Significance
In Latin America, vampire bat rabies constrains livestock production and is the main cause of lethal human rabies outbreaks. Despite knowledge that bat dispersal prevents viral extinction and compromises control campaigns, the movement patterns of infected bats are unknown. Using large host and virus datasets, we illustrate a genetic approach to link population level patterns of host dispersal to pathogen spatial spread that overcomes logistical limitations of tracking animal movement in the wild. The results implicate male vampire bats as contributing disproportionately to rabies spatial spread and offer opportunities to forecast and prevent rabies. The ubiquity of sex-biased dispersal in animals suggests sex-biased pathogen spread could widely influence the distribution and invasion dynamics of emerging diseases.
Keywords: Desmodus, zoonotic disease, forecasting, sex bias, spatial dynamics
Abstract
Anticipating how epidemics will spread across landscapes requires understanding host dispersal events that are notoriously difficult to measure. Here, we contrast host and virus genetic signatures to resolve the spatiotemporal dynamics underlying geographic expansions of vampire bat rabies virus (VBRV) in Peru. Phylogenetic analysis revealed recent viral spread between populations that, according to extreme geographic structure in maternally inherited host mitochondrial DNA, appeared completely isolated. In contrast, greater population connectivity in biparentally inherited nuclear microsatellites explained the historical limits of invasions, suggesting that dispersing male bats spread VBRV between genetically isolated female populations. Host nuclear DNA further indicated unanticipated gene flow through the Andes mountains connecting the VBRV-free Pacific coast to the VBRV-endemic Amazon rainforest. By combining Bayesian phylogeography with landscape resistance models, we projected invasion routes through northern Peru that were validated by real-time livestock rabies mortality data. The first outbreaks of VBRV on the Pacific coast of South America could occur by June 2020, which would have serious implications for agriculture, wildlife conservation, and human health. Our results show that combining host and pathogen genetic data can identify sex biases in pathogen spatial spread, which may be a widespread but underappreciated phenomenon, and demonstrate that genetic forecasting can aid preparedness for impending viral invasions.
Knowledge of the mechanisms governing the spread of pathogens across landscapes is vital to predict disease emergence in humans, domestic animals, and wildlife (1). The spread of directly transmitted pathogens is intimately linked to host dispersal, but for most wildlife hosts, actively tracking the movements of infected individuals is logistically impractical at pertinent temporal and spatial scales (2). Indirect population genetic methods overcome this limitation by characterizing historical patterns of dispersal in host genomes, raising the possibility that host genetic structure could forecast the pathways of invading pathogens. However, host genetic structure may not correlate with pathogen spread because of the different timescales reflected in host and pathogen genomes or if infection alters host dispersal behavior (3, 4). Differences among genomes in modes of inheritance might also influence genetic forecasts of pathogen spread. Biparentally inherited host nuclear DNA (nuDNA) reveals the joint population structure of both sexes, whereas maternally inherited mitochondrial DNA (mtDNA) indicates female population structure. Incongruence between host genomes is commonly attributed to sex biases in dispersal and/or differences in rates of lineage sorting (5, 6). In contrast, pathogen genomes are transmitted horizontally between hosts and often evolve on rapid timescales, providing high-resolution markers of the contemporary movements of infected individuals of both sexes (4). Therefore, congruence of pathogen population structure to host nuDNA or mtDNA population structure, but incongruence to the other, could identify which host genome is most relevant to epidemic spread while signaling a potentially sex-linked mechanism of pathogen dispersal.
Here, we apply nuclear, mitochondrial, and viral genetic markers to explore the spatial spread of vampire bat (Desmodus rotundus) rabies virus (VBRV) (Lyssavirus, Rhabodviridae), a directly transmitted zoonosis that causes universally fatal encephalitis when infected bats feed on livestock and humans (7). VBRV is a constant impediment to public health and agriculture in Latin America, with annual costs exceeding US$30 million in livestock mortality alone (8). Control programs focus on reducing the size of bat populations by using topical anticoagulant poisons (7). However, because long-term viral maintenance depends more strongly on viral dispersal between vampire bat colonies than on colony size, decades of culling have failed to eliminate VBRV (9, 10). Instead, human and livestock VBRV mortality are increasing and the virus is emerging in historically VBRV-free areas throughout Latin America (7, 11). Linking patterns of bat dispersal to viral spread is therefore essential to improve disease control in endemic areas and to predict pathways of ongoing invasions.
Prior population genetic studies concluded that the low vagility and small home range sizes of vampire bats generate high genetic differentiation among populations (12, 13). However, genetic lineages of VBRV are geographically widespread, implying that the virus overcomes the genetic isolation of its host through a currently unidentified dispersal mechanism (14, 15). In carnivores, nonresident, nomadic individuals and seasonal variation in contact networks are thought to influence pathogen prevalence within populations, but the role of host social structure on pathogen spread at larger spatial scales is less understood (16, 17). Like most mammals, male vampire bats disperse upon sexual maturity, whereas females retain strong fidelity to the natal roost (18, 19). We therefore hypothesized that male-biased dispersal could spread VBRV between colonies, giving males a pivotal role in the regional persistence and spatial propagation of outbreaks across landscapes. As male dispersal contributes to nuclear but not mitochondrial gene flow, we predicted that bat nuclear population structure would explain historical invasions of VBRV. Further, because male dispersal may be prompted by the subsequent year’s annual birth pulse (19), we hypothesized that expansions could be seasonal. Finally, we tested whether linking simple landscape resistance models with viral phylogeography can forecast rates and routes of viral invasion to currently VBRV-free regions. We realize these objectives by using datasets representing hundreds of host nuclear, host mitochondrial, and viral genomes from Peru along with a 13-y spatially explicit time series on VBRV outbreaks in sentinel livestock.
Results
Viral Genetic Structure.
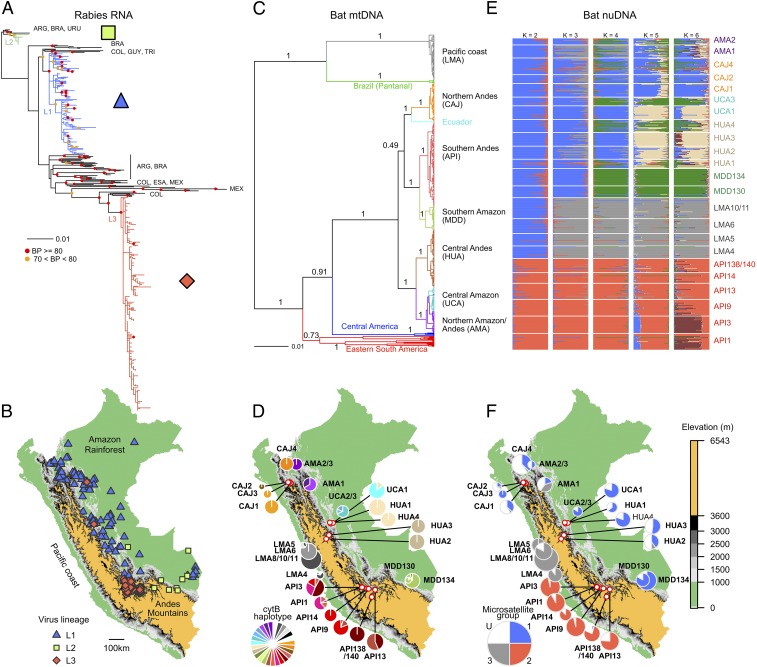
We sequenced the complete nucleoprotein (N) gene from 264 rabies isolates collected from Peruvian livestock between 1997 and 2012 and compared these to representative sequences from throughout the Americas. Because livestock infect neither each other nor bats, each isolate represents a single transmission event from bat to livestock, providing a window of insight into locally circulating bat viruses (14, 15). A maximum likelihood (ML) phylogenetic tree revealed three viral lineages in Peru, each of which shared a most recent common ancestor (MRCA) with viruses from other South American countries, consistent with multiple, independent introductions of VBRV into Peruvian bats (Fig. 1A). Two viral lineages, L1 and L2, circulated exclusively east of the Andes mountains and overlapped in the southern and central Amazon (Fig. 1B). Viral lineage 3 (L3) was isolated in inter-Andean valleys in southern Peru, with the exception of two samples found well outside that range (Fig. 1B). The outlier viruses were paraphyletic to each other (indicating distinct introductions) and were found in rarely infected species (dog and horse), suggesting human-mediated translocations of companion animals, rather than infections acquired from indigenous bats at the sampling locality.
Fig. 1.
Genetic and geographic structure of host and viral markers with distinct inheritance mechanisms. (A) The ML tree of VBRV, using 434 complete N sequences from Peru (colored branches) and other representative countries in the Americas (black branches; ARG, Argentina; BRA, Brazil; COL, Colombia; ESA, El Salvador; GUY, French Guiana; MEX, Mexico; TRI, Trinidad; URU, Uruguay). Colored symbols show bootstrap support from 1,000 replicate ML searches. Two outgroup sequences from a rabies variant circulating in Peruvian dogs were excluded for visualization. (B) Geographic distributions of viral lineages in Peru. Areas above 3,600 m (the upper limit to vampire bats in Peru) are colored gold. (C) Bayesian phylogenetic tree of cytB sequences from vampire bats from Peru (n = 442) and other countries in the Americas (n = 26). Branches are colored by geographic region. Node values are posterior probabilities. (D) Distribution of CytB haplotypes across vampire bat colonies in Peru. Sites with <8 sequenced individuals were grouped with other colonies surveyed within 10 km. Pie charts are proportionate to sample size (range = 8–30). (E) Estimates from STRUCTURE analyses assuming K = 2–6 populations using 9 microsatellites (n = 480 bats). Each bar represents the probability of membership assignment to each of K groups. (F) Pie charts show the distribution of microsatellite groups [K = 3, threshold probability for group membership = 0.85, unassigned individuals (U) in white].
Contrasting the Population Structure of Vampire Bat Genomes to Rabies Virus.
A Bayesian phylogenetic analysis of mitochondrial cytochrome B (cytB) sequences from 468 vampire bats showed marked geographic structure. Bats captured west of the Andes mountains in the department of Lima (LMA sites) were highly divergent from other Peruvian bats and were more closely related to bats from southwestern Brazil (Fig. 1C). Bats east of the Andes formed a separate monophyletic group (with a single sequence from Ecuador) that shared an MRCA with populations in Central America [posterior probability (PP) = 0.91]. Most mitochondrial lineages were found exclusively within single departments (equivalent to US states) of Peru (Fig. 1C). In some cases [e.g., sites from Amazonas (AMA) and sites from Cajamarca (CAJ)], colonies separated by short distances were comprised exclusively of individuals from distinct and paraphyletic mitochondrial lineages, indicating a lack of female gene flow among nearby colonies since lineages diverged. A lineage found in both the central Amazon in Ucayali (UCA sites) and the northern Amazon (AMA sites) was a potential exception; however, the well-supported subclades within this lineage were restricted to either UCA or AMA, but not both (Fig. 1C and SI Appendix, Fig. S6). Spatial isolation was even stronger at the haplotype level. Of 27 haplotypes, 16 were exclusive to single bat colonies and the mean distance occupied by haplotypes was only 22 km (range = 0–258.1 km). Minimal sharing of haplotypes between distant colonies implied the absence of contemporary mitochondrial gene flow between regions (Fig. 1D).
We next compared the population structure of biparentally inherited nuclear markers to that of mtDNA and viral RNA. Nine nuclear microsatellites from 480 vampire bats indicated 2–3 genetic groups (K), with admixture among all colonies east of the Andes (microsatellite group 1, MG1; Fig. 1 E and F) and genetic isolation of vampire bat colonies found in inter-Andean valleys in southern Peru in the department of Apurimac (API sites, microsatellite group 2, MG2). At K > 2, bats from the Pacific coast (LMA sites) formed a distinct group (MG3), which was also detected at low to moderate frequency in the northern Andes and Amazon (Fig. 1E). These findings were robust to analysis of only six loci without null alleles, two alternative methods of statistical inference [STRUCTURE and discriminant analysis of principal components (DAPC)], and to relaxing priors on the spatial locations of sampled bat populations (SI Appendix, Figs. S2–S4). Larger (less plausible) values of K revealed additional geographic clusters but implied extensive gene flow among them (Fig. 1E), an expected pattern given the isolation by distance in our microsatellite data (SI Appendix, Fig. S5). Importantly, even at the highest levels of K, microsatellite groups spanned areas east of the Andes, providing a potential corridor for viral spread through the Amazon and eastern slopes of the Andes.
We assessed individual-level genomic mismatches between nuDNA and mtDNA by using 352 bats for which we had both microsatellite and mitochondrial data. Bats east of the Andes carried highly divergent mtDNA lineages, but belonged to the same microsatellite group (SI Appendix, Fig. S6). Therefore, viral lineages L1 and L2 were maintained by MG1 bats with diverse and paraphyletic mitochondrial haplotypes, whereas L3 exclusively infected MG2 bats with API mitochondrial haplotypes, creating viral genetic congruence to nuDNA, but not mtDNA. The atypical MG3 bats found in the Andes and Amazon had locally prevalent mtDNA haplotypes, suggesting that male immigrants from the coast reproduce with resident females despite the long branch separating these populations in the cytB phylogeny (Fig. 1C).
Microsatellite Confirmation of Male-Biased Dispersal.
Greater structure in maternally inherited markers relative to nuclear markers signaled the expected pattern of female philopatry and male-biased dispersal (19, 20). Additional tests using microsatellites found a nearly 14-fold increase in the FIS (the inbreeding coefficient of an individual relative to the subpopulation) of male vampire bats relative to females (female FIS = −0.0003, male FIS = 0.1363, P < 0.0004), consistent with the expectation that the dispersing sex should have heterozygote deficiency because samples represent a mixture of populations (i.e., the Wahlund effect) (21). Other genetic comparisons of males and females were not statistically significant (SI Appendix, Table S7), but detection by any one of the above methods implies intense sex-biased dispersal (21). Similarly, using the assignment probabilities from our STRUCTURE analysis, we found that putative recent migrants (PP < 0.2 for belonging to the locally abundant genotype group) tended to be male (59.3% for all Peru, 65.2% for bats within the range of MG1).
Seasonal Expansions of Rabies Across the Landscape.
We estimated the monthly area of Peru infected by VBRV from 2003 to 2014 using a database of 1,146 laboratory-confirmed livestock rabies outbreaks (11). Time series decomposition revealed significant seasonality in the area infected (generalized additive model: deviance explained = 27.7%, P < 0.001) with peak spatial expansions in November and December (SI Appendix, Fig. S10).
Reconstructing and Forecasting Viral Invasion Dynamics.
Having identified congruence between bat nuDNA and viral geographic distributions as a potential by-product of male-biased dispersal, we sought to reconstruct and forecast the dynamics of viral spread across the landscape. We applied Bayesian continuous phylogenetic ancestral state estimation to L1 and L3 viruses with precisely known collection dates and Global Positioning System (GPS) locations (NL1 = 81, NL3 = 179). To enhance phylogenetic resolution, we added sequence data from the hypervariable, noncoding region between the glycoprotein and polymerase genes (G-L, 510 bp) to the complete N gene sequences. Similar analyses of lineage L2 were precluded by low sample size and the disappearance of this virus after 2009 (SI Appendix, Table S8). However, the lack of genetic isolation by geographic distance in available data suggests relatively unconstrained dispersal of that virus during its tenure in Peru (SI Appendix, Fig. S7). Phylogeographic models showed viral invasions occurred within the past 40 y [L1: 95% highest posterior density (HPD) on MRCA = 20.65–35.02; L3: 95% HPD = 16.27–28.0]. Although some geographic clustering was apparent in both viral lineages, jumps between the Peruvian departments within the range of each lineage were common (Fig. 2 A and B). Both viruses underwent decelerating invasions, with initially rapid increases in geographic extent followed by gradual expansions in the last 10–15 y (Fig. 2D). The greater spatial scale of historical expansions of L1 produced a higher median velocity in L1 compared with L3 [61.5 (95% HPD = 26.2–194.5) vs. 25.4 (95% HPD = 14.9–50.1) km/y]. However, contemporary velocities calculated from tip branches (1997–2012) were similar between lineages (L1: 33.9 vs. L3: 28.4 km/y, Fig. 2E).
Fig. 2.
Dynamics of historical viral dispersal within Peru. Bayesian phylogenetic trees of VBRV L1 (A) and L3 (B), with tip symbols colored according to department of Peru (C). Inner node symbols are PPs of clades. (D) Spatial expansions of each viral lineage, depicted as the cumulative geographic distance from the inferred outbreak origin through time. (E) Posterior distributions of the epidemic velocity and diffusion coefficient of each viral lineage. Points are parameter estimates from the tips of one randomly sampled tree from the posterior distribution of each Bayesian phylogeographic analysis. Solid diamonds and lines are the median and 95% HPDs on parameter estimates, respectively. Black diamonds are median statistics calculated across all branches. Vertical dashed lines are the 95% bounds of the wavefront velocity estimated from time series data in northern Peru (Fig. 3 and SI Appendix, Fig. S9).
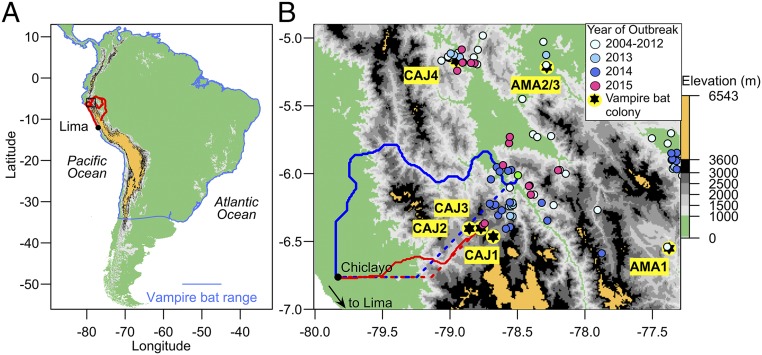
Unexpected gene flow of MG3 from the VBRV-free Pacific coast to the VBRV-endemic Andes and Amazon prompted us to explore corridors for viral invasion to the Pacific coast of South America (Fig. 1F and SI Appendix, Fig. S6). A landscape resistance model describing the well-known pattern of VBRV spread along valleys (7, 11) showed least-cost routes from Andean and Amazonian bat colonies with MG3 individuals to the coast passed through a corridor in the north of Peru that forms the lowest pass throughout the length of the Andes (the Huancabamba Depression; Fig. 3A). We projected the invasion of VBRV by combining phylogeographic estimates of viral dispersal velocities with least cost distances to the Pacific coast. Assuming velocities inferred from the tips of L1 and L3 phylogenies, respectively, we forecast the arrival of VBRV to Chiclayo (a major city on the coast) by July 2019 (95% HPD: 2016.85–2023.25) or June 2020 (95% HPD: 2017.12–2024.93). Rabies mortality data from livestock during the 3 y after our viral sequence data were collected (2013–2015) confirmed ongoing viral invasion along the routes projected by landscape resistance models (Fig. 3B). These nongenetic data show that VBRV traveled 50.1 km southwest from June 2012 to April 2015 at 16.1 km/y (95% CI = 14.6–17.6, Fig. 2E and SI Appendix, Fig. S9), leaving less than 145 km to the Pacific coast of South America.
Fig. 3.
Forecasting invasion of VBRV to the Pacific coast of South America. (A) The South American range of vampire bats, colored as in Fig. 1. Red lines are least-cost pathways from bat colonies in the Andes and Amazon with MG3 individuals (typical of the coast) to Lima using the “valley” resistance model. The black box indicates the region in B. (B) Blue lines are least-cost routes from the westernmost VBRV outbreak in 2012 (green point) to Chiclayo (a reference point for the Pacific coast) according to the valley (solid) and threshold (dotted) resistance models. Red lines are routes from the western front of the epidemic in 2015. Light blue points (2004–2012) are outbreaks that were included in phylogenetic analyses.
Discussion
By combining large host and virus genetic datasets, we show female philopatry and male-biased dispersal in vampire bats likely creates a disproportionate role for male bats in the spatial spread of VBRV. Using these insights on host and virus dispersal, we forecast routes and rates of an ongoing viral invasion that we predict will cause an historic and damaging first invasion of VBRV to the Pacific coast of South America. Independent epidemiological data support our genetic predictions.
Reduced host population structure in nuclear relative to mitochondrial markers and heterozygote deficiency in males are consistent with male dispersal and female philopatry in vampire bats, as is known from field studies and is the general expectation for mammals (18, 19). We suggest that a by-product of sex-biased dispersal is that the spatial spread and geographic distribution of VBRV will also be male-driven. Biologically, the long incubation period (2–4 wk) and the short infectious period of rabies (2–3 d) means that most viral dispersal will occur before the onset of disease (22). Moreover, the debilitating clinical signs of rabies (ataxia, lethargy, and death) during most of the infectious period make it unlikely that infection could induce unusually long-distance dispersal in infected females, thereby spreading VBRV without leaving a mtDNA signature of dispersal through reproduction. Therefore, the sex that disperses most while incubating rabies (males) will naturally dominate viral spread, and barriers to male dispersal will delimit the boundaries of viral distributions, as we observed. The seasonality in the geographic area infected by VBRV was also consistent with male-biased spatial spread. Viral expansions peaked at the start of the wet season (SI Appendix, Fig. S10), when a new cohort of births is expected to initiate dispersal of males from the previous year (19, 23). This concurrence provides indirect evidence for pulses of VBRV expansion driven by seasonal male bat dispersal, although other seasonal and nonseasonal factors could also influence spatial expansions.
In principle, incongruence between host genomes could arise from limitations of genetic data examined here; however, several lines of evidence argue against this interpretation. First, rates of lineage sorting and mutation differ between nuDNA and mtDNA and imply different timescales of population structure (6). Given that viral invasions occurred within the last 40 y, we are most concerned with contemporary bat population structure as revealed by microsatellites (which matched the viral distribution) and the landscape distribution of mtDNA haplotypes. If contemporary dispersal were equal among sexes and the patterns we observed arose from different genetic timescales, we would have expected mtDNA haplotypes to span epidemiologically connected regions, rather than being most often restricted to single bat colonies. The number or power of microsatellites also cannot explain their weaker population structure relative to mtDNA. Both six and nine microsatellites differentiated API bats from bats captured in Madre de Dios (MDD) sites, despite their close relatedness in the mtDNA phylogeny (Fig. 1C), and simulations showed 98–100% power to detect population structure (SI Appendix, Fig. S8).
The ubiquity of sex-biased dispersal in nature [typically female biased in birds and male biased in mammals (18)] could make sex-biased pathogen spread a widespread determinant of epidemic propagation at the landscape level. Sex-biased pathogen spread is difficult to detect by using traditional methods for studying animal movement such as radio telemetry or GPS tags because rare dispersal events that can be critical for disease spread will be missed in studies carried out over small spatiotemporal scales. This study shows that contrasting host and pathogen genetic markers with different inheritance modes provides a framework to begin to evaluate sex-biased pathogen dispersal. Other approaches, such as theoretical modeling of pathogen transmission within host contact networks, incorporate similar concepts, but typically lack corresponding data from pathogens to verify how host dispersal heterogeneity affects disease spread, require extensive field datasets on host contacts, and offer limited guidance for managing pathogens emerging at the landscape level (17, 24). Identifying the sex responsible for pathogen spread carries practical implications for the prevention and control of VBRV because blocking viral movement between colonies is predicted to cause viral extirpation (10). Future work should quantify the scales at which males and females contribute to intercolony viral spread to evaluate the efficacy of targeting dispersing males in rabies control campaigns.
We also show that combining host population genetics, pathogen phylogeography, and landscape ecology can predict rates and routes of pathogen invasion to disease-free areas. A similar approach could be useful to forecast other emerging pathogen invasions where the lack of long-term infection data precludes traditional epidemiological analyses. Most importantly, we forecast viral invasion to the historically VBRV-free Pacific coast of South America (25) via a previously undetected corridor of vampire bat gene flow across the Peruvian Andes. Three years of independent livestock rabies mortality data confirmed viral expansion along comparable routes and velocity to model forecasts (Figs. 2E and 3). We caution invasions could accelerate or decelerate closer to the coast where landscapes are less complex than the Andes or Amazon and that alternate routes have not been excluded. However, at present, we foresee no significant barriers to continued invasion. Vampire bats occur continuously from the leading edge of the wavefront to the Pacific coast (26), and VBRV summited the highest remaining peak in 2015 (Fig. 3B). The evolutionary divergence of coastal from Andean subpopulations is also unlikely to stop VBRV because rabies host shifts are common within bat genera (27), whereas the subpopulations in question are interbreeding (SI Appendix, Fig. S6). More work is needed to determine whether the current invasion was triggered by recent changes in bat population structure, gradual viral invasion to the fringes of the vampire bat distribution, or a combination. Regardless of the initiating mechanism, the arrival of VBRV to the coastal regions of Peru and subsequent potential spread to Ecuador and northern Chile would be profoundly damaging for agriculture. The presence of VBRV would also create new risks to humans that interact with bats or infected livestock and to wildlife such as sea lions that constitute an important food source for coastal vampire bats (28). Culling vampire bats failed to stop advancing VBRV epidemics in Argentina and could conceivably exacerbate viral spread if culls promote bat dispersal, as was observed in badger culls aiming to control bovine tuberculosis in the United Kingdom (10, 29, 30). We therefore advocate heightened surveillance, preventative livestock vaccination, and educational campaigns to reduce the burden of impeding epidemics.
Previous comparisons of host and pathogen genetic data have exploited pathogens as a high resolution marker of host demography and dispersal or have studied coevolutionary dynamics over longer timescales (4, 31). Our study shows that similar data can verify key host demographic groups for pathogen spatial spread and forecast epidemic invasions to disease-free areas. As the abundance and resolution of host and pathogen genomic data increase, similar approaches could test the generality of sex-biased pathogen dispersal while providing important foresights into the landscape dynamics of emerging pathogen invasions.
Materials and Methods
Vampire Bat and Rabies Virus Data.
Biopsies of bat wing membranes were collected in 2008–2013 from 29 vampire bat colonies across 7 Peruvian departments in the Pacific coast, Andes mountains, and Amazon rainforest (SI Appendix, Table S1). In addition, we acquired 264 rabies-infected livestock brains from 13 departments of Peru, collected between 1997 and 2012 by the passive surveillance system of the National Service of Agrarian Health of Peru (SENASA). Further data details and laboratory procedures for sequencing and genotyping are described in SI Appendix. All sequences have been deposited to GenBank (cytB: KU937964–KU938399; rabies virus: KU938400–KU938924).
Viral Phylogenetic Analyses.
ML phylogenetic trees were estimated with Garli 2.01 by using Peruvian samples with complete N gene sequences and geographic information at least to the district level (32). We included up to five sequences per lineage for VBRVs from other regions of Latin America and two dog rabies sequences as outgroups. The topology was selected as the best of 10 replicate ML searches with random starting trees by using the GTR+I+G substitution model. Bootstrap values were calculated from 1,000 additional ML searches by using the best topology from the first set of searches as a starting tree. Phylogeographic analyses were carried out in BEAST v.1.8 (33). Preliminary runs indicated use of a lognormal relaxed molecular clock for N and a strict molecular clock for G-L data for both lineages. Nucleotide substitution models were selected for codon positions one and two (together) and for codon position 3 by using Akaike’s Information Criterion (AIC) corrected for small sample size in jModeltest 2 (34). Triplicate MCMC chains were run for 50 million generations, with trees and parameters sampled every 10,000 steps. We evaluated three relaxed random walk models of spatial diffusion by using Bayes Factors (BF) (35, 36) and present results from the gamma model in Fig. 2 (BF > 8.5 for gamma vs. other models). Convergence within and across runs and appropriate burn-in periods were checked in Tracer. For L3, the overall likelihood converged, but tree likelihoods for individual data partitions swapped between competing values throughout runs regardless of chain lengths. Demographic parameters (MRCA, diffusion rate) were uncorrelated with swaps. Diffusion coefficients and dispersal velocities were estimated from 1,000 randomly sampled trees from each posterior distribution using the Seraphim package of R (37).
Host Population Genetic Analyses.
Bayesian phylogenetic analysis of mitochondrial Cytb sequences was performed with BEAST by using the HKY+I model of nucleotide substitution selected by AIC in jModeltest 2, assuming constant effective population size. One sequence of Diphylla ecaudata (Hairy-legged vampire bat, GenBank accession no.: DQ077399) was included as an outgroup. Trees were sampled every 1,000 states for 20 million generations, and the first 2,001 trees were removed before generating a maximum clade credibility tree. CytB haplotypes were designated by using the pegas package of R after removing the first 5 bp from sequences because of missing data in some samples (38). Nuclear population structure was assessed with two classification methods: STRUCTURE (a Bayesian clustering method) minimizes Hardy–Weinberg and linkage disequilibrium, and DAPC identifies genetic clusters that maximize between-group variance and minimize within-group variance (39, 40). SI Appendix provides summary statistics on microsatellites, additional checks performed, and details of analyses of host population structure.
Forecasts of Viral Invasion.
We applied the velocities estimated from continuous phylogeographic analyses to least-cost distances from landscape models of bat dispersal to predict pathways and dates of VBRV spread. Distances were calculated with two landscape models in the gdistance package of R: first, a path through any elevation under 3,600 m (“threshold model”) and second, assuming exponentially increasing costs from 1 to 3,600 m, with no dispersal above 3,600 m (“valley model”). These models qualitatively and quantitatively capture the observed spread of VBRV epidemics through river valleys (7, 11). Distances from the leading edge of a westward-expanding VBRV epidemic (April 2015, La Colca, Department of Cajamarca) to Chiclayo differed by only 5.7 km between models (136.37 versus 143.32 km); we therefore use the second, more conservative scenario in forecasts. To confirm ongoing VBRV invasions along the projected route, we analyzed 31 laboratory confirmed rabies outbreaks in livestock that were reported after our sequence data were collected (2013–2015) by using the linear regression technique of ref. 11.
Supplementary Material
Acknowledgments
We thank Heather Danaceau, Jennifer Towner, and Victoria Estacio for laboratory assistance and Roman Biek, Barbara Mable, and Mafalda Viana for comments on the manuscript. SENASA contributed surveillance data and livestock samples under an International Cooperative Agreement with the University of Georgia (30-11-2012). The Peruvian Government authorized collection, exportation, and use of genetic resources (RD-222-2009-AG-DGFFS-DGEFFS, 003851-AG-DGFFS, RD-273-2012-AG-DGFFS-DGEFFS, and RD-054-2016-SERFOR-DGGSPFFS). The University of Georgia Advanced Computing Resource Center provided computational resources. Funding was provided by National Science Foundation Grant DEB-1020966 (to D.G.S. and S.A.) and the Pan American Health Organization (W.V.). D.S. was funded by a Sir Henry Dale Fellowship, jointly funded by Wellcome Trust and Royal Society Grant 102507/Z/13/Z. The findings and the conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.J.B. is a Guest Editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KU937964–KU938399 (cytB) and KU938400–KU938924 (rabies virus)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606587113/-/DCSupplemental.
References
- 1.Plowright RK, et al. Ecological dynamics of emerging bat virus spillover. Proc Biol Sci. 2015;282(1798):20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altizer S, Bartel R, Han BA. Animal migration and infectious disease risk. Science. 2011;331(6015):296–302. doi: 10.1126/science.1194694. [DOI] [PubMed] [Google Scholar]
- 3.Mazé-Guilmo E, Blanchet S, McCoy KD, Loot G. Host dispersal as the driver of parasite genetic structure: A paradigm lost? Ecol Lett. 2016;19(3):336–347. doi: 10.1111/ele.12564. [DOI] [PubMed] [Google Scholar]
- 4.Biek R, Drummond AJ, Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311(5760):538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- 5.Turmelle AS, Kunz TH, Sorenson MD. A tale of two genomes: Contrasting patterns of phylogeographic structure in a widely distributed bat. Mol Ecol. 2011;20(2):357–375. doi: 10.1111/j.1365-294X.2010.04947.x. [DOI] [PubMed] [Google Scholar]
- 6.Zink RM, Barrowclough GF. Mitochondrial DNA under siege in avian phylogeography. Mol Ecol. 2008;17(9):2107–2121. doi: 10.1111/j.1365-294X.2008.03737.x. [DOI] [PubMed] [Google Scholar]
- 7.Johnson N, Aréchiga-Ceballos N, Aguilar-Setien A. Vampire bat rabies: Ecology, epidemiology and control. Viruses. 2014;6(5):1911–1928. doi: 10.3390/v6051911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization WHO Expert Consultation on Rabies. Second report. World Health Organ Tech Rep Ser. 2013;982(982):1–139. [PubMed] [Google Scholar]
- 9.Linhart SB, Mitchell GC, Crespo RF. Control of vampire bats by topical application of an anticoagulent (chlorophacinone) Bol La Of Sanit Panam. 1972;73(2):31–38. [PubMed] [Google Scholar]
- 10.Blackwood JC, Streicker DG, Altizer S, Rohani P. Resolving the roles of immunity, pathogenesis, and immigration for rabies persistence in vampire bats. Proc Natl Acad Sci USA. 2013;110(51):20837–20842. doi: 10.1073/pnas.1308817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benavides JA, Valderrama W, Streicker DG. Spatial expansions and travelling waves of rabies in vampire bats. Proc R Soc B Biol Sci. 2016;283(1832) [Google Scholar]
- 12.Martins FM, Templeton AR, Pavan ACO, Kohlbach BC, Morgante JS. Phylogeography of the common vampire bat (Desmodus rotundus): Marked population structure, Neotropical Pleistocene vicariance and incongruence between nuclear and mtDNA markers. BMC Evol Biol. 2009;9(294):294. doi: 10.1186/1471-2148-9-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romero-Nava C, León-Paniagua L, Ortega J. Microsatellites loci reveal heterozygosis and population structure in vampire bats (Desmodus rotundus) (Chiroptera: Phyllostomidae) of Mexico. Rev Biol Trop. 2014;62(2):659–669. [PubMed] [Google Scholar]
- 14.Condori-Condori RE, Streicker DG, Cabezas-Sanchez C, Velasco-Villa A. Enzootic and epizootic rabies associated with vampire bats, peru. Emerg Infect Dis. 2013;19(9):1463–1469. doi: 10.3201/eid1909.130083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres C, et al. Phylodynamics of vampire bat-transmitted rabies in Argentina. Mol Ecol. 2014;23(9):2340–2352. doi: 10.1111/mec.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loveridge AJ, Macdonald DW. Seasonality in spatial organization and dispersal of sympatric jackals (Canis mesomelas and C. adustus): Implications for rabies management. J Zool (Lond) 2001;253:101–111. [Google Scholar]
- 17.Craft ME, Volz E, Packer C, Meyers LA. Disease transmission in territorial populations: The small-world network of Serengeti lions. J R Soc Interface. 2011;8(59):776–786. doi: 10.1098/rsif.2010.0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenwood PJ. Mating systems, philopatry and dispersal in birds and mammals. Anim Behav. 1980;28(4):1140–1162. [Google Scholar]
- 19.Wilkinson GS. The social organization of the common vampire bat: II Mating system, genetic structure and relatedness. Behav Ecol Sociobiol. 1985;17:123–134. [Google Scholar]
- 20.Melnick DJ, Hoelzer GA. Differences in male and female macaque dispersal lead to contrasting distributions of nuclear and mitochondrial DNA variation. Int J Primatol. 1992;13(4):379–393. [Google Scholar]
- 21.Goudet J, Perrin N, Waser P. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol Ecol. 2002;11(6):1103–1114. doi: 10.1046/j.1365-294x.2002.01496.x. [DOI] [PubMed] [Google Scholar]
- 22.Moreno JA, Baer GM. Experimental rabies in the vampire bat. Am J Trop Med Hyg. 1980;29(2):254–259. doi: 10.4269/ajtmh.1980.29.254. [DOI] [PubMed] [Google Scholar]
- 23.Lord RD. Seasonal reproduction of vampire bats and its relation to seasonality of bovine rabies. J Wildl Dis. 1992;28(2):292–294. doi: 10.7589/0090-3558-28.2.292. [DOI] [PubMed] [Google Scholar]
- 24.Nunn CL, Thrall PH, Stewart K, Harcourt AH. Emerging infectious diseases and animal social systems. Evol Ecol. 2008;22(4):519–543. [Google Scholar]
- 25.Navarro AM, Bustamante J, Sato A. Situación actual y control de la rabia en el Perú [Current status and control of rabies in Peru] Rev Peru Med Exp Salud Publica. 2007;24(1):46–50. [Google Scholar]
- 26.Quintana H, Pacheco V. Identificación y distribución de los murciélagos vampiros del Perú [Identification and distribution of vampire bats in Peru] Rev Peru Med Exp Salud Publica. 2007;24:81–88. [Google Scholar]
- 27.Streicker DG, et al. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329(5992):676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- 28.Catenazzi A, Donnelly MA. Sea lion (Otaria flavescens) as host of the common vampire bat (Desmodus rotundus) Mar Ecol Prog Ser. 2008;360:285–289. [Google Scholar]
- 29.Fornes A, et al. Control of bovine rabies through vampire bat control. J Wildl Dis. 1974;10(4):310–316. doi: 10.7589/0090-3558-10.4.310. [DOI] [PubMed] [Google Scholar]
- 30.Pope LC, et al. Genetic evidence that culling increases badger movement: Implications for the spread of bovine tuberculosis. Mol Ecol. 2007;16(23):4919–4929. doi: 10.1111/j.1365-294X.2007.03553.x. [DOI] [PubMed] [Google Scholar]
- 31.Hafner MS, et al. Disparate rates of molecular evolution in cospeciating hosts and parasites. Science. 1994;265(5175):1087–1090. doi: 10.1126/science.8066445. [DOI] [PubMed] [Google Scholar]
- 32.Zwickl DJ, Hills DM. 2006. Genetic algorithim approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Dissertation (University of Texas, Austin)
- 33.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baele G, et al. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 2012;29(9):2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemey P, Rambaut A, Welch JJ, Suchard MA. Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol. 2010;27(8):1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dellicour S, Rose R, Pybus OG. Explaining the geographic spread of emerging viruses: A new framework for comparing viral genetic information and environmental landscape data. BMC Bioinformatics. 2016;17(82):1–12. doi: 10.1186/s12859-016-0924-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paradis E. pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26(3):419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 39.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010;11(1):94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.