Significance
Lignocellulosic biomass is a potential major resource for renewable energy production. Plant cell-wall deconstruction, however, remains an inefficient process, mainly due to the recalcitrant nature of the lignin and cellulosic components, that requires chemical pretreatment methods prior to degradation. This study aims to overcome this barrier by combining two paradigms into a single system, by using a synthetic biology approach. The designed system integrates an engineered laccase (an oxidizing enzyme that acts on lignin) into a multienzyme cellulosome complex, thereby producing enhanced decomposition of wheat straw. These findings demonstrate the potential of introducing complementary enzymes that fail to occur together in nature into designer cellulosomes for improved lignocellulose conversion.
Keywords: lignocellulose, multienzyme complex, catalysis, biomass conversion
Abstract
Efficient breakdown of lignocellulose polymers into simple molecules is a key technological bottleneck limiting the production of plant-derived biofuels and chemicals. In nature, plant biomass degradation is achieved by the action of a wide range of microbial enzymes. In aerobic microorganisms, these enzymes are secreted as discrete elements in contrast to certain anaerobic bacteria, where they are assembled into large multienzyme complexes termed cellulosomes. These complexes allow for very efficient hydrolysis of cellulose and hemicellulose due to the spatial proximity of synergistically acting enzymes and to the limited diffusion of the enzymes and their products. Recently, designer cellulosomes have been developed to incorporate foreign enzymatic activities in cellulosomes so as to enhance lignocellulose hydrolysis further. In this study, we complemented a cellulosome active on cellulose and hemicellulose by addition of an enzyme active on lignin. To do so, we designed a dockerin-fused variant of a recently characterized laccase from the aerobic bacterium Thermobifida fusca. The resultant chimera exhibited activity levels similar to the wild-type enzyme and properly integrated into the designer cellulosome. The resulting complex yielded a twofold increase in the amount of reducing sugars released from wheat straw compared with the same system lacking the laccase. The unorthodox use of aerobic enzymes in designer cellulosome machinery effects simultaneous degradation of the three major components of the plant cell wall (cellulose, hemicellulose, and lignin), paving the way for more efficient lignocellulose conversion into soluble sugars en route to alternative fuels production.
Plant biomass is one of the most abundant sources of organic material on earth. Given its widespread availability and renewability, it is considered to be a promising resource for alternative and sustainable energy production (1). The plant cell wall is composed of lignocellulose, a heterogeneous amalgamation of cellulose, hemicellulose, and lignin. Cellulose and hemicellulose are attractive components for the production of biofuels or synthons, because these polysaccharides can be biologically hydrolyzed to simple sugars, which, in turn, can be converted into ethanol or other high-value chemicals (2, 3). On the other hand, lignin is mostly considered to be a hindrance in bioethanol production processes (4). Indeed, although lignin-derived aromatic compounds are valuable in the green chemistry sector, this complex organic heteropolymer encases the cellulose/hemicellulose fibers, and thus limits the accessibility of enzymes or chemicals. As a result, and to increase the amount of fermentable sugars produced from plant biomass, lignin must be removed (5).
Currently, the techniques available to remove lignin from the biomass efficiently are limited. For example, chemical pretreatment of biomass, with acids or organic solvents, before enzymatic digestion can generate lignocellulose-derived byproducts such as phenolic compounds that inhibit the enzymatic biocatalysts (6). More favorable techniques, based on the use of lignin-degrading enzymes, such as laccase and lignin peroxidase, have long been identified; however, their utilization in the biofuel production process is currently limited (7). Laccases (EC 1.10.3.2) are well-studied blue multicopper oxidases (type 1) found in fungi, plants, and microorganisms that catalyze the oxidation of a wide variety of phenolic and nonphenolic compounds, with concomitant reduction of molecular oxygen to water (8–10). The broad substrate range of laccases allows their use in numerous types of industrial and biotechnological fields, such as the paper, textile, and food industries (11–14). Known functions of fungal (15) and bacterial (16) laccases include roles in morphogenesis, sporulation, and pigmentation. Both bacterial and fungal forms can effect lignin catalysis and lignocellulose degradation, whereas plant laccases are involved in lignin synthesis in the plant cell wall.
Recently, a new laccase-like enzyme (Tfu1114) was characterized in the model cellulolytic bacterium Thermobifida fusca (17). Tfu1114 is able to catalyze the oxidation of several phenolic and nonphenolic lignin-related compounds, such as 2,6-dimethoxyphenol (2,6-DMP), veratryl alcohol (VA), and guaiacol. It is, however, unable to oxidize 2,2-azobis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS), a classical substrate for laccases, and only exhibits one copper atom per protein instead of the four found in canonical laccases. Interestingly, it was shown that Tfu1114 can significantly increase the hydrolysis of bagasse polysaccharides when combined with a cellulase or a xylanase from the same organism (17). This enhancement is linked to the oxidation of phenolic subunits of lignin and the generation of free radicals, which could create cleavage sites in the network structure of the lignin–carbohydrate complex, consequently exposing more polysaccharide fibers to the glycoside hydrolases (18).
In previous studies, our group has extensively studied the enzymes produced by T. fusca to degrade lignocellulose, particularly in the context of their conversion to the cellulosomal mode (19–22). Indeed, in this bacterium, like in the large majority of aerobic microorganisms, the cellulolytic system is based on the secretion of individual, free enzymes. By contrast, a restricted number of anaerobic bacteria perform efficient degradation of plant biomass through the secretion of unique, large multienzyme complexes termed cellulosomes (23–26). These complexes are based on a noncatalytic subunit called scaffoldin that binds to the insoluble substrate via a cellulose-specific carbohydrate-binding module (CBM). The scaffoldin also contains a set of subunit-binding modules termed cohesins that mediate the specific incorporation and organization of catalytic subunits through the dockerin, a complementary binding module carried by each enzymatic subunit (27, 28). The existence of cellulosome complexes in anaerobic fungi, which differ by the absence of true cohesins and dockerins, has also been reported (29, 30).
Previous studies have demonstrated the high efficiency of cellulosomes for the degradation of cellulose and hemicellulose, due to the spatial proximity of synergistically acting enzymes and to the locally increased concentrations in enzymes and substrate (31). However, cellulosomal systems discovered to date all appear to be lacking some of the key oxidative enzymes produced by the majority of aerobic lignocellulolytic microbes involved in the reduction of cellulose crystallinity and in lignin breakdown.
Two decades ago, the concept of designer cellulosomes was proposed, based on the modular nature of the cellulosome complex. Designer cellulosomes were devised as a tool to manipulate cellulosomal architecture and to incorporate noncellulosomal enzymes into these artificial complexes (32). This incorporation is accomplished by taking advantage of the high specificity between cohesins and dockerins originating from the same species. A chimeric scaffoldin, bearing several cohesin modules from multiple origins, can thus be designed, as well as chimeric enzymes bearing the corresponding dockerin, thus enabling self-assembly of a tailor-made enzymatic complex. The chimeric scaffoldin enables control of the location of each cellulolytic enzyme in the cellulosomal complex, as well as its molar ratio. This Lego-like complex has been used to test the effects of enzymatic compositions, the relative positioning of the enzymes within the complex, and their spatial proximity, which, together, generate the synergistic action of the cellulosomal components (33–35). The majority of the T. fusca biomass-degrading enzymes have been converted previously to the cellulosomal mode (19, 20, 36–41), including a pair of lytic polysaccharide monooxygenases (LPMOs) involved in the oxidative cleavage of crystalline cellulose fibers (22). Interestingly, in most cases, their inclusion into designer cellulosomes proved to be more efficient than the equivalent mixture of wild-type enzymes (20–22, 37, 40, 42).
In this study, we examined the conversion to the cellulosomal mode of the laccase-like enzyme Tfu1114 by fusion to a dockerin-xylanase chimera and the impact of the conversion on its enzymatic activity. This chimeric bifunctional enzyme was incorporated into a designer cellulosome alongside two cellulases and a xylanase, and was used to deconstruct a wheat straw substrate so as to assess the benefits of the simultaneous degradation by cellulosome action of the three components of lignocellulose (i.e., cellulose, hemicellulose, lignin).
Results
Conversion of the Selected Enzymes to the Cellulosomal Mode.
T. fusca enzymes involved in lignocellulose degradation have been extensively studied because they are industrially relevant due to their high thermostability, broad pH range, and high activity (19, 37, 42). In previous studies, the majority of these enzymes have been converted to the cellulosomal mode by fusion of a dockerin module, without adverse effects on their activity. In this study, we used two well-characterized T. fusca cellulases, the family 5 endoglucanase Cel5A and exoglucanase Cel48A, together with Clostridium thermocellum xylanase Xyn11V, and integrated them in a designer cellulosome. This combination of enzymatic activities was shown to be highly synergistic and efficient for the simultaneous hydrolysis of cellulose and hemicellulose (20–22, 37, 39). This complex was used as a base to evaluate the benefits of the incorporation in designer cellulosomes of a lignin-active enzyme: the recently characterized T. fusca laccase-like Tfu1114 (hereafter termed Lac).
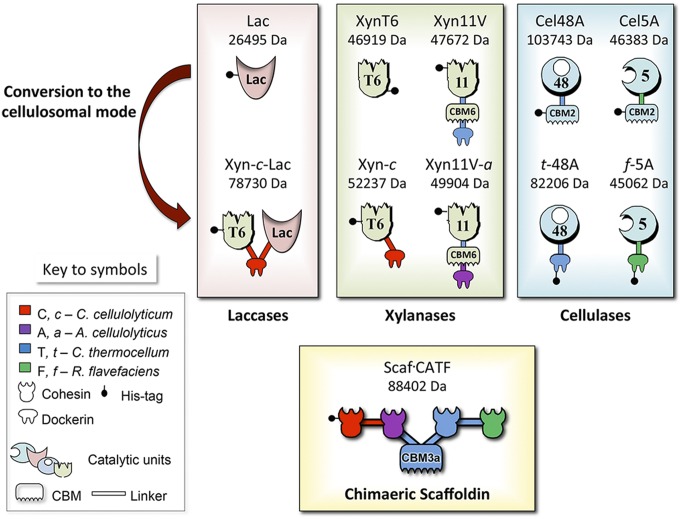
To convert T. fusca enzymes to the cellulosomal mode, dockerin domains originating from different bacterial species and showing different binding specificities were used. T. fusca Cel48A and Cel5A were fused at their N termini with dockerins originating from C. thermocellum and Ruminococcus flavefaciens, and termed t-48A and f-5A, respectively (39). In both cases, the original N-terminal CBM2 was replaced by the dockerin module. C. thermocellum xylanase Xyn11V was fused to an Acetivibrio cellulolyticus dockerin (termed a). This enzyme displays the same modular arrangement as T. fusca Xyn11A, a GH11 module, and a CBM2 (specific for xylan binding and cellulose to a lesser extent) (43), but it is advantageous over the latter because the recombinant form is expressed very efficiently in Escherichia coli expression systems. Similar to the chimeric form of T. fusca Xyn11A, the CBM was maintained in C. thermocellum Xyn11V to preserve enzymatic activity in designer cellulosomes as reported previously (39) and the dockerin module was added at the C terminus of the protein. The enzymatic activity of the resultant Xyn11V-a was comparable to the wild-type Xyn11V on xylan substrates (Table S1).
Table S1.
Specific activities of the recombinant enzymes on various xylan substrates
| Substrate | Xyn11V | Xyn11V-a |
| Birchwood xylan | 619 ± 13 | 490 ± 34 |
| Oat spelled xylan | 658 ± 1 | 921 ± 19 |
| Beechwood xylan | 977 ± 8 | 1,300 ± 20 |
Specific activities are measured as the catalytic activity (katal) that raises the rate of a chemical reaction by one mole per second per enzyme.
The conversion of the laccase Tfu1114 to the cellulosomal mode proved more difficult, because the chimeric enzyme bearing the dockerin from Clostridium cellulolyticum at either the N or C terminus failed to express in E. coli cells under various growth conditions. To overcome this barrier, a highly expressed GH10 xylanase, XynT6, from the thermophile Geobacillus stearothermophilus, was used as a solubility tag (44, 45) and fused at the N terminus of the dockerin-bearing laccase. The resulting bifunctional chimera was successfully expressed and purified. The XynT6 solubility tag was kept fused to the converted Tfu1114, because its thermophilic xylanase activity is expected to promote xylan degradation further within the framework of our experiments. As a control, a variant of XynT6 fused only to the dockerin (c) was also produced, and termed Xyn-c.
Construction of the Tetravalent Scaffoldin.
To drive the assembly of the above-described dockerin-bearing enzymes into a defined designer cellulosome, a tetravalent scaffoldin, Scaf⋅CATF, containing the appropriate matching cohesins was produced. It includes four different cohesin types, originating from C. cellulolyticum (C), A. cellulolyticus (A), C. thermocellum (T), and R. flavefaciens (F). To target the scaffoldin to the cellulose-containing substrate, a cellulose-binding CBM3a from C. thermocellum was incorporated between A and T, as shown in Fig. 1.
Fig. 1.
Schematic representation of the recombinant proteins used in this study. The numbers 5, 11, and 48 refer to the corresponding GH family (GH5, GH48, and GH11) of the catalytic module; uppercase characters (A, C, F, T) indicate the source of the cohesin module; and lowercase, italicized fonts (a, c, f, t) indicate the source of the dockerin module.
Functionality of the Chimeric Bifunctional Enzyme Xyn-c-Lac.
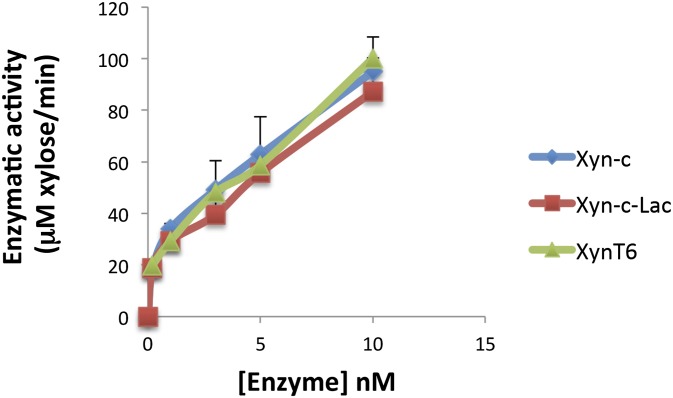
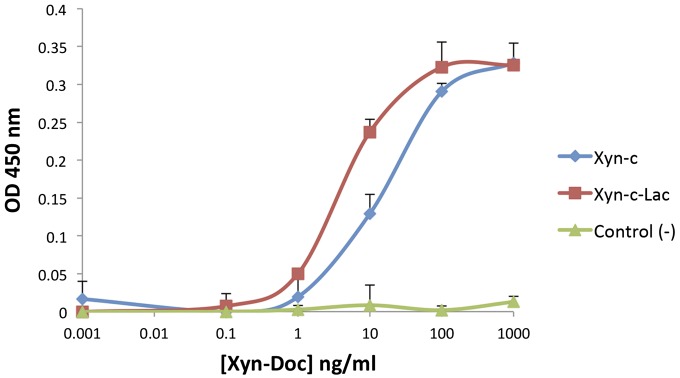
To assess whether the laccase and xylanase components of Xyn-c-Lac were affected by the translational fusion, enzymatic activities of the chimera were compared with the corresponding wild-type moieties. Xylanase activity was assessed by dinitrosalicylic acid (DNS) assay using beechwood xylan as a substrate and utilizing previously reported reactions parameters for the wild-type xylanase XynT6 (46). The xylanase moiety retains enzymatic activity, and its activity profile was similar to the wild-type and Xyn-c xylanases (Fig. S1). The functionality of the dockerin module was confirmed by affinity-based ELISA that measured the binding between the chimeric Xyn-c-Lac and the cohesin partner. The dockerin-bearing enzyme specifically bound the respective cohesin in an exclusive manner (Fig. S2).
Fig. S1.
Comparative xylanase activity of XynT6, Xyn-c, and Xyn-c-Lac on beechwood xylan at 50 °C. The assay was repeated twice, and the error bars indicate the SD from the mean of triplicate samples from one experiment.
Fig. S2.
Cohesin-dockerin binding measured by ELISA. Xyn-c and Xyn-c-Lac interacted with the cohesin 1 from C. cellulolyticum scaffoldin CipC as expected. As a negative control, Xyn-c-Lac failed to interact with C. thermocellum cohesin 3 from CipA. Error bars indicate the SD from the mean of triplicate samples from one experiment.
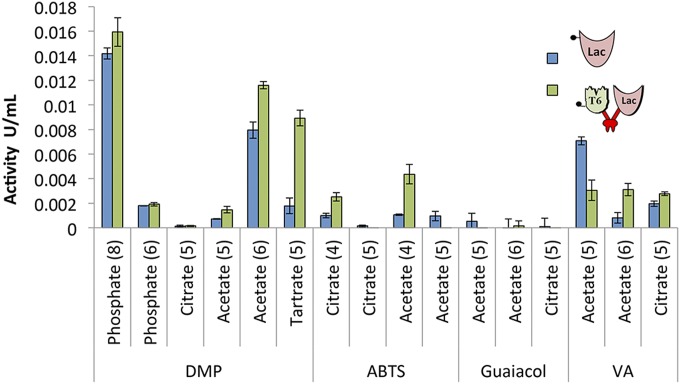
To ensure that the addition of xylanase and dockerin does not affect the laccase activity in the chimeric Xyn-c-Lac, its activity was assayed and compared with the activity of the wild-type laccase on a variety of laccase substrates. In the first characterization of the wild-type laccase (17), enzymatic assays were conducted with the 2,6-DMP substrate within a pH range of 6.5–9.0. The activity for oxidation was assayed by 15-min incubation of the enzymes at 50 °C with 20 mM ABTS, 2,6-DMP, guaiacol, and VA substrates in various buffers and pH conditions. As can be seen in Fig. 2, the addition of the xylanase and dockerin moieties to the chimeric Xyn-c-Lac did not affect its activity in comparison to the wild-type enzyme under most of the conditions examined. Both the chimeric enzyme and the wild-type enzyme exhibited the highest activity toward 2,6-DMP using phosphate buffer at pH 8. This result is in agreement with a previous study that characterized the wild-type enzyme activity (17). The substrate 2,6-DMP was oxidized by both the wild-type and chimeric enzymes in tartrate buffer (pH 5) and citrate buffer (pH 6). The second-highest oxidized substrate was VA. Surprisingly, the chimeric enzyme exhibited moderate activity for ABTS at pH 4, whereas the activity of the wild-type enzyme was very low in our experiments and negligible in previous studies (17). We identified only minor activity of both the wild-type and chimeric enzymes toward guaiacol. Because the optimal pH of the laccase varied with the substrate and buffering agent, and the T. fusca enzymes cellulases and xylanases in the designer cellulosome machinery are known to be active at pH ranges of 5–6 (20–22), we therefore elected pH 5 as the working pH in our experiments.
Fig. 2.
Laccase activity of the wild-type Lac and chimeric Xyn-c-Lac toward different substrates. Enzymatic activity toward 2,6-DMP, ABTS, guaiacol, and VA was determined with different buffers in various pH conditions (pH value is given in parentheses under each bar pair). The oxidation rate was calculated based on the extinction coefficient of the oxidized product of each substrate. Each reaction was performed three times. Error bars represent SDs.
Analysis of Designer Cellulosome Complex Formation.
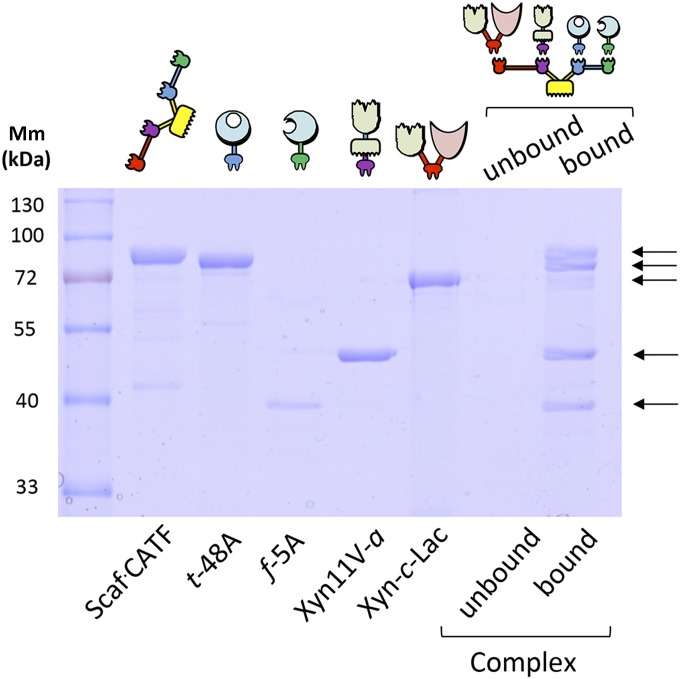
To examine the incorporation of the various components into the designer cellulosome complex, we used the ability of the complex to bind microcrystalline cellulose by virtue of its resident CBM to perform an affinity pull-down assay. The designer cellulosome complex was first incubated with cellobiose before its incubation with cellulose to prevent nonspecific binding of the catalytic modules to cellulose. The bound and unbound fractions were examined by SDS/PAGE. Dockerin-bearing enzymes that failed to interact properly with the matching cohesin on the scaffoldin would appear in the unbound fraction. The four enzymes were incorporated together at their optimal ratio into the scaffoldin. As seen in Fig. 3, complex formation seems to be complete because all five proteins appear in the bound fraction, whereas the amount of protein observed in the unbound fraction is negligible. In addition, the assembly of the complex and its components was further confirmed by nondenaturing PAGE electrophoresis (Fig. S3).
Fig. 3.
Assessment of cellulosome complex formation by cellulose pull-down. The designer cellulosome preparations were subjected to interaction with cellulose by using the ability of its resident CBM to bind microcrystalline cellulose. Bound and unbound fractions were examined by SDS/PAGE. Dockerin-bearing enzymes interacting properly with matching cohesins of the scaffoldin protein will appear in the bound fraction. Bands in the unbound fraction (none visible) indicate enzymes that failed to interact properly with the matching cohesins of the scaffoldin. Mm, molecular mass.
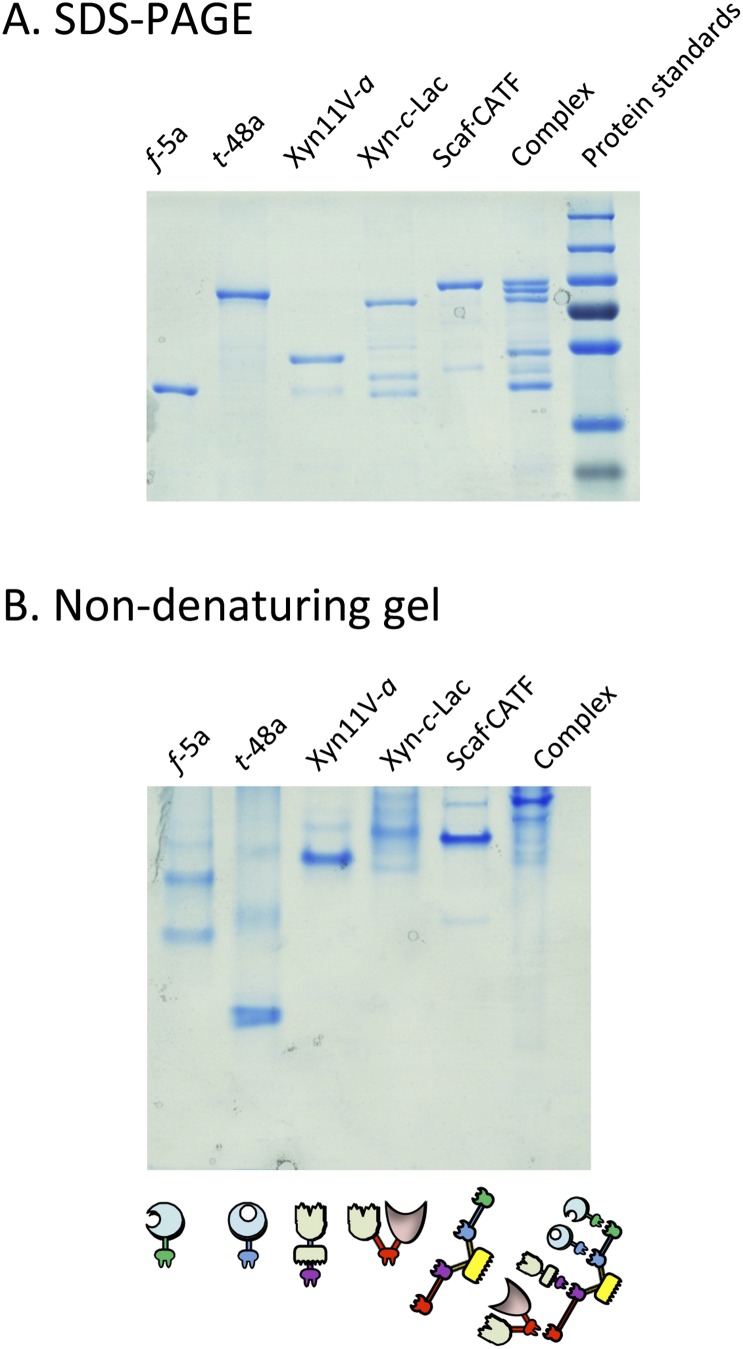
Fig. S3.
Verification of designer cellulosome complex formation by native gel. Electrophoretic mobility of components and assembled complexes on denaturing (A) and nondenaturing (B) gels. Equimolar concentrations of the chimeric enzymes and their matching scaffoldin were combined. Analysis of the mixture of the matching components by nondenaturing PAGE indicates their near-complete interaction as a single major band formed.
Degradation of Unpretreated Wheat Straw.
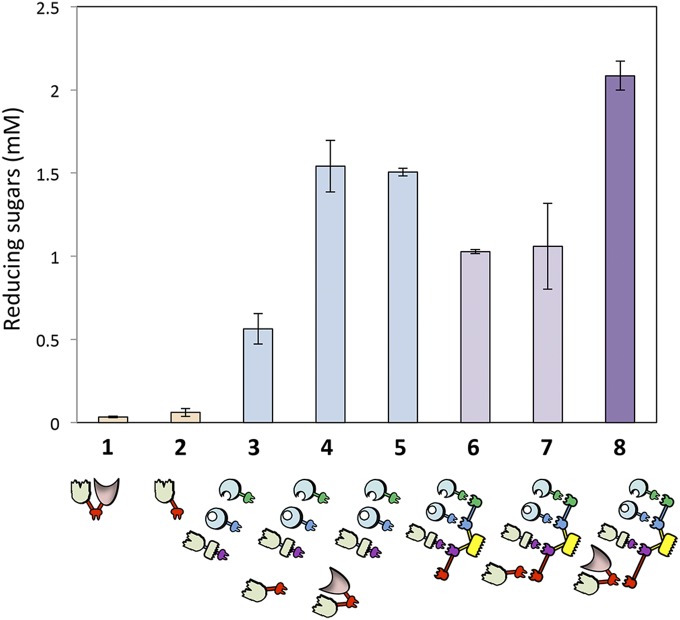
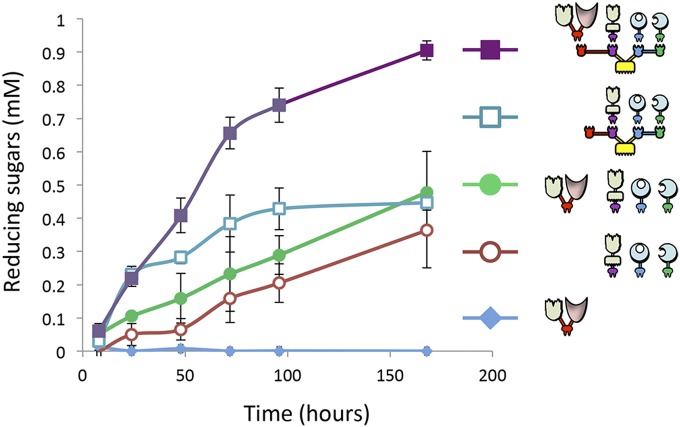
The Xyn-c-Lac and the Xyn-c produced negligible amounts of reducing sugars after incubation with unpretreated wheat straw. The combination of the three enzymes f-5A, t-48A, and Xyn11V-a into a designer cellulosome resulted in a 1.8-fold increase in enzymatic activity compared with the mixtures of the free enzymes (Fig. 4). Xyn-c-Lac served to enhance the amount of reducing sugars dramatically by 2.7-fold when combined with the free enzymes. This activity could, in fact, be attributed to the xylanase solubility tag (Xyn), because similar levels of reducing sugars were obtained while adding Xyn-c to the mixture of free enzymes. In contrast, the incorporation of the Xyn-c-Lac into the designer cellulosome machinery generated an increase of 1.4-fold in the amount of reducing sugar production compared with the mixture of the chimeric enzymes in their free state, and a twofold increase compared with the trivalent designer cellulosomes lacking the laccase (with or without the control Xyn-c) following 72 h of incubation with unpretreated wheat straw (Fig. 4). The fact that the combination of Xyn-c with the designer cellulosomal system did not serve to increase the amount of reducing sugar production demonstrates that the increase in activity caused by addition of Xyn-c-Lac to the designer cellulosome machinery can be attributed to the laccase moiety. Our results suggest a strong proximity effect between the laccase and the other enzymatic (glycoside hydrolase) partners and the necessity of the laccase to be located in the cellulosome complex to achieve efficient degradation (Fig. 4). Interestingly the Xyn-c enzyme contributed to the enzymatic activity in combination with the other enzymes in the free state only; as part of the designer cellulosome, Xyn-c reduced the activity compared with the free enzyme mixture (Fig. 4). Kinetics studies over 7 d of the tetravalent designer cellulosome bearing the Xyn-c-Lac enzyme compared with selected controls are shown in Fig. 5. After 7 d, the tetravalent designer cellulosome caused a 50% increase in the amount of reducing sugars in comparison to the trivalent designer cellulosome lacking the laccase (Xyn-c-Lac). Together, these results demonstrate the ability of laccase to enhance cellulolytic and hemicellulolytic degradation when integrated into the designer cellulosome machinery.
Fig. 4.
Comparative degradation of unpretreated wheat straw by laccase in tetravalent designer cellulosomes and free-enzyme systems. The unpretreated wheat straw substrate was incubated for 72 h at 50 °C. Bars: 1, bifunctional chimeric xylanase-tagged laccase (Xyn-c-Lac); 2, xylanase tag alone (Xyn-c); 3, three free dockerin-bearing GHs (Xyn11V-a, t-48A, and f-5A); 4, the three free GHs with the additional xylanase tag (Xyn-c); 5, the three free GHs with the addition of the xylanase-tagged laccase (Xyn-c-Lac); 6, the three scaffoldin-complexed GHs; 7, scaffoldin-complexed GHs + xylanase tag; 8, scaffoldin complexed GHs + laccase. Enzymes were used at 0.5 μM with a substrate concentration of 7 g/L. Reaction yields from 1 to 8 were as follows: 0.14%, 0.3%, 2.4%, 6.7%, 6.5%, 4.5%, 4.6%, and 9% using the predetermined maximum sugar release of 3.3 mmol of reducing sugars per gram of dry matter (57). Each reaction was performed three times. Error bars represent SDs.
Fig. 5.
Kinetics studies of unpretreated wheat straw hydrolysis by the trivalent and tetravalent designer cellulosomes (with or without Xyn-c-Lac) versus the free enzyme mixtures. Enzymes were used at 0.5 μM with a substrate concentration of 3.5 g/L. Each reaction was performed three times. Error bars represent SDs.
Discussion
Recent efforts have focused on the development of renewable alternatives to fossil fuels, and cellulosic biomass has great potential to contribute to the demand for liquid fuel (5, 47). The production of lignocellulosic biofuel initially involves the deconstruction of cell-wall polymers into simple sugars. This critical step has been studied extensively, although an efficient, inexpensive process has yet to be developed. Lignin acts as a physical barrier that restricts the access of hydrolytic enzymes to cellulose and hemicellulose components of the plant cell wall. Moreover, it has been shown that cellulases have a tendency to bind lignin in a nonreversible manner (48–51), thus limiting their availability in solution. Thermochemical pretreatment processes that are used for removal of the lignin component before cellulose degradation tend to be expensive, slow, and relatively inefficient. Furthermore, some of these pretreatments produce inhibitors that decrease the overall yield of the fermentation process (7). In this context, laccases have drawn much attention in past decades as an ecological alternative for many processes in the biotechnology industries (11–14). Laccases, which facilitate the degradation of lignin, can largely contribute to current efforts to overcome limitations of enzymatic degradation of lignocellulosic biomass. In parallel, several studies have shown that highly efficient plant cell-wall degradation can be achieved by the use of the designer cellulosome complexes (20, 21, 37, 52). Designer cellulosome technology, which harnesses the proximity effect of ordered hydrolytic enzymes to maximize their combined synergistic effects, was used in this work in combination with integration of a nonconventional enzyme that degrades lignin. The goal of this study was to assess whether this laccase could be converted to the cellulosomal mode and integrated efficiently into the designer cellulosome machinery, and whether it would enhance the degradation of lignocellulose.
For this purpose, the recently characterized laccase-like Tfu1114 from T. fusca, which exhibited synergistic activity in combination with hydrolytic enzymes on biomass, was selected (17). The effect of the Lac enzyme in combination with T. fusca endoglucanase Tfu_1074 (Cel6A) and endoxylanase Tfu_1213 (Xyn11A) on degrading cellulosic biomass was previously demonstrated (17). In this study, a mixture of T. fusca endo- and exo-acting cellulases, together with an endoxylanase, was selected for integration into the designer cellulosome complex alongside the laccase for maximizing their synergism. Interestingly, members of the family-6 glycoside hydrolase enzymes have never been found to be part of any known cellulosome system. However, endocellulase Tfu_1074 (Cel6A) was demonstrated to confer enhanced cellulose-degrading activity in simple designer cellulosomes (38, 53). In contrast, Tfu_0620 (Cel6B) is an exoglucanase, which was shown to cause an antiproximity effect in designer cellulosomes, and was therefore deemed incompatible with the cellulosomal mode (38). Therefore, in this study, we chose to work with the well-characterized family-48 exocellulase Cel48A, with GH family-48 enzymes being a major component in the natural secreted cellulosomes (54–57) together with the family-5 endocellulase Cel5A, whose synergistic effect with Cel48A has been demonstrated previously and which were extensively used in their dockerin-bearing chimeric forms as successful components of designer cellulosomes (20–22, 37, 58). Endoxylanase Tfu_1213 is part of the family-11 glycoside hydrolase enzymes. Therefore, C. thermocellum Xyn11V, which is also a thermophilic family-11 enzyme and has the same modular arrangement of T. fusca Xyn11A, was used as a suitable replacement (43), and its chimeric form was expressed very efficiently in E. coli.
As previously discussed, the insertion of noncellulosomal enzymes into a designer cellulosome complex is far from trivial (59). In most cases, when a free cellulolytic enzyme bears a CBM, the native CBM can be replaced with a dockerin, therefore preserving the overall modular architecture of the protein. However, in a protein that contains a single catalytic module and lacks any CBM, the dockerin can be added at either the N or C terminus via an appropriate linker regardless of any former knowledge of its effect on the functionality of the chimeric enzyme (catalytic module and dockerin). In this study, the conversion of the enzyme Lac into the cellulosomal mode was complex, because upon inclusion of the dockerin, the chimeric enzyme failed to be expressed in soluble form under various expression conditions. This issue was solved by the addition of the thermophilic G. stearothermophilus xylanase, which was previously shown to be expressed at high levels and in a soluble form; we were able to express the chimeric cellulosomal form of the laccase successfully. The resultant chimeric bifunctional enzyme, Xyn-c-Lac, retains both the xylanase and laccase activity, and its dockerin module incorporated properly into the designer cellulosome in combination with the cellulases Cel5A and Cel48A and the xylanase Xyn11V (Figs. 2 and 3 and Fig. S1).
We validated proper activity of both native Lac and the chimeric enzyme Xyn-c-Lac at pH 5–6 compatible with the cellulases and xylanase optimal pH condition. In addition, we observed activity of the laccase enzymes with the ABTS substrate at low pH, whereas in a previous study, the laccase failed to exhibit any activity on that substrate at pH 8 (17). In fact, the optimal pH range for laccases is highly variable and depends on the substrate tested; in that context, fungal laccases have also been reported to be active in the pH range of 3–5 on the ABTS substrate (12). Additional results on the other substrates were in concordance with the previous characterization efforts (17). By demonstrating the activity of the enzyme at low pH on different substrates, we established the possibility of combining the chimeric laccase into the designer cellulosome machinery in combination with other cellulases and xylanases.
In contrast to the results of Chen and coworkers (17), in our study, the addition of the laccase to the free dockerin-containing cellulases and xylanase failed to increase enzymatic activity. However, incorporation of the laccase into the designer cellulosome machinery, together with the cellulases and the xylanase, resulted in a twofold increase in the amount of reducing sugars compared with the trivalent designer cellulosomes lacking the laccase. Moreover, because the Xyn solubility tag was not effective in the cellulosomal mode [the activity of the cellulosome containing only Xyn-c was significantly lower than the mixture of free enzymes (Fig. 4, bar 7 versus bar 4)], it is likely that an enzyme tag that would contribute to the activity of the cellulosomal complex would further increase the proximity effect of the laccase with the glycoside hydrolase components.
Successful incorporation of a laccase into a designer cellulosome offers new potential for enzymatic degradation of lignocellulosic biomass and can contribute to future efforts for production of alternative fuels. The incorporation of nonconventional enzymes into designer cellulosomes, such as β-glucosidases, LPMOs, and expansins, has proved an efficient strategy for enhancement of biomass degradation by glycoside hydrolases (22, 60–62). These accessory enzymes act in concert with glycoside hydrolases, either by releasing product inhibition or by acting directly on the substrate. Designer cellulosomes that have contained either LPMOs or the laccase have enabled combination of both hydrolytic and oxidative activities in the same reaction, which would not have been possible in natural cellulosomes, which are restricted to anaerobic environments.
The nonconventional enzymes are part of the miniature toolbox that can serve to enhance the value of designer cellulosome technology and contribute synergistically to biomass degradation, either by assisting the glycoside hydrolases in their methodic degradation of polysaccharide substrates or by attacking different components of the plant cell-wall matrix.
Materials and Methods
Cloning.
Recombinant laccases were cloned by using a two-step restriction-free procedure (63). The plasmids and primers used for the restriction-free cloning are listed in Table S2. The other enzymes and scaffoldin were cloned using restriction enzymes. Additional details can be found in SI Cloning.
Table S2.
Plasmids and primers used for the restriction-free cloning of the recombinant enzymes
| Vector name | Forward primer (5′–3′) | Reverse primer (5′–3′) | Primary PCR template | Secondary PCR template |
| pETduet-Lac | tataccatggtgacgggcaccgt | ggtgctcgagtgggaccctccag | T. fusca YX genomic DNA | pETduet |
| pETduet-c-Lac | cggttcgccggatatgtctggagggtcccaactagtcctgtaattgtat | ggtggcagcagcctaggttaattaagctgcttaaggtagcttacttacc | pET21a-Xyl43-c (21) | pETduet-Lac |
| pETduet-Xyn-c-Lac | atgggcagcagccatcaccatcatcaccacaagaatgcagattcctatgcgaaaaaacct | accctggaagtacaggttttcacccgcggatttgtggtcgataatagcccaatatgcggg | pET9d-Xyn-c (44) | pETduet-c-Lac |
| pET28a t-48A | TATACCATGGcacatcaccatcaccatcacgcagttgaaagcagttccac | gtcacctcggccgagtcgtggccgGGTACCtctgtaataatgcggagtat | C. thermocellum ATCC genomic DNA | pET28a b-48A (37) |
Expression and Purification.
All recombinant proteins were expressed in E. coli BL21(DE3), grown in autoinduction media (64), and supplemented with the appropriate antibiotics. Protein purification was performed by immobilized metal-ion affinity chromatography on a nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen). Scaf⋅CATF was purified on phosphoric acid-swollen cellulose, 7.5 mg⋅mL−1 (pH 7), as previously described (36), followed by an additional purification step using the Ni-NTA column.
Designer Cellulosome Assembly.
Designer cellulosome assembly was examined by both affinity pull-down assay and electrophoretic mobility in native and denaturing conditions as described earlier (34).
Cohesin–Dockerin Interaction.
The procedure of Barak et al. (44) was followed to test the binding activity of the cohesin and dockerin components of the designer cellulosome.
Enzymatic Activity Assays.
The laccase activity assay was carried out in vertical shaker incubator at 50 °C for 15 min, using 1.5 μM enzyme and 2 mM substrate (17). Xylanase activity was assayed on xylan substrates by the DNS method (65). Hatched wheat straw [blended at 0.2–0.8 mm and containing 32% (wt/wt) cellulose, 30% hemicellulose, and 21% lignin (21)] was washed for 24 h to remove residual reducing sugars. The enzymes and the scaffoldin at optimal molar ratios (as determined in Designer Cellulosome Assembly) were incubated for 2 h at 37 °C with 20 mM CaCl2. Then, the complex or enzyme mixtures (at 0.5 μM, ∼25 mg of protein per gram of wheat straw for the largest complex) were assayed for wheat straw degradation in a 200-μL reaction [50 mM acetate buffer (pH 5.0), 12 mM CaCl2, 2 mM EDTA, 7 g/L wheat straw]. Reaction mixtures were incubated in a vertical shaker incubator for 72 h at 50 °C. All assays were performed in triplicate. To evaluate the concentration of reducing sugars, the DNS method was used (65). The 7-d activity assay was conducted similarly with 3.5 g/L unpretreated wheat straw (instead of 7 g/L, ∼49 mg of protein per gram of wheat straw for the largest complex).
SI Cloning
The plasmid pETduet-Lac was obtained as previously described (17). The pETduet-c-Lac plasmid was obtained by inserting a sequence coding for the dockerin module from Clostridium cellulolyticum Cel5A (c) (40), amplified from the vector pET21a-Xyl43-c (21), in pETDuet plasmid (Novagen, Inc.). The Geobacillus stearothermophilus xylanase was obtained from a previous study (44) and was inserted at the 5′ of the c-Lac coding sequence to produce the plasmid pETduet-Xyn-c-Lac. Similarly, the t-48A chimera was obtained by inserting a sequence coding for the dockerin module from Clostridium thermocellum XynZ (t) amplified from strain American Type Culture Collection (ATCC) 27405 genomic DNA to replace the dockerin from Bacteroides cellulosovens in pET28a b-48A (37).
The Xyn11V-a chimera, was constructed by ligating sequentially the two modules into the linearized form of pET21a (Novagen, Inc.). Xyn11V was first amplified using C. thermocellum ATCC 27405 genomic DNA utilizing the primers 5′-ATTATGCATATGCACCATCACCATCACCACGATGTAGTAATTACGTCAAACCAGAC-3′ and 5′-ATTCTACTCGAGATTATCACTAGTAGGTGTAGGTGTAGGATTTACA-3′ (NdeI, SpeI, and XhoI sites are shown in boldface) and inserted into pET21a linearized with NcoI and XhoI. Then, the dockerin from scaffoldin B was cloned from Acetivibrio cellulolyticus genomic DNA using the primers 5′-CTACAACTAGTACTACAACACCAACGCCTAAAT-3′ and 5′-GGTGGTCTCGAGTTATTCTTCTTTCTCTTCAA-3′ (SpeI and XhoI sites are shown in boldface) and inserted into pXyn11V linearized with SpeI and XhoI. Wild-type enzymes (XynT6, Cel5A, and Cel48A) were cloned as described previously (45, 66, 67). The plasmid encoding the chimeric glycoside hydrolase pET28a f-5A was obtained from previous studies (39). All of the enzyme constructs were designed to contain a His tag for subsequent purification.
The recombinant scaffoldin pScaf⋅CATF was derived from pETscaf6 (52). pETscaf6, originally pET9d- (Novagen, Inc.), is composed of cohesin C (cohesin 1 from scaffoldin C from C. cellulolyticum), CBM-T (CBM3a and cohesin 3 from the cellulosomal scaffoldin subunit C. thermocellum YS), and cohesin F (cohesin 1 from Ruminococcus flavefaciens strain 17 scaffoldin B). To construct Scaf⋅CATF, cohesin A (cohesin 3 from A. cellulolyticus scaffoldin C) was amplified from the A. cellulolyticus genomic DNA using 5′-TATCGGGTACCGCGGCCGCATTTACAGGTTGACATTGGAAGT-3′ and 5′-TACGTGGTACCGATGCAATTACCTCAATTTT-3′ primers (KpnI and KpnI sites are shown in boldface) and ligated (T4 DNA ligase; Fermentas UAB) to pETscaf6-linearized using KpnI (all of the restriction enzymes were purchased from New England Biolabs, Inc.). Correct orientation of the cloned fragment was checked by restriction analysis of the resulting plasmid, pScaf⋅CATF.
PCR assays were performed using Phusion High Fidelity DNA polymerase (New England Biolabs, Inc.), PCR products were purified using a HiYieldTM Gel/PCR Fragments Extraction Kit (Real Biotech Corporation), and plasmids were extracted using a Qiagen Miniprep Kit (Qiagen). Plasmids were maintained and propagated in Escherichia coli DH5α.
Acknowledgments
We thank Rachel Salama and Yuval Shoham (Technion) for the valuable help with Dionex analysis of the soluble sugars. We appreciate the support of the European Union (Area NMP.2013.1.1-2: Self-assembly of naturally occurring nanosystems: CellulosomePlus Project 604530), the Israel Science Foundation (Grant 1349), and the Israeli Center of Research Excellence (I-CORE Center Grant 152/11). E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608012113/-/DCSupplemental.
References
- 1.Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol. 2009;60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 2.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311(5760):484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 3.Lynd LR, et al. How biotech can transform biofuels. Nat Biotechnol. 2008;26(2):169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 4.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454(7206):841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 5.Himmel ME, Bayer EA. Lignocellulose conversion to biofuels: Current challenges, global perspectives. Curr Opin Biotechnol. 2009;20(3):316–317. doi: 10.1016/j.copbio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Jönsson LJ, Martín C. Pretreatment of lignocellulose: Formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol. 2016;199:103–112. doi: 10.1016/j.biortech.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Kudanga T, Le Roes-Hill M. Laccase applications in biofuels production: Current status and future prospects. Appl Microbiol Biotechnol. 2014;98(15):6525–6542. doi: 10.1007/s00253-014-5810-8. [DOI] [PubMed] [Google Scholar]
- 8.Pollegioni L, Tonin F, Rosini E. Lignin-degrading enzymes. FEBS J. 2015;282(7):1190–1213. doi: 10.1111/febs.13224. [DOI] [PubMed] [Google Scholar]
- 9.Claus H. Laccases: Structure, reactions, distribution. Micron. 2004;35(1-2):93–96. doi: 10.1016/j.micron.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 10.Giardina P, et al. Laccases: A never-ending story. Cell Mol Life Sci. 2010;67(3):369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pezzella C, Guarino L, Piscitelli A. How to enjoy laccases. Cell Mol Life Sci. 2015;72(5):923–940. doi: 10.1007/s00018-014-1823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madhavi V, Lele SS. Laccase: Properties and applications. BioResources. 2009;4:1694–1717. [Google Scholar]
- 13.Riva S. Laccases: Blue enzymes for green chemistry. Trends Biotechnol. 2006;24(5):219–226. doi: 10.1016/j.tibtech.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Virk AP, Sharma P, Capalash N. Use of laccase in pulp and paper industry. Biotechnol Prog. 2012;28(1):21–32. doi: 10.1002/btpr.727. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, et al. Lignin engineering through laccase modification: A promising field for energy plant improvement. Biotechnol Biofuels. 2015;8:145. doi: 10.1186/s13068-015-0331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins LO, Durão P, Brissos V, Lindley PF. Laccases of prokaryotic origin: Enzymes at the interface of protein science and protein technology. Cell Mol Life Sci. 2015;72(5):911–922. doi: 10.1007/s00018-014-1822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C-Y, Hsieh ZS, Cheepudom J, Yang CH, Meng M. A 24.7-kDa copper-containing oxidase, secreted by Thermobifida fusca, significantly increasing the xylanase/cellulase-catalyzed hydrolysis of sugarcane bagasse. Appl Microbiol Biotechnol. 2013;97(20):8977–8986. doi: 10.1007/s00253-013-4727-y. [DOI] [PubMed] [Google Scholar]
- 18.Du X, et al. Understanding pulp delignification by laccase-mediator systems through isolation and characterization of lignin-carbohydrate complexes. Biomacromolecules. 2013;14(9):3073–3080. doi: 10.1021/bm4006936. [DOI] [PubMed] [Google Scholar]
- 19.Caspi J, et al. Conversion of Thermobifida fusca free exoglucanases into cellulosomal components: Comparative impact on cellulose-degrading activity. J Biotechnol. 2008;135(4):351–357. doi: 10.1016/j.jbiotec.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Stern J, Moraïs S, Lamed R, Bayer EA. Adaptor scaffoldins: An original strategy for extended designer cellulosomes, inspired from nature. MBio. 2016;7(2):e00083–e16. doi: 10.1128/mBio.00083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morais S, Morag E, Barak Y, et al. Deconstruction of lignocellulose into soluble sugars by native and designer cellulosomes. MBio. 2012;3(6):e00508-12. doi: 10.1128/mBio.00508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arfi Y, Shamshoum M, Rogachev I, Peleg Y, Bayer EA. Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc Natl Acad Sci USA. 2014;111(25):9109–9114. doi: 10.1073/pnas.1404148111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamed R, Setter E, Bayer EA. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983;156(2):828–836. doi: 10.1128/jb.156.2.828-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayer EA, Belaich J-P, Shoham Y, Lamed R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 25.Bayer EA, Lamed R, White BA, Flint HJ. From cellulosomes to cellulosomics. Chem Rec. 2008;8(6):364–377. doi: 10.1002/tcr.20160. [DOI] [PubMed] [Google Scholar]
- 26.Bayer EA, Shoham Y, Lamed R. Lignocellulose-decomposing bacteria and their enzyme systems. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Springer; Berlin Heidelberg: 2013. pp. 215–265. [Google Scholar]
- 27.Stahl SW, et al. Single-molecule dissection of the high-affinity cohesin-dockerin complex. Proc Natl Acad Sci USA. 2012;109(50):20431–20436. doi: 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pagès S, et al. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: Prediction of specificity determinants of the dockerin domain. Proteins. 1997;29(4):517–527. [PubMed] [Google Scholar]
- 29.Nagy T, Tunnicliffe RB, Higgins LD, et al. Characterization of a Double dockerin from the cellulosome of the anaerobic fungus Piromyces equi. J Mol Biol. 2007;373(3):612–622. doi: 10.1016/j.jmb.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Solomon KV, et al. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science. 2016;351(6278):1192–1195. doi: 10.1126/science.aad1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Himmel ME, et al. Microbial enzyme systems for biomass conversion: Emerging paradigms. Biofuels. 2010;1:323–341. [Google Scholar]
- 32.Bayer EA, Morag E, Lamed R. The cellulosome--a treasure-trove for biotechnology. Trends Biotechnol. 1994;12(9):379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 33.Mingardon F, Chanal A, Tardif C, Bayer EA, Fierobe HP. Exploration of new geometries in cellulosome-like chimeras. Appl Environ Microbiol. 2007;73(22):7138–7149. doi: 10.1128/AEM.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stern J, et al. Significance of relative position of cellulases in designer cellulosomes for optimized cellulolysis. PLoS One. 2015;10(5):e0127326. doi: 10.1371/journal.pone.0127326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vazana Y, et al. A synthetic biology approach for evaluating the functional contribution of designer cellulosome components to deconstruction of cellulosic substrates. Biotechnol Biofuels. 2013;6(1):182. doi: 10.1186/1754-6834-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caspi J, et al. Thermobifida fusca family-6 cellulases as potential designer cellulosome components. Biocatalysis Biotransform. 2006;24:3–12. [Google Scholar]
- 37.Caspi J, et al. Effect of linker length and dockerin position on conversion of a Thermobifida fusca endoglucanase to the cellulosomal mode. Appl Environ Microbiol. 2009;75(23):7335–7342. doi: 10.1128/AEM.01241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caspi J, et al. Thermobifida fusca exoglucanase Cel6B is incompatible with the cellulosomal mode in contrast to endoglucanase Cel6A. Syst Synth Biol. 2010;4(3):193–201. doi: 10.1007/s11693-010-9056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moraïs S, et al. Contribution of a xylan-binding module to the degradation of a complex cellulosic substrate by designer cellulosomes. Appl Environ Microbiol. 2010;76(12):3787–3796. doi: 10.1128/AEM.00266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moraïs S, et al. Assembly of xylanases into designer cellulosomes promotes efficient hydrolysis of the xylan component of a natural recalcitrant cellulosic substrate. MBio. 2011;2(6):1–11. doi: 10.1128/mBio.00233-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moraïs S, et al. Paradigmatic status of an endo- and exoglucanase and its effect on crystalline cellulose degradation. Biotechnol Biofuels. 2012;5(1):78. doi: 10.1186/1754-6834-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraïs S, et al. Enhanced cellulose degradation by nano-complexed enzymes: Synergism between a scaffold-linked exoglucanase and a free endoglucanase. J Biotechnol. 2010;147(3-4):205–211. doi: 10.1016/j.jbiotec.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 43.Fernandes AC, et al. Homologous xylanases from Clostridium thermocellum: Evidence for bi-functional activity, synergism between xylanase catalytic modules and the presence of xylan-binding domains in enzyme complexes. Biochem J. 1999;342(Pt 1):105–110. [PMC free article] [PubMed] [Google Scholar]
- 44.Barak Y, et al. Matching fusion protein systems for affinity analysis of two interacting families of proteins: The cohesin-dockerin interaction. J Mol Recognit. 2005;18(6):491–501. doi: 10.1002/jmr.749. [DOI] [PubMed] [Google Scholar]
- 45.Lapidot A, Mechaly A, Shoham Y. Overexpression and single-step purification of a thermostable xylanase from Bacillus stearothermophilus T-6. J Biotechnol. 1996;51(3):259–264. doi: 10.1016/s0168-1656(96)01604-5. [DOI] [PubMed] [Google Scholar]
- 46.Khasin A, Alchanati I, Shoham Y. Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl Environ Microbiol. 1993;59(6):1725–1730. doi: 10.1128/aem.59.6.1725-1730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson DB. Cellulases and biofuels. Curr Opin Biotechnol. 2009;20(3):295–299. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Heiss-Blanquet S, Zheng D, Lopes Ferreira N, Lapierre C, Baumberger S. Effect of pretreatment and enzymatic hydrolysis of wheat straw on cell wall composition, hydrophobicity and cellulase adsorption. Bioresour Technol. 2011;102(10):5938–5946. doi: 10.1016/j.biortech.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Pareek N, Gillgren T, Jönsson LJ. Adsorption of proteins involved in hydrolysis of lignocellulose on lignins and hemicelluloses. Bioresour Technol. 2013;148:70–77. doi: 10.1016/j.biortech.2013.08.121. [DOI] [PubMed] [Google Scholar]
- 50.Le Costaouëc T, Pakarinen A, Várnai A, Puranen T, Viikari L. The role of carbohydrate binding module (CBM) at high substrate consistency: Comparison of Trichoderma reesei and Thermoascus aurantiacus Cel7A (CBHI) and Cel5A (EGII) Bioresour Technol. 2013;143:196–203. doi: 10.1016/j.biortech.2013.05.079. [DOI] [PubMed] [Google Scholar]
- 51.Kumar L, Arantes V, Chandra R, Saddler J. The lignin present in steam pretreated softwood binds enzymes and limits cellulose accessibility. Bioresour Technol. 2012;103(1):201–208. doi: 10.1016/j.biortech.2011.09.091. [DOI] [PubMed] [Google Scholar]
- 52.Fierobe H-P, et al. Action of designer cellulosomes on homogeneous versus complex substrates: Controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J Biol Chem. 2005;280(16):16325–16334. doi: 10.1074/jbc.M414449200. [DOI] [PubMed] [Google Scholar]
- 53.Moraïs S, Shterzer N, Lamed R, Bayer EA, Mizrahi I. A combined cell-consortium approach for lignocellulose degradation by specialized Lactobacillus plantarum cells. Biotechnol Biofuels. 2014;7:112. doi: 10.1186/1754-6834-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morag E, Halevy I, Bayer EA, Lamed R. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J Bacteriol. 1991;173(13):4155–4162. doi: 10.1128/jb.173.13.4155-4162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zverlov VV, Kellermann J, Schwarz WH. Functional subgenomics of Clostridium thermocellum cellulosomal genes: Identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics. 2005;5(14):3646–3653. doi: 10.1002/pmic.200401199. [DOI] [PubMed] [Google Scholar]
- 56.Artzi L, Morag E, Barak Y, Lamed R, Bayer EA. Clostridium clariflavum: Key cellulosome players are revealed by proteomic analysis. MBio. 2015;6(3):e00411–e00415. doi: 10.1128/mBio.00411-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ravachol J, et al. Combining free and aggregated cellulolytic systems in the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Biotechnol Biofuels. 2015;8:114. doi: 10.1186/s13068-015-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moraïs S, et al. Cellulase-xylanase synergy in designer cellulosomes for enhanced degradation of a complex cellulosic substrate. MBio. 2010;1(5):e00285–e10. doi: 10.1128/mBio.00285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vazana Y, Moraïs S, Barak Y, et al. Designer cellulosomes for enhanced hydrolysis of cellulosic substrates. Methods Enzymol. 2012;510:249–252. doi: 10.1016/B978-0-12-415931-0.00023-9. [DOI] [PubMed] [Google Scholar]
- 60.Gefen G, Anbar M, Morag E, Lamed R, Bayer EA. Enhanced cellulose degradation by targeted integration of a cohesin-fused β-glucosidase into the Clostridium thermocellum cellulosome. Proc Natl Acad Sci USA. 2012;109(26):10298–10303. doi: 10.1073/pnas.1202747109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen C, et al. Integration of bacterial expansin-like proteins into cellulosome promotes the cellulose degradation. Appl Microbiol Biotechnol. 2016;100(5):2203–2212. doi: 10.1007/s00253-015-7071-6. [DOI] [PubMed] [Google Scholar]
- 62.Artzi L, Morag E, Shamshoum M, Bayer EA. Cellulosomal expansin: Functionality and incorporation into the complex. Biotechnol Biofuels. 2016;9:61. doi: 10.1186/s13068-016-0474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Unger T, Jacobovitch Y, Dantes A, Bernheim R, Peleg Y. Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J Struct Biol. 2010;172(1):34–44. doi: 10.1016/j.jsb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 64.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 65.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Biochem. 1959;31:426–428. [Google Scholar]
- 66.Ghangas GS, Wilson DB. Expression of a Thermomonospora fusca cellulase gene in Streptomyces lividans and Bacillus subtilis. Appl Environ Microbiol. 1987;53(7):1470–1475. doi: 10.1128/aem.53.7.1470-1475.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irwin DC, Zhang S, Wilson DB. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur J Biochem. 2000;267(16):4988–4997. doi: 10.1046/j.1432-1327.2000.01546.x. [DOI] [PubMed] [Google Scholar]