Significance
Box C/D RNAs are a large family of noncoding RNAs that guide 2′-O-methylation of RNAs. These RNAs associate with three or four proteins into C/D ribonucleoproteins (RNPs). The guide region of C/D RNAs is variable in length, particularly in eukaryotes, and by prediction, it can form 10–21 bp with substrates. Crystallographic and biochemical analyses revealed that the guide recognizes only a maximum of 10 nt in a substrate. Longer guide–substrate duplexes need to be unwound to fit into a size-limiting protein channel for modification. Our study reveals an aspect of the substrate recognition mechanism of C/D RNA. This mechanism is incompatible with the RNA-swapped model for dimeric C/D RNP.
Keywords: crystal structure, box C/D snoRNA, RNA modification, RNA–protein complex
Abstract
Box C/D RNAs guide site-specific 2′-O-methylation of RNAs in archaea and eukaryotes. The spacer regions between boxes C to D′ and boxes C′ to D contain the guide sequence that can form a stretch of base pairs with substrate RNAs. The lengths of spacer regions and guide-substrate duplexes are variable among C/D RNAs. In a previously determined structure of C/D ribonucleoprotein (RNP), a 12-nt-long spacer forms 10 bp with the substrate. How spacers and guide–substrate duplexes of other lengths are accommodated remains unknown. Here we analyze how the lengths of spacers and guide-substrate duplexes affect the modification activity and determine three structures of C/D RNPs assembled with different spacers and substrates. We show that the guide can only form a duplex of a maximum of 10 bp with the substrate during modification. Slightly shorter duplexes are tolerated, but longer duplexes must be unwound to fit into a capped protein channel for modification. Spacers with <12 nucleotides are defective, mainly because they cannot load the substrate in the active conformation. For spacers with >12 nucleotides, the excessive unpaired sequences near the box C/C′ side are looped out. Our results provide insight into the substrate recognition mechanism of C/D RNA and refute the RNA-swapped model for dimeric C/D RNP.
Box C/D RNAs are a large family of noncoding RNAs conserved in archaea and eukaryotes (1–3). The majority of these RNAs function as guides in site-specific 2′-O-methylation of ribosomal RNAs (rRNAs) and small nuclear RNAs (4–7). A few special members, including U3 and U14, small nucleolar RNAs are required for pre-rRNA processing and ribosome assembly. One of the most abundant modifications in rRNAs, 2′-O-methylated nucleotides cluster in functionally important regions and are believed to fine-tune ribosome structure and function (8–11).
Box C/D RNAs have a bipartite structure and contain the C (RUGAUGA; R is a purine) and D (CUGA) box motifs at two ends and the related C′ and D′ box motifs in the internal regions. Each pair of boxes C and D combine into a characteristic kink turn (K-turn) or K-loop structure (12–15). The spacer regions located between boxes D and C′ (D spacer) and between boxes D′ and C (D′ spacer) harbor the guide sequence. The guide sequence can potentially form 10–21 bp with complementary sequences in substrate RNAs and select the fifth nucleotide apart from box D′/D for modification (5).
To form a functional ribonucleoprotein (RNP) enzyme, each C/D RNA associates with four core proteins [Snu13, fibrillarin (Nop1 in yeast), Nop56, and Nop58] in eukaryotes (16–23) and with three proteins (L7Ae, fibrillarin, and Nop5) in archaea (24). The homologous eukaryotic Nop56 and Nop58 proteins are replaced by a single protein, Nop5, in archaea. Fibrillarin is the catalyst that transfers a methyl group from S-adenosylmethionine to the target ribose (21, 25). The RNA-guided methylation activity has been reconstituted for archaeal C/D RNPs (24), but not yet for eukaryotic complexes (26).
Structural studies have revealed the organization of archaeal C/D RNPs. Nop5 dimerizes through its central coiled-coil domain and binds fibrillarin with its N-terminal domain (NTD) (27). The K-turn/K-loop structure of C/D RNA is sandwiched between L7Ae and the C-terminal domain (CTD) of Nop5 (28, 29). The linker between the NTD and the coiled-coil domain of Nop5 is flexible, allowing fibrillarin to access the bound substrate in a dynamic manner (27, 29–31).
We have previously determined a crystal structure of substrate-bound C/D RNP in the active conformation (30). The structure illustrates the organization of a monomeric C/D RNP (mono-RNP), in which a single bipartite C/D RNA is anchored by its two K-turn motifs onto the Nop5 dimer platform. In the structure, the spacer region of the crystallized C/D RNA, previously known as CD45, is 12 nt long and forms a 10-bp duplex with the substrate. The guide-substrate duplex is placed on the dimerized coiled-coil domains of Nop5 and capped at two ends by the CTDs of Nop5. The spacer sequence located between boxes D and C′ and boxes D′ and C is of variable length in C/D RNAs. In archaeal C/D RNAs, the spacer is most frequently 12 nt long (32); however, in eukaryotes, spacer length varies widely and can potentially form 10–21 bp with the substrate (5, 32, 33). This raises an important question of how C/D RNPs can accommodate differently sized spacers and guide-substrate duplexes while still maintaining the specificity of modification.
In vitro reconstitution of archaeal C/D RNPs also yielded a dimeric RNP (di-RNP) (30, 34). The structural organization and biological relevance of di-RNP is currently a matter of debate. C/D RNA has proposed to be swapped between two different protein complexes in the di-RNP (34–38), rather than associating with the same Nop5 dimer as shown in the mono-RNP structure (30). It also has been argued that the RNA-swapped di-RNP model is advantageous for accommodating long guide-substrate duplexes compared with the mono-RNP model.
In this study, we investigated the mechanism by which C/D RNPs recognize differently sized spacers and guide-substrate duplexes. We varied the length of spacers and guide-substrate duplexes and examined how the modification activity of C/D RNP was affected. We also determined three additional structures of C/D RNPs assembled with different spacers and substrates. Contrary to what was previously thought, we found that C/D RNAs recognize only a maximum of 10 nt of a substrate. The protein channel that accommodates the guide-substrate duplex is rather rigid and restricts the size of the duplex.
Results
C/D RNP Activities Are Affected by the Length of Spacers and Guide-Substrate Duplexes.
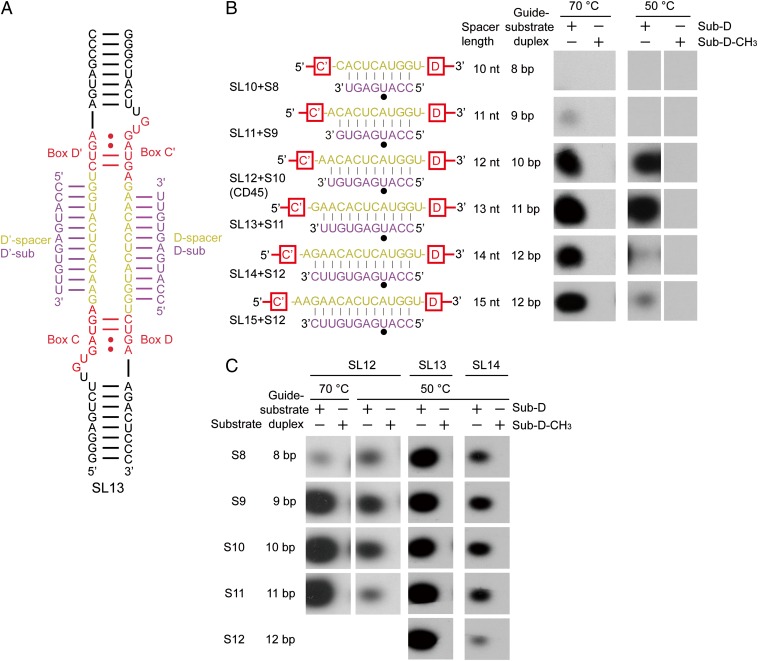
We first asked how the modification activity of C/D RNP is affected by the length of spacers and guide-substrate duplexes. We varied the spacer length by deleting or inserting nucleotides at the box C/C′ side based on a two-piece C/D RNA backbone (Fig. 1 A and B). A series of C/D RNAs was constructed with spacer lengths (SL) ranging from 10–15 nt. For clarity, the constructed C/D RNAs are named based on their spacer length (SL10–SL15), and the CD45 RNA is renamed SL12. These C/D RNAs were assembled with purified recombinant proteins Nop5, fibrillarin, and L7Ae from Sulfolobus solfataricus (24, 30). The modification activities of C/D RNPs were measured on cognate substrates at multiple-turnover conditions (30-fold excess of substrate over enzyme) by monitoring incorporation of 3H-methyl.
Fig. 1.
C/D RNP activities depend on the length of spacers and guide-substrate duplexes. (A) Secondary structure of the two-piece C/D RNA SL13. Other C/D RNAs differ only in the spacer regions. Boxes C, D, C′, and D′ are shown in red, the spacer regions are in yellow, and the bound substrates are in purple. (B and C) Activity of C/D RNPs. C/D RNPs were reconstituted with guide RNAs SL10–SL15 (1 μM) and incubated with substrates S8–S12 or their premethylated versions (30 μM), [methyl-3H] SAM, and cold SAM (30 μM) at 70 °C or 50 °C for 20 min. RNAs were separated by denaturing PAGE and visualized by 3H autoradiography. Base pairing interactions between the guide and the substrate are displayed. The target site is marked by a black circle.
The SL12 RNA was highly active in guiding modification of a 10-nt substrate S10 (Fig. 1B), as shown previously (30). No modification was observed for the substrate premethylated at the target site, indicating that the modification was site-specific. The activity was significantly reduced when the spacer and the substrate were both shortened by 1 nt (SL11 + S9), and was completely abolished when they were further shortened by 1 nt (SL10 + S8). The loss of activity likely occurred because the shorter substrates were unable to form a thermodynamically stable duplex with the shorter spacers, or because the associated substrates cannot adopt an active conformation to be modified. To distinguish the two possibilities, we measured the modification of substrates S8 and S9 by C/D RNPs with a spacer length of 12–14 nt (Fig. 1C). Significant activities were detected, indicating that a guide-substrate duplex as short as 8 bp is tolerated. Nevertheless, the S8 substrate was less modified than longer substrates, particularly at high temperature of 70 °C (Fig. 1C), as expected for reduced binding of short substrates. In addition, the guiding activity of SL10 and SL11 RNAs was not restored at a lower temperature of 50 °C (Fig. 1B). We can conclude that short spacers (<12 nt long) are defective primarily because the substrate cannot be loaded into a modification-competent conformation.
Modification activity was not noticeably affected at 70 °C and 50 °C when the spacer was lengthened to 13 nt and the guide-substrate duplex was lengthened to 11 bp (Fig. 1B) (SL13 + S11). Further increases of the spacer length to 14 nt and 15 nt and of the substrate length to 12 nt slightly reduced the modification at 70 °C, but dramatically decreased the modification at 50 °C (Fig. 1B) (SL14 + S12, SL15 + S12). The data show that extensive guide-substrate pairing (12 bp in this case) inhibits the modification in a temperature-dependent manner. An archaeal C/D RNA with spacers shorter or longer than 12 nt was previously shown to be defective in guiding modification (32). We confirmed that short spacers are not functional, but did not find that long spacers (up to 15 nt) are defective.
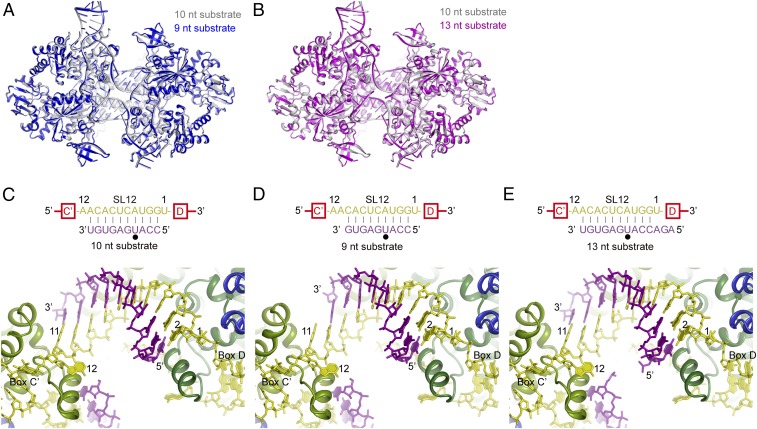
Structures of SL12 RNP Bound with Different Substrates.
To provide structural insight into the guiding mechanism of C/D RNA, we determined two crystal structures of SL12 RNP bound to a 9-nt substrate at 3.3-Å resolution and a 13-nt substrate at 3.6-Å resolution (Fig. 2 and Table S1). The two structures were crystallized in the same space group as the previously determined structure of SL12 RNP bound to 10-nt substrates. The three SL12 RNP structures bound with different substrates are nearly identical, with an rmsd of 0.195–0.324 Å (Fig. 2 A and B). In the previous structure, the 10-nt substrate pairs with the spacer at positions 2–11 (counted from box D/D′) (Fig. 2C).
Fig. 2.
Structures of substrate-bound SL12 C/D RNPs. (A) Alignment of SL12 RNP structures bound to 10-nt (silver) and 9-nt (blue) substrates. (B) Alignment of SL12 RNP structures bound to 10-nt (silver) and 13-nt (magenta) substrates. (C–E) Conformation of guide-substrate duplexes in SL12 RNP structures bound to 10-nt (C), 9-nt (D), and 13-nt (E) substrates. These structures share the same orientation. Proteins are shown as ribbons; RNAs, as sticks. Fibrillarin is omitted for clarity. The two Nop5 subunits are colored in dark green and light green, L7Ae is in blue, C/D RNA is in yellow, and the substrates are in purple. Base pairing interactions between the guide and the substrate are displayed at the top. The target site is marked by a black circle. Nucleotides in spacers and substrates are numbered by their distances to box D.
Table S1.
Data collection and refinement statistics
| Crystal form | SL12 RNP/9-nt substrate | SL12 RNP/13-nt substrate | SL13 RNP/11-nt substrate |
| Data collection | |||
| Space group | P41212 | P41212 | P1 |
| Cell dimensions | |||
| a, b, c (Å) | 242.676, 242.676, 146.898 | 241.781, 241.781, 145.002 | 97.916, 97.718, 128.793 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 108.98, 104.42, 89.86 |
| Wavelength (Å) | 0.9789 | 0.9789 | 0.97915 |
| Resolution range (Å) | 20–3.3 (3.36–3.3) | 20–3.6 (3.66–3.6) | 25–3.12 (3.17–3.12) |
| Unique reflections | 63,174 (2,486) | 49,349 (2,376) | 74,336 (3,707) |
| Redundancy | 7.2 (3.1) | 7.6 (4.0) | 2.3 (2.4) |
| <I>/<σ(I)> | 12.0 (1.2) | 12.4 (2.1) | 22.5 (4.1) |
| Completeness (%) | 96.5 (77.2) | 99.7 (97.4) | 97.3 (98.1) |
| Rmerge | 0.131 (0.729) | 0.165 (0.528) | 0.087 (0.514) |
| Structure refinement | |||
| Resolution range (Å) | 20–3.3 (3.36–3.3) | 20–3.6 (3.67–3.6) | 25–3.13 (3.21–3.13) |
| No. of reflections | 62,833 (2,130) | 49,034 (2,346) | 74,281 (5,061) |
| No. of atoms | 19,986 | 20,055 | 26,608 |
| Rwork | 0.250 (0.305) | 0.256 (0.305) | 0.178 (0.245) |
| Rfree | 0.303 (0.354) | 0.298 (0.340) | 0.234 (0.319) |
| Mean B (Å2) | 91.89 | 74.19 | 66.22 |
| rmsd bond length (Å) | 0.013 | 0.005 | 0.013 |
| rmsd bond angles (°) | 1.711 | 0.885 | 1.479 |
| Ramachandran plot | |||
| Favored (%) | 95.59 | 95.91 | 95.44 |
| Allowed (%) | 4.36 | 4.09 | 4.46 |
| Outliers (%) | 0.05 | 0 | 0.10 |
Values in parentheses are for the data in the highest-resolution shell.
The 9-nt substrate lacks one nucleotide at the 3′ end, resulting in the absence of 1 bp at position 11 (Fig. 2D and Fig. S1A). Nevertheless, the missing base pair causes no structural change in the remainder of the complex, including the now-unpaired guide nucleotide at position 11. The structure illustrates how a 9-bp guide-substrate duplex is accommodated in C/D RNP and accounts for the efficient modification of 9-nt substrates by SL12 RNP (Fig. 1C).
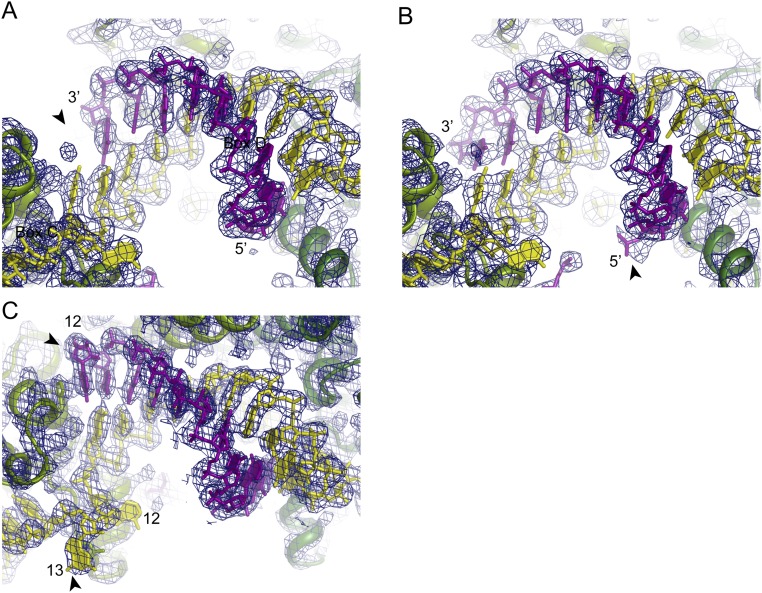
Fig. S1.
Electron density map of C/D RNP structures. The 2Fo–Fc electron density map contoured at the 1.5σ level is displayed for the SL12 RNP structure bound to the 9-nt (A) or 13-nt (B) substrates and the SL13 RNP structure bound to the 11-nt substrate (C). The determined structures are overlaid on the map. Substrate RNAs are shown in purple, C/D RNAs are in yellow, and two Nop5 subunits are in light green and dark green. The SL12 RNP structures in A and B have the same orientation. Important features in each map are marked with arrowheads. In A, the arrowhead indicates a missing density at position 11 for the 9-nt substrate. In B, the arrowhead indicates extra density for a phosphate group from the 5′ extension of the 13-nt substrate. In C, the arrowheads indicate the unpaired substrate nucleotide at position 12 and the unpaired spacer nucleotide at position 13.
The 13-nt substrate contains three additional nucleotides at the 5′ end (Fig. 2E). The extension is largely disordered in the structure, with only a phosphate group visible (Fig. S1B). The extension was designed to pair with the spacer at position 1; however, this base pair was not formed in the structure, confirming that the first nucleotide in the spacer is not involved in binding substrate.
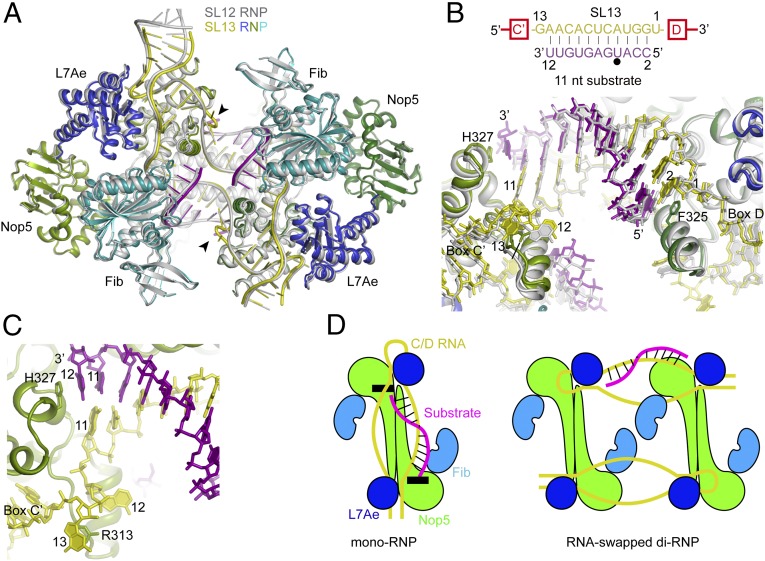
Structure of SL13 RNP.
We further determined the crystal structure of SL13 RNP bound with the 11-nt substrate S11 at 3.1-Å resolution (Fig. 3 A–C). The overall structure of SL13 RNP is highly similar to that of the SL12 RNP structure despite the different spacers and substrates used. The two structures can be aligned with an rmsd of 0.976 Å over 1,441 Cα atom pairs (Fig. 3A). Of note, the SL12 and SL13 RNPs are crystallized in different space groups (P41212 vs. P1), excluding the possibility that their structural similarity is imposed by the same crystal packing environment.
Fig. 3.
Structure of SL13 RNP bound to 11-nt substrates. (A) Structural alignment of SL13 RNP bound with 11-nt substrates and SL12 RNP bound with 10-nt substrates. The SL12 RNP structure is colored in silver. The SL13 RNP structure is colored by molecule; the two Nop5 subunits are in dark green and light green, L7Ae is in blue, fibrillarin (Fib) is in cyan, C/D RNA is in yellow, and substrate RNA is in purple. The looped-out guide nucleotide 13 is marked with an arrowhead. (B) Conformation of guide-substrate duplexes in the aligned SL12 RNP and SL13 RNP structures. Fibrillarin is omitted for clarity. Proteins are shown as ribbons; RNAs, as sticks. Base pairing interactions between the guide and the substrate are displayed at the top. The target site is marked by a black circle. Nucleotides in spacers and substrates are numbered by their distances to box D. (C) Close-up view of the SL13 RNP structure at the box C' side. (D) Cartoons of C/D mono-RNP and RNA-swapped di-RNP. Black boxes indicate the structural elements that cap the guide-substrate duplex in the mono-RNP model.
The spacer region of SL13 RNA is 13 nt long and is expected to form a 11-bp duplex with the S11 substrate (Fig. 3B); however, in the S11–SL13 RNP structure, the substrate forms only a 10-bp duplex with the guide at positions 2–11. The 3′ end base of S11 is unpaired and sandwiched between its 5′ base and the imidazole ring of Nop5 His327 (Fig. 3C). The nucleotide at position 12 of the spacer is also unpaired and adopts a similar conformation as its counterpart in the SL12 RNP structure. The extra spacer nucleotide at position 13 is looped out and stacks on the guanidine group of Nop5 Arg313. The structure shows that C/D RNP is unable to accept an 11-bp guide-substrate duplex. For even longer spacers, the structure also suggests that additional spacer sequences between position 12 and box C′/C can be looped out in a similar manner as for nucleotide 13.
The size of the RNA-binding channel is preserved as well. For example, the Cα–Cα distance between His327 and Phe325 from the opposing Nop5 subunits, which are two aromatic residues capping the two ends of the guide-substrate duplex, is 37.1 ± 0 Å for the SL13 RNP structure and 38.0 ± 0.2 Å for the SL12 RNP structure (Fig. 3B).
Discussion
In principle, the CTDs of the Nop5 dimer could adjust their positions somewhat to accommodate the guide-substrate duplex of various sizes. Our structural and biochemical data reveal a different scenario, in which the substrate-binding channel formed by the Nop5 dimer is rather rigid and exactly fits a 10-bp duplex. Slightly shorter duplexes (8–9 bp) are tolerated as long as they are thermodynamically stable. Longer duplexes must be partially unwound at the C/C′ side to fit into the size-limiting channel. Consequently, fibrillarin can approach the guide-substrate duplex in the same manner, ensuring the efficiency and specificity of modification. The substrate recognition mechanism of C/D RNA explains that excessive guide-substrate pairing inhibits the modification in a temperature-dependent manner (Fig. 1B), because duplex unwinding slows at low temperatures.
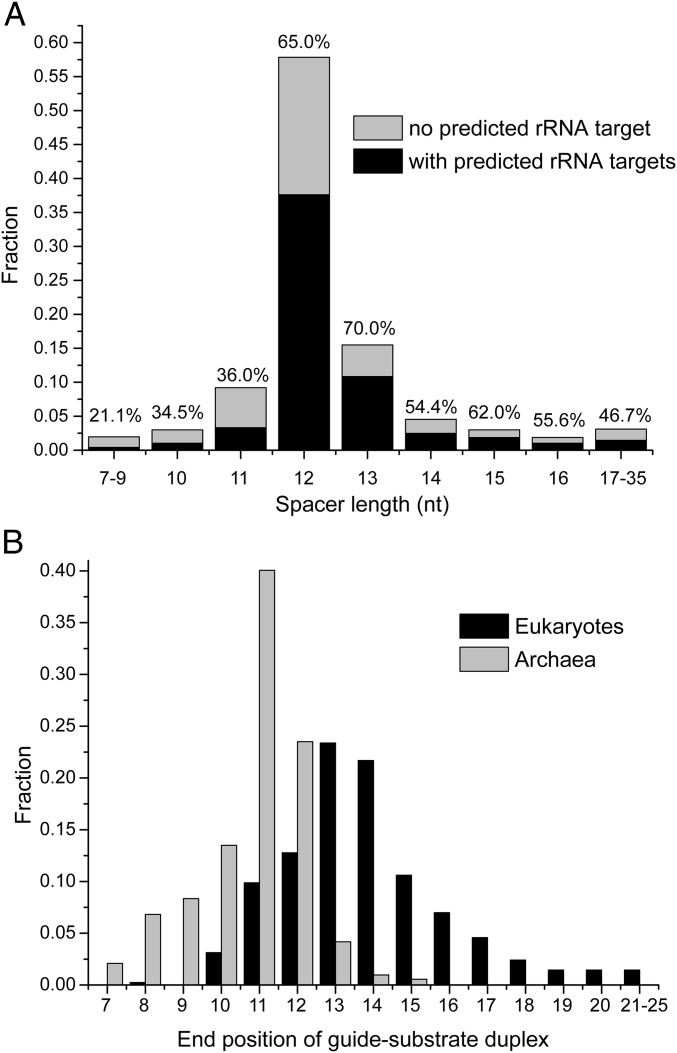
Archaeal C/D RNAs have a compact structure, with the spacer region length constrained to 12 nt (32). Recently, 489 C/D RNAs were compiled from the RNA-Seq data from seven archaea, and 719 guide–rRNA interactions were predicted (39). Analysis of this large dataset showed that 57.8% of the spacers are 12 nt long (Fig. 4A), confirming a previous report (32). Only 14.2% of spacers are <12 nt long; however, these short spacers have significantly lower percentages (20.1–36%) of predicted rRNA targets compared with the spacers of ≥12 nt (46.7–70%). This is consistent with the finding that short spacers are defective in guiding modification (Fig. 1B). The predicted guide-substrate duplexes terminate mainly at positions 10–12 relative to box D or D′ (Fig. 4B). A guide-substrate duplex that ends at or before position 11 can be directly loaded onto the substrate-binding channel and modified efficiently. Although longer spacers are functional (Fig. 1), they potentially can make extra base pairs with the substrate and are not optimal. Archaeal C/D RNAs appear to be under strong evolutionary pressure to maintain high efficiency in modification by restraining the spacer length to 12 nt.
Fig. 4.
Spacers and guide-substrate duplexes in archaeal and eukaryotic C/D RNAs. (A) Distribution of spacer length in archaeal C/D RNAs. A total of 968 spacer sequences from 489 archaeal C/D RNAs were counted (39). Ten spacers could be determined owing to missing box C' or/and D'. The percentage of spacers with predicted rRNA targets is indicated for each length group. (B) Distribution of end positions of guide-rRNA duplexes in archaeal and eukaryotic C/D RNAs. Distances to box D or D' are shown on the x-axis. The archaeal dataset include 719 predicted guide-rRNA duplexes from 489 archaeal C/D RNAs (39). The eukaryotic dataset includes 415 predicted guide-rRNA duplexes from Homo sapiens, Mus musculus, Arabidopsis thaliana, Oryza sativa, Saccharomyces cerevisiae, and Schizosaccharomyces pombe (33). The end of the duplex must be followed by a mismatch that must not be followed by a minimum of 2 Watson–Crick bp or G:U bp.
Although our data are based on archaeal C/D RNPs, recognition of a maximum of 10 nt in substrates is likely a general principle that holds for eukaryotic C/D RNPs as well. The size of the substrate-binding channel is determined by the Nop5 structure in archaeal C/D RNP. Nop5 is highly conserved in sequence with its eukaryotic counterparts Nop56 and Nop58 (30), suggesting that a similar-sized channel exists in eukaryotic complexes.
The spacer region in eukaryotic C/D RNAs is much longer and of more variable size compared with archaeal C/D RNAs. Analysis of 415 predicted guide-rRNA duplexes from six distantly related eukaryotes shows that 86.7% of them are longer than the optimal duplex that ends at position 11 (33) (Fig. 4B). These duplexes most often end at positions 13 and 14. The long guide sequence may load substrates in two steps. At the first step, it binds the substrate with full range of complementarity; however, the resulting >10-bp duplex is placed out of the substrate-binding channel and cannot be modified. After unwinding of a few terminal base pairs to position 11 by breathing or other mechanisms, the substrate is then loaded and subsequently modified and released. The long guide sequence may be advantageous for enhancing the specificity and rate of target recognition at the first binding step. In addition, the unwinding of long guide-substrate duplexes takes more time, allowing C/D RNP to bind the rRNA transcript for longer times. We propose that the extent of guide-substrate pairing provides a mechanism to control the association time of C/D RNP with rRNA. In eukaryotes, the precursor rRNA is cotranscriptionally modified by C/D RNPs and H/ACA RNPs and assembled into preribosomes (40–43). The association of guide RNAs could suppress misfolding of the target site sequence when other sequences required for assembling native structures have not been transcribed. In this way, eukaryotic C/D RNPs regulate the folding and assembly of rRNA by sequestering the target sequence. In contrast, the primary function of archaeal C/D RNP is apparently to efficiently modify rRNA.
We demonstrate that only the base pair interactions from positions 2–11 are effective for loading the substrate to the active site. Previous mutational studies have shown that the pairing interactions at the core region are mostly important for efficient modification of the substrate, whereas the pairing interactions outside the core region are dispensable (44, 45). Moreover, the pairing at the core region has been shown to impose strong constraints on the evolution of guide sequences of eukaryotic C/D RNAs. A previous analysis of conservation in eukaryotic guide-rRNA duplexes found that the duplex region from positions 3–11 is composed of most conserved Watson–Crick pairs (33). The other paired regions are less conserved than the core region (33), although they may aid in target recognition and rRNA folding.
Our reconstituted C/D RNP for activity measurement consists of a mixture of mono-RNP, di-RNP, and other higher-order species (30). Both mono-RNP and di-RNP are active in site-specific modification (30). The inhibitory effect of long guide-substrate duplexes was observed for the mixture, indicating that the di-RNP also recognizes only 10 nt of substrate as the mono-RNP.
The di-RNP likely contains two C/D RNAs and four copies of each of Nop5, L7Ae, and fibrillarin. In the proposed RNA-swapped model (34, 35), the spacer regions of C/D RNAs are placed between two Nop5 dimers and mediate dimerization of the RNP (Fig. 3D). As a result, the guide-substrate duplex cannot be loaded onto the substrate-binding channel present in the mono-RNP structure. The RNA-swapped model is also unlikely to limit the size of the guide-substrate duplex, because the ends of the guide-substrate duplex are now attached to two freely placed Nop5 molecules. Thus, the RNA-swapped di-RNP model cannot account for the recognition of 10 nt of substrate by C/D RNA.
The di-RNP structure has been analyzed by low-resolution approaches (34, 35), but the key information about the organization of C/D RNA cannot be determined directly from these studies. Without invoking the unconventional RNA-swapped model, the di-RNP could be accounted for simply by self-association of the mono-RNP through protein–protein interaction. In this way, each C/D RNA would still be bound to an Nop5 dimer as in the mono-RNP, and the di-RNP would adopt the same mechanism of action as the mono-RNP.
Methods
Activity Assay.
Substrate RNAs and premethylated substrate RNAs were purchased from Takara. The new C/D RNAs were constructed from the CD45 RNA by QuikChange mutagenesis (30). The D and D′ spacer sequences were different for the SL12/CD45 RNA and the same for other C/D RNAs. The modification reactions (10 μL) contained 10 pmol C/D RNA, 30 pmol L7Ae, 20 pmol Nop5-Fib dimer, and 300 pmol D site substrate or premethylated substrate in buffer containing 20 mM Hepes-Na (pH 8.0), 150 mM NaCl, and 1.5 mM MgCl2. The reactions were prepared in PCR tubes and incubated at 20 °C for 10 min. To start the modification, the tubes were added with 300 pmol unlabeled S-adenosylmethionine (SAM) and 0.25 µCi [methyl-3H] SAM (83 Ci mmol−1; PerkinElmer) and incubated at 70 °C or 50 °C for 20 min. The reactions were stopped by freezing in liquid nitrogen. Each sample was mixed with an equal volume of 8 M urea in Tris-borate-EDTA (TBE) buffer and heated at 95 °C for 2 min. The samples were then loaded onto a 10% (wt/vol) polyacrylamide gel containing 8 M urea and then separated in TBE buffer at a constant power of 13 W for 1 h. The gel was fixed in a solution containing 10% (vol/vol) acetic acid, 25% (vol/vol) isopropanol, and 65% (vol/vol) water for 30 min, then soaked in Amplify solution (GE Healthcare) with agitation for 15–30 min and dried by vacuum at 80 °C. For autoradiography, the gel was exposed to X-ray films for 2–3 d at −80 °C.
Crystallization.
Purification of Nop5, fibrillarin, and L7Ae proteins; preparation of C/D RNAs; and assembly of C/D RNPs were done as described previously (29, 30). Crystallization was performed by the hanging-drop vapor diffusion method at 20 °C. The SL12 RNP was loaded with a 1.5 molar equivalent of the 9-nt substrates (5′-UCCAGUACU-3′ and 5′-CCAUGAGUG-3′) or the 13-nt substrates (5′-AGAUCCAGUACUU-3′ and 5′-AGACCAUGAGUGU-3′). The two SL12 RNPs were crystallized from 2.0 M (NH4)2SO4, 2% (wt/vol) PEG-400, 10 mM MgCl2, and 0.1 M Hepes-Na (pH 6.0), the same condition as the previous SL12 RNP bound to the 10-nt substrates.
The SL13 RNP (4 mg/mL in 5 mM Hepes-K, pH 8.0) bound to the 11-nt substrate (5′-CCAUGAGUGUU-3′) was crystallized in 1.7 M DL-malic acid (pH 7.0) and 0.1 M potassium sodium tartrate tetrahydrate. The crystal was cryoprotected in 30% (vol/vol) glycerol created in the well solution and flash-frozen in liquid nitrogen.
Structure Determination.
Diffraction data were collected at the BL17U beamline of the Shanghai Synchrotron Radiation Facility and processed with the HKL-2000 program package (46). All structures were solved by molecular replacement and refined with PHENIX (47). Models were built in COOT (48). Structural figures were prepared with PyMol (49).
The two SL12 RNP structures bound to the 9-nt and 13-nt substrates contain 1.5 copies of C/D RNP in the asymmetric unit and have the same space group of P41212 as the previously determined SL12 RNP structure bound to the 10-nt substrates. The 9-nt substrate-bound structure was refined to 3.3 Å, and the 13 nt substrate-bound structure was refined to 3.6 Å. The SL13 RNP structure bound with the 11-nt substrate is in the P1 space group with two copies of C/D RNPs in the asymmetric unit and was refined to 3.1 Å.
Acknowledgments
We thank the staff at beamline BL17U of the Shanghai Synchrotron Research Facility for assistance in diffraction data collection. This work was supported by the National Natural Science Foundation of China (Grants 31325007, 31430024, and 91540201), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB08010203), and the Beijing Municipal Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.L.J.L. is a Guest Editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5GIN, 5GIO, and 5GIP).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604872113/-/DCSupplemental.
References
- 1.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20(14):3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3(3):397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 3.Liang B, Li H. Structures of ribonucleoprotein particle modification enzymes. Q Rev Biophys. 2011;44(1):95–122. doi: 10.1017/S0033583510000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavaillé J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383(6602):732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 5.Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85(7):1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 6.Tycowski KT, Smith CM, Shu MD, Steitz JA. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93(25):14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omer AD, et al. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288(5465):517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 8.Baudin-Baillieu A, et al. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009;37(22):7665–7677. doi: 10.1093/nar/gkp816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15(9):1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esguerra J, Warringer J, Blomberg A. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA. 2008;14(4):649–656. doi: 10.1261/rna.845808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27(7):344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 12.Watkins NJ, et al. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103(3):457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 13.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20(15):4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidovic I, Nottrott S, Hartmuth K, Lührmann R, Ficner R. Crystal structure of the spliceosomal 15.5-kD protein bound to a U4 snRNA fragment. Mol Cell. 2000;6(6):1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 15.Nolivos S, Carpousis AJ, Clouet-d’Orval B. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005;33(20):6507–6514. doi: 10.1093/nar/gki962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman DR, Kuhn JF, Shanab GM, Maxwell ES. Box C/D snoRNA-associated proteins: Two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 2000;6(6):861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lafontaine DL, Tollervey D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol Cell Biol. 2000;20(8):2650–2659. doi: 10.1128/mcb.20.8.2650-2659.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafontaine DL, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5(3):455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu P, Brockenbrough JS, Metcalfe AC, Chen S, Aris JP. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J Biol Chem. 1998;273(26):16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17(12):7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72(3):443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 22.Schimmang T, Tollervey D, Kern H, Frank R, Hurt EC. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8(13):4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galardi S, et al. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol Cell Biol. 2002;22(19):6663–6668. doi: 10.1128/MCB.22.19.6663-6668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omer AD, Ziesche S, Ebhardt H, Dennis PP. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci USA. 2002;99(8):5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Boisvert D, Kim KK, Kim R, Kim SH. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 2000;19(3):317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Y, Yu G, Tian S, Li H. Co-expression and co-purification of archaeal and eukaryal box C/D RNPs. PLoS One. 2014;9(7):e103096. doi: 10.1371/journal.pone.0103096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aittaleb M, et al. Structure and function of archaeal box C/D sRNP core proteins. Nat Struct Biol. 2003;10(4):256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- 28.Xue S, et al. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Mol Cell. 2010;39(6):939–949. doi: 10.1016/j.molcel.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye K, et al. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc Natl Acad Sci USA. 2009;106(33):13808–13813. doi: 10.1073/pnas.0905128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J, et al. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature. 2011;469(7331):559–563. doi: 10.1038/nature09688. [DOI] [PubMed] [Google Scholar]
- 31.Oruganti S, et al. Alternative conformations of the archaeal Nop56/58-fibrillarin complex imply flexibility in box C/D RNPs. J Mol Biol. 2007;371(5):1141–1150. doi: 10.1016/j.jmb.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 32.Tran E, Zhang X, Lackey L, Maxwell ES. Conserved spacing between the box C/D and C′/D′ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA. 2005;11(3):285–293. doi: 10.1261/rna.7223405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CL, Perasso R, Qu LH, Amar L. Exploration of pairing constraints identifies a 9 base-pair core within box C/D snoRNA-rRNA duplexes. J Mol Biol. 2007;369(3):771–783. doi: 10.1016/j.jmb.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 34.Bleichert F, et al. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science. 2009;325(5946):1384–1387. doi: 10.1126/science.1176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lapinaite A, et al. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013;502(7472):519–523. doi: 10.1038/nature12581. [DOI] [PubMed] [Google Scholar]
- 36.Graziadei A, Masiewicz P, Lapinaite A, Carlomagno T. Archaea box C/D enzymes methylate two distinct substrate rRNA sequences with different efficiency. RNA. 2016;22(5):764–772. doi: 10.1261/rna.054320.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bower-Phipps KR, Taylor DW, Wang HW, Baserga SJ. The box C/D sRNP dimeric architecture is conserved across domain Archaea. RNA. 2012;18(8):1527–1540. doi: 10.1261/rna.033134.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bleichert F, Baserga SJ. Dissecting the role of conserved box C/D sRNA sequences in di-sRNP assembly and function. Nucleic Acids Res. 2010;38(22):8295–8305. doi: 10.1093/nar/gkq690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dennis PP, Tripp V, Lui L, Lowe T, Randau L. C/D box sRNA-guided 2′-O-methylation patterns of archaeal rRNA molecules. BMC Genomics. 2015;16:632. doi: 10.1186/s12864-015-1839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osheim YN, et al. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004;16(6):943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Wu C, Cai G, Chen S, Ye K. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev. 2016;30(6):718–732. doi: 10.1101/gad.274688.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol. 2015;22(11):920–923. doi: 10.1038/nsmb.3111. [DOI] [PubMed] [Google Scholar]
- 43.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37(6):809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Appel CD, Maxwell ES. Structural features of the guide:target RNA duplex required for archaeal box C/D sRNA-guided nucleotide 2′-O-methylation. RNA. 2007;13(6):899–911. doi: 10.1261/rna.517307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavaillé J, Bachellerie JP. SnoRNA-guided ribose methylation of rRNA: Structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 1998;26(7):1576–1587. doi: 10.1093/nar/26.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.DeLano WL. The PyMOL User’s Manual. Delano Scientific; San Carlos, CA: 2002. [Google Scholar]