In a whole genome-based phylogeny, clinical and fecal isolates of Escherichia coli sequence type 131 (H30R1 and H30Rx subclones) from six households formed household-specific clusters, interspersed among reference ST131 genomes. This supported the fecal-urethral hypothesis and confirmed within-household strain sharing.
Keywords: colonization, Escherichia coli infections, ST131, transmission, whole genome sequence
Abstract
Background. Within-household sharing of strains from the resistance-associated H30R1 and H30Rx subclones of Escherichia coli sequence type 131 (ST131) has been inferred based on conventional typing data, but it has been assessed minimally using whole genome sequence (WGS) analysis.
Methods. Thirty-three clinical and fecal isolates of ST131-H30R1 and ST131-H30Rx, from 20 humans and pets in 6 households, underwent WGS analysis for comparison with 52 published ST131 genomes. Phylogenetic relationships were inferred using a bootstrapped maximum likelihood tree based on core genome sequence polymorphisms. Accessory traits were compared between phylogenetically similar isolates.
Results. In the WGS-based phylogeny, isolates clustered strictly by household, in clades that were distributed widely across the phylogeny, interspersed between H30R1 and H30Rx comparison genomes. For only 1 household did the core genome phylogeny place epidemiologically unlinked isolates together with household isolates, but even there multiple differences in accessory genome content clearly differentiated these 2 groups. The core genome phylogeny supported within-household strain sharing, fecal-urethral urinary tract infection pathogenesis (with the entire household potentially providing the fecal reservoir), and instances of host-specific microevolution. In 1 instance, the household's index strain persisted for 6 years before causing a new infection in a different household member.
Conclusions. Within-household sharing of E coli ST131 strains was confirmed extensively at the genome level, as was long-term colonization and repeated infections due to an ST131-H30Rx strain. Future efforts toward surveillance and decolonization may need to address not just the affected patient but also other human and animal household members.
Escherichia coli, a major opportunistic colonizing pathogen, is the most common facultative member of the human gut microbiota and the commonest cause of urinary and bloodstream infections in humans [1, 2]. Extraintestinal pathogenic E coli (ExPEC) are a group of E coli lineages characterized by an enhanced ability to transit from their usual gut habitat to normally sterile body compartments and cause infections [3]. Several ExPEC lineages account for most extraintestinal E coli infections [4–7]. Some of these, especially E coli sequence type 131 (ST131) and its H30R1 and H30Rx subclones (aka, clades C1 and C2 [8]), are associated with antimicrobial resistance and have spread extensively, creating a global epidemic of drug-resistant E coli infections [8–11].
The cause of such widespread dissemination of antimicrobial-resistant ExPEC lineages is unclear. One possibility, host-to-host transmission, is supported by evidence showing colonization of multiple household members with putatively the same ExPEC strain, with sporadic infection episodes occurring in colonized individuals [12, 13]. This suggests that household transmission may precede the classic fecal-urethral pathway of urinary tract infection (UTI) pathogenesis, in which the immediate reservoir for an infecting ExPEC strain is the host's own intestinal and genital microbiota [14–16].
The growing availability of whole genome sequence (WGS) analysis for high-resolution strain typing obliges reconsideration of traditional assumptions regarding strain identity and transmission. For example, the first WGS-based ST131 phylogeny showed that pulsed-field gel electrophoresis (PFGE), historically preferred for E coli strain typing, misrepresents the genetic similarity of many ST131 strains [11]. Only 2 previous studies applied WGS analysis to household colonization patterns. One examined a single non-ST131 ExPEC strain, in 6 hosts [17], and the other documented sharing of a single H30Rx strain between 2 sisters, 1 with recurrent UTI, the other with fatal urosepsis [11].
In this study, we applied WGS analysis to 33 clinical or commensal E coli ST131 isolates from 20 members of 6 households, within each of which household the ST131 isolates (hereafter, household isolates) putatively represented a single H30R1 or H30Rx strain. We sought to place these household isolates within the ST131 phylogeny [11] and to assess the degree of genetic similarity between (1) isolates from the same versus different households and (2) clinical versus commensal isolates within individuals and households. We also compared PFGE and WGS analysis for assessing clonal relationships within ST131.
SUBJECTS AND METHODS
Isolates
The 52 ST131 comparison isolates, which were epidemiologically unlinked to the present household isolates, were selected from the 104 ST131 genomes reported by Price et al [11], which had been selected to maximize diversity of PFGE profiles, host species, and resistance phenotypes. Because conventional typing (as described below) placed all the present household isolates within the H30R subclone (Table 1), as comparison isolates we used 46 H30R isolates from Price et al [11] plus the 6 next-most-basal isolates in the phylogram. The 52 comparison isolates were predominantly North American human clinical isolates, although some were from other locales (Asia, Australia, Europe) and hosts (dog, cat, monkey, dolphin).
Table 1.
Characteristics of 33 Household-Source Isolates of Escherichia coli ST131
| Household | Datea | Isolate | Host | Sampleb | ESBLc | PFGE | Subcloned | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 2007 | CU758 | Child (index) | Abscessb | 968 | H30R1 | [18] | |
| CU761 | Mother | Fecal | 968 | H30R1 | [18] | |||
| 2 | 2008 | CD364 | Dog (index) | Urine | 968 | H30R1 | [19] | |
| CU797 | Dog (index) | Fecal | 968 | H30R1 | [19] | |||
| CU790 | Cat no. 1 | Fecal | 968 | H30R1 | [19] | |||
| CU799 | Cat no. 2 | Fecal | 968 | H30R1 | [19] | |||
| 3 | 2007 | JJ1887 | Sister 1 (index) | Urine | Pos | 968 | H30Rx | [11, 20] |
| JJ1886 | Sister 2 | Blood | Pos | 968 | H30Rx | [11, 20] | ||
| 4 | 2008 | JJ2546 | Father (index) | Urine | Pos | 812 | H30Rx | [21] |
| JJ2547 | Daughter | Abscessb | Pos | 812 | H30Rx | [21] | ||
| JJ2548 | Daughter | Blood | Pos | 812 | H30Rx | [21] | ||
| 2014 | JJ2963 | Mother | Urine | Pos | 812 | H30Rx | This study | |
| JJ2974 | Mother | Fecal | Pos | 812 | H30Rx | This study | ||
| 5 | 2013 | JJ2888 | Child (index) | Urine | Pos | 806 | H30Rx | This study |
| HVAST501 | Child (index) | Fecal | Pos | 806 | H30Rx | This study | ||
| HVAST502 | Mother | Fecal | Pos | 806 | H30Rx | This study | ||
| HVAST503 | Other adult | Fecal | Pos | 1883 | H30Rx | This study | ||
| HVAST504 | Pet | Fecal | Pos | 806 | H30Rx | This study | ||
| 6 | January 2014 | JJ2913 | Child (index) | Urine | Pos | 903 | H30Rx | [12] |
| HVAST656 | Child (index) | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST695 | Mother | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST697 | Father | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST702 | Sibling no. 1 | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST709 | Sibling no. 3 | Fecal | Pos | 903 | H30Rx | [12] | ||
| March 2014 | HVAST736 | Child (index) | Fecal | Pos | 903 | H30Rx | [12] | |
| HVAST737 | Mother | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST738 | Sibling no. 1 | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST740 | Sibling no. 3 | Fecal | Pos | 903 | H30Rx | [12] | ||
| May 2014 | HVAST762 | Child (index) | Fecal | Pos | 903 | H30Rx | [12] | |
| HVAST757 | Mother | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST758 | Father | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST760 | Sibling no. 2 | Fecal | Pos | 903 | H30Rx | [12] | ||
| HVAST761 | Sibling no. 3 | Fecal | Pos | 903 | H30Rx | [12] |
Abbreviations: ESBL, extended-spectrum β-lactamase; PFGE, pulsed-field gel electrophoresis pulsotype; Pos, positive; ST131, sequence type 131.
a Dates (shown as year, or month and year) apply to the indicated isolate and subsequent same-household isolates without a listed date, until the next isolate with a listed date.
b Isolate CU758 was from a surgically drained bone abscess (acute osteomyelitis). Isolate JJ2547 was from a catheter-drained renal gas abscess (emphysematous pyelonephritis).
c Pos for both ESBL phenotype and blaCTX-M-15; blank, negative for both traits.
d The H30R and H30Rx subclones correspond with clades C1 and C2 of Petty et al [8].
The 33 household isolates were clinical or fecal isolates from 20 members of 6 households, each in a different city in Maine [20], Minnesota [12], Missouri [18], Pennsylvania [21], or Virginia [19] (Table 1). The 6 study households came to the investigators' attention because, within each household, 1 or multiple members had developed 1 or more infection episodes, some of unusual severity, involving fluoroquinolone-resistant E coli, with or without extended-spectrum β-lactamase (ESBL) production. These striking clinical and microbiological features prompted the patient's clinicians to contact the investigators for molecular strain typing, to determine whether there was something special about the causative strain(s). The investigators then instigated household fecal surveillance, given the known tendency of E coli strains to circulate within households. Six of the 10 clinical isolates from these households (CU758, household 1; CU799 and CD364, household 2; JJ1886 and JJ1887, household 3; JJ2547, household 4) appeared in Price et al [11], but here they were analyzed as household isolates rather than comparison isolates. (In Price et al [11], CD364 was designated CD449.)
For each household, the investigators obtained and typed the index clinical isolate(s) and, when possible, arranged for fecal surveillance of all available household members, 1 or more times. For this, antibiotic-supplemented media were used to recover fluoroquinolone-resistant E coli from self-collected fecal swabs [12]. Fluoroquinolone-resistant fecal E coli isolates were typed similarly to the clinical isolates (described below). Some of these household-based screenings have been reported previously, in full for households 1–3 and 6 [11, 12, 18–20] and partially for household 4 [21]. In this study, novel extended follow-up data are provided for household 4, and household 5 is newly reported.
Conventional Strain Typing
Putative E coli isolates were identity-confirmed by API 20E (bioMérieux) and/or uidA PCR [22]. Susceptibility profiles were determined by disk diffusion using procedures, reference strains, and interpretive criteria as specified by the Clinical Laboratory Standards Institute [23]. Detection of E coli ST131, its H30 and H30Rx subclones, and blaCTX-M-15 (encoding the ST131-associated ESBL CTX-M-15) used established polymerase chain reaction (PCR) assays targeting key single-nucleotide polymorphisms (SNPs) [24]. Because H30R1 and H30Rx (or clades C1 and C2 [8]) are sister subclones that constitute the ST131-H30R clade [11], fluoroquinolone-resistant ST131-H30 isolates that screened negative for H30Rx were regarded as H30R1. XbaI PFGE analysis was done using a standardized protocol [25]. Profiles were captured digitally, analyzed, and used to infer a similarity dendrogram based on Dice coefficients using the unweighted pair group method (UPGMA) within BioNumerics (BioRad). Pulsotype designations were assigned based on ≥94% similarity to reference profiles [25].
Whole Genome Sequence Analysis
For households other than household 4, from 1 to 5 µg deoxyribonucleic acid (DNA) per isolates in 200 µL was sheared to 200–1000 base pairs (bp) with the SonicMAN sonicator (Matrical BioScience, Spokane, WA), then purified using the QIAGEN QIAquick PCR purification kit (QIAGEN, Valencia, CA). The DNA was processed enzymatically according to Illumina's recommendations (Illumina, Inc., San Diego, CA) using enzymes from New England Biolabs (Ipswich, MA) and oligonucleotides and adaptors from Illumina.
After adaptor ligation, DNA was separated electrophoretically on a 2% agarose gel for 2 hours. A gel slice containing 500- to 600-bp fragments of each DNA sample was isolated and purified using the QIAGEN QIAquick Gel Extraction kit (QIAGEN). Individual libraries underwent quantitative PCR using a Kapa Library Quantification kit (Kapa Biosystems, Woburn, MA). Equimolar pools of ≤12 indexed E coli libraries were prepared at a concentration of ≥1 nM using 10 mM Tris-HCl (pH 8.0) with 0.05% Tween 20. The pooled paired-end libraries were sequenced on an Illumina MiSeq and Genome Analyzer IIx to a read length of ≥100 bp.
The genomes were sequenced at an average depth of 58.27× (standard deviation [SD] = 35.4, using the 5 129 934-base JJ1886 chromosome as a reference [26]). On average, 5 058 582 bases (SD = 12 993) were sequenced at ≥10× coverage for each genome.
For the household 4 isolates, DNA libraries were prepared using the Nextera XT DNA library preparation kit (Illumina), and after normalization they underwent sequencing on a Miseq platform using the Miseq 150 cycle kit.
Single-Nucleotide Polymorphism Identification
Basic quality checks were used to confirm that for each sample >80% of bases had a sequencing quality score ≥30 (regarded as high) and the mean quality score for all bases was ≥30. For qualifying samples, Illumina short reads of sufficient length (≥100 bp) were mapped to the strain JJ1886 chromosome [26] using the Burrows-Wheeler aligner [27]. Each alignment was analyzed for SNPs using the genome analysis tool kit software [28]. Single-nucleotide polymorphism loci were included if they were present in all the sample genomes, had a minimum coverage of 10×, were present in >90% of base calls for that position, and were not from duplicated or repetitive regions on the reference genome. Recombinant sites were identified and removed from the SNP matrix using Gubbins [29].
Phylogenetic Analyses
Model selection was performed in MEGA version 6.06 [30] using the matrix of nonrecombinant SNPs as input; the substitution model with the lowest Bayesian information criterion score (a criterion for model selection among a finite set of models, based partly on the likelihood function) was used for phylogenetic analyses. A phylogeny was estimated using the maximum-likelihood (ML) method, and node support was determined based upon 1000 bootstrap replicates. Phylogenetic analyses were conducted in MEGA version 6.06 [30].
Accessory Genome Analysis
Deoxyribonucleic acid sequences were assembled into contigs using CLC Genomics Workbench version 8.5.1 (QIAGEN, Waltham, MA). Contigs were screened for resistance determinants, virulence genes, and plasmid replicons using ResFinder [31], VirulenceFinder [32], and PlasmidFinder [33]. All searches were conducted using default parameters. Two-group comparisons of proportions were tested using Fisher's exact text (2-tailed).
RESULTS
Household Histories
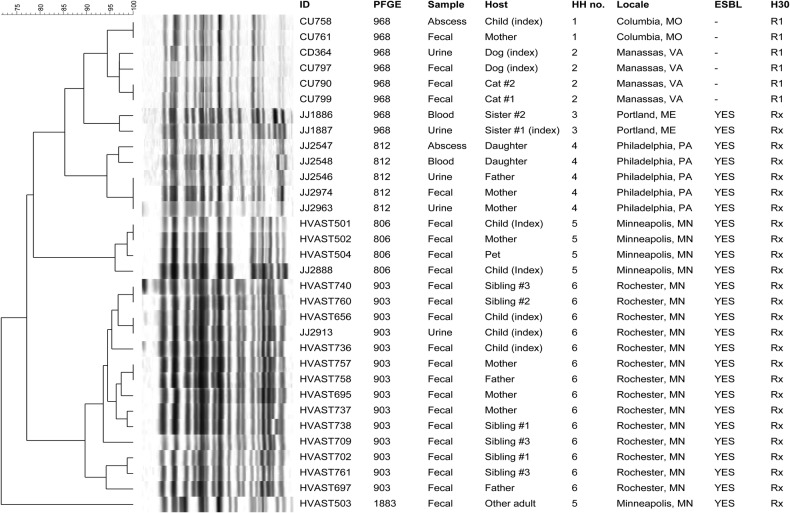
Household 1 included an otherwise healthy 8-month-old girl with E coli septic arthritis and bone abscess due to a fluoroquinolone-resistant ST131-H30R1 strain, plus her parents [18]. Fecal screening suggested that the mother (but not the father; data not shown) carried the girl's ST131-H30R1 clinical strain (Figure 1).
Figure 1.
Pulsed-field gel electrophoresis profiles of 33 clinical and fecal isolates of Escherichia coli sequence type 131 (ST131) from the members of 6 households. Scale is % profile similarity. Only ST131 isolates and the corresponding subjects are included; other isolates, and subjects without an ST131 isolate, are not shown. All isolates were fluoroquinolone resistant. H30, clonal subset within the ST131-H30 clade (R1 = H30R1, Rx = H30Rx). Abbreviations; ESBL, extended-spectrum β-lactamase production; HH, household; ID, identifier; PFGE, pulsotype.
Household 2 included a dog with recurrent bacteriuria due to a fluoroquinolone-resistant ST131-H30R1 strain and multiple other pets [19]. Fecal screening suggested that the index dog and 2 cats carried the dog's ST131-H30R1 clinical strain (Figure 1).
Household 3 comprised 2 sisters, 1 with recurrent UTI, 1 with fatal urosepsis after caring for her sister. Both PFGE (Figure 1) and WGS analysis [11] indicated that their multidrug-resistant E coli clinical isolates represented the same ST131-H30Rx strain (Table 1).
Household 4 included an elderly man and his adult daughter, who in 2008 both developed severe E coli kidney infections, seemingly caused by the same multidrug-resistant ST131 H30-Rx strain [21]. Six years later (2004), after the father's death, the daughter's mother also developed UTI due to a multidrug-resistant ST131 H30-Rx strain. Fecal screening suggested that the mother carried the daughter's UTI strain, which matched the father and daughter's shared 2008 clinical strain (Figure 1).
Household 5 included a 6-year-old girl with recurrent multidrug-resistant E coli UTI, her mother and godmother (both of whom had recently experienced multidrug-resistant E coli UTI), and 2 pets. Fecal screening suggested that the girl, the mother, and a pet carried the girl's ST131-H30Rx urine strain (Figure 1). The godmother also carried an ST131-H30Rx strain, but it differed markedly by PFGE from the household's other ST131 isolates (Figure 1).
Household 6 included a female infant with recurrent urosepsis due to a multidrug-resistant ST131-H30Rx strain, her parents, a pet dog, and 3 siblings, 1 of whom also recently experienced a multidrug-resistant E coli UTI [12]. Fecal screening suggested that at 1 or more of 3 samplings (weeks 1, 10, and 19), all individuals except the dog (data not shown) carried the infant's index ST131-H30Rx clinical strain (Figure 1).
Genome Data
Alignment of the short-read genomic libraries from the present 33 household ST131 isolates and 52 published ST131 genomes, followed by identification of conserved sequences and removal of putatively recombined regions, yielded a total core genome of 4 031 345 bp. This core genome yielded an SNP matrix containing 6526 SNPs, which were used to infer an ML tree (data not shown). In this tree, the minimal clade containing all the household isolates, which corresponded with H30R, included 46 comparison genomes, with the next-most-basal comparison isolate being CD306 (data not shown).
For higher resolution, the above process was repeated using the 33 household isolates plus the 46 corresponding comparison H30R genomes and CD306. The new core genome contained 4 050 617 nucleotides, and the new SNP matrix 2782 nucleotides. Removal of recombinant regions left 588 nucleotides for phylogenetic analysis.
Consensus Maximum-Likelihood Tree
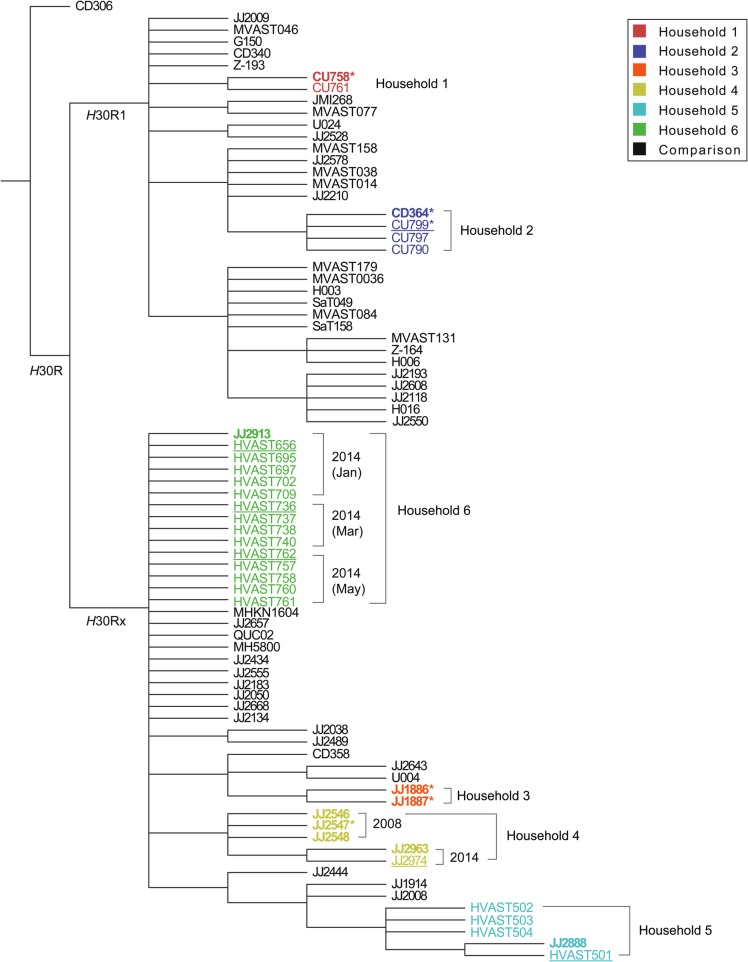
This SNP matrix was used to generate a bootstrapped 70% consensus ML tree based on a Kimura 2-parameter substitution model [34], using 1000 iterations (Figure 2). In this tree, the H30R1 and H30Rx subclades were resolved as distinct sister groups within the H30R clade. The household isolates were grouped as 6 well separated household-specific clusters, distributed between the H30R1 and H30Rx subclades as predicted by PCR-based typing, ie, households 1 and 2 were H30R1 and households 3–6 were H30Rx (Figure 2).
Figure 2.
Bootstrapped consensus core genome phylogeny for the 33 present household isolates and 47 comparison isolates of Escherichia coli sequence type 131 (ST131). The tree was based on 1000 bootstrapped maximum likelihood trees, retaining only those nodes that appeared in >70% of trees, and was rooted with strain CD306, which in the all-isolate phylogeny (data not shown) was basal. Clades H30R1 and H30Rx correspond, respectively, with clades C1 and C2 of Petty et al [8]. Branch lengths are meaningless. Household isolates are color-coded by household (1–6); comparison isolates, from Price et al [11], are shown in black. For the household isolates: boldface indicates clinical isolates; regular font indicates fecal isolates; underlining indicates fecal isolates from a clinical isolate's source host; and asterisks indicate the 6 household isolates that were included in Price et al [11], ie, JJ1886, JJ1887, JJ2547, CU758, CU799, and CD364 (which in Price et al was labeled as CD449). Dates are shown for the 2 households that underwent serial sampling.
For 4 households (1–3 and 6), the corresponding isolates were statistically indistinguishable at the core genome level, without evidence of meaningful within-household differentiation, whether across individuals or between fecal and clinical isolates (Figure 2). In contrast, households 4 and 5 exhibited within-household variation. Specifically, within household 4 the father and daughter's (indistinguishable) 2008 clinical isolates were distinct from the mother's (indistinguishable) 2014 clinical and fecal isolates. Likewise, within household 5 the index girl's (indistinguishable) clinical and fecal isolates were distinct from the other household members' (indistinguishable) fecal isolates, including the godmother's isolate HVAST503, with its unique pulsotype (Figure 1).
For 5 of the 6 households, the associated isolates were separated in the consensus tree from all epidemiologically unlinked comparison isolates. In contrast, the 15 isolates from household 6 (1 clinical, 14 fecal) shared an indistinguishable core genome not only with one another but also with 10 comparison isolates, which derived from diverse locales, hosts, and years [11] (Figure 2).
To test whether accessory genome content might differentiate the 15 household 6 isolates from the 10 epidemiologically unrelated but genomically similar comparison isolates, accessory traits were analyzed. The household isolates differed categorically from the comparison isolates by uniformly possessing plasmid-borne enterotoxin gene senB and 2 plasmid replicons, Col156 and IncI1 (Table 2). They also differed significantly for the prevalence of multiple other accessory traits (Table 2).
Table 2.
Accessory Traits of 25 Sequence Type 131 (ST131)-H30Rx Household and Comparison Isolates With Highly Similar Core Genomes
| Traita |
Prevalence of Trait, no. (Column %) |
P Valueb | ||
|---|---|---|---|---|
| Category | Specific Trait | Household 6 (n = 15) | Comparison (n = 10) | |
| Resistance gene | aac(3′)-IIa | 13 (87) | 2 (20) | .002 |
| aadA5 | 15 (100) | 4 (40) | .001 | |
| blaCTX-M-15 | 15 (100) | 5 (50) | .005 | |
| blaTEM-1B | 15 (100) | 4 (40) | .001 | |
| dfrA17 | 15 (100) | 5 (50) | .005 | |
| mph(A) | 15 (100 | 2 (20) | <.001 | |
| strA | 15 (100) | 1 (10) | <.001 | |
| sul1 | 15 (100) | 4 (40) | .001 | |
| sul2 | 15 (100) | 1 (10) | <.001 | |
| tetA | 15 (100) | 3 (30) | <.001 | |
| Virulence gene | senB | 15 (100) | 0 (0) | <.001 |
| Plasmid marker | ColI156 | 15 (100) | 0 (0) | <.001 |
| IncFIA | 2 (13) | 8 (80) | .002 | |
| IncFIB(AP001918) | 15 (100) | 1 (10) | <.001 | |
| IncI1 | 15 (100) | 0 (0) | <.001 | |
| ≥1 of 6 markersc | 0 (0) | 6 (60) | .001 | |
a Total number of traits sought: resistance-associated, 2084; virulence-associated, 102; plasmid-associated, 109. Traits listed are those that yielded P < .05 in between-group comparisons. Additional traits detected in ≥ 1 isolate each, but that yielded P ≥ .05, included (overall prevalence): aac(3)-IId (24%), blaCTX-M-19 (4%), blaOXA-1 (28%), catB3 (28%), dfrA14 (4%), sat (96%), ccl (4%), cnf1 (4%), gad (100%), iha (96%), nfaE (12%), ColI(BS512) (16%), ColI(MGS31) (4%), and colI(MG828) (20%).
b By Fisher's exact test (2-tailed).
c The 6 plasmid markers, which were present in 1–2 isolates each, included ColRNAI, ColI(MGS31), ColI (RNAI), IncB/O/K/Z, IncN, IncX1, and IncY.
DISCUSSION
This study, the first large-scale WGS analysis of household-source E coli ST131 isolates, provides robust confirmation of within-household strain sharing involving ST131, as described previously with less reliable typing methods such as PFGE [18-19]. It also strongly supports the fecal-urethral hypothesis of UTI pathogenesis, host-specific microevolution of ST131 variants within a household, and long-term within-household persistence of a virulent ST131 strain. Thus, our WGS analysis provides a more robust understanding of ST131 strain dynamics within households.
Consistent with previous evidence that PFGE can misrepresent genomic similarity relationships between different ST131 strains [11], here PFGE and WGS disagreed within 4 of the 6 households. For across-household strain differences, WGS analysis clearly resolved the isolates from households 1–3 into distinct household-specific clades, whereas PFGE assigned them all to the same pulsotype, albeit with clustering by household in the PFGE dendrogram. For within-household strain similarity, whereas WGS analysis identified all household 5 isolates as representing a single strain, PFGE identified the godmother's isolate (HVAST503) as representing a unique pulsotype that was quite distant in the dendrogram. This discrepancy likely derives from the WGS phylogram being based only on non-recombined core genome regions (<20% of the genome), whereas PFGE considers the entire genome, including accessory traits and recombined core regions, which can obscure underlying clonal relationships. These findings suggest that WGS analysis provides more accurate clonal discrimination and could supersede PFGE for molecular epidemiology once it becomes sufficiently available, affordable, and simple [35, 36].
Our findings strongly support the fecal-urethral hypothesis [14, 15] by documenting multiple instances in which an individual's clinical and fecal isolates shared indistinguishable core genomes. Moreover, the index clinical isolate often was as similar to other household members' fecal isolates as to the index host's fecal isolate. Thus, WGS analysis supports extending the fecal-urethral hypothesis to include the entire household. These data suggest that the fecal microbiota of multiple household members conceivably could have been the source for the index infection episode. Compared with the 2 previous studies that provided WGS support for the fecal-urethral hypothesis [17, 37], ours is much larger and uniquely addresses ST131 [17, 37].
Our findings also confirm that for ST131, the household is a critical ecological unit. Household clustering of E coli strains was documented previously in a large PFGE-based cross-sectional survey [13]. However, unlike the present study, that study suggested strain sharing across households, possibly because it predated the ST131 emergence, was based in 1 locale, and relied on PFGE. In this study, ST131 strains were non-overlapping across households, and accessory genome analysis disproved the only instance of seeming commonality between household isolates versus comparison isolates.
It is interesting to note that in 3 of 6 households the index subject was a young child. This may simply reflect sampling bias, with pediatricians perhaps being more likely than other clinicians to refer cases of multidrug-resistant E coli infection for strain typing due to their comparative rarity in children. However, it also may indicate that young children, who ingest and excrete microorganisms freely, are especially likely to share multidrug-resistant E coli with other household members.
The 6-year persistence of an ST131-H30Rx strain in household 4 equals or exceeds previous persistence records for commensal E coli, ie, 6 years [38], 3.9 years (https://opac.library.usyd.edu.au:443/record=b4803147~S4), and 3 years [17]. Some evidence supports that ST131-H30 strains may persist longer than other antimicrobial-resistant E coli [39], possibly promoting the ST131 pandemic. It is notable that the core genome of household 4's ST131-H30Rx strain was quite stable over 6 years, consistent with the low background mutation rate of E coli, including ST131 [17, 40].
The study has limitations. First, the nature and functional significance of the observed within-household core genome variation, including between clinical and fecal isolates, remains undefined. Second, accessory genome content was not compared between clinical and fecal isolates to identify possible niche-adaptive traits. Third, the primarily cross-sectional design and minimal within-household core genome variation precluded inferences regarding transmission. Fourth, the comparison isolates represented diverse locales, time periods, and host species; more closely matched comparison isolates might yield more evidence of commonality with household isolates. Study strengths include the unique set of 10 clinical and 23 fecal ST131 isolates from 20 members of 6 households; longitudinal follow-up in 2 households; comparison of conventional typing and WGS analysis; and leveraging of a large set of comparison ST131 genomes.
CONCLUSIONS
In summary, WGS analysis provided strong evidence that ST131 strains cluster by household, within which they are shared extensively among different individuals, behave both as commensals and pathogens, and sometimes persist for extended periods, causing recurrent infections. The findings support and extend the fecal-urethral hypothesis for UTI pathogenesis and establish the household as an important functional unit in the ecology of H30R1 and H30Rx. This suggests that future risk assessment, surveillance, and decolonization efforts may need to consider household members as well as patients.
Acknowledgments
We thank Nicole Stoesser for providing helpful editorial suggestions.
Disclaimer. The opinions expresses here are strictly those of the authors and do not necessarily reflect those of the author's respective institutions, the Department of Veterans Affairs, or the National Institutes of Health.
Financial support. This material is based in part on work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs (grant nos. 1 I01 CX000192 01 and 1 I01 CX000920-01; to J. R. J.); National Institutes of Health (NIH) grant no. R01AI106007 (to E. V. S.); and NIH grant no. 2R21-AI117654 (to L. B. P. and J. R. J.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect 2003; 5:449–56. [DOI] [PubMed] [Google Scholar]
- 2.Gerver S, Mihalkova M, Abernethy J et al. . Escherichia coli bacteraemia mandatory reports, 2014/15. Summary of the Mandatory Surveillance Annual Epidemiological Commentary, 2014/15. London: Public Health England, 2015: pp 4–5. [Google Scholar]
- 3.Russo TA, Johnson JR. A proposal for an inclusive designation for extraintestinal pathogenic Escherichia coli: ExPEC. J Infect Dis 2000; 181:1753–4. [DOI] [PubMed] [Google Scholar]
- 4.Manges AR, Tabor H, Tellis P et al. . Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg Infect Dis 2008; 14:1575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tchesnokova V, Billig M, Chattopadhyay S et al. . Predictive diagnostics for Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J Clin Microbiol 2013; 51:2991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibreel TM, Dodgson AR, Cheesbrough J et al. . Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 2012; 67:346–56. [DOI] [PubMed] [Google Scholar]
- 7.Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 2014; 20:380–90. [DOI] [PubMed] [Google Scholar]
- 8.Petty NK, Ben Zakour N, Stanton-Cook M et al. . Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 2014; 111:5694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson JR, Tchesnokova V, Johnston B et al. . Abrupt emergence of a single dominant multi-drug-resistant strain of Escherichia coli. J Infect Dis 2013; 207:919–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colpan A, Johnston B, Porter S et al. . Escherichia coli sequence type 131 (ST131) as an emergent multidrug-resistant pathogen among U.S. veterans. Clin Infect Dis 2013; 57:1256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price LB, Johnson JR, Aziz M et al. . The epidemic of ESBL-producing Escherichia coli ST131 is driven by a single highly virulent subclone, H30-Rx. MBio 2013; 6:e00377–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madigan T, Johnson JR, Clabots C et al. . Extensive household outbreak of urinary tract infection and intestinal colonization due to extended-spectrum beta lactamase (ESBL)-producing Escherichia coli sequence type 131 (ST131). Clin Infect Dis 2015; 61:e5–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JR, Owens K, Gajewski A, Clabots C. Escherichia coli colonization patterns among human household members and pets, with attention to acute urinary tract infection. J Infect Dis 2008; 197:218–24. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto S, Tsukamoto T, Terai A et al. . Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol 1997; 157:1127–9. [PubMed] [Google Scholar]
- 15.Grüneberg RN. Relationship of infecting urinary organism to the faecal flora in patients with symptomatic urinary infection. Lancet 1969; 1:766–8. [DOI] [PubMed] [Google Scholar]
- 16.Ulleryd P, Sandberg T, Scheutz F et al. . Colonization with Escherichia coli strains among female sex partners of men with febrile urinary tract infection. J Clin Microbiol 2015; 53:1947–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeves PR, Liu B, Zhou Z et al. . Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 2011; 6:e26907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson JR, Anderson JT, Clabots C et al. . Within-household sharing of a fluoroquinolone-resistant Escherichia coli sequence type ST131 strain causing pediatric osteoarticular infection. Pediatr Infect Dis J 2010; 29:474–5. [DOI] [PubMed] [Google Scholar]
- 19.Johnson JR, Miller S, Johnston B et al. . Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household . J Clin Microbiol 2009; 47:3721–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owens RC, Johnson JR, Stogstill P et al. . Community transmission in the United States of a CTX-M-15-producing sequence type ST131 Escherichia coli strain resulting in death. J Clin Microbiol 2011; 49:3406–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ender PT, Gajanana D, Johnston B et al. . Transmission of extended-spectrum beta-lactamase-producing Escherichia coli (sequence type ST131) between a father and daughter resulting in septic shock and emphysematous pyelonephritis. J Clin Microbiol 2009; 47:3780–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walk ST, Alm E, Gordon DM et al. . Cryptic lineages of the genus Escherichia. Appl Environ Microbiol 2009; 75:6534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 20th Informational Supplement. CLSI document M100-S20: Wayne, PA: Clinical and Laboratory Standards Institute; 2010. [Google Scholar]
- 24.Banerjee R, Robicsek A, Kuskowski M et al. . Molecular epidemiology of Escherichia coli sequence type ST131 and its H30 and H30-Rx subclones among extended-spectrum beta-lactamase-positive and -negative E. coli clinical isolates from the Chicago region (2007-2010). Antimicrob Agents Chemother 2013; 57:6385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JR, Nicolas-Chanoine M, Debroy C et al. . Comparison of Escherichia coli sequence type ST131 pulsotypes by epidemiologic traits, 1967–2009. Emerg Infect Dis 2012; 18:598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen PS, Stegger M, Aziz M et al. . Complete genome sequence of the epidemic and highly virulent CTX-M-15-producing H30-Rx subclone of Escherichia coli ST131. Genome Announc 2013; 1:e00988–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A, Hanna M, Banks E et al. . The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croucher M, Page A, Connor T et al. . Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D et al. . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zankari E, Hasman H, Cosentino S et al. . Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67:2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joensen K, Scheutz F, Lund O et al. . Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 2014; 52:1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carattoli A, Zankari E, García-Fernández A et al. . In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58:3895–03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980; 16:111–20. [DOI] [PubMed] [Google Scholar]
- 35.Salipante SJ, Roach DJ, Kitzman J et al. . Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 2015; 15:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snitkin E, Zelazny A, Thomas P et al. . Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 2012; 4:148ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weissman SJ, Beskhlebnaya V, Chesnokova V et al. . Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin. Infect Immun 2007; 75:3548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clermont O, Lescat M, O'Brien CL et al. . Evidence for a human-specific Escherichia coli clone. Environ Microbiol 2008; 10:1000–6. [DOI] [PubMed] [Google Scholar]
- 39.Overdevest I, Haverkate M, Veenemans J et al. . Prolonged colonization with Escherichia coli O25:ST131 versus other extended-spectrum β-lactamase-producing E. coli in a long-term care facility with a high endemic level of rectal colonization, the Netherlands, 2013–2014. Euro Surveill (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoesser N, Sheppard AE, Pankhurst L et al. . Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio 2016; 7:e02162–15. [DOI] [PMC free article] [PubMed] [Google Scholar]