Abstract
We report the case of a 60-year-old man with septic shock due to Capnocytophaga canimorsus that was diagnosed in 24 hours by a novel whole-genome next-generation sequencing assay. This technology shows great promise in identifying fastidious pathogens, and, if validated, it has profound implications for infectious disease diagnosis.
Keywords: high-throughput nucleotide sequencing, microbiological techniques, sepsis/diagnosis
Mortality due to sepsis is high worldwide and further increased in the 20% of patients who receive inappropriate, mismatched antimicrobial therapy [1, 2]. In up to 40% of cases, no organism is identified [3], highlighting the need for more sensitive pathogen identification technologies. Next-generation sequencing (NGS) has the potential to detect a broad range of pathogens with exceptional sensitivity [4, 5] and provide clinically actionable results [6]. In this study, we report a case of septic shock due to Capnocytophaga canimorsus diagnosed by a novel NGS method prior to and confirmed by results of conventional blood cultures. This NGS method, if validated, has the potential to improve (1) the identification of causative organisms in patients with sepsis and (2) the delivery of appropriate, pathogen-directed antibiotic therapy.
CASE PRESENTATION
A 60-year-old man presented to an outside hospital with fever, hypotension, and altered mental status. He had been well until the day of presentation when he developed low back pain, epigastric pain, chills, and vomiting. Several hours later, his wife found him disoriented with dusky discoloration of his face and extremities. The patient's history was notable for splenectomy in 1974 after trauma and a complicated cholecystectomy in 2014 requiring multiple laparotomies. He took no medications and denied recent travel and sick contacts, but within the last week, he had sustained several bites and scratches from the family German shepherd. In the emergency department, he was given intravenous fluids, 2 units of packed red blood cells, and levofloxacin. A computed tomography (CT) scan of the chest, abdomen, and pelvis without contrast demonstrated patchy bilateral lung parenchymal opacities consistent with acute respiratory distress syndrome. Shortly after presentation, the patient was intubated for worsening confusion and hypoxemia and transferred to our hospital.
Upon arrival to our hospital, the patient was heavily sedated and mechanically ventilated. Vital signs were notable for a blood pressure of 80/58 mmHg, heart rate of 111 beats per minute, and temperature of 38.5°C. Physical exam revealed coarse breath sounds, absent peripheral pulses with cold, dusky extremities, mottling of the skin throughout, and rare violaceous purpura on the thighs. Superficial linear scratches with surrounding erythema were noted on the upper extremities and attributed by the patient's wife to their dog. Intravenous fluids and vasopressors were administered. Two blood cultures were collected. Laboratory studies were notable for white blood cell count 18.0 × 109/L (50% bands), platelet count 14 × 109/L, international normalized ratio 1.7, fibrinogen 178 mg/dL, creatinine 3.8 mg/dL (baseline 0.9 mg/dL), lactate 5.6 mmol/L, and PaO2 60 mmHg (FiO2 0.5 and positive end-expiratory pressure 10 cm H2O). Urinalysis showed 3 erythrocytes and 3 leukocytes per high-power field, negative leukocyte esterase, positive nitrite, 2+ protein, 1+ bilirubin, and >50 bacteria.
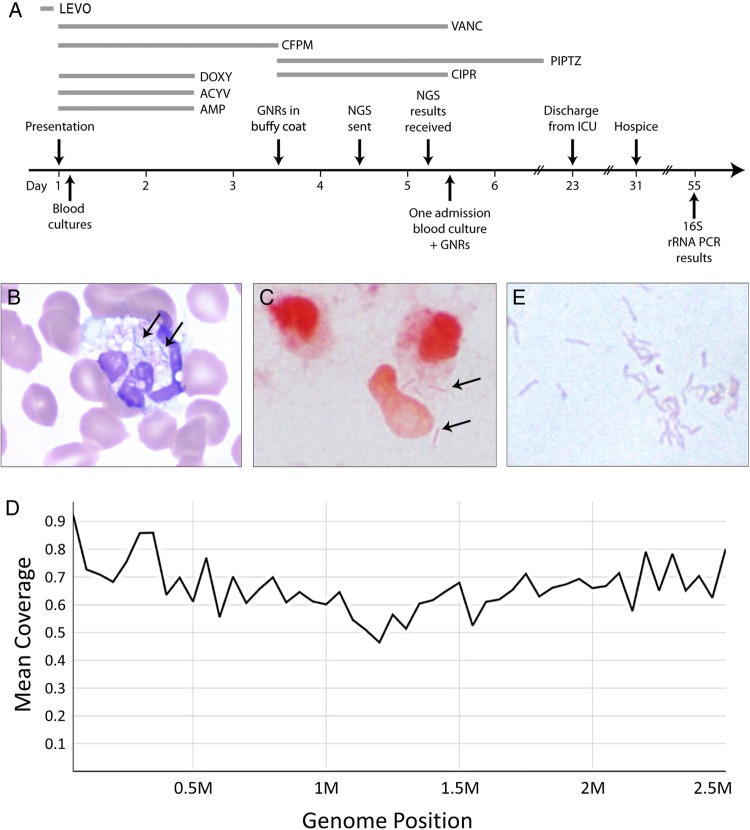
Given the concern for a severe infection, empiric broad-spectrum antibiotics consisting of vancomycin, cefepime, ampicillin, doxycycline, and acyclovir were administered to cover typical bacterial pathogens, atypical organisms (eg, Rickettsia), and bacterial and viral central nervous system infections (Figure 1A). A CT scan of the head without contrast was negative for acute abnormalities. A lumbar puncture was performed and revealed 11 nucleated cells/µL (51% neutrophils, 30% lymphocytes, 14% monocytes/macrophages), 531 erythrocytes/µL, glucose 81 mg/dL (finger stick, 101 mg/dL), and protein 100 mg/dL. Cerebrospinal fluid Gram stain and herpes simplex virus polymerase chain reaction (PCR) were negative, and ampicillin and acyclovir were discontinued. Continuous renal replacement therapy was initiated due to oliguria. A peripheral blood Wright's stain showed neutrophils with blue inclusions resembling bacterial rods (Figure 1B), which were confirmed as Gram-negative rods by buffy coat Gram stain (Figure 1C). Given the history of dog bites, cefepime was changed to piperacillin-tazobactam and ciprofloxacin for better coverage of Pasteurella multocida and Capnocytophaga canimorsus. Despite finding Gram-negative rods in the peripheral blood buffy coat, admission blood cultures remained negative. Because of the uncertainty of the pathogen responsible for the patient's overwhelming sepsis, on hospital day 4 a sample of the patient's plasma collected upon hospital admission was sent for analysis by a novel investigational assay to screen for circulating microbial deoxyribonucleic acid (DNA).
Figure 1.
(A) Timeline of events. Piperacillin-tazobactam (PIPTZ) was administered for a total of 14 days. (B) Wright-Giemsa stain of peripheral blood smear demonstrating blue bacilli (arrows) within neutrophils (original magnification, 1000×). (C) Gram stain of buffy coat demonstrating Gram-negative rods ([GNRs] arrows) within and around neutrophils (original magnification, 1000×). (D) Average alignment coverage over the entire Capnocytophaga canimorsus genome. Overall coverage is 0.71×. (E) Gram-negative rods cultured from blood (original magnification, 1000×) later confirmed by 16S sequencing to be C canimorsus. Abbreviations: ACYV, acyclovir; AMP, ampicillin; CFPM, cefepime; CIPR, ciprofloxacin; DOXY, doxycycline; ICU, intensive care unit; LEVO, levofloxacin; NGS, next-generation sequencing; PCR, polymerase chain reaction; rRNA, ribosomal ribonucleic acid; VANC, vancomycin.
METHODS
After obtaining emergency approval from the Institutional Review Board (IRB) at Duke University Medical Center (IRB no. Pro00070628) and informed consent from the patient's wife, a plasma sample from the day of hospital admission was prepared and sent to Karius, Inc. (Menlo Park, CA) for analysis. Deoxyribonucleic acid libraries for NGS were prepared as previously described [7, 8]. Sequencing was performed on an Illumina NextSeq instrument and analysis was performed by Karius. In brief, after removing low-quality reads, reads were mapped to the human reference genome (hg19). Remaining reads were mapped to a curated reference database of viral, bacterial, fungal, and other eukaryotic pathogens. Further analysis was performed to identify sequences known to confer resistance to β-lactams [9]. Additional details are provided in the Supplementary Methods.
RESULTS
Within 24 hours, the NGS assay detected high levels of microbial DNA (851 306 genome copies/mL plasma) with reads that aligned along the entirety of the C canimorsus genome (Figure 1D). Because the sequencing results fit with the buffy coat Gram stain, the recent history of dog bites, and the clinical picture of septic shock in an asplenic patient, and because no other convincing hits for other pathogens were identified by sequencing, the presumptive diagnosis was C canimorsus infection. The analysis did not detect any β-lactamase DNA sequences (eg, cfxA2 and cfxA3) [9]. The antibiotic regimen was narrowed to piperacillin-tazobactam monotherapy. Subsequently, at the end of hospital day 5, 1 of 2 admission blood cultures grew Gram-negative rods (Figure 1E). Gram stain morphology showed thin, faintly staining Gram-negative fusiform bacilli. Definitive identification of the organism using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF) was attempted but failed because of an inadequate database of prior Capnocytophaga isolates at our institution. The specimen was sent to a reference laboratory for 16S ribosomal ribonucleic acid (rRNA) PCR sequencing, which, despite 2 attempts, was unsuccessful due to poor growth of the organism.
The patient's clinical course was further complicated by Clostridium difficile colitis and Candida tropicalis fungemia. On hospital day 26, the patient developed recurrent sepsis due to progressive gangrene of both upper and lower extremities. Rather than proceed with multiple amputations, and because of the patient's poor prognosis, persistent dialysis dependence, and previously stated end-of-life wishes, the patient's wife elected to transition to comfort care only. The patient subsequently died on day 40. Postmortem, a third attempt at 16S sequencing was performed on the blood culture isolate and this time successfully identified the organism as C canimorsus (Figure 1E), 55 days after initial blood culture collection.
DISCUSSION
We report a case of septic shock and multiple organ failure due to C canimorsus that was diagnosed by plasma whole-genome NGS and confirmed by 16S rRNA sequencing. Capnocytophaga canimorsus is a fastidious Gram-negative bacteria found in the saliva of dogs and cats. Human infection can occur after animal bites or scratches, especially in those with anatomic or functional asplenia or heavy alcohol use [10]. The organism gains access to the blood stream and proliferates rapidly, especially in asplenic patients, causing high-grade bacteremia, septic shock, multiple organ failure, and death in up to one quarter of patients [10].
Although C canimorsus bacilli can be seen on peripheral blood smear or buffy coat Gram stain, as was the case in our patient, identification of the organism by conventional microbiological techniques is difficult and can take days to weeks owing to slow growth [11]. Presumptive identification can occasionally be made based on colony morphology and gliding motility [12]; however, definitive identification often requires 16S rRNA sequencing, which can take several additional days to weeks. In our patient, confirmation was delayed due to the fastidious nature of the pathogen and technical difficulties with the 16S sequencing, highlighting the shortcomings of this technique.
Next-generation sequencing is a powerful tool with the potential to revolutionize infectious disease diagnosis [4–6] and offers several distinct advantages over existing pathogen identification techniques. Unlike Sanger sequencing, which lacks sufficient throughput to detect microbial DNA directly from patient samples with high human DNA background, or MALDI-TOF, which requires positive cultures and well validated reference databases [5], or 16S rRNA sequencing, which is not available directly from blood, or PCR-based assays, which identify only a limited set of bacterial species, the novel NGS approach described herein is as follows: (1) it combines NGS, molecular biology techniques, and informatics to filter human sequences and identify pathogen sequences directly from patient plasma; (2) it is unbiased and detects virtually any microorganism; (3) it is high throughput and returns results within a clinically actionable timeframe; (4) it is culture-independent and allows identification of fastidious organisms; and (5) it can potentially screen for known antibiotic resistance genes. To our knowledge, no other validated tools exist with these characteristics.
CONCLUSIONS
This case is the first to report the use of whole-genome NGS to identify a pathogen in plasma and the first to report its use in a septic critically ill patient. This case highlights the great promise of NGS to provide data within a clinically actionable timeframe and has implications for ensuring delivery of pathogen-targeted antibiotics and improving antibiotic stewardship. Although our NGS findings in this case were confirmed by conventional blood culture and 16S rRNA sequencing, this NGS assay requires further validation in a clinical trial.
Supplementary Data
Supplementary material is available online at Open Forum Infectious Diseases online (http://OpenForumInfectiousDiseases.oxfordjournals.org/).
Acknowledgments
Author contributions. M. K. A., A. S. B., K. W., and B. D. K. drafted the manuscript, which was approved by all authors. M. K. A., A. S. B., K. W., E. F., A. S., B. M. H., B. A. B., A. C. S., S. F. C., B. D. A., C. M. M., M. O. A., V. G. F., and B. D. K. provided clinical care to the patient and/or collected/interpreted clinical data. B. S.-K. and F. R. prepared the plasma samples. D. K. H., T. A. B., and M. K. performed the DNA sequencing, analysis, and interpretation.
Financial support. This work was supported by the National Institutes of Health (grant K24-AI093069; to V. G. F).
Potential conflicts of interest. B. D. A. receives royalties from Up-To-Date; she is the recipient of research grants from Cidara, Astellas, and Synexis; and she is a paid consultant for Astellas, Cidara, and Shire pharmaceuticals. C. M. M. is a paid consultant for Almac Diagnostics, outside the context of the submitted work. D. K. H., T. A. B, and M. K. are employees at Karius, Inc. V. G. F. is the Chair of the Scientific Advisory Board for Merck V710; he is a paid consultant for Pfizer, Novartis, Galderma, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, Medicines Co., Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Cubist, Basilea, Affinergy; he has grants pending from Karius, Contrafect, MedImmune, Actavis/Forest/Cerexa, Pfizer, Merck/Cubist, Advanced Liquid Logics, Theravance, and Novartis; he receives royalties (Up-To-Date) and personal fees for development or presentation of educational presentations outside the submitted work (Green Cross, Cubist, Cerexa, Durata, Theravance); and he has a patent pending related to sepsis diagnostics. B. D. K. reports a working relationship with 12th Man Technologies outside the scope of the submitted work and receives research funding from the National Institutes of Health and Defense Advanced Research Projects Agency. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Valles J, Rello J, Ochagavia A et al. Community-acquired bloodstream infection in critically ill adult patients: impact of shock and inappropriate antibiotic therapy on survival. Chest 2003; 123:1615–24. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Ellis P, Arabi Y et al. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest 2009; 136:1237–48. [DOI] [PubMed] [Google Scholar]
- 3.Vincent JL, Sakr Y, Sprung CL et al. Sepsis in European intensive care units: results of the SOAP study. Crit Care Med 2006; 34:344–53. [DOI] [PubMed] [Google Scholar]
- 4.Dunne WM Jr, Westblade LF, Ford B. Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis 2012; 31:1719–26. [DOI] [PubMed] [Google Scholar]
- 5.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. Next-generation sequencing for infectious disease diagnosis and management: a report of the Association for Molecular Pathology. J Mol Diagn 2015; 17:623–34. [DOI] [PubMed] [Google Scholar]
- 6.Wilson MR, Naccache SN, Samayoa E et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 2014; 370:2408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Vlaminck I, Khush KK, Strehl C et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 2013; 155:1178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Vlaminck I, Martin L, Kertesz M et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A 2015; 112:13336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jolivet-Gougeon A, Sixou JL, Tamanai-Shacoori Z, Bonnaure-Mallet M. Antimicrobial treatment of Capnocytophaga infections. Int J Antimicrob Agents 2007; 29:367–73. [DOI] [PubMed] [Google Scholar]
- 10.Butler T. Capnocytophaga canimorsus: an emerging cause of sepsis, meningitis, and post-splenectomy infection after dog bites. Eur J Clin Microbiol Infect Dis 2015; 34:1271–80. [DOI] [PubMed] [Google Scholar]
- 11.Lion C, Escande F, Burdin JC. Capnocytophaga canimorsus infections in human: review of the literature and cases report. Eur J Epidemiol 1996; 12:521–33. [DOI] [PubMed] [Google Scholar]
- 12.Meybeck A, Aoun N, Granados D et al. Meningitis due to Capnocytophaga canimorsus: contribution of 16S RNA ribosomal sequencing for species identification. Scand J Infect Dis 2006; 38:375–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.