Abstract
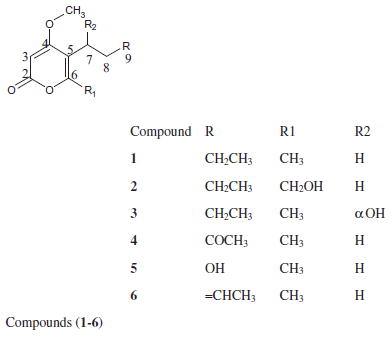
Four new (1–4) and two known (5 and 6) α-pyrone derivatives have been isolated from Alternaria phragmospora, an endophytic fungus from Vinca rosea, leaves. The isolated compounds were chemically identified to be 5-butyl-4-methoxy-6-methyl-2H-pyran-2-one (1), 5-butyl-6-(hydroxymethyl)-4-methoxy-2H-pyran-2-one (2), 5-(1-hydroxybutyl)-4-methoxy-6-methyl-2H-pyran-2-one (3), 4-methoxy-6-methyl-5-(3-oxobutyl)-2H-pyran-2-one (4), 5-(2-hydroxyethyl)-4-methoxy-6-methyl-2H-pyran-2-one (5), and 5-[(2E)-but-2-en-1-yl]-4-methoxy-6-methyl-2H-pyran-2-one (6). Compounds 2 and 4 showed moderate antileukemic activities against HL60 cells with IC50 values of 2.2 and 0.9 μM and against K562 cells with IC50 values of 4.5 and 1.5 μM, respectively.
Keywords: Antileukemic, α-Pyrone derivatives, Alternaria phragmospora, Endophytic fungi
Introduction
The endophytic fungi can be considered as an unexplored source for chemically novel and biologically active secondary metabolites in a wide variety of medical and agricultural areas.1 The genus Alternaria was established in 1817, and contains 44 different species. In the last few decades more than 268 metabolites from different species of Alternaria fungi have been reported.2 The isolated metabolites from Alternaria fungi were found to be belonging to several chemical classes including nitrogen-containing compounds,3 steroids,4 terpenoids,5 pyranones,6 quinones,7 phenolics,8 and various other classes.2 The isolated metabolites showed different biological activities: antitumor, herbicide, antimicrobial, antimalarial, and antileishmanial.2 However the endophytic fungus Alternaria phragmospora has not been examined phytochemically or biologically before. The crude ethyl acetate extract of the cultured endophytic fungus A. phragmospora showed antileukemic activities against Leukemic K562 cells and HL60 cells with IC50 values of 0.035 and 0.045 μg/ml, respectively. Consequently, it was chosen for further investigations seeking for isolation and identification of the secondary metabolites which may be responsible for this activity. Leukemic chronic K562 cells and acute HL60 cells have been chosen as models in this research. The K562 cell line is composed of undifferentiated blast cells that are rich in glycophorin and may be induced to produce fetal and embryonic hemoglobin in the presence of hemin, while the HL-60 is a promyelocytic leukemia cell line which in the presence of DMSO, matures into granulocytes and when exposed to phorbol esters, differentiate into nondividing mononuclear phagocytes.9,10
Results and discussion
In this study, phytochemical and biological investigations for the endophytic fungus are reported. The ethyl acetate extract was subjected to different chromatographic techniques which led to the isolation and structural elucidation of four new (1–4) and two known (5 and 6) α-pyrone derivatives. We herein report the isolation, structural elucidation, including relative and absolute configuration, and the results of some bioactivity tests of these compounds.
Compound 1 was isolated as white crystalline needles. The molecular formula was determined to be C11H16O3 by HR-ESI-MS (+ve mode) showing molecular ion peak [M+H]+ at m/z 197.1176 (calcd for C11H17O3, 197.1178) indicating four degrees of unsaturation. The IR spectrum of 1 showed absorption bands at 1720, 1647, and 1563 cm−1 indicating the presence of a lactone group and α-pyrone ring. The 13C NMR spectrum (Table 2) of compound 1 displayed 11 carbon resonances, while the DEPT 135 experiment sorted these signals into two methyl groups at (δC 13.8) and (δC 17.097), one methoxy group at (δC 55.9), three methylenes at (δC 22.4, 23.8, and 31.3), one methine at (δC 87.6), and four quaternary carbons signals, one of them was found to be an ester carbonyl at (δC 164.5) while the other three were found to be resonating at (δC 170.8, 157.8, and 111.6). By analysis of 1H NMR (Table 1) and HMQC spectral data, compound 1 exhibited two signals for methyl groups [δ 0.86 (3H, t, (6.8), H-10) and δ 2.11 (s, 3H, H-11)], a signal of aromatic proton at δ 5.34 (s, H-3), a methoxy group signal at δH 3.72 (s), and three methylene group signals [δ 1.25 (m, 2H, H-9), 1.26 (m, 2H, H-8), and 2.21 (t, 7.6, 2H, H-7)]. The α-pyrone ring structure was confirmed by several HMBC correlations between H-3 and both of C-4 and C-5. HMBC spectrum confirmed the presence of a butyl side chain by showing some correlations between H-10 to both of C-9 and C-8, H-7 to both of C-8 and C-9. The attachment of butyl side chain at C-5 was confirmed from HMBC correlations of H-7 to C-4, C-5, and C-6. The attachment of the methoxy group was confirmed by a HMBC correlation between the methoxy group and C-4. The structure of 1 was authenticated by 2D NMR experiments, giving pertinent COSY and HMBC correlations (Fig. 2). Compound 1 was suggested to be 5-butyl-4-methoxy-6-methyl-2H-pyran-2-one. The assigned chemical structure of 1 was confirmed by X-ray diffraction analysis. Crystals of 1 suitable for X-ray diffraction were obtained by slow evaporation of a solution of 1 in methanol–water (30:70). The final X-ray crystallographic model of 1 (Fig. 1) confirmed the structure of 1 as shown. The asymmetric unit contains two independent molecules, one of which (not shown) is disordered, with two conformations of the n-butyl substituent.
Table 2.
13C NMR spectroscopic data (400 MHz in CDCl3) for (1–4)
| Position |
δC
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| 2 | 164.5 | 164.8 | 163.8 | 167.0 |
| 3 | 87.6 | 89.1 | 89.0 | 88.2 |
| 4 | 170.8 | 170.9 | 170.4 | 172.8 |
| 5 | 111.6 | 113.4 | 113.1 | 112.0 |
| 6 | 157.8 | 158.1 | 159.0 | 160.1 |
| 7 | 23.8 | 23.3 | 68.6 | 19.4 |
| 8 | 31.3 | 32.0 | 39.2 | 42.6 |
| 9 | 22.4 | 22.4 | 19.4 | 210.1 |
| 10 | 13.8 | 13.8 | 13.4 | 29.5 |
| 11 | 17.0 | 58.5 | 17.8 | 17.0 |
| O–CH3 | 55.9 | 56.3 | 56.2 | 57.0 |
Table 1.
1H NMR spectroscopic data (400 MHz in CDCl3) for (1–4)
| Position |
δH (J in Hz)
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| 3 | 5.34, s | 5.48, s | 5.49, s | 5.57, s |
| 7 | 2.21, t (7.6) | 2.35, t (7.6) | 4.63, m | 2.62, m |
| 8 | 1.26, m | 1.34, m | 1.65, m | 2.64, m |
| 8a: 1.84, m | ||||
| 9 | 1.25, m | 1.28, m | 8b: 1.27, m | – |
| 10 | 0.86, t (6.8) | 0.86, t (7.2) | 0.93, t (7.2) | 2.14, s |
| 11 | 2.11, s | 4.41, s | 2.27, s | 2.28, s |
| O–CH3 | 3.72, s | 3.80, s | 3.85, s | 3.88, s |
Figure 2.
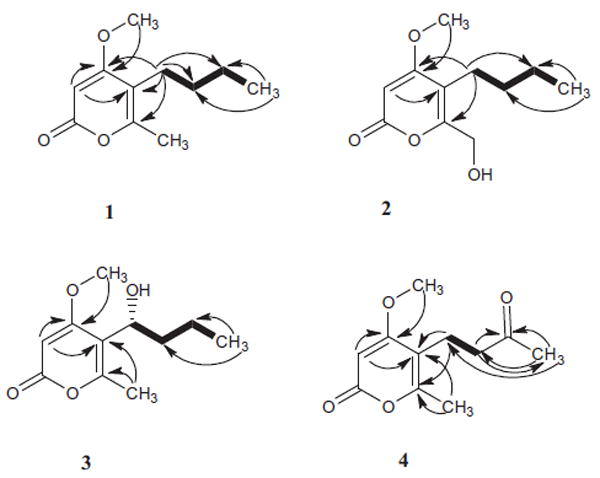
Key HMBC
 and H–H cosy
and H–H cosy
 correlations for (1–4).
correlations for (1–4).
Figure 1.
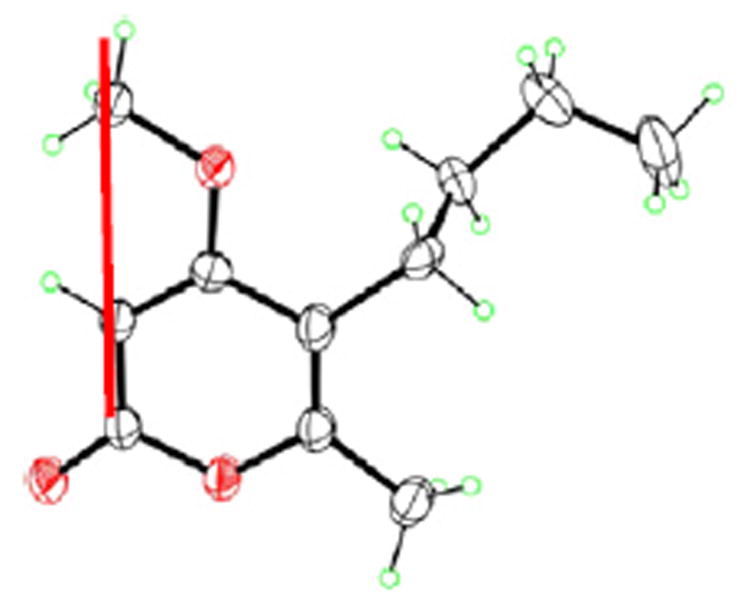
X-ray structure of 1.
Compound 2 was isolated as yellowish needles. The molecular formula was determined to be C11H16O4 by HR-ESI-MS (+ve mode) showing molecular ion peak [M+H]+ at m/z 213.1133 (calcd for C11H17O4, 213.1127) indicating four degrees of unsaturation. The IR spectrum of 2 absorption bands at 1720, 1647, and 1563 cm−1 indicated the presence of a lactone group and α-pyrone ring.
The 1H and 13C NMR spectral data (Tables 1 and 2) of compound 2 and those for 1 were found to be similar, with a difference observed in the chemical shift of carbon 11, the methyl group 11 in 1 of (δH 2.11, s, and δC 17.0) was replaced with oxygenated methylene (δH 4.41, s, and δC 58.5) in 2. The structure of 2 was authenticated by 2D NMR experiments, giving pertinent COSY and HMBC correlations (Fig. 2). Compound 2 was identified to be 5-butyl-6-(hydroxymethyl)-4-methoxy-2H-pyran-2-one.
Compound 3 was isolated as white amorphous powder. The molecular formula was determined to be C11H16O4 by HR-ESI-MS (+ve mode) showing molecular ion peak [M+H]+ at m/z 213.1123 (calcd for C11H17O4, 213.1127) indicating four degrees of unsaturation. The IR spectrum of 3 showed absorption bands at 1720, 1647, and 1563 cm−1 indicating the presence of a lactone group and α-pyrone ring. The 1H and 13C NMR spectral data (Tables 1 and 2) of compound 3 and those for 1 were found to be similar, with a difference observed in the chemical shift of carbon 7, The methylene group 7 in 1 of (δH 2.21, t, J 7.6, and δC 23.8) was replaced by oxygenated methine (δH 4.63, m and δC 68.6) in compound 3 indicating the oxygenation of C-7. The structure of 3 was authenticated by 2D NMR experiments, giving pertinent COSY and HMBC correlations (Fig. 2). The absolute configuration of 3 was assigned by application of the modified Mosher method.11-13 Treatment of 3 with (S)- and (R)-MTPA Cl afforded the (R)-MTPA ester (3a) and (S)-MTPA ester (3b), respectively. The difference in chemical shift values (Δδ) (δS – δR) for the diastereomeric esters 3b and 3a was calculated in order to assign the absolute configuration at C-4. Calculation for all relevant signals suggested the (R) absolute configuration at C-7, as shown in Figure 3. Here in this research we report the first isolation for compound 3 from a natural source and the identification of its absolute configuration. Compound 3 was obtained before by a chemical reduction reaction for citrepyrone, an α-pyrone derivative isolated from Penicillium citreoviride14 and another time by a chemical reduction reaction for pyrenocine C, a phytotoxin-related metabolite produced by onion pink root fungus, Pyrenochaeta terrestris.15
Figure 3.
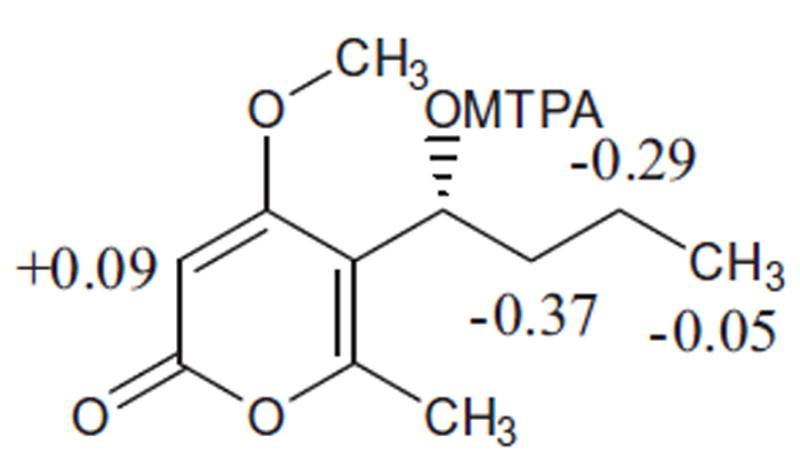
Δδ (δS – δR) values (in ppm) for the MTPA ester of 3.
Compound 3 was identified to be (R)-5-(1-hydroxybutyl)-4-methoxy-6-methyl-2H-pyran-2-one.
Compound 4 was isolated as white amorphous powder. The molecular formula was determined to be C11H14O4 by HR-ESI-MS (+ve mode) showing molecular ion peak [M+H]+ at m/z 211.0966 (calcd for C11H15O4, 211.0970) indicating four degrees of unsaturation. The IR spectrum of 4 absorption bands at 1720, 1647, and 1563 cm−1 indicated the presence of a lactone group and α-pyrone ring. The 1H and 13C NMR spectral data (Tables 1 and 2) of compound 4 were found to be similar to those of 1, with a difference observed in the chemical shift of carbon 9. The methylene group 9 in 1 of (δH 1.25, m and δC 22.4) was replaced by a carbonyl group (δC 210.1) in compound 4. The structure of 4 was authenticated by 2D NMR experiments, giving pertinent COSY and HMBC correlations (Fig. 2). Compound 4 was identified to be 4-methoxy-6-methyl-5-(3-oxobutyl)-2H-pyran-2-one.
Compound 5 was isolated as white amorphous powder and characterized by analysis of its spectroscopic data and by comparison with data reported in the literature16 was found to be macommelinol [5-(2-hydroxyethyl)-4-methoxy-6-methyl-2H-pyran-2-one].
Compound 6 was isolated as white amorphous powder and identified by analysis of its spectroscopic data and by comparison with data reported in the literature17 was found to be novae-zelandin A [5-[(2E)-but-2-en-1-yl]-4-methoxy-6-methyl-2H-pyran-2-one].
Compounds 2 and 4 showed moderate antileukemic activities against HL60 cells with IC50 values of 0.45 and 0.18 μg/ml and against K562 cells with IC50 values of 0.9 and 0.3 μg/ml, respectively, as shown in Table 3 and Figure 4
Table 3.
Inhibitory effects of compounds 2 and 4 on the growth of human leukemia cells in vitro
| Compounds | HL60 cells IC50 (μg/ml) | K562 cells IC50 (μg/ml) |
|---|---|---|
| 2 | 0.45 | 0.9 |
| 4 | 0.18 | 0.3 |
| Taxol | 0.0003 | 0.0023 |
Figure 4.
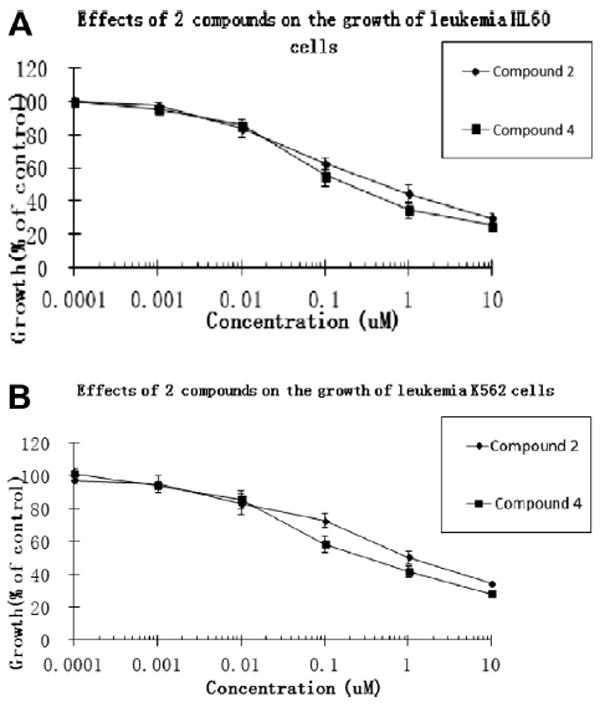
The concentration-dependent effects of compounds 2 and 4 on the growth of acute leukemia HL60 (A) and chronic leukemia K562 (B) cells.
All the isolated compounds have been examined for antimicrobial, antimalarial, and antileishmania activities, but did not show any promising effects.
Supplementary Material
Acknowledgments
We are grateful to the Egyptian Government and National Center for Natural Products Research, School of Pharmacy, University of Mississippi, for their financial support. We are also thankful to Dr. Baharthi Avula for the HRMS. This investigation was conducted in part in a facility constructed with support from the research facilities improvement Program C06RR14503 from the NIH NCRR. This work is supported in part by the United States Department of Agriculture ARS Specific Cooperative Agreement No. 58-6408-2-0009.
Footnotes
Supplementary data
Supplementary data (1H, 13C, and 2D NMR, spectra, and HR-ESI-MS of compounds 1–4, X-ray supporting data for 1, and the experimental section) associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tetlet.2014.04.085.
References and notes
- 1.Schulz B, Boyle C, Draeger S, Rommert A-K, Krohn K. Mycol Res. 2002;106:996. [Google Scholar]
- 2.Lou J, Fu L, Peng Y, Zhou L. Molecules. 2013;18:5891. doi: 10.3390/molecules18055891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottini AT, Gilchrist DG. Tetrahedron Lett. 1981;22:2719. [Google Scholar]
- 4.Gu W. World J Microb Biotechnol. 2009;25:1677. [Google Scholar]
- 5.Liebermann B, Nussbaum R-P, Günther W. Phytochemistry. 2000;55:987. doi: 10.1016/s0031-9422(00)00268-5. [DOI] [PubMed] [Google Scholar]
- 6.Sheridan H, Canning A-M. J Nat Prod. 1999;62:1568. doi: 10.1021/np990154w. [DOI] [PubMed] [Google Scholar]
- 7.Suemitsu R, Yamada Y, Sano T, Yamashita K. Agric Biol Chem. 1984;48:2383. [Google Scholar]
- 8.Gamboa-Angulo MM, García-Sosa K, Alejos-González F, Escalante-Erosa F, Delgado-Lamas G, Peña-Rodríguez LM. J Agric Food Chem. 2001;49:1228. doi: 10.1021/jf000872k. [DOI] [PubMed] [Google Scholar]
- 9.Koeffler H, Golde D. Blood. 1980;56:344. [PubMed] [Google Scholar]
- 10.Ritke MK, Rusnak JM, Lazo JS, Allan WP, Dive C, Heer S, Yalowich JC. Mol Pharmacol. 1994;46:605. [PubMed] [Google Scholar]
- 11.Dale JA, Mosher HS. J Am Chem Soc. 1973;95:512. [Google Scholar]
- 12.Ohtani I, Kusumi T, Kashman Y, Kakisawa H. J Am Chem Soc. 1991;113:4092. [Google Scholar]
- 13.Su B-N, Park EJ, Mbwambo ZH, Santarsiero BD, Mesecar AD, Fong HH, Pezzuto JM, Kinghorn AD. J Nat Prod. 2002;65:1278. doi: 10.1021/np0202475. [DOI] [PubMed] [Google Scholar]
- 14.Niwa M, Ogiso S, Endo T, Furukawa H, Yamamura S. Tetrahedron Lett. 1980;21:4481. [Google Scholar]
- 15.Sparace SA, Mudd JB, Burke BA, Aasen A. J Phytochemistry. 1984;23:2693. [Google Scholar]
- 16.Sakurai I, Shimizu S, Yamamoto Y. Chem Pharm Bull. 1988;36:1328. [Google Scholar]
- 17.Christensen KB, Van Klink JW, Weavers RT, Larsen TO, Andersen B, Phipps RK. J Agric Food Chem. 2005;53:9431. doi: 10.1021/jf0513213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


