Abstract
Within a large clonal population, such as cancerous tumor entities, cells are not identical, and the differences between intracellular pH levels of individual cells may be important indicators of heterogeneity that could be relevant in clinical practice, especially in personalized medicine. Therefore, the detection of the intracellular pH at the single-cell level is of great importance to identify and study outlier cells. However, quantitative and real-time measurements of the intracellular pH of individual cells within a cell population is challenging with existing technologies, and there is a need to engineer new methodologies. In this paper, we discuss the use of nanopipette technology to overcome the limitations of intracellular pH measurements at the single-cell level. We have developed a nano-pH probe through physisorption of chitosan onto hydroxylated quartz nanopipettes with extremely small pore sizes (~100 nm). The dynamic pH range of the nano-pH probe was from 2.6 to 10.7 with a sensitivity of 0.09 units. We have performed single-cell intracellular pH measurements using non-cancerous and cancerous cell lines, including human fibroblasts, HeLa, MDA-MB-231 and MCF-7, with the pH nanoprobe. We have further demonstrated the real-time continuous single-cell pH measurement capability of the sensor, showing the cellular pH response to pharmaceutical manipulations. These findings suggest that the chitosan-functionalized nanopore is a powerful nano-tool for pH sensing at the single-cell level with high temporal and spatial resolution.
Personalized medicine holds great potential, especially in treating cancer, which remains a major medical challenge due to both intrinsic and acquired resistance to conventional chemotherapeutics.1–3 In the last decade, advances have been made in the development of personalized cancer therapeutics to increase the efficacy of chemotherapy.4 Despite every effort to tailor drugs to the individual, results vary.5 This fact has been correlated with the presence of genetically distinct cells within an individual tumor.6 In recent studies genome sequencing technology has been employed to identify these genetic alterations in a large population of cells.7–9 While genetic aspects of cancer cell heterogeneity and the relationship between mutations and drug resistance have been studied extensively, development of pre-screening technologies to detect heterogeneity, that is, to find cancer cells that differ in their cellular metabolism and physiology within large cell populations, is under-investigated.
Evaluation of cells heterogeneity can be performed through the measurement of cytoplasmic ions and molecules. Accumulation of metal ions,10 changes in reactive oxygen (ROS) and nitrogen species (RNS) levels,11 and protein expression12 are important markers of cancerous cells within cell populations. Although less recognized, pH is also a distinctive factor of cancer cells. pH is one of the most intriguing features in initiating and regulating a myriad of cellular events, such as multi-drug resistance in tumor,13 protein processing,14 endocytosis15 and apoptosis.16 Due to its vital importance, the pH of the intracellular environment is strictly regulated through various ion channels and intracellular weak acids and bases, such as alkali cation-H+ exchangers, bicarbonate and acid loading transporters. In mammalian cells, subcellular compartments have different pH values in order to sustain optimum operational conditions for certain metabolic functions.17 In normal physiological conditions, the resting intracellular pH of mammalian cells is maintained between 6.8 and 7.3.18 On the other hand, extracellular pH values are slightly alkaline with the range of 7.2 to 7.4. A dysregulation of intracellular pH is ofen associated with altered cell functions, proliferation and drug resistance, and is observed in cancerous tumors.19 Moreover pH has a great effect on tumor growth and cancer cell migration and therefore the potential for metastases.20,21
Carcinogenic tumors are heterogeneous and widely assumed to be acidic due to the high metabolic rate of cancer cells coupled with poor blood supply. This regional high metabolism and lack of perfusion triggers an anaerobic metabolism which leads extracellular pH levels to decrease to ~6.0.22 Additionally, aerobic metabolism can increase the intracellular concentration of carbon dioxide (CO2), which results in a decrease of local pH levels. These two mechanisms of acidification are commonly accepted in cancer research. Little is known, however, about whether intracellular pH levels contribute to intratumoral heterogeneity, and if it is an indicator of preexisting metabolic heterogeneity in cancer cells in a large cell population. Greater granularity of pH data will be of great importance not only for the development of new anti-cancer drugs and carriers, as most of new drug delivery systems propose to use pH sensitive polymers or pH sensitive polymeric nanoparticles, but also to ascertain how effectively anti-cancer drug work over the course of treatment. Therefore, real-time quantitative measurement of intracellular pH may be crucial to link intratumoral cell heterogeneity, drug resistance and drug delivery systems for effective treatment.
pH can be used as a marker for the identiffication of variants of cancer cells in a tumor tissue. Once identified, these cells can be tagged and followed over the course of drug treatment. Then samples can be collected from the tagged cells to sequence their RNA and DNA to illuminate what makes these cells drug-resistant.
Detecting pH at the cellular level is not only important to investigate single cancer cells and cell heterogeneity in a tumor environment but also to understand neurodegeneration and aging. Neurodegenerative diseases, such as Parkinson's and Alzheimer's diseases, create heterogeneous physico-chemical environments due to mitochondrial oxidative phosphorylation, and therefore it is important to measure pH and understand its effect on neural recovery at the damaged site of brain.23 Additionally, cerebral pH has been found to be one of the major markers of metabolic disturbance and lethality after brain injury.24 Many of these studies have suffered the lack of an appropriate analytical tool.
Commonly utilized analytical techniques to measure intracellular pH values include nuclear magnetic resonance (NMR),25 electrochemistry,26,27 confocal microscopy,28 and absorbance and fluorescence spectroscopy.29–31 Of these, fluorescence spectroscopy and imaging are the most widely used techniques. However fluorescence intensity is hard to quantify directly and suffers from the experimental factors such as dye localization, photobleaching, excitation wavelength and cellular uptake and release rate. Additionally, fluorescence intensity can be affected by autofluorescence. Moreover, fluorescence probes do not allow continuous and site-specific detection of intracellular pH levels.
Here, we report the development of a nano-sized pH probe based on ionic conductance technique to measure pH at the single-cell level. This new nano-pH probe can be used as an analytical tool to illuminate the relationship between pH and a variety of diseases. To solve the aforementioned issues we propose the use of nanopipette platform that utilizes scanning ion conductance microscopy (SICM) principles.32 Nanopipettes are electrical devices that can measure the differences in ionic current at a nanopore. Their small sizes enables direct, real-time in vitro measurements with high spatial resolution and reduced invasiveness, allowing the monitoring of intracellular changes of an individual cell over the course of drug treatment. Recently, nanopipettes have gained importance as novel sensing tools and have been investigated for the detection of proteins,33,34 metal cations,32,35 DNA36 and carbohydrates.37 Quartz nanopipettes can be functionalized with various recognition materials. In this work chitosan, a biopolymer, is used as a pH-sensitive surface coating of internal surface of nanopipettes. Chitosan is biocompatible and has low-toxicity which makes it ideal for biological purposes. It possesses unique film-forming ability, high adherence to surfaces and remarkable mechanical strength. In addition, chitosan has been shown as a selective coating for biosensor fabrication.38–40
In this work, we demonstrate the development and characterization of chitosan-modified quartz nanopipettes for pH measurements in physiological buffers and cell media. We then used the chitosan-modified nanopipettes for the direct measurement of intracellular pH at four different cells types, including human fibroblast, HeLa, MCF-7 and MDA-MB-231. We evaluated the in vitro specificity of chitosan-modified nano-pH probes using a chloride channel blocker. The nano-pH probe is a powerful candidate not only to investigate cell heterogeneity in a variety of pathologic states, including cancerous tumors, but also neurodegenerative states and aging.
Materials and methods
Reagents and materials
Chitosan (low molecular weight), 5-nitro-2-(3-phenylpropylamino)-benzoate (NPPB), sodium phosphate dibasic and monobasic were purchased from Sigma Aldrich. Sodium chloride (ACS grade), hydrochloric acid, sodium hydroxide and hydrogen peroxide were obtained from Fisher Scientific. Acetic acid (glacial) was supplied from Riedel-de-Haen. 2-Propanol was obtained from Spectrum Chemicals. 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxymethyl ester (BCECF,AM) was bought from Invitrogen. Dimethyl sulfoxide (anhydrous) was supplied from Fluka. Minimum essential medium eagle (MEM), Dulbecco's modified eagle medium (DMEM) and trypsin were purchased from CellGro while fetal bovine serum (FBS) and penicillin-streptomycin from Gibco. All aqueous solutions are prepared in distilled, deionized water (Millipore, Synthesis System) with a resistivity of 18.2 Ω cm.
Preparation of nano-pH probe
Nanopipettes were fabricated from quartz capillaries with filament (QF100-70.7.5, Sutter Instrument). Prior to pulling, capillaries were treated with piranha solution (sulfuric acid-: hydrogen peroxide, 3 : 1 v/v) (Caution: ‘piranha solution’ reacts violently with organic materials and may become extremely hot when prepared.) and rinsed thoroughly with distilled water and 2-propanol. Treated capillaries were kept in 2-propanol until use to prevent contamination. Capillaries were pulled using a P-2000 laser puller (Sutter Instrument) with a two-line program with following parameters; line 1: heat 700, fil 4, vel 20, del 170, pull 0 and line 2: heat 680, fil 4, vel 40, del 170, pull 200. The resulting nanopipettes had a pore diameter of ~97 nm detected by a FEI Quanta 3D field emission microscope. Nanopipettes were stored in a sealed box until modification. Nanopipettes were functionalized by backfilling 10 μl of 0.25% chitosan solution and centrifuged at 4000 rpm to assure the coverage of the nanopipette tip with chitosan matrix. After centrifugation excess of chitosan was aspirated and nanopipettes were left airdry overnight. Dried nanopipettes were backfilled with 10 mM phosphate buffer saline (PBS) solution at pH 7.0, then centrifuged to remove residual air bubbles trapped at the tip of nanopipettes. Once filled all nanopipettes were kept in 10 mM PBS (pH 7.0) until pH measurements to prevent clogging of the nanopore.
Sensing setup
To carry out analytical characterization experiments of chitosan-modified nanopipette sensors, a two-electrode setup connected to a potentiostat (1030C, CH Instruments Inc.) was used for sensing. A 125 μm platinum wire (Goodfellow Corporation) placed into nanopipettes filled with electrolyte served as working electrode while a pseudo Ag/AgCl electrode placed in bulk solution (PBS or cell media) served as reference electrode. Linear sweep voltammetry was utilized for all in vitro measurements with a scan rate of 0.1 V s−1.
Intracellular measurements were performed by combining the potentiostat and scanning ion conductance microscope (SICM) with a low-noise mechanical switch. The SICM setup consisted of an Axopatch 200B amplifier (Molecular Devices) for current feedback measurements, a MP-285 motorized micro-manipulator (Sutter Instrument) for coarse positioning of nano-pH probe, a piezo stage (NanoCube, Physik Instrumente) for fine positioning and insertion of the nano-pH probe sensors, and a programmable interface for hardware control of the setup. This system is run by custom software written in Lab-VIEW (National Instruments). All experiments with cells were conducted on an inverted fluorescence microscope (Olympus IX 70) equipped with an eyepiece camera (Dino-Eye, Big C).
Cell culture
HeLa, MCF-7, MDA-MB-231 and human fibroblast cells were cultured in a conditioned environment with 5% CO2 and 90% humidity at 37 °C. HeLa, MCF-7 and MDA-MB-231 cells were cultured in 1× MEM, while human fibroblasts in 1× DMEM. All media were supplemented with 10% FBS and 1% Penicillin-Streptomycin.
Fluorescence microscopy
MDA-MB-231 cell cultures were exposed to a pH-sensitive fluorescent indicator, BCECF,AM. The working solution was prepared to a concentration of 1 μM in Hank's Buffered Salt Solution (HBSS) and incubated at 37 °C for 15 min before fluorescent imaging. Cells were washed with Dulbecco's phosphate-buffered saline (DPBS) before loading of 1 μM BCECF,AM solution. After incubation, excess fluorescent dye was rinsed off the cells with HBSS was loaded on the culture for imaging.
For intracellular pH buffer calibration, cell cultures were exposed to complete pH calibration buffer prepared according to the protocol supplied with the Intracellular pH Calibration Buffer Kit (Life Technologies, P35379), and were incubated at 37 °C for 10 min before imaging. Intracellular pH calibration was done in three replicates. All fluorescence microscopy analyses were carried out with a Leica SP5 confocal microscope using the Leica Application Suite Advance Fluorescence (LAS AF 3) software. Further image analyses were performed with Fiji-ImageJ software.
Results & discussion
Characterization of pH-responsive quartz nanopipette sensors
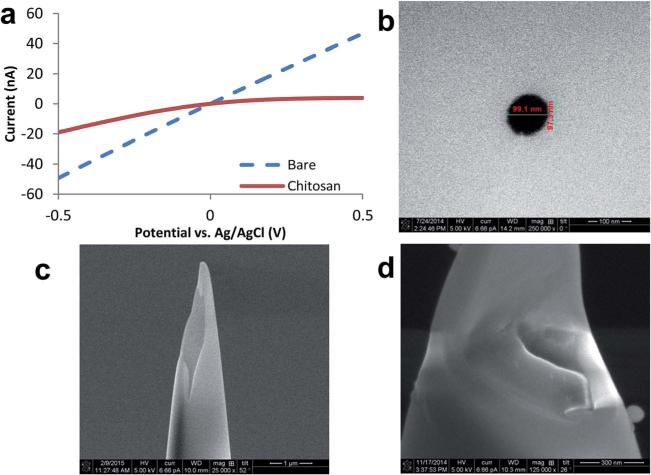
The measurement principle of nanopipettes is based on the ionic current at tip. This ionic current is highly dependent on the pore size and surface charge of nanopipette.34 The surface charge of a quartz nanopipette is negative due to dissociation of silanol groups at the glass–liquid interface. Quartz undergoes protonation at extremely acidic pH values.41 These surface properties of quartz reduce pH sensing capabilities, making bare nanopipettes inappropriate for measuring very small pH changes. Limitations associated with the low sensitivity of bare quartz surfaces can be overcome through the incorporation of pH responsive polymeric entities onto nanopipette surfaces. Here, we employed chitosan as the pH sensitive surface coating. Chitosan, with a strong positive charge at acidic pH, is attracted to hydroxyl moieties on the negatively charged quartz surface through electrostatic interactions. In addition to alterations of the surface charge, the thickness of the chitosan layer has been shown to change with pH which may enhance the sensitivity of the nanopipette.42,43 To evaluate the presence and impact of the chitosan layer on the nanopipette surface, we monitored the changes in current responses as a result of surface modification. Fig. 1A demonstrates the electrochemical traces of the bare and chitosan-modified quartz nanopipettes filled with 10 mM PBS (pH 7.0) in the potential range of −0.5 to 0.5 V (vs. Ag/AgCl reference electrode). The recorded current response significantly decreases after chitosan modification. To optimize the sensitivity of the nano-pH sensor, various concentrations of chitosan were studied (ESI Fig. SI1†). Due to viscous properties and limited tip-geometry, high concentrations of chitosan solutions did not reach to the nanopore. Typical geometric shape of a nanopipette tip is conical (Fig. SI2A†), and the pore size of quartz nanopipettes were determined by SEM and found to be ~97 nm (Fig. 1B). Additional SEM micrographs were taken to further confirm the presence of the chitosan layer (Fig. SI2B†). Because the chitosan modification was done on the inside of the nanopipette, a focused ion beam was used to vertically etch the nanopipette and expose internal surface. The cross-section image shows chitosan residues inside of the nanopipette surface when compared to that of bare nanopipette (Fig. 1C and D).
Fig. 1.
(A) Comparison of ionic current rectifications of a bare and chitosan-modified quartz nanopipette. Both measurements were carried out with quartz nanopipettes filled with 10 mM PBS (pH 7.0). Scanning electron micrographs demonstrating (B) a typical nanopipette pore opening. SEM images of focused ion beam cut (C) bare nanopipette tip and (D) chitosan-modified nanopipette, showing the chitosan layer onto the inner surface of nanopipette.
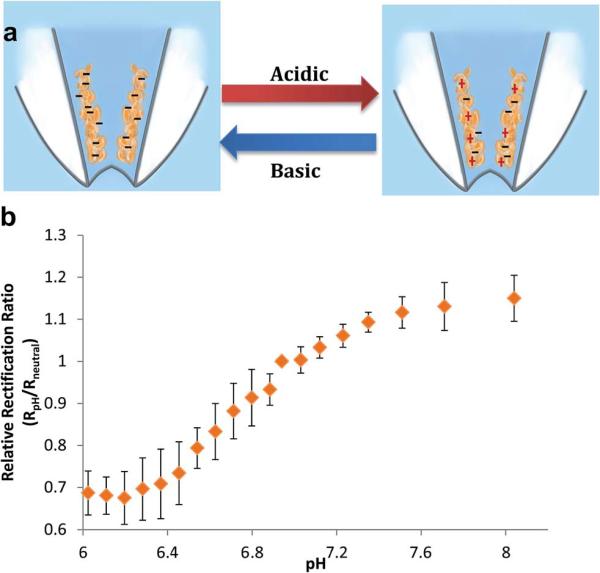
Once the presence of the chitosan layer was confirmed with SEM and electrochemistry, analytical characterization of the functionalized nanopipettes was conducted using linear sweep voltammetry. The potential range spanned from −0.5 to 0.5 V with a scan rate of 0.1 V s−1. The modulation of pH was achieved by a conventional acid–base titration approach. Calibration of chitosan-modified nanopipettes was performed by consecutive additions of 20 μl of first 1 M NaOH and second HCl into 0.1 M PBS (pH 7.0). Current rectification of modified nanopipettes at +/−0.5 V changed in response to the changing pH of the buffer solution, as expected with the alteration charge on the chitosan layer. Chitosan contains a glucosamine residue on its polysaccharide backbone (pKa ~6.5) making chitosan pH-responsive.38 pH values below the pKa protonate the chitosan layer making the nanopipette surface positively charged, whereas basic conditions deprotonate chitosan's amine functional group, increasing the net negative charge at the surface (Fig. 2A). For quantitation of pH, a relative rectification ratio (RR) has been defined as RRR = RRpH/RRneutral where RRpH and RRneutral are RR at a specific pH and at pH 7.0 respectively. Fig. 2B displays the calibration curve obtained by acid–base titrations using the chitosan-modified nanopipette within the physiologically relevant pH range from 6.02 to 8.04. The trend observed in pH calibration curve is typical of isoelectric point determination experiments. A slight shift in the isoelectric point of chitosan may be due to the nanoscale conical geometry of the nanopipette tip, which can impede the uniform diffusion of ions. The sensitivity of the chitosan-functionalized pH-nanoprobe was 0.09 pH units. This high sensitivity to pH makes the nanoprobe a powerful tool for intracellular pH measurements. Current-potential curves of individual pH as well as larger range pH calibration are given in ESI Fig. SI3.† Bare nanopipettes were tested for pH sensing; as expected; these nanopipettes demonstrated low sensitivity towards pH changes (Fig. SI4†). The chitosan-modified nanopipettes showed excellent storage stability, at dry-state, up to week having 98% of the initial sensitivity; however after one and a half month the sensitivity of the nano-pH sensors went down to 47% of the initial sensitivity (Fig. SI5†).
Fig. 2.
(A) Schematic of the reversible changes in surface charge as a result of pH. (B) Calibration of chitosan-modified nanopipettes within the physiologically relevant pH range from 6.02 to 8.04. All data points are represented as relative rectification ratios at +/− 0.5 V vs. Ag/AgCl reference electrode. The error bars represent standard deviations for n = 4 replicate measurement. 0.1 M PBS was used as supporting electrolyte.
pH sensing in cell culture media
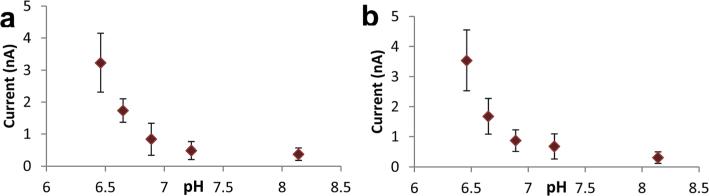
Our motivation for developing a solid nanopore pH probe is to measure intracellular pH at the single-cell level and to identify cancer cells with their distinctive metabolic characters. To perform intracellular pH measurements, chitosan-modified nanopipettes were further calibrated in cell culture media, MEM and DMEM. As cell media contain various amino acids, vitamins and other ingredients, optimum working parameters were different from those determined for PBS. The scanned potential range was from −0.2 to 0.6 V with a scan rate of 0.1 V s−1. The sensitivity of chitosan-modified nanopipettes for pH changes was the highest at 0.6 V. Fig. 3 shows the calibration of nano-pH probes in 1× MEM and DMEM solution. Calibration of nano-pH probes in the media was carried out by consecutive additions of 20 μl of 0.1 M HCl. The measurements were done 15 s after the addition of acid solution to cell culture media to obtain a homogeneous solution. Representative linear sweep voltammograms are demonstrated in Fig. SI6† for acid titration of MEM and DMEM media. As ingredients of these media are different, their buffering capacities are slightly different with DMEM being more resistant to pH changes compared to MEM.
Fig. 3.
Calibration of chitosan-modified nanopipettes in cell culture media: (A) 1× MEM and (B) DMEM. Current responses were measure at a fixed bias potential of 0.6 V. The error bars represent standard deviations for n = 4 replicate measurement.
Measurement of intracellular pH of cancerous and non-cancerous cells
Direct measurement of intracellular pH is challenging due to the small size of cells and the complexity of physiological matrix. While physiological pH level is marginally alkaline, intracellular pH level of individual cells in a large population and subcellular compartments is unknown. Conventionally, fluorescence dyes (e.g. BCECF,AM, oregon green) are utilized for indirect detection of pH in cells.30 Although these pH indicators reveal an approximation of pH over a large cell population, there are several disadvantages of using fluorescence dyes: (i) low sensitivity due to short pH range, (ii) fast photobleaching, (iii) cytotoxicity. Additionally, accumulation of these dyes in certain organelles and their rate of leakage can result in incorrect interpretations. Our studies using a conventional pH indicator, BCECF,AM, to measure intracellular pH of MDA-MB-231 cells have proven the drawbacks of using fluorescence for accurate and sensitive evaluation of intracellular events (for details see ESI Fig. SI7 & SI8†). In addition, continuous interrogation of a single-cell over the course of time for evaluating the cellular impact of therapeutics, channel activators, or toxins cannot be carried out with conventional fluorescence probes.
In order to directly and accurately measure intracellular pH, chitosan-modified nanopipettes were inserted in the cytoplasm of the cells in culture. We used this sensing technology, for the first time, for the direct monitoring of intracellular pH of human cancerous and non-cancerous cell lines, including human fibroblast, HeLa, MCF-7 and MDA-MB-231. Human fibroblast cells are selected as a non-cancerous model to investigate intracellular pH levels at normal cytoplasmic conditions. HeLa cell lines are the most commonly used human cancer type due to their rapid and continuous growth in cell culture. Additionally, because of reports of contamination and heterogeneity of HeLa cells determination of the intracellular pH levels of these cells may allow us to evaluate the cell heterogeneity.44 MCF-7 and MDA-MB-231 are distinct breast cancer cell lines. MCF-7 is a hormone-responsive cell line and its growth is stimulated with estrogen; MDA-MB-231 derives from an invasive breast cancer which was found to be highly metastatic.45 We chose to interrogate these two breast cancer cell lines because they exhibit different drug sensitivities and we sought to determine whether this could be correlated with differences in the intracellular pH levels.
Chitosan-modified nanopipettes were inserted to individual cells using a customized scanning ion conductance microscope which detects current feedback for positioning the nanopipettes. Recently we have demonstrated that this custom-built platform can perform nanobiopsies at the single-cell level for genomic investigations.46 Fig. SI9A† demonstrates a representative feedback signal recorded during the approach-penetration-retraction process of chitosan-modified nanopipettes. After the insertion of nano-pH probe to a cell, linear sweep voltammograms were recorded and the current-responses at a bias potential of 0.6 V were used to calculate the intracellular pH levels of single-cells.
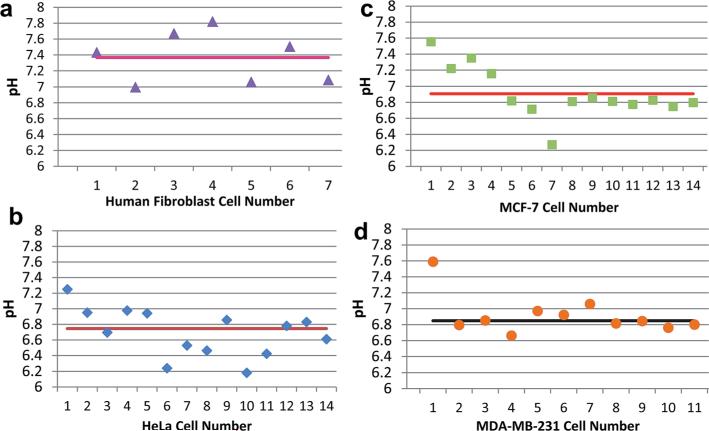
From voltammetric current responses at 0.6 V versus Ag/AgCl, the calculated intracellular pH levels of individual cells and average pH values for all cell lines are shown in the Fig. 4. Seven human fibroblast cells were interrogated for intracellular pH and the average pH was 7.37 ± 0.29 (Fig. 4A). The observed intracellular pH level in these human fibroblasts is in line with previous reports estimating pH levels through indirect and destructive approaches, including monitoring of ion exchangers (NHEs and NBCs) and acid transporters (AEs).17
Fig. 4.
Intracellular pH levels of individual cells determined by chitosan-modified nanopipettes. pH levels recorded for (A) human fibroblast, (B) HeLa, (C) MCF-7 and (D) MDA-MB-231 cells. Horizontal lines represent the average intracellular pH measured with the nano-pH probe.
We also used the nano-pH probes to investigate the metabolic differences between non-cancerous and cancerous cells. As cancer cells have a faster metabolic rate compared to non-cancerous cells, production of acidic species and CO2 in cancer cells is higher as well. Using the chitosan-modified nano-pH probe in 14 individual HeLa cells for intracellular pH measurements, we found the average pH for HeLa cells to be 6.75 ± 0.27 (Fig. 4B).
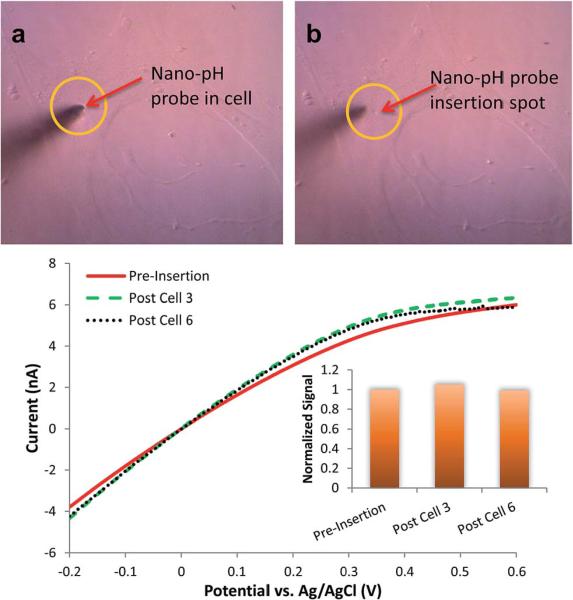
To compare whether a similarly acidic intracellular environment is present in other cancer cell lines, we performed pH measurements on breast cancer lines. Using the nano-pH probe, was observed an average intracellular pH level for 14 individual MCF-7 cells of 6.91 ± 0.20 (Fig. 4C). The average intracellular pH was found to be 6.85 ± 0.11 for MDA-MB-231 using 11 individual cells (Fig. 4D). Representative linear sweep voltammograms of individual cell measurements are given in Fig. SI10.† Our data demonstrate that the intracellular environment can differ from cell to cell in a way that is detectible by pH. These differences can be attributed to different metabolic speeds of individual cells and may be used for the identification of heterogeneous cells in a large population, such as tumors. The small tip size of nano-pH probe reduces the damage during insertion (Fig. 5, compare micrograph A and B) and measurement. This aspect enables continuous or intermittent interrogation of the same cell over the course of pharmaceutical manipulations and drug therapies (see next section). Fig. 5C illustrates regeneration and reusability of the same nano-pH probes for consecutive in vitro measurements, specifically after the third and sixth cell. pH probes were tested after several cell interrogations in 0.1 M PBS (pH 7.0). Additionally, this test is important to control the integrity of the probe after use for in vitro measurement.
Fig. 5.
Representative micrographs of (A) a nano-pH probe inserted to a MDA-MB-231 and (B) after retraction of the probe. Cells did not show any morphological changes and stayed intact over the course of insertion and measurement, and survived after retraction. (C) In vitro reusability of the nano-pH probes. Linear sweep voltammograms of regenerating baseline of the same nano-pH probe after the third and sixth cell interrogation in 0.1 M PBS (pH 7.0).
To fully deploy the pH nano-probe, we intend to build a fully-automated high-throughput robotic system that will allow us to interrogate hundreds of cells in a range of minutes. Cells having lower or higher pH values compared to the general population of cells will be identified and then tagged with a molecular marker to nanobiopsy for DNA and RNA sequencing.
Pharmaceutical manipulation of intracellular pH
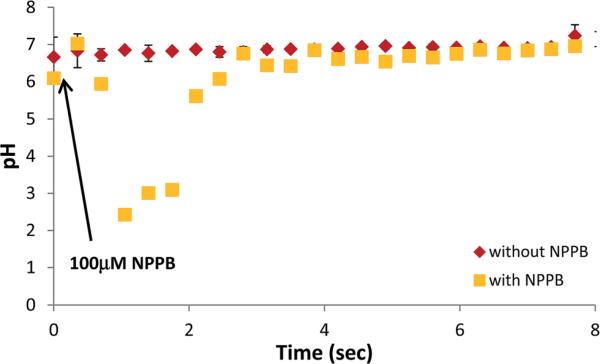
Of its many possible uses, we hope that the nano-pH probe can be used to monitor intracellular pH changes during drug therapy. To this end, we deployed the nano-pH probe for continuous monitoring at a single-cell during the addition of a known chloride channel blocker, 5-nitro-2-(3-phenylpropylamino)-benzoate (NPPB). NPPB has been shown previously to block chloride channels in renal epithelial and macrophage cells, with a resulting increase in acidity of the intracellular environment. Conventionally, the change in pH has been measured indirectly by introduction of fluorescent dye (BCECF,AM).47,48 (Thus this pharmaceutical manipulation test not only serves to demonstrate the capability of real-time measurement of nano-pH probes but also the specificity towards pH detection.) To obtain a baseline, nano-pH probes were inserted in MDA-MB-231 cells and consecutive pH measurements were performed for every 21 second for 7 min. This real-time pH monitoring in MDA-MB-231 cells showed minimal drift over the course of measurement (Fig. 6, red diamonds). To study the effect of NPPB, a nano-pH probe was inserted into MDA-MB-231 cells and intracellular pH recording was initiated just prior to the addition of 100 μM of NPPB (freshly prepared in anhydrous DMSO) to the cell media. The yellow squares in Fig. 6 display the pH changes as a result of NPPB exposure over a 7 min time period. Intracellular pH level dropped significantly within the first 2 min after the introduction of NPPB and went as low as 2.5. Measured pH levels stabilized by 4 min post NPPB introduction. This increase in pH can be due to apoptosis resulting in the shrinkage of cell body, which would expose the tip of the nano-pH probe to the cell media. Intracellular pH measurements of three individual MDA-MB-231 cells with nano-pH probe not only showed the real-time pH changes after NPPB exposure but also the variations from cell to cell in terms of drug-response (Fig. SI11†).
Fig. 6.
Real-time intracellular pH measurements with nano-pH probes. The pH measurements were performed on MDA-MB-231 cells in the absence (red diamonds) and presence (yellow cubes) of 100 μM NPPB (Cl− channel blocker). Arrow in the figure shows the addition time of NPPB. Readings are obtained every 21 s for 7 min post channel blocker exposure. Error bars represent standard deviation for n = 3 replicates.
Conclusions
Direct measurement of intracellular pH has been investigated for more than forty years.26,27 We have achieved in a new way via simple physisorption of chitosan into a quartz nanopipette. Our approach takes advantage of a pH-responsive chitosan polymeric layer and the small size of a nanopipette for intracellular pH measurement at the single-cell level. Leveraging a scanning ion conductance microscope customized for single-cell navigation, we were able to insert nano-pH probes into individual cells. In vitro results showed that chitosan-functionalized nanopipettes measure intracellular pH selectively with high temporal resolution. The average intracellular pH levels were 7.37 ± 0.29, 6.75 ± 0.27, 6.91 ± 0.20 and 6.85 ± 0.11 for human fibroblast, HeLa, MCF-7 and MDA-MB-231, respectively. These results show good separation between fibroblast and cancerous cells, which have a more acidic cytoplasmic environment than non-cancerous cells. Additionally, our findings reveal that individual cells within a population may differ in their intracellular pH. Finally, NPPB exposure experiment demonstrates that nano-pH probe enables real-time, continuous interrogation of single-cell upon biochemically induced changes in intracellular pH.
Our data show that chitosan-modified nanopipette sensing technology is a powerful approach for interrogating single-cell pH levels with high spatial and temporal resolution with high selectivity and sensitivity. Further application of this nano-pH probe technology may provide a deeper understanding of cell heterogeneity and drug resistance. To achieve this aim, we are working on the development of a fully automated system for high-throughput screening of cell populations over the course of drug treatment. Additionally, we will use nano-pH probes to investigate pH changes and differences in tumorous microenvironments (e.g. tumor tissues).
Supplementary Material
Acknowledgements
This work was supported in part by grants from the National Institutes of Health [P01-35HG000205], and National Institute of Neurological Disorders and Stroke [R21NS082927]. We acknowledge Dr Benjamin Abrams for technical support and UCSC Life Sciences Microscopy Center for the use of confocal microscope, and Dr Tom Yuzvinsky for image acquisition and the W.M. Keck Center for Nanoscale Optofluidics for use of the FEI Quanta 3D Dualbeam microscope.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra06721a
References
- 1.Higgins CF. Nature. 2007;446:749–757. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 2.Videira M, Reis RL, Brito MA. Biochim. Biophys. Acta. Rev. Canc. 2014;1846:312–325. doi: 10.1016/j.bbcan.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Lee M-CW, Lopez-Diaz FJ, Khan SY, Tariq MA, Dayn Y, Vaske CJ, Radenbaugh AJ, Kim HJ, Emerson BM, Pourmand N. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4726–E4735. doi: 10.1073/pnas.1404656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Workman P. Mol. BioSyst. 2005;1:17–26. doi: 10.1039/b501751n. [DOI] [PubMed] [Google Scholar]
- 5.Garraway LA, Jänne PA. Cancer Discovery. 2012;2:214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 6.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Sci. Transl. Med. 2012;4:127ps110. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 7.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow C-W, Cao Y, Gumbs C, Gold KA, Kalhor N, Little L, Mahadeshwar H, Moran C, Protopopov A, Sun H, Tang J, Wu X, Ye Y, William WN, Lee JJ, Heymach JV, Hong WK, Swisher S, Wistuba II, Futreal PA. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, Jarosz M, Lipson D, Tagawa ST, Nanus DM, Stephens PJ, Mosquera JM, Cronin MT, Rubin MA. Eur. Urol. 2013;63:920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu T, Sempos CT, Freudenheim JL, Muti P, Smit E. Ann. Epidemiol. 2004;14:195–201. doi: 10.1016/S1047-2797(03)00119-4. [DOI] [PubMed] [Google Scholar]
- 11.Wiseman H, Halliwell B. Biochem. J. 1996;313:17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat. Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 13.Raghunand N, Gillies RJ. Drug Resist. Updates. 2000;3:39–47. doi: 10.1054/drup.2000.0119. [DOI] [PubMed] [Google Scholar]
- 14.Paroutis P, Touret N, Grinstein S. The pH of the Secretory Pathway: Measurement, Determinants, and Regulation. 2004 doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- 15.Weisz OA. Traffic. 2003;4:57–64. doi: 10.1034/j.1600-0854.2003.40201.x. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb RA, Nordberg J, Skowronski E, Babior BM. Proc. Natl. Acad. Sci. U. S. A. 1996;93:654–658. doi: 10.1073/pnas.93.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casey JR, Grinstein S, Orlowski J. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 18.Mahnensmith RL, Aronson PS. Circ. Res. 1985;56:773–788. doi: 10.1161/01.res.56.6.773. [DOI] [PubMed] [Google Scholar]
- 19.Damaghi M, Wojtkowiak JW, Gillies RJ. Front. Physiol. 2013;4:370. doi: 10.3389/fphys.2013.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stock C, Schwab A. Pflugers Arch–Eur J Physiol. 2009;458:981–992. doi: 10.1007/s00424-009-0677-8. [DOI] [PubMed] [Google Scholar]
- 21.Parks SK, Chiche J, Pouyssegur J. J. Cell. Physiol. 2011;226:299–308. doi: 10.1002/jcp.22400. [DOI] [PubMed] [Google Scholar]
- 22.Gatenby RA, Gillies RJ. Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 23.Rango M, Bonifati C, Bresolin N. J. Cereb. Blood Flow Metab. 2005;26:283–290. doi: 10.1038/sj.jcbfm.9600192. [DOI] [PubMed] [Google Scholar]
- 24.Timofeev I, Nortje J, Al-Rawi PG, Hutchinson PJA, Gupta AK. J. Cereb. Blood Flow Metab. 2013;33:422–427. doi: 10.1038/jcbfm.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashim AI, Zhang X, Wojtkowiak JW, Martinez GV, Gillies RJ. NMR Biomed. 2011;24:582–591. doi: 10.1002/nbm.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter NW. Kidney Int. 1972;1:341–346. doi: 10.1038/ki.1972.45. [DOI] [PubMed] [Google Scholar]
- 27.Mabe H, Blomqvist P, Siesjo BK. J. Cereb. Blood Flow Metab. 1983;3:109–114. doi: 10.1038/jcbfm.1983.13. [DOI] [PubMed] [Google Scholar]
- 28.Lucien F, Harper K, Pelletier P-P, Volkov L, Dubois CM. 2014:e51395. doi: 10.3791/51395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talley CE, Jusinski L, Hollars CW, Lane SM, Huser T. Anal. Chem. 2004;76:7064–7068. doi: 10.1021/ac049093j. [DOI] [PubMed] [Google Scholar]
- 30.McNamara KP, Nguyen T, Dumitrascu G, Ji J, Rosenzweig N, Rosenzweig Z. Anal. Chem. 2001;73:3240–3246. doi: 10.1021/ac0102314. [DOI] [PubMed] [Google Scholar]
- 31.Peng H.-s., Stolwijk JA, Sun L-N, Wegener J, Wolfbeis OS. Angew. Chem. 2010;122:4342–4345. doi: 10.1002/anie.200906926. [DOI] [PubMed] [Google Scholar]
- 32.Actis P, Vilozny B, Seger RA, Li X, Jejelowo O, Rinaudo M, Pourmand N. Langmuir. 2011;27:6528–6533. doi: 10.1021/la2005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Actis P, Mak A, Pourmand N. Bioanal Rev. 2010;1:177–185. doi: 10.1007/s12566-010-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umehara S, Karhanek M, Davis RW, Pourmand N. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4611–4616. doi: 10.1073/pnas.0900306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilozny B, Actis P, Seger RA, Vallmajo-Martin Q, Pourmand N. Anal. Chem. 2011;83:6121–6126. doi: 10.1021/ac201322v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karhanek M, Kemp JT, Pourmand N, Davis RW, Webb CD. Nano Lett. 2005;5:403–407. doi: 10.1021/nl0480464. [DOI] [PubMed] [Google Scholar]
- 37.Vilozny B, Wollenberg AL, Actis P, Hwang D, Singaram B, Pourmand N. Nanoscale. 2013;5:9214–9221. doi: 10.1039/c3nr02105j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozel RE, Wallace KN, Andreescu S. Anal. Chim. Acta. 2011;695:89–95. doi: 10.1016/j.aca.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozel RE, Hayat A, Wallace KN, Andreescu S. RSC Adv. 2013;3:15298–15309. doi: 10.1039/C3RA41739E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozel RE, Ispas C, Ganesana M, Leiter JC, Andreescu S. Biosens. Bioelectron. 2014;52:397–402. doi: 10.1016/j.bios.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behrens SH, Grier DG. J. Chem. Phys. 2001;115:6716–6721. [Google Scholar]
- 42.Claesson PM, Ninham BW. Langmuir. 1992;8:1406–1412. [Google Scholar]
- 43.Lee H-S, Eckmann DM, Lee D, Hickok NJ, Composto RJ. Langmuir. 2011;27:12458–12465. doi: 10.1021/la202616u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masters JR. Nat. Rev. Cancer. 2002;2:315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- 45.Holliday D, Speirs V. Breast Cancer Res. 2011;13:215. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Actis P, Maalouf MM, Kim HJ, Lohith A, Vilozny B, Seger RA, Pourmand N. ACS Nano. 2013;8:546–553. doi: 10.1021/nn405097u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown CDA, Dudley AJ. Br. J. Pharmacol. 1996;118:443–444. doi: 10.1111/j.1476-5381.1996.tb15422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukacs GL, Nanda A, Rotstein OD, Grinstein S. FEBS Lett. 1991;288:17–20. doi: 10.1016/0014-5793(91)80992-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.