Abstract
Salmonella enterica subsp. enterica serovar Kentucky is frequently isolated from healthy poultry and dairy cows and is occasionally isolated from people with clinical disease. A genomic analysis of 119 isolates collected in the United States from dairy cows, ground beef, poultry and poultry products, and human clinical cases was conducted. Results of the analysis demonstrated that the majority of poultry and bovine-associated S. Kentucky were sequence type (ST) 152. Several bovine-associated (n = 3) and food product isolates (n = 3) collected from the United States and the majority of human clinical isolates were ST198, a sequence type that is frequently isolated from poultry and occasionally from human clinical cases in Northern Africa, Europe and Southeast Asia. A phylogenetic analysis indicated that both STs are more closely related to other Salmonella serovars than they are to each other. Additionally, there was strong evidence of an evolutionary divergence between the poultry-associated and bovine-associated ST152 isolates that was due to polymorphisms in four core genome genes. The ST198 isolates recovered from dairy farms in the United States were phylogenetically distinct from those collected from human clinical cases with 66 core genome SNPs differentiating the two groups, but more isolates are needed to determine the significance of this distinction. Identification of S. Kentucky ST198 from dairy animals in the United States suggests that the presence of this pathogen should be monitored in food-producing animals.
Introduction
Salmonella enterica subsp. enterica is a major cause of human and animal salmonellosis worldwide. The majority of human salmonellosis in the United States is caused by several serovars, namely Enteritidis, Typhimurium, Newport, and Javiana [1]. Although it is currently assumed that all serovars are potentially pathogenic to humans, the association between S. enterica and non-human animal illness as well as aymptomatic carriage is not as clear and frequently animals shedding several serovars such as Kentucky, Enteritidis, and Seftenberg are asymptomatic [1][2][3]. S. Kentucky is frequently isolated from both asymptomatic cattle, poultry and poultry products in the United States, but has been isolated from other sources such as the environment and domesticated dogs [2][3][4][5][6];;. Recently, the global spread of multi-drug resistant S. Kentucky ST198 has been described, indicating this serovar is an emerging public health threat[7]. In North America, human clinical cases of S. Kentucky ST198 infections have been associated with travel to the Middle East, Southeast Asia or Africa [7][8][9], and clinical cases caused by S. Kentucky ST152 are relatively rare.
Salmonella Kentucky has been identified as the most frequently isolated serovar from non-human non-clinical cases in the United States [1] and until recently, has mostly been considered a concern with poultry due to its high prevalence in broilers as well as the established link between poultry products and human salmonellosis. Further, S. Kentucky isolated from poultry and poultry products in the United States has been reported to be resistant to multiple antibiotics [4][10][11][12][13]. This serovar is currently the most frequently isolated serovar from poultry in the United States, recently supplanting Enteritidis and Heidelberg in poultry flocks [2]. In recent years, research on dairy farms has demonstrated that S. Kentucky is frequently isolated from dairy cows and dairy production operations in the United States and the isolation rates of this serovar in dairy cows appear to be increasing [3][14][15]). What remains unclear is how S. Kentucky is capable of colonizing these two distantly related hosts and if there are genomic differences between poultry-associated and bovine-associated S. Kentucky.
Both S. Kentucky ST152 and ST198 have been isolated globally, but the degree of relatedness among S. Kentucky isolates recovered on a global scale is not yet known. Multi-Locus Sequence Typing (MLST) analyses have demonstrated that there are no common alleles between these two STs, indicating that they are distantly related [7]. However, comprehensive description of the genomic differences between these two STs has not yet been conducted, and a comparison of the two may identify genomic factors involved in host-specificity and virulence potential. In a whole-genome phylogenetic analysis of S. enterica Timme et al. [16] demonstrated that S. Kentucky was polyphyletic; a phenomenon identified in other serovars [17]. For S. enterica and other bacteria, polyphyly has been associated with lateral gene transfer (LGT) of the antigenic coding regions [17][18]. The historical application of serology to strain naming and description has thus been misleading in that distantly related strains that have acquired an antigenic coding region from the same source through LGT have been presumed to be similar based on this scheme, when in fact their genomes may be highly diverged.
The objective of this study was to infer the phylogeny of S. Kentucky, identify the genomic features associated with the apparent specificity of S. Kentucky ST152 to bovine and poultry hosts/products, and to identify differences between S. Kentucky ST152 and ST198 genomes. To accomplish this we sequenced the genomes of 49 dairy-cow associated isolates and two poultry-associated isolates that were collected from across the United States, and coupled these data with the genomes of 68 isolates previously collected from poultry, poultry products and production environments and clinical cases in the United States, Canada, and the Middle East.
Materials and Methods
Salmonella Kentucky isolates from dairy cows were recovered from previously collected National Animal Health Monitoring System (NAHMS) samples as well as an eight-year dairy farm monitoring program conducted in south-central Pennsylvania [3][19] (two poultry-associated isolates were supplied by S. Parveen). Isolates were serogrouped following the methods of Herrera-Leon et al. [20]and Karns et al. [21]and serotypes were identified by National Veterinary Services Laboratories (NVSL; Ames, IA). Isolates were streaked onto tryptic soy agar, and a single colony was inoculated in tryptic soy broth overnight at 37°C. This inoculum was centrifuged, decanted, and then processed for DNA extraction using a Qiagen DNeasy Kit (Qiagen, Valencia, CA). Nextera XT libraries were made for each sample and pooled into equimolar concentrations following the manufacturer’s instructions (Illumina, San Diego, CA). Paired-end sequencing (2 X 151 bp) was conducted on an Illumina NextSeq 500 sequencing platform with a High-Output flow cell. Data were demultiplexed and trimmed to remove adaptor sequences using the BCL2FastQ program and PhiX reads were removed using DeconSeq [22]. Reads were further cleaned using Trimmomatic [23] and assembled using SPAdes 3.6.2 [24]. ST56 complex genomes were downloaded from NCBI GenBank prior to February 2016 and ST15 complex genomes were downloaded prior to December 2015 (PRJNA242614, PRJNA273513, PRJNA78335, PRJNA78339, PRJNA78337, PRJNA225734, PRJNA225734, PRJNA66693, PRJNA20069, PRJNA19457, PRJEB6491, PRJNA337914, and PRJNA186035) (S1 Table). Genomes from BioProject PRJEB6491 [25] were labeled as 915c and 917c in this analysis. Bovine-associated S. Kentucky isolates used in this study will be provided upon request.
Three methods were used to identify SNPs among the S. Kentucky genomes. To identify high quality SNPs based on read coverage, Lyve-SET [26] [27]and CFSAN SNP Pipeline [28] [29] were used. For the Lyve-SET analysis SNP identification was conducted with a minimum 10X coverage requirement and the CGP read cleaner. When fastq files were not available, assembled genomes were included in the project/asm directory prior to SNP detection. Default settings were used for the CFSAN SNP Pipeline analysis. For this analysis only genomes for which fastq files or.sff files were available could be used for the SNP search and therefore one ST152 (strain CDC 191) genome and several ST198 genomes were excluded (strains CVM 43824, CVM 43780, CVM 43756). To identify SNPs in assembled genomes, Parsnp from the Harvest package was used [30][31]. Regions undergoing high levels of recombination were removed using the–x option (PhiPack) and all genomes were included using the–c option. The chromosome of S. Kentucky CVM21988 was selected as the reference genome for all analyses. Parsnp was used to determine the core-genome SNP differences between S. Kentucky ST152 and ST198 genomes and other subclade A1 serovars identified by Timme et al. [16]. SNPs were annotated using snpEff [32].
To infer the phylogeny of S. Kentucky ST152/318/2132 isolates the multi-fasta SNP matrices from each of the three SNP detection methods were imported into MEGA6 [33] and RAxML [34]. Maximum Parsimony (MP) trees were inferred for all S. Kentucky isolates (ST198, ST152/318/2132, and unknown STs) as well as S. Kentucky isolates with non-S. Kentucky isolates using MEGA6. For phylogenetic inference of ST152/318/2132 isolates a Maximum Likelihood phylogeny was inferred using RAxML version 8.2.8 with the General Time Reversible (GTR) selected as the model of nucleotide substitution and all other parameters set to default settings. MP and ML analyses were each conducted with 1000 bootstrap replicates. The bootstrapped MP tree inferred from the Parsnp method of SNP detection was used to determine the genealogical sorting index (gsi) [35] for bovine and poultry isolates using the GSI webserver [36]. MLST data were retrieved from the University of Warwick Salmonella enterica MLST database [6]and through the Center for Genomic Epidemiology server [37].
The resulting SNP matrices were also analyzed using the Bayesian clustering program STRUCTURE v2.3.4 [38]. This program clusters individuals based on patterns of SNP differences without enforcing a bifurcating tree-like structure and, thus, may reveal additional details about the genomic content, similarity, and differences among isolates. In particular, STRUCTURE is suitable for detecting evidence for admixture (i.e., individuals whose genomes appear to be a combination of SNPs associated with distinct groups). STRUCTURE analyses were run with default settings and 60,000 generations, the first 10,000 of which served as the burnin. STRUCTURE results were visualized using DISTRUCT v1.1 [39].
Putative plasmid sequences were identified with PlasmidFinder [40] using the Enterobacteriaceae database with the detection thresholds set to 95% sequence identity. The identified HSP fragments were compared to the NCBI database (BLASTN analysis) to identify similar plasmids. The presence/absence of protein coding genes was conducted using BLASTP. Putative genomic islands in S. Kentucky CVM29188 (ST152) and S. Kentucky CVM N51290 (ST198) were identified using IslandViewer 3 [41].
Forty nine bovine-associated S. Kentucky isolates, and two poultry-associated isolates (including type strain ATCC 9263) were evaluated for their susceptibility to 25 antimicrobials (azithromycin, ciprofloxacin, gentamicin, tetracycline, nalidixic acid, cefoxitin, chloramphenicol, ceftriaxone, amoxicillin/clavulanic acid, ceftiofur, sulfisoxazole, trimethoprim/sulfamethoxazole, ampicillin, streptomycin, cephalothin, cefotaxime, cefotaxime/clavulanic acid, ceftazidime, ceftazidime/clavulanic acid, imipenem, cefepime, cefpodoxime, piperacillin/tazobactam, meropenem, and cefazolin) using an automated microdilution procedure (Sensititre, ThermoFisher, Lenexa, KS) and specialty plates CVM3AGNF and ESB1F. Antimicrobial minimum inhibitory concentrations (MICs) were interpreted based upon the epidemiological cut-off values (ECOFFs) used by the National Antimicrobial Resistance Monitoring System (NARMS).
Results and Discussion
Description of Isolates and Antibiotic Susceptibility Testing of Bovine-associated Isolates
In total 119 S. Kentucky genomes were utilized in the in silico analyses of this study. Of these, 49 dairy cow-associated isolates were selected from a collection of isolates that have been recovered from routine analysis of milk, milk filters, fecal samples from dairy cows, and the dairy cow farm environment in the United States over a 13-year period (Table 1). Two poultry isolates recovered from a broiler operation in the Eastern Shore of Maryland were used in this study, and the remaining poultry-associated genomes (n = 59) were gathered as assembled genomes and raw sequencing reads from the NCBI GenBank database and the SRA database, respectively. All human clinical isolates (n = 6) were all gathered from NCBI (two as raw sequencing reads and four as assembled genomes).
Table 1. Kentucky genomes used in this study.
S.
| Genome | Source | Location of Isolation | Year of Isolation | Sequence Type (ST)1 | ST Complex 2 | Phylogenetic Cluster 3 |
|---|---|---|---|---|---|---|
| ARS-CC97 | Fecal composite (dairy cow farm) | Pennsylvania (USA) | 2004 | 318 | 15 | 1.2.2 |
| 0253 | Dairy cow feces | Pennsylvania (USA) | 2004 | 318 | 15 | 1.2.2 |
| 5349 | Dairy cow feces | Pennsylvania (USA) | 2006 | 152 | 15 | 1.2.3 |
| ARS-CC444 | Milk filter (dairy cow farm) | Wisconsin (USA) | 2007 | 152 | 15 | 1.1 |
| ARS-CC457 | Milk filter (dairy cow farm) | Texas (USA) | 2007 | 152 | 15 | 1.1 |
| ARS-CC515 | Bulk tank milk (dairy cow farm) | Wisconsin (USA) | 2007 | 152 | 15 | 1.1 |
| ARS-CC521 | Milk filter (dairy cow farm) | Wisconsin (USA) | 2007 | 152 | 15 | 1.1 |
| ARS-CC572 | Milk filter (dairy cow farm) | Wisconsin (USA) | 2007 | 152 | 15 | 1.1 |
| ARS-CC913 | Dairy cow feces | Virginia (USA) | 2007 | 152 | 15 | 1.2.1 |
| ARS-CC526 | Bulk tank milk (dairy cow farm) | Pennsylvania (USA) | 2007 | 152 | 15 | 1.2.1 |
| ARS-CC496 | Milk filter (dairy cow farm) | New York (USA) | 2007 | 152 | 15 | 1.2.3 |
| CFSAN011775 | Dairy cow feces | Pennsylvania (USA) | 2008 | 152 | 15 | 1.2.3 |
| CFSAN011776 | Dairy cow feces | Pennsylvania (USA) | 2009 | 152 | 15 | 1.2.3 |
| CFSAN011777 | Dairy cow feces | Pennsylvania (USA) | 2009 | 152 | 15 | 1.2.3 |
| CFSAN011778 | Dairy cow feces | Pennsylvania (USA) | 2009 | 152 | 15 | 1.2.3 |
| CFSAN011779 | Dairy cow feces | Pennsylvania (USA) | 2010 | 152 | 15 | 1.2.3 |
| CFSAN011780 | Dairy cow feces | Pennsylvania (USA) | 2010 | 152 | 15 | 1.2.3 |
| CFSAN011782 | Dairy cow feces | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC6181 | Milk filter (dairy cow farm) | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC6183 | Milk filter (dairy cow farm) | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC6204 | Milk filter (dairy cow farm) | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC6329 | Milk filter (dairy cow farm) | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC6333 | Milk filter (dairy cow farm) | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC6340 | Milk filter (dairy cow farm) | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC8561 | Dairy cow feces | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC8574 | Fecal composite (dairy cow farm) | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC8601 | Dairy cow feces | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC8619 | Dairy cow feces | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC8624 | Dairy cow feces | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC8625 | Dairy cow feces | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| ARS-CC8633 | Fecal composite (dairy cow farm) | Pennsylvania (USA) | 2011 | 152 | 15 | 1.2.3 |
| CFSAN011781 | Dairy cow feces | Pennsylvania (USA) | 2012 | 152 | 15 | 1.2.3 |
| ARS-CC7487 | Dairy cow hide swab | Pennsylvania (USA) | 2013 | 152 | 15 | 1.2.3 |
| ARS-CC8078 | Fecal composite (dairy cow farm) | Pennsylvania (USA) | 2013 | 152 | 15 | 1.2.3 |
| ARS-CC8289 | Milk filter (dairy cow farm) | Ohio (USA) | 2014 | 152 | 15 | 1.2.2 |
| ARS-CC8294 | Milk filter (dairy cow farm) | Pennsylvania (USA) | 2014 | 152 | 15 | 1.2.3 |
| ARS-CC8297 | Milk filter (dairy cow farm) | Ohio (USA) | 2014 | 152 | 15 | 1.2.3 |
| ARS-CC8417 | Trough water (dairy cow farm) | Pennsylvania (USA) | 2014 | 152 | 15 | 1.2.3 |
| ARS-CC8446 | Milk filter (dairy cow farm) | Ohio (USA) | 2014 | 152 | 15 | 1.2.3 |
| ARS-CC353 | Milk filter (dairy cow farm) | New York (USA) | 2007 | 152 | 15 | 2.1 |
| ARS-CC469 | Bulk tank milk (dairy cow farm) | Vermont (USA) | 2007 | 152 | 15 | 2.1 |
| ARS-CC661 | Milk filter (dairy cow farm) | Vermont (USA) | 2007 | 152 | 15 | 2.1 |
| ARS-CC690 | Milk filter (dairy cow farm) | Wisconsin (USA) | 2007 | 152 | 15 | 2.1 |
| ARS-CC912 | Dairy cow feces | Wisconsin (USA) | 2007 | 152 | 15 | 2.1 |
| ARS-CC917 | Dairy cow feces | Vermont (USA) | 2007 | 152 | 15 | 2.1 |
| CDC 191 | Human clinical | Wisconsin (USA) | 2002 | 152 | 15 | 2.1 |
| ARS-CC621 | Milk filter (dairy cow farm) | Texas (USA) | 2007 | 152 | 15 | 2.2 |
| CVM N50435 | Chicken breast | New Mexico (USA) | 2013 | 152 | 15 | 2.3.1 |
| CVM N51252 | Chicken wings | Colorado (USA) | 2013 | 152 | 15 | 2.3.1 |
| CVM N51981 | Chicken breast | Louisiana (USA) | 2013 | 152 | 15 | 2.3.1 |
| CVM N43447 | Chicken breast | California (USA) | 2013 | 152 | 15 | 2.3.1 |
| 13562 | Chicken breast | Iowa (USA) | 2001 | 152 | 15 | 2.3.1 |
| CVM N43450 | Chicken breast | California (USA) | 2013 | 152 | 15 | 2.3.2 |
| CVM N43478 | Chicken breast | Oregon (USA) | 2013 | 152 | 15 | 2.3.2 |
| CVM N43820 | Chicken breast | Georgia (USA) | 2013 | 152 | 15 | 2.3.2 |
| SA20030505 | Chicken cecal contents | Ontario (Canada) | 2002 | 152 | 15 | 2.3.2 |
| ABBSB1008-2 | Chicken feces | British Columbia (Canada) | 2006 | 152 | 15 | 2.3.2 |
| CVM N45412 | Chicken breast | Washington (USA) | 2013 | 152 | 15 | 2.3.3.1 |
| CVM N47718 | Chicken breast | New Mexico (USA) | 2013 | 152 | 15 | 2.3.3.1 |
| CVM N47730 | Chicken breast | New Mexico (USA) | 2013 | 152 | 15 | 2.3.3.1 |
| CVM N51241 | Chicken breast | California (USA) | 2013 | 152 | 15 | 2.3.3.1 |
| CVM N47729 | Chicken breast | Washington (USA) | 2013 | unknown | 15 | 2.3.3.1 |
| CVM N45934 | Chicken breast | Maryland (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N45937 | Chicken breast | Maryland (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N45939 | Chicken breast | Maryland (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N45944 | Chicken breast | Maryland (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N46849 | Chicken breast | Pennsylvania (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N47721 | Chicken breast | New Mexico (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N47723 | Ground turkey | New Mexico (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N48688 | Chicken breast | Maryland (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N48710 | Chicken breast | Tennessee (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N47722 | Ground turkey | New Mexico (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N48687 | Chicken breast | Maryland (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N48705 | Chicken breast | Louisiana (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N44693 | Chicken wings | California (USA) | 2013 | 152 | 15 | 2.3.3.2 |
| CVM N43448 | Chicken wings | California (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM N43455 | Chicken breast | Colorado (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM N43835 | Chicken breast | Washington (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM N46820 | Chicken breast | Colorado (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM N46857 | Chicken breast | Pennsylvania (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM N50419 | Chicken breast | Connecticut (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM N50421 | Chicken wings | Georgia (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM N51294 | Chicken breast | New Mexico (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM N51313 | Chicken breast | Tennessee (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| ABB07-SB3057-2 | Chicken feces | British Columbia (Canada) | 2005 | 152 | 15 | 2.4.1.1 |
| ABB1087-1 | Chicken feces | British Columbia (Canada) | 2005 | 152 | 15 | 2.4.1.1 |
| 22694 | Chicken breast | Massachusetts (USA) | 2002 | 152 | 15 | 2.4.1.1 |
| CVM N51982 | Chicken wings | Louisiana (USA) | 2013 | 152 | 15 | 2.4.1.1 |
| CVM29188 | Chicken breast | Georgia (USA) | 2003 | 152 | 15 | 2.4.1.1 |
| CVM N43465 | Chicken breast | Minnesota (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N43466 | Chicken wings | Minnesota (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N44708 | Chicken breast | New York (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N47720 | Chicken breast | New Mexico (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N48707 | Chicken breast | New York (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N48711 | Chicken breast | Tennessee (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N50437 | Chicken wings | New York (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N51256 | Chicken wings | Connecticut (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N51273 | Chicken breast | Maryland (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| ARS-CC5795 | Poultry | Maryland (USA) | 2010 | 152 | 15 | 2.4.1.2 |
| ARS-CC5805 | Poultry | Maryland (USA) | 2010 | 152 | 15 | 2.4.1.2 |
| CVM N50429 | Chicken breast | New Mexico (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N51277 | Chicken breast | Maryland (USA) | 2013 | 152 | 15 | 2.4.1.2 |
| CVM N51249 | Chicken breast | Colorado (USA) | 2013 | 2132 | 15 | 2.4.1.2 |
| CVM N43471 | Chicken breast | New Mexico (USA) | 2013 | 152 | 15 | 2.4.1.3 |
| SALC-205-3 | Chicken feces | British Columbia (Canada) | 2004 | 152 | 15 | 2.4.2 |
| 20793 | Chicken breast | Georgia (USA) | 2002 | 152 | 15 | 2.4.2 |
| N312 | Chicken breast | Minnesota (USA) | 2004 | 152 | 15 | 2.4.2 |
| ARS-CC938 | Milk filter (dairy cow farm) | Florida (USA) | 2003 | 198 | 56 | 198.1 |
| ATCC 9263 | unknown | unknown | Unknown | 198 | 56 | 198.1 |
| CVM N51290 | Ground beef | New Mexico (USA) | 2013 | 198 | 56 | 198.1 |
| ARS-CC273 | Dairy calf feces | Florida (USA) | 2011 | 198 | 56 | 198.1 |
| ARS-CC274 | Dairy calf feces | Florida (USA) | 2011 | 198 | 56 | 198.1 |
| CVM N41913 | Ground beef | Minnesota (USA) | 2012 | 198 | 56 | 198.1 |
| CVM N42453 | Ground turkey | California (USA) | 2012 | unknown | 56 | 198.1 |
| 915c | Human clinical (sacral wound) | Kuwait | 2012 | 198 | 56 | 198.2 |
| 917c | Human clinical (stool) | Kuwait | 2012 | 198 | 56 | 198.2 |
| CVM 43824 | Human clinical | unknown | 2012 | 198 | 56 | 198.2 |
| CVM 43780 | Human clinical | unknown | 2011 | 198 | 56 | 198.2 |
| CVM 43756 | Human clinical | unknown | 2012 | 198 | 56 | 198.2 |
For in-house isolates (bovine-associated isolates and two poultry isolates, and ATCC 9263) antibiotic susceptibility tests (AST) were conducted. All bovine-associated isolates and ATCC 9263 were susceptible to all tested antibiotics. Poultry isolate ARS-CC5795 was resistant to amoxicillin/clauvulanic acid, ampicillin, cefitoxin, ceftiofur, and ceftriaxone, while ARS-CC5805 (poultry isolate) was resistant to streptomycin and tetracycline. The absence of antibiotic resistance in a diverse collection of S. Kentucky recovered from dairy cows is consistent with other studies and is notable due to the reported prevalence of antibiotic resistance among poultry-associated S. Kentucky within the United States [4][10][11][12][13][42][43]. Antibiotic resistance conferring plasmids are frequently identified in poultry-associated S. Kentucky ST152 isolates [12] while antibiotic resistance in human clinical S. Kentucky ST198 is associated with acquisition of the Salmonella Genomic Island 1 (SGI1), plasmids, and core genome polymorphisms [7][9][44][45].
Phylogenetic Inference
To date at least 15 S. Kentucky sequence types (ST) have been described MLST studies [7][46]. In these assays, each ST differs from another ST by at least one of seven alleles with some sequence types of the same serovar sharing no alleles. Based on the number of deposited sequences in the MLST database, ST152 and ST198 are among the most frequently isolated S. Kentucky sequence types. However, they share no common alleles indicating they are highly diverged from each other [7]. These two STs are part of two larger groups of “ST complexes” which consist of closely related STs. ST152 is a member of the ST15 complex which to date also includes ST151, 212, 318, and 723 among several others. ST198 is a member of ST56 complex, which also includes ST727, 835, and 1680. Outside of these defined complexes there are other closely related STs sharing one or more alleles with members of each complex. A pairwise comparison of seven MLST loci of ST152 and ST198 identified 42 SNPs resulting in a 1.25% sequence divergence (data not shown). When applying the same MLST criteria to the S. Kentucky genomes used in this study, three STs were identified (Table 1). All of the poultry-associated isolates were ST152 with one identified as 2132 (one allele difference with ST152), while two bovine-associated isolates were ST318 (one allele difference with ST152 and ST2132), 42 were ST152, and three were ST198. Two strains could not be accurately typed due to abbreviated genes at the ends of contigs. S. Kentucky CVM N47729, isolated from a chicken breast, shared six of seven alleles with ST152 and S. Kentucky CVM N42453 shared six of seven alleles with ST198. Eight S. Kentucky genomes gathered from the NCBI database were identified as ST198. These included five human clinical isolates, two ground beef isolates, and the S. Kentucky type strain ATCC 9263. These results are consistent with those of the MLST database in that ST152 from the United States are commonly isolated from poultry and cattle. A single ST318 isolate is included in the MLST database and, consistent with our study, was isolated from cattle in the mid-Atlantic region of the United States. Based on data entries in the MLST database ST198 isolates were recovered on an apparently broader global scale and were more frequently associated with human infections and poultry, cattle, and occasionally other organisms, and the environment.
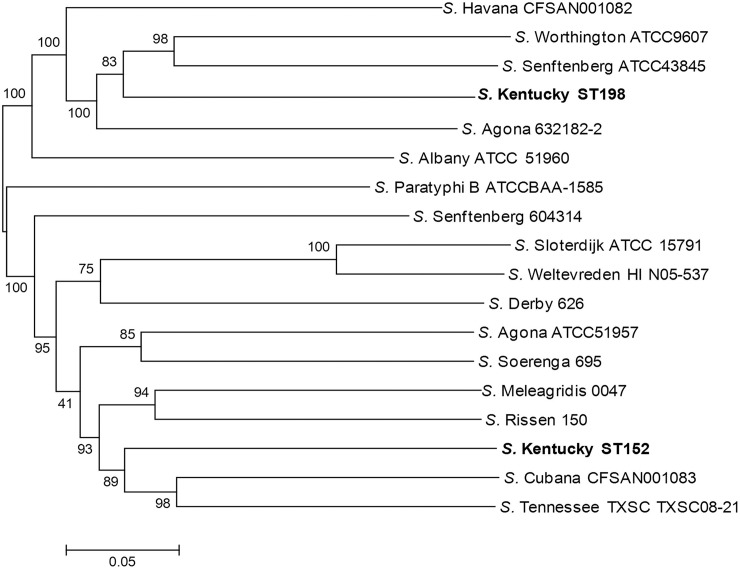
To investigate the relatedness among the S. Kentucky isolates and infer their evolutionary history on a genome-wide scale, SNPs across the genomes of all S. Kentucky and closely related serovars were identified. Using a core-genome SNP matrix determined from representative genomes of S. enterica subclade A1 as described by Timme et al. [16], two distant lineages of S. Kentucky were identified (Fig 1). These two S. Kentucky lineages correspond to the ST15 complex (ST152, 318 and 2132) and ST56 complex (ST198) of S. Kentucky sequence types described above (Table 1).
Fig 1. Phylogenetic relationships of S. Kentucky ST152 and ST198 with representatives of subclade A1 serovars as described by Timme et al.
[16] inferred using the Maximum Likelihood method with the General Time Reversible model of nucleotide substitution. Bar length represents number of substitutions per site.
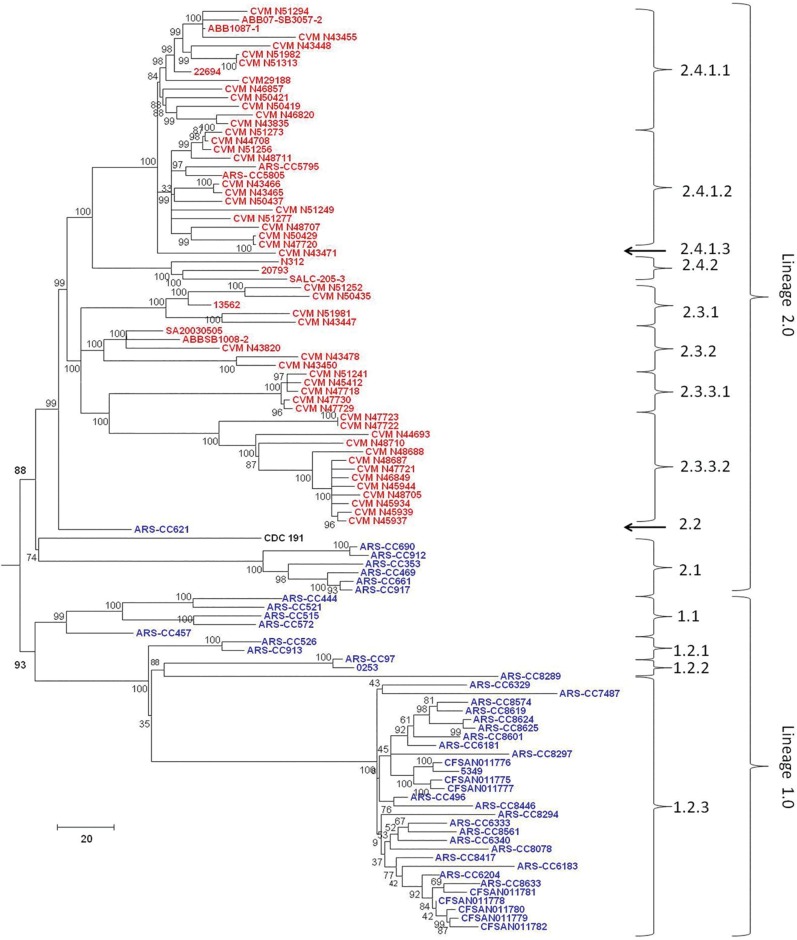
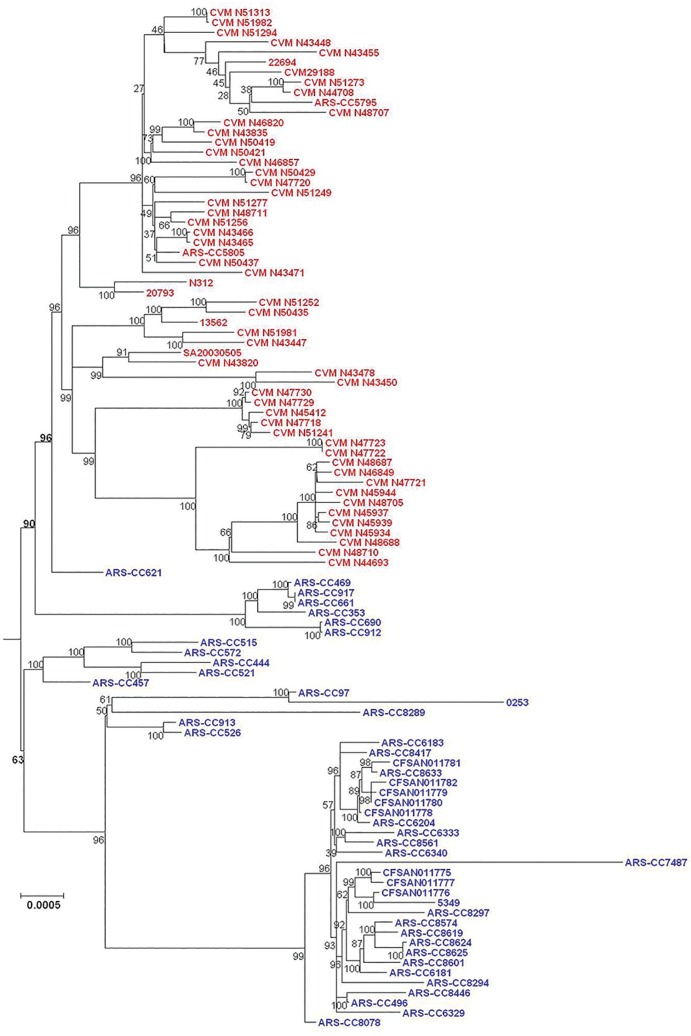
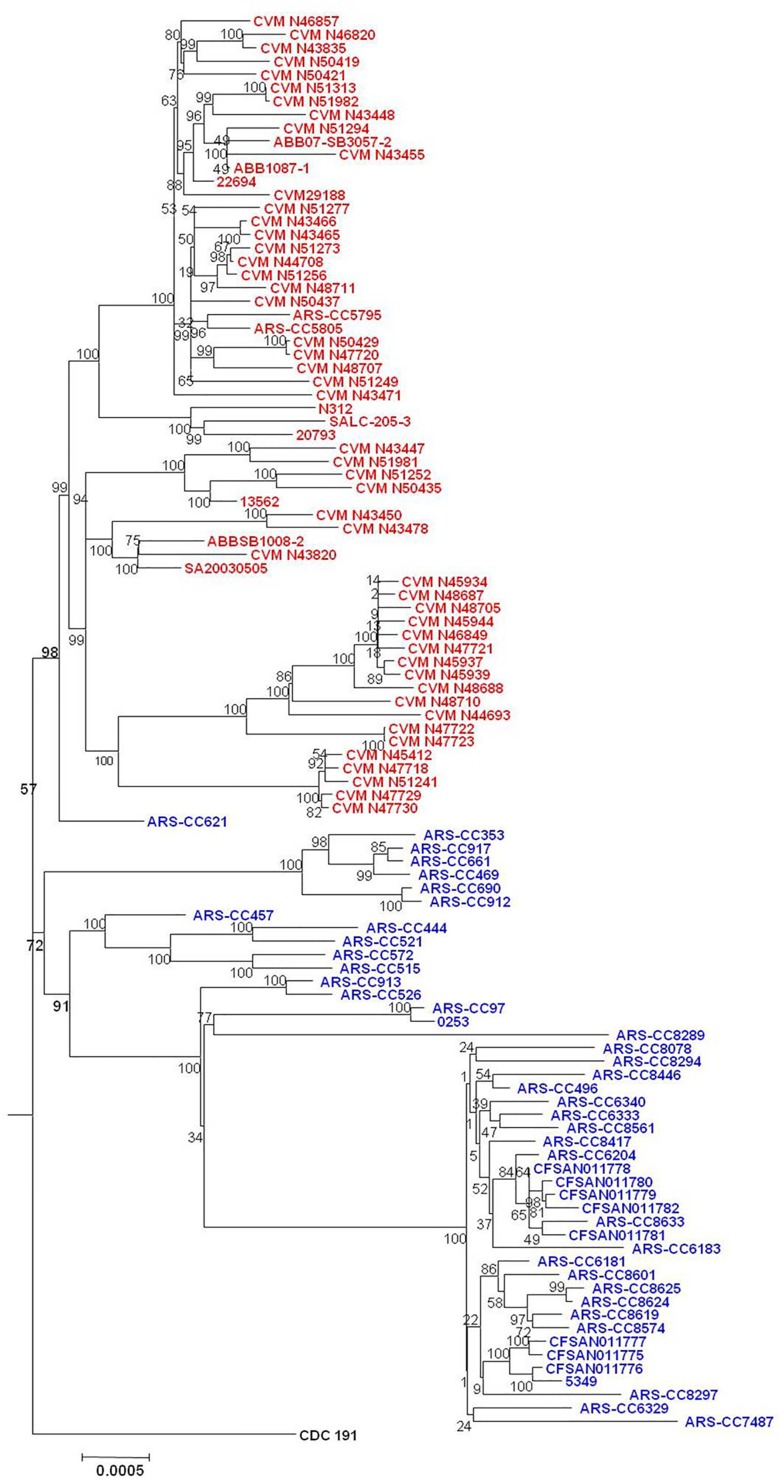
SNPs within the S. Kentucky genomes (excluding other subclade A1 serovars) were identified using three methods and phylogenetic trees were inferred from these data. Phylogenies inferred from data derived from the three SNP-detection methods were, for the most part, approximately similar in topology. For all SNP detection methods and both methods of tree inference the ST15 complex was observed to have two major sublineages (labeled as Lineages 1.0 and 2.0 in this analysis) (Figs 2–7). For both MP and ML analyses there was weak support for the placement of some sublineages in the ancestral node of the Lineage 1.0 and for the Parsnp ML analysis the human clinical isolate (S. Kentucky CDC 191) is rooted in the ancestral node of the tree while it is placed in Lineage 2.0 in the other phylogenetic analyses of this study (Figs 2, 4, and 5).
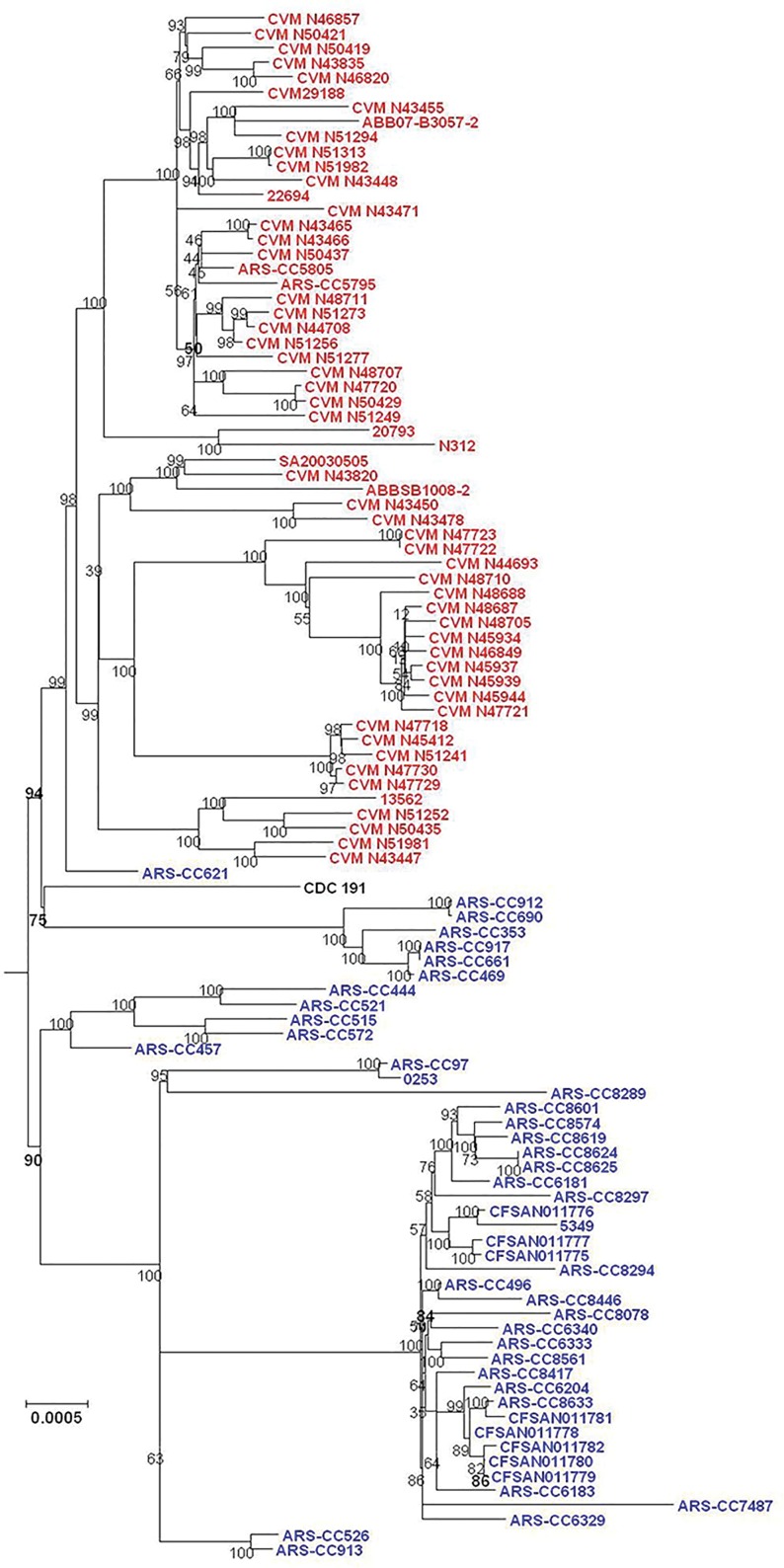
Fig 2. Maximum Parsimony tree of S. Kentucky ST152/318/2132 genomes based on 40,795 SNPs detected by Parsnp and rooted in twelve S. Kentucky ST198 isolates.
Bovine-associated strains are labeled in blue, poultry-associated strains are labeled in red, and the human clinical isolate CDC 191 is labeled in black. Clusters of genomes are labeled on right of tree. Tree was inferred with 1000 bootstrap replicates. Bar length = number of substitutions.
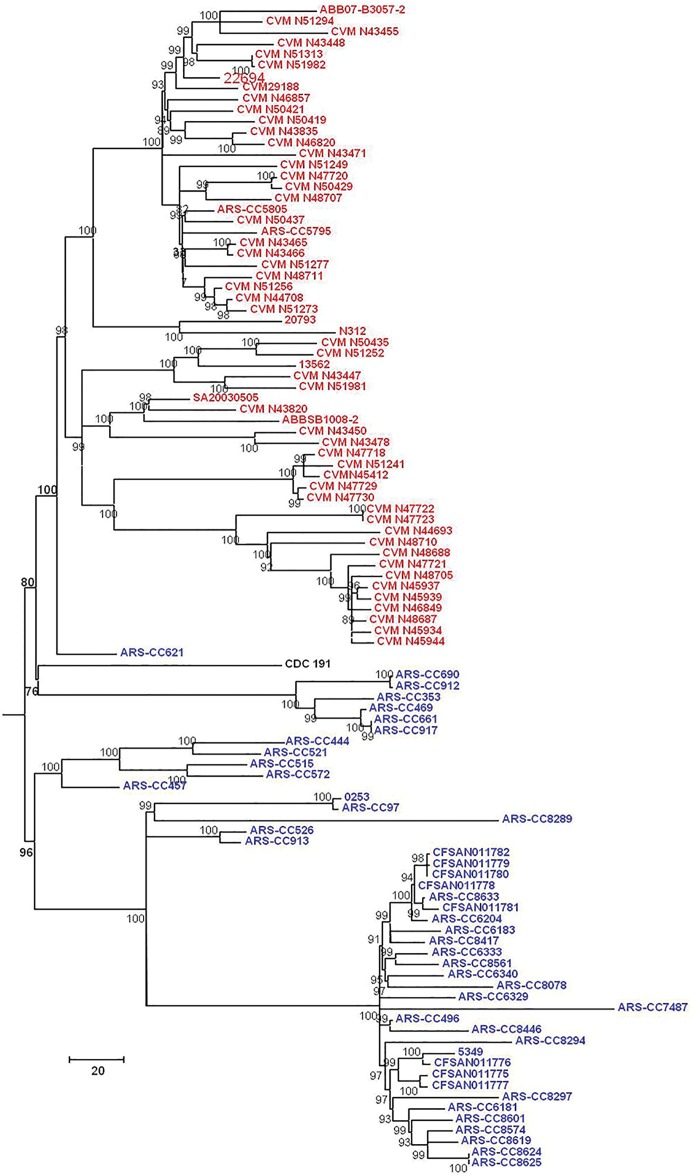
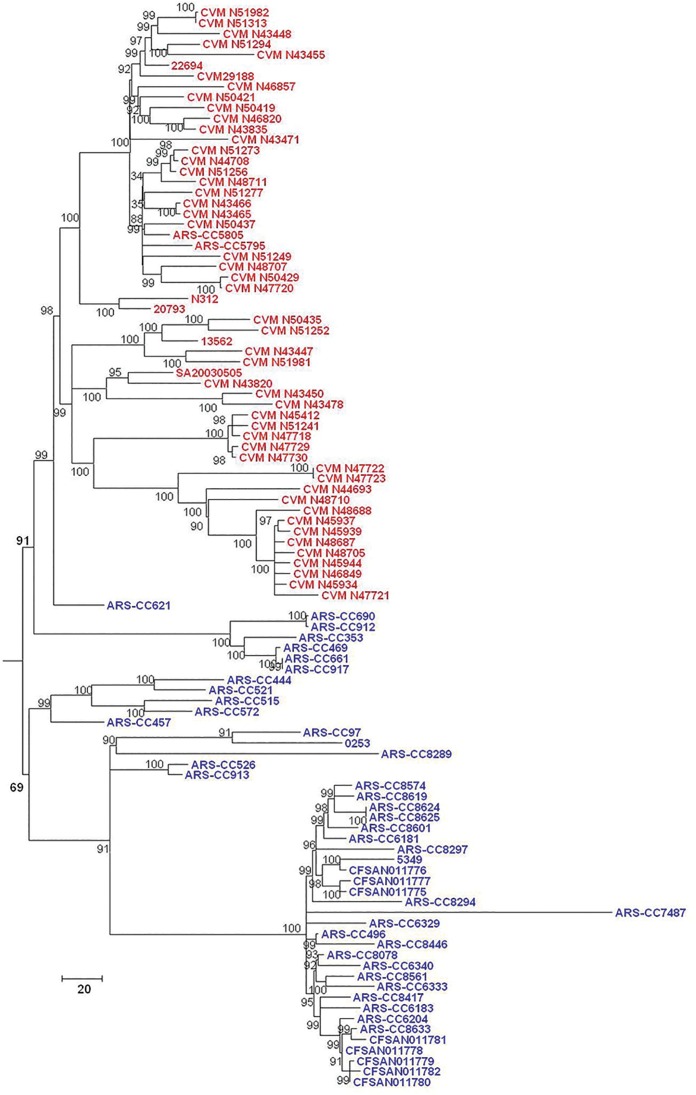
Fig 7. Maximum Likelihood tree of S. Kentucky ST152/318/2132 genomes based on SNPs detected by CFSAN SNP Pipeline.
Bovine-associated strains are labeled in blue and poultry-associated strains are labeled in red. Tree was inferred using the General Time Reversible model of nucleotide substitution in RAxML with 1000 bootstrap replicates and rooted in seven S. Kentucky ST198 genomes. Bar = number of substitutions per site.
Fig 4. Maximum Parsimony tree of S. Kentucky ST152/318/2132 genomes based on 45,999 SNPs detected by Lyve-SET and rooted in twelve S. Kentucky ST198 genomes.
Bovine-associated strains are labeled in blue, poultry-associated strains are labeled in red, and the human clinical isolate CDC 191 is labeled in black. Tree was inferred with 1000 bootstrap replicates. Bar length = number of substitutions.
Fig 5. Maximum Likelihood tree of S. Kentucky ST152/318/2132 genomes based on SNPs detected by Lyve-SET.
Bovine-associated strains are labeled in blue, poultry-associated strains are labeled in red, and the human clinical isolate CDC 191 is labeled in black. Tree was inferred using the General Time Reversible model of nucleotide substitution in RAxML with 1000 bootstrap replicates and rooted in twelve S. Kentucky ST198 genomes. Bar = number of substitutions per site.
Fig 6. Maximum Parsimony tree of S. Kentucky ST152/318/2132 genomes based on 48,269 SNPs detected by CFSAN SNP Pipeline and rooted in seven S. Kentucky ST198 isolates.
Bovine-associated strains are labeled in blue and poultry-associated strains are labeled in red. Tree was inferred with 1000 bootstrap replicates. Bar length = number of substitutions.
For five of the six analyses Lineage 1.0 is composed to several clusters of bovine isolates (n = 39) labeled as clusters 1.1, 1.2.1, 1.2.2 and 1.2.3. Cluster 1.1 consists of five ST152 isolates, one recovered from Texas and four from Wisconsin in 2007. The Wisconsin isolates grouped together, separately from the single Texas isolate. Cluster 1.2.1 consisted of one isolate from Pennsylvania and one from Virginia. Cluster 1.2.2 consisted of three isolates, two of them were typed as ST318 and collected from the same farm in Pennsylvania while the third isolate was a highly diverged ST152 isolate collected from Ohio. Cluster 1.2.3 consisted of 29 isolates, mainly from Pennsylvania with one isolate from New York, and two from Ohio. These results, however, are not consistent across all tree inference methods as the ML analysis of the SNPs detected with Parsnp place all bovine isolates except ARS-CC621 in Lineage 1.0 (Fig 3), albeit with low bootstrap support, and consistent with other methods place all poultry isolates in Lineage 2.0.
Fig 3. Maximum Likelihood tree of S. Kentucky ST152/318/2132 genomes based on SNPs detected by Parsnp.
Bovine-associated strains are labeled in blue, poultry-associated strains are labeled in red, and the human clinical isolate CDC 191 is labeled in black. Tree was inferred using the General Time Reversible model of nucleotide substitution in RAxML with 1000 bootstrap replicates and rooted in twelve S. Kentucky ST198 genomes. Bar = number of substitutions per site.
The topology of Lineage 2.0 is moderately to strongly supported for all analyses and is composed of three to four clusters of strains. For the Parsnp MP and Lyve-SET and CFSAN SNP Pipeline MP and ML analyses the ancestral Lineage 2.0 node moderately to strongly supported (80 to 94%) with much stronger support for the node from which the bovine-associated isolate ARS-CC621 and all poultry-associated isolates descended for all analyses.
For all analyses excluding the Parsnp ML analysis the Lineage 2.0 clusters included one group of bovine/human-associated genomes, (cluster 2.1), one bovine-associated genome (cluster 2.2) and two groups of poultry-associated genomes (clusters 2.3.1, 2.3.2, 2.3.3.1, 2.3.3.2 and 2.4.1.1, 2.4.1.2, 2.4.1.3, and 2.4.2) which is consistent with the two dominant poultry-associated pulsotypes described by Ladely et al. [13]. Cluster 2.1 consists of seven isolates; three from Vermont, one from New York two from Wisconsin, and a distantly related human clinical isolate (CDC 191) from Wisconsin in 2002. Within this group the Vermont isolates are more closely related to each other than they are to the New York and Wisconsin isolates, and the Wisconsin isolates are more closely related to each other than they are to the New York or Vermont isolates. Cluster 2.2 consists of a single bovine-associated isolate (ARS-CC621) recovered in Texas in 2007.
Clusters 2.3.1 to 2.4.2 consist solely of isolates recovered from poultry, poultry products or processing plants and chicken feces/cecum samples collected across North America over a 12 year period. These clusters are monophyletic (geneaological sorting index = 1, P <0.0001) and do not include any bovine isolates indicating complete lineage sorting from the most recent common ancestor that gave rise to poultry-associated isolates. All poultry-associated clusters were moderately to strongly supported for all MP analyses, while ML analyses resulted in lower bootstrap support for clusters 2.4.1.1 to 2.4.2. Within the latter tree, the topologies of cluster 2.4.1 and 2.4.2 are not consistent with those of all other MP and ML trees.
Within the paraphyletic bovine-associated groups presence of different clusters in the same region (Wisconsin, Texas, and Pennsylvania) indicates multiple evolutionary sublineages of S. Kentucky ST152 are circulating in dairy cow herds of the same region. For example, clusters 1.1 and 2.1 isolates were recovered from dairy cows in Wisconsin, cluster 1.1 and 2.2 isolates were recovered from Texas, and clusters 1.2.1, 1.2.2 and 1.2.3 isolates were detected in Pennsylvania. Thus, broad geographic dispersal of S. Kentucky strains may have occurred repeatedly over time. Dairy farms are open environments in that there is a potential of wildlife intrusions on the farm and interactions with dairy cows [47]. For example, migratory birds are known vehicles of enteric bacteria and have been suggested to be a transmission route to livestock [48] and some wild bird species such as European starlings (Sturnus vulgaris) are known to concentrate in areas of cattle operations where their presence has been associated with increased S. enterica contamination of feed and water [49].
Within the poultry-associated clusters, a clear geographic pattern of distribution was not observed. For example, isolates from New Mexico, Tennessee, California, and British Columbia were identified in both clusters 2.3 and 2.4. Along with the presence of isolates from the same state in multiple clusters, geographically disparate isolates grouped together as well. For instance, some isolates from British Columbia (ABBSB1008-2) are sister taxa on the same phylogenetic sublineage with one from Georgia (CVM N43820), two regions that are separated by over 4500 km. Similarly, isolates from California also clustered with those from Tennessee, Massachusetts, New York, Colorado, and Washington State in cluster 2.4. This lack of geographic clustering among the poultry isolates is most likely due to the fact that the majority of these isolates were not collected on-farm, but rather after poultry product processing. Sequencing of isolates collected from farms would help identify any possible geographic signatures present in the currently circulating ST152 populations.
Based on the genomes analyzed in this study there is strong evidence to support the hypothesis that bovine-associated ST152 isolates are phylogenetically distinct from poultry-associated ST152 isolates. However, these data only reflect those S. Kentucky ST152 isolates collected in North America in recent years. It is known that ST152 isolates have been recovered from various sources worldwide and as far back as the 1950s, but what is not well documented in the literature is how frequently ST152 are recovered from cattle and poultry in other continents, or if this ST152-Bovine/Poultry relationship occurs in other regions globally, i.e, if the ST152 strains have entered bovine and/or poultry populations outside of North America or if they are circulating through the populations of other animals. At present, the metadata of only five ST152 strains from outside of North America are deposited in the MLST database and none of these were recovered from poultry or bovine sources, but rather fish meal and human clinical cases. Addition of other ST152 genomes from a variety of sources may result in a somewhat different phylogenetic relationship among strains than what is presented here, as well as the presence of other evolutionary lineages not detected in this study.
Genomic Polymorphisms within the ST152/ST318/2132 Isolates (ST complex 15)
In total there were 2662 SNPs identified in the core genome analysis by Parsnp, and 3353 and 3336 identified by Lyve-SET and CFSAN SNP Pipeline, respectively. We further identified Lineage and host-associated SNPs. The Lineage 1.0/2.0 divergence event for all trees excluding the Parsnp ML tree is marked by eight identified SNPs; four in intergenic regions and four in protein coding genes resulting in three synonymous mutations and one non-synonymous mutation (SeKA_A1002, SeKA_A1027, SeKA_A1948, SeKA_A4700) (Table 2).
Table 2. Identified SNPs defining the Lineage 1.0/2.0 divergence (Top) and the Bovine-associated/Poultry-associated divergence (Bottom).
S = synonymous mutation. NS = non-synonymous mutation.
| Lineage 1.0 / 2.0 | Locus Tag a | Annotation b | Genome Position a | nt Lineage 2.0 | nt Lineage 1.0 | Substitution c | aa Lineage 2.0 | aa Lineage 1.0 | aa Position a | Method |
| intergenic | 528068 | C | T | 1, 2, 3 | ||||||
| SeKA_A1002 | LysR substrate binding domain protein | 953413 | A | G | S | Leu | Leu | 223 | 1, 2, 3 | |
| SeKA_A1027 | nitrite extrusion protein 2 | 984718 | C | T | S | Pro | Pro | 288 | 1, 2, 3 | |
| intergenic | 1032798 | A | G | 2, 3 | ||||||
| SeKA_A1948 | amidophosphoribosyltransferase | 1876321 | G | A | S | Ala | Ala | 288 | 1, 2, 3 | |
| intergenic | 2432705 | T | G | 1, 2, 3 | ||||||
| intergenic | 3169966 | T | G | 1, 2, 3 | ||||||
| SeKA_A4700 | propionate—CoA ligase | 4587482 | A | C | NS | Asn | Thr | 240 | 2, 3 | |
| Bovine / Poultry | Locus Tag a | Annotation b | Genome Position a | nt Poultry-associated | nt Bovine-associated | Substitution c | aa Poultry | aa Bovine | aa Position | Method |
| SeKA_A1094 | protein YdcF | 1055798 | T | C | NS | Thr | Ala | 43 | 1, 2, 3 | |
| SeKA_A2591 | hemolysin-3 | 2519134 | A | C | NS | Phe | Val | 32 | 3 | |
| SeKA_A2812 | methyltransferase family protein | 2713142 | G | A | NS | Ser | Leu | 39 | 1, 2, 3 | |
| SeKA_A4467 | carbonate dehydratase | 4347265 | G | T | NS | Ala | Asp | 22 | 1, 2, 3 | |
a = Locus/Position in S. Kentucky CVM29188
b = Annotation in S. Kentucky CVM29188 (accession no. ABAK02000001.1)
c = S(synonymous), NS(nonsynonymous)
1 = Parsnp
2 = Lyve-SET
3 = CFSAN SNP Pipeline
In a comparison of bovine-associated isolates to poultry-associated isolates an average of 217, 247, and 281 SNP differences were identified between the two by the Parsnp, Lyve-SET, and CFSAN SNP Finder analyses, respectively (Parsnp range = 62 to 308 SNPs, Lyve-SET range = 72 to 377 SNPs, and CFSAN SNP Pipeline range = 72 to 489). Within these SNP matrices there were four SNPs in protein coding genes, all resulting in non-synonymous mutations, that defined the bovine-poultry divergence for all strains (SeKA_A1094, SeKA_A2591, SeKA_A2812, SeKA_A4467) (Table 2). No evidence of the role of these genes in host-specificity, colonization of the gut by S. enterica or persistence of S. enterica in cows, poultry, poultry and bovine products, or the environment could be found in the literature. However, hemolysin-3 (SeKA_A2591) has been shown in Gram-negative and Gram-positive organisms to be involved in the infection process of other mammals, particularly Vibrio vulnificus and Bacillus cereus [50][51]. The hemolysin-3 in S. Kentucky demonstrates 71% and 45% amino acid identity to that of V. vulnificus and B. cereus. In vitro and in vivo assays need to be conducted to further elucidate the potential roles of these protein-coding genes in persistence in the poultry and bovine hosts or specificity to either environment.
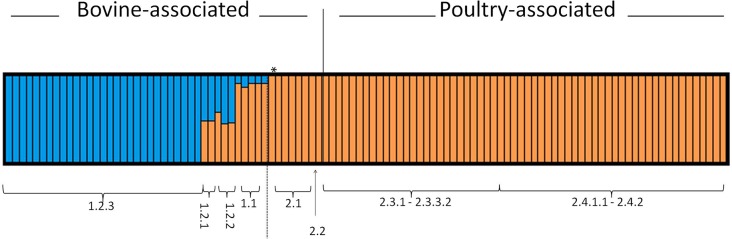
A Bayesian analysis of SNP characters using the STRUCTURE v 2.3.4 program was consistent with the inferred ML and MP phylogenies (Fig 8) in showing that there were primarily two distinct groups. However, these results provided additional details that help explain the placement of certain isolates within the phylogenetic analysis. For example, STRUCTURE showed that isolates within Cluster 1.2.3 have SNP profiles that are, for the most part, distinct from all others (blue profiles). In contrast, clusters 1.1, 1.2.1, and 1.2.2 showed evidence of admixture (blue/orange profiles) where those isolates had some proportion of SNPs that are indicative of clusters 1.2.3 (blue profiles) and Lineage 2.0 isolates (orange profiles). Cluster 1.1 isolates had SNP profiles more closely related to clusters 2.1 to 2.4 than to 1.2.3. All poultry isolates all show approximately similar SNP profiles with the basal cow isolates in clusters 2.1 and 2.2 indicating a high level of similarity among the cow and poultry isolates within evolutionary Lineage 2.0.
Fig 8. STRUCTURE analysis of the proportion of each isolate’s SNP profile attributed to each of the different groups.
Dashed line = Lineage 1.0/2.0 divergence. Profiles to the left of the solid black line = bovine-associated isolates, while profiles to the right of the solid black line = poultry-associated isolates. Asterisk = human clinical isolate CDC 191.
Plasmid Detection
Using the assembled genomes, multiple plasmids were detected in the S. Kentucky ST152 isolates while several were identified as plasmid-free (Table 3 and S2 Table). Thirty three poultry-associated isolates encoded sequences similar to the ColV plasmid pCVM29188_146 plasmid (IncFIB), which is somewhat consistent with previous studies of poultry-isolated S. Kentucky in North America that have shown that the majority of these strains (72.9%) harbored markers of ColV plasmids [52]. However, this study also demonstrated that S. Kentucky isolates collected on farms were more likely to harbor this plasmid than those collected from retail meats [52], while the majority of isolates analyzed here were recovered from the latter. This plasmid has been shown to be involved in enhanced survival of S. Kentucky in poultry and is highly similar in gene content and nucleotide sequence similarity to the ColV plasmid of avian pathogenic Escherichia coli (APEC) [52]. The presence of this plasmid was restricted to poultry-associated isolates, providing further evidence that it is involved in interactions with the poultry host. However, it should be noted that Fricke et al. [12] identified six bovine-associated isolates with PCR markers indicative of the presence of APEC-like plasmids similar to those of pCVM29188_146 suggesting that more bovine-specific S. Kentucky need to be evaluated for the presence of this or highly similar plasmids. Although, Johnson et al. [52]demonstrated the role of this plasmid in enhanced extracellular survival in poultry, its absence from poultry-associated S. Kentucky suggests there are other factors involved in specificity of these strains to the poultry host. Similar to Ladely et al. [13] the IncFIB plasmid type was not evenly distributed across poultry-associated strains as it was identified in 81% of cluster 2.4.1.1 to 2.4.2 isolates but only in 25% of cluster 2.3.1 to 2.3.3.2 isolates.
Table 3. Plasmids identified by PlasmidFinder in genomes analyzed in this study.
| ST | Poulty/Bovine | Plasmid Replicon | Number of Plasmids Detected |
|---|---|---|---|
| ST152 | Poultry-associated | IncFIB | 33 |
| IncX1 | 58 | ||
| IncHI2 | 12 | ||
| Col156 | 2 | ||
| IncI1 | 29 | ||
| Bovine-associated | IncI1 | 34 | |
| IncI2 | 3 | ||
| IncA/C2 | 4 | ||
| ColpVC | 1 | ||
| Col8282 | 3 | ||
| No plasmid detected | 12 | ||
| ST198 | IncI1 | 1 | |
| No plasmid detected | 11 | ||
The most frequently detected plasmid sequences in poultry-associated isolates were those similar to the canonical pCVM29188_46 (IncX1) plasmid, which was also not detected in any of the bovine-associated isolates (Table 3 and S2 Table). The biological role of this plasmid has not been well-elucidated, but its high prevalence among these strains indicates it may play a significant role in the survival of S. Kentucky within the poultry host, poultry production environment, or in transmission of S. Kentucky between animals. However, this would need to be further evaluated in vivo. Coupling the plasmid presence/absence data with the inferred phylogeny suggests that poultry-associated S. Kentucky acquired this plasmid, as well as the IncFIB plasmid after diverging from the most recent common ancestor shared with the bovine-associated isolates.
Within the bovine-associated S. Kentucky 12 isolates were identified as plasmid-free and these isolates belonged to clusters 1.1, 1.2.1, 2.1, and 2.2 (Table 3 and S2 Table). Isolates from several clusters harbored a variety of plasmids, including those belonging to IncI1 (34 isolates), IncI2 (3 isolates), and IncA/C2 (4 isolates) replicon types, and plasmids identified by PlasmidFinder as Col8282 (3 isolates) and one as ColpVC. The IncI1 plasmid, represented by sequence AOYZ01000068.1 in GenBank of S. Kentucky 5349, was the most frequently detected plasmid sequence among the bovine-associated isolates. This sequence is ca. 92 kb, encodes 100 ORFs and is somewhat similar to plasmid sequences available in the NCBI database. These include plasmid pSTY1-1898 of S. Typhimurium str. USDA-ARS-USMARC-1898 (coverage = 81%, similarity = 99%) and S. Kentucky plasmid pCS0010A_95 (coverage = 79%, similarity = 99%), among several others.
The majority of isolates that harbored plasmids were from the mid-Atlantic region of the United States. The absence of plasmids in many bovine-associated isolates (25%) suggests they are not necessary for survival or persistence within the bovine gastrointestinal tract or the dairy farm environment. However, the high level prevalence of IncI1 plasmids in cluster 1.2.3 isolates indicates that they may provide an advantage in these environments, and their restriction to isolates from the mid-Atlantic and Northeast United States may be indicative of their importance to the ecology of these strains in this region.
Phylogeny and Genomic Polymorphisms within the ST198 Isolates (ST complex 56)
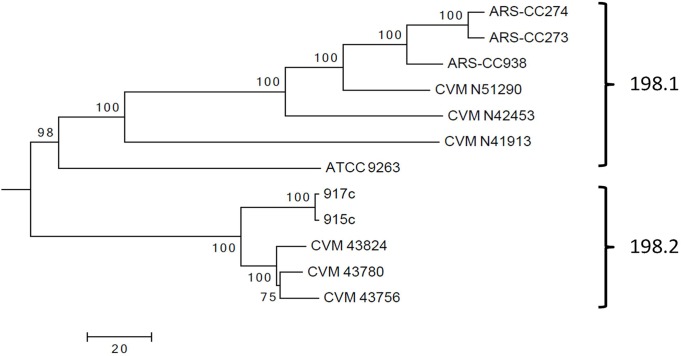
Within the S. Kentucky ST198 lineage two major clusters were identified (labeled here as clusters 198.1 and 198.2) (Fig 9). Cluster 198.1 consists of isolates recovered from agricultural sources in the United States (dairy cow feces, dairy cow milk, ground beef, and ground turkey) and the S. Kentucky type strain ATCC 9263. Cluster 198.2 consists of five human clinical isolates, two of these collected from the same patient in Kuwait in 2012 (915c from a sacral wound and 917c from a stool sample) [25], and three were collected from human clinical cases in the United States. It is not known if these three infections were acquired abroad or within the United States, as metadata for these isolates are lacking in the public database. Source attribution would need to be verified before the locations of infections can be confirmed. S. Kentucky ST198 infections have been reported in Africa, Eastern Europe, and Southeast Asia and travel-acquired infections with these organisms has been reported in people traveling to these regions [7][8]. Until a more comprehensive geophylogeny of S. Kentucky ST198 strains is conducted assumptions about strain traceback are speculative.
Fig 9. Maximum Parsimony tree of S. Kentucky ST198 isolates collected from human clinical cases (cluster 198.2) and dairy cows and food products in North America (198.1).
The tree was inferred from an alignment of SNPs detected among all S. Kentucky ST198 genomes and using the chromosome of S. Kentucky CVM29188 as a reference. The tree is based on 42,958 SNPs (Parsnp analysis), of these, 532 SNPs were identified among the ST198 genomes. The tree displayed here is rooted in the distantly related S. Kentucky CVM29188. Tree was inferred with 1000 bootstrap replicates. Bar length = number of substitutions.
Using Parsnp, with S. Kentucky CVM29188 as a reference genome, there were 532 SNPs identified in the core-genomes of the ST198 isolates with an average of 209 SNP differences between the clusters 198.1 and 198.2 genomes (range = 169 to 228 SNPs). Cluster 198.1 isolates collected from agricultural sources in the United States had, for the most part, fewer SNPs between each other (mean = 138, range = 10 to 210) than they did with the human clinical isolates (cluster 198.2) (Table 4). Similarly, the human clinical isolates (cluster 198.2) had fewer SNP differences between each other than they did with the cluster 198.1 isolates (mean = 32, range = 2 to 48). Interestingly, in a genome-genome comparison fewer SNPs were detected between ATCC 9263 and 198.2 genomes than between ATCC 9263 and 198.1 genomes. However, there were fewer SNPs between ATCC 9263 and the 198.1 cluster than ATCC 9263 and the 198.2 cluster when the average base pair differences over all of the sequence pairs were calculated per cluster.
Table 4. SNP differences among S. Kentucky ST198 genomes.
| Cluster / Genome | ATCC 9263 | CVM N51290 | CVM N42453 | CVM N41913 | ARS-CC274 | ARS-CC273 | ARS-CC938 | CVM 43780 | CVM 43756 | CVM 43824 | 915c | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 198.1 | ATCC 9263 | |||||||||||
| CVM N51290 | 195 | |||||||||||
| CVM N42453 | 201 | 94 | ||||||||||
| CVM N41913 | 200 | 193 | 197 | |||||||||
| ARS-CC274 | 210 | 69 | 107 | 206 | ||||||||
| ARS-CC273 | 210 | 71 | 111 | 210 | 10 | |||||||
| ARS-CC938 | 197 | 58 | 98 | 197 | 35 | 33 | ||||||
| 198.2 | CVM 43780 | 169 | 204 | 210 | 209 | 219 | 219 | 206 | ||||
| CVM 43756 | 178 | 211 | 215 | 214 | 224 | 228 | 215 | 19 | ||||
| CVM 43824 | 174 | 207 | 211 | 210 | 220 | 224 | 211 | 17 | 22 | |||
| 915c | 177 | 208 | 214 | 213 | 223 | 225 | 212 | 41 | 48 | 44 | ||
| 917c | 179 | 210 | 214 | 213 | 223 | 227 | 214 | 43 | 48 | 44 | 2 | |
There were 66 conserved SNP differences identified between clusters 198.1 and 198.2 (Table 5). Of particular interest are the DNA gyrase subunit A (gyrA) (AEX15_13770) substitutions Ser83 → Phe in 198.2; Asp87 → Tyr in CVM 43780, CVM 43756, and CVM 43824; and Asp87 → Asn in 915c and 917c. These substitutions confer resistance to nalidixic acid, an attribute that is presumed to have emerged in ST198 isolates in the early 2000s in Egypt[7]. Substitution Ser80 → Ile in DNA topoisomerase IV subunit A (parC) (AEX15_04910) conferring resistance to ciprofloxacin, was also observed in 198.2 isolates. These substitutions are characteristic of ST198 strains circulating in North and East Africa and the Middle East [7] suggesting that the infections caused by these strains (S. Kentucky CVM 43780, CVM 43756, and CVM 43824) may have been acquired outside of the United States or from exposure to imported products.
Table 5. SNP differences between 198.1 (non-clinical North American agricultural isolates) and 198.2 (human clinical isolates).
The reference genome for this analysis is the chromosome of S. Kentucky CVM29188.
| Locus Tag in CVM N51290 | Locus Tag in CVM29188 | Annotation a | Genome Position a | nt 198.1 | nt 198.2 | Substitution b | aa 198.1 | aa 198.2 | aa Position |
|---|---|---|---|---|---|---|---|---|---|
| AEX15_00985 | SeKA_A0118 | putative cytoplasmic protein | 112694 | A | G | NS | Asn | Ser | 215 |
| AEX15_01465 | SeKA_A0272 | ABC transporter ATP-binding protein | 259861 | G | A | S | Asn | Asn | 352 |
| AEX15_07125 | SeKA_A0450 | outer membrane protein F | 449431 | G | A | S | Ala | Ala | 310 |
| intergenic | 511489 | G | A | ||||||
| intergenic | 528058 | C | G | ||||||
| intergenic | 768380 | G | T | ||||||
| intergenic | 932324 | T | C | ||||||
| AEX15_09820 | SeKA_A1028 | Nitrate reductase alpha subunit | 986092 | C | A | NS | Ser | Tyr | 249 |
| AEX15_09905 | SeKA_A1046 | virulence factor SrfB | 1006926 | C | T | S | Asn | Asn | 389 |
| AEX15_10005 | SeKA_A1065 | PTS system, IIc component | 1028557 | T | G | NS | Val | Gly | 232 |
| AEX15_10580 | SeKA_A1172 | phosphatidylglycerophosphatase B | 1132142 | T | C | NS | Thr | Ala | 184 |
| AEX15_10645 | SeKA_A1188 | anthranilate synthase component I | 1148000 | C | T | NS | Pro | Ser | 225 |
| AEX15_11230 | SeKA_A1320 | hypothetical protein | 1268383 | G | C | NS | Gly | Ala | 47 |
| AEX15_11825 | SeKA_A1435 | aspartyl-tRNA synthetase | 1376156 | C | T | NS | Ala | Thr | 273 |
| AEX15_11880 | SeKA_A1448 | flagellar protein FlhE | 1388508 | T | C | S | Leu | Leu | 624 |
| AEX15_11945 | SeKA_A1461 | transcriptional activator FlhD | 1402136 | G | T | S | Ser | Ser | 4 |
| AEX15_12480 | SeKA_A1563 | arsenical pump-driving ATPase | 1479380 | A | G | NS | Leu | Ser | 578 |
| AEX15_13315 | SeKA_A1750 | beta-D-glucoside glucohydrolase | 1662011 | G | A | S | Thr | Thr | 560 |
| AEX15_13615 | SeKA_A1814 | nucleoid-associated protein NdpA | 1730145 | T | C | NS | Asn | Asp | 136 |
| AEX15_13770 | SeKA_A1846 | DNA gyrase subunit A (gyrA) | 1764089 | G | A | NS | Ser | Phe | 83 |
| AEX15_13940 | SeKA_A1884 | O-succinylbenzoate synthase | 1805065 | C | A | NS | Glu | Asp | 44 |
| AEX15_15785 | SeKA_A2190 | DNA repair protein RecO | 2129001 | T | C | NS | Lys | Glu | 156 |
| AEX15_21540 | SeKA_A2229 | phospho-2-dehydro-3-deoxyheptonate aldolase | 2169354 | G | A | S | Asn | Asn | 149 |
| AEX15_21995 | SeKA_A2343 | murein hydrolase effector LrgB | 2282024 | G | A | S | Gly | Gly | 38 |
| AEX15_22185 | SeKA_A2383 | putative type III secretion system effector protein OrgC | 2317497 | G | A | NS | Ser | Leu | 98 |
| AEX15_22530 | SeKA_A2451 | sulfate adenylyltransferase subunit 1 | 2379190 | A | C | NS | Ile | Ser | 339 |
| intergenic | 2476378 | G | A | ||||||
| AEX15_04160 | SeKA_A2564 | nickel/cobalt efflux protein RcnA | 2496525 | C | T | NS | Pro | Ser | 31 |
| intergenic | 2497451 | C | G | ||||||
| AEX15_04380 | SeKA_A2619 | D-erythrose-4-phosphate dehydrogenase | 2542815 | G | A | NS | Pro | Ser | 224 |
| AEX15_04910 | SeKA_A2749 | DNA topoisomerase IV subunit A (parC) | 2654521 | C | A | NS | Ser | Ile | 80 |
| intergenic | 2760667 | C | A | ||||||
| AEX15_03530 | SeKA_A3124 | cytoplasmic protein | 2998220 | G | A | S | Thr | Thr | 395 |
| AEX15_02625 | SeKA_A3321 | putative glucarate transporter | 3208206 | T | A | NS | Leu | Gln | 355 |
| AEX15_02555 | SeKA_A3336 | ADP-heptose—LPS heptosyltransferase | 3221877 | G | A | S | Pro | Pro | 122 |
| AEX15_02175 | SeKA_A3395 | PTS sorbose transporter subunit IIC | 3280995 | C | T | NS | Gly | Asp | 241 |
| AEX15_14010 | SeKA_A3451 | trimethylamine N-oxide reductase I catalytic subunit | 3334071 | G | A | S | Cys | Cys | 320 |
| AEX15_14230 | SeKA_A3482 | phosphate transporter permease subunit PstC | 3369116 | G | T | NS | Pro | His | 74 |
| AEX15_14255 | SeKA_A3487 | glucosamine—fructose-6-phosphate aminotransferase | 3375283 | T | C | NS | Ile | Val | 490 |
| AEX15_20905 | SeKA_A3529 | threonine dehydratase | 3421216 | G | A | S | Ala | Ala | 446 |
| AEX15_21030 | SeKA_A3553 | putative common antigen polymerase | 3445490 | G | A | S | Leu | Leu | 125 |
| AEX15_21080 | SeKA_A3565 | porphobilinogen deaminase | 3453638 | G | T | NS | Asp | Glu | 117 |
| AEX15_21220 | SeKA_A3595 | chondroitin sulfate/heparin utilization regulation protein | 3480727 | C | T | S | Asn | Asn | 103 |
| AEX15_21260 | SeKA_A3603 | sec-independent translocase | 3489186 | C | T | NS | Pro | Ser | 142 |
| AEX15_21305 | SeKA_A3612 | proline dipeptidase | 3500226 | C | T | S | Ser | Ser | 179 |
| intergenic | 3575800 | C | A | ||||||
| AEX15_21420 | SeKA_A3805 | thiamine-phosphate pyrophosphorylase | 3686081 | C | A | NS | Ala | Ser | 57 |
| AEX15_18585 | SeKA_A3887 | LexA repressor | 3764969 | C | T | S | Ser | Ser | 60 |
| intergenic | 3770533 | C | A | ||||||
| AEX15_18425 | SeKA_A3921 | Na+/H+ antiporter | 3813232 | G | A | S | Ser | Ser | 30 |
| intergenic | 3829881 | C | T | ||||||
| intergenic | 3832063 | T | A | ||||||
| intergenic | 3896075 | C | T | ||||||
| AEX15_17850 | SeKA_A4040 | putative cytoplasmic protein | 3929069 | T | C | S | Thr | Thr | 54 |
| intergenic | 4002925 | G | A | ||||||
| AEX15_20245 | SeKA_A4293 | chitinase | 4165915 | A | G | S | Val | Val | 533 |
| AEX15_20105 | SeKA_A4321 | isoleucyl-tRNA synthetase | 4192511 | C | A | S | Thr | Thr | 62 |
| AEX15_17125 | SeKA_A4346 | carnitine operon protein CaiE | 4219742 | C | T | NS | Glu | Lys | 128 |
| intergenic | 4233585 | C | A | ||||||
| AEX15_16055 | SeKA_A4409 | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase | 4286504 | T | C | NS | Ser | Pro | 265 |
| AEX15_16220 | SeKA_A4445 | hypothetical protein | 4325107 | A | G | S | Gly | Gly | 137 |
| AEX15_16390 | SeKA_A4481 | putative fimbrial outer membrane usher protein | 4359640 | G | T | NS | Leu | Met | 704 |
| intergenic | 4463447 | T | C | ||||||
| AEX15_22970 | SeKA_A4665 | fimbrial outer membrane usher protein StbC | 4548312 | A | C | NS | Val | Gly | 119 |
| AEX15_06235 | SeKA_A4762 | 2-aminoethylphosphonate transport system permease PhnU | 4645904 | A | G | NS | Ile | Thr | 92 |
| AEX15_06585 | SeKA_A4840 | copper exporting ATPase | 4725203 | A | G | S | Ser | Ser | 111 |
a = Position/annotation in S. Kentucky CVM29188 (accession no. ABAK02000001.1)
b = S(synonymous), NS(nonsynonymous)
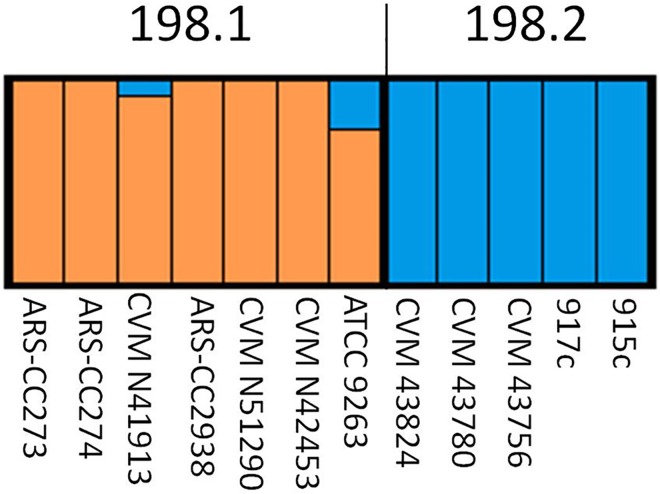
A Bayesian analysis of SNP differences among isolates using the STRUCTURE program indicated that for the most part 198.1 and 198.2 isolates were distinct with the exception of ATCC 9263 and CVM N41913, which were basal within the cluster 198.1 lineage (Fig 10). For these two isolates a proportion of SNPs detected in their genomes (ATCC 9263 = 0.24 and CVM N41913 = 0.078) were characteristic of those identified in 198.2.
Fig 10. STRUCTURE analysis of the proportion of each isolate’s SNP profile attributed to each of the different clusters (198.1 and 198.2).
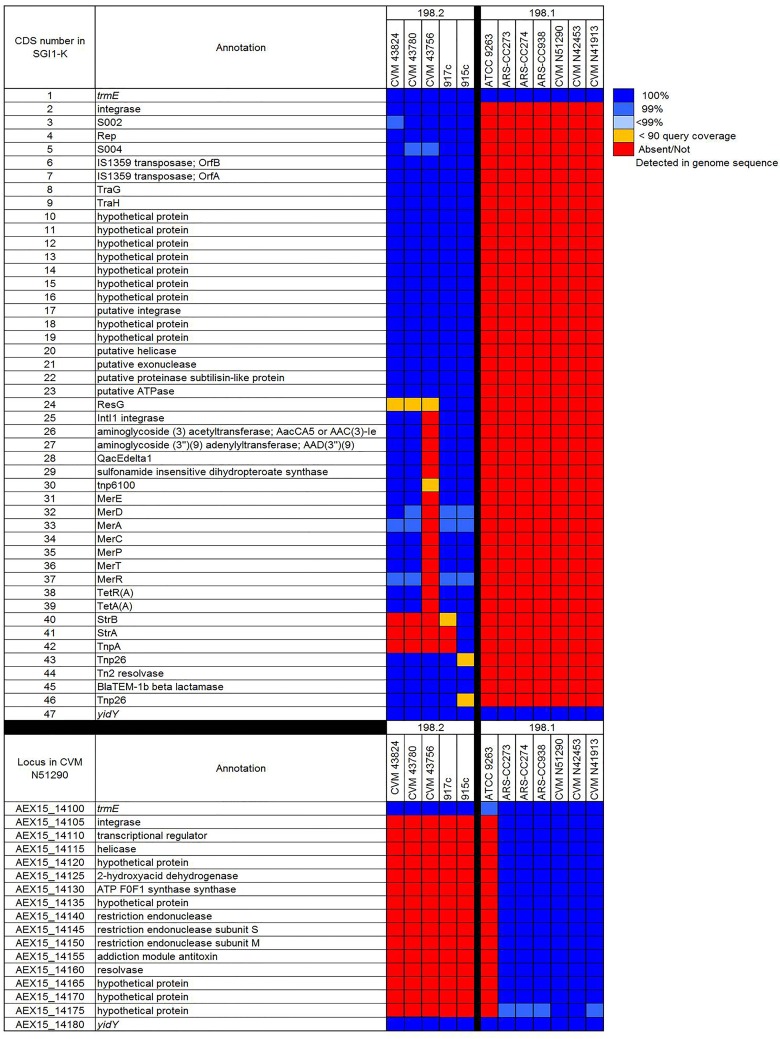
All cluster 198.2 genomes were identified as encoding ORFs homologous to the complete sequence of Salmonella Genomic Island 1 inserted at the trmE-yidY locus (SGI-K used as a reference; NCBI accession AY463797) (Fig 11). However, the structure of this island was difficult to discern due to the presence of SGI1 ORFs on multiple contigs in the 198.2 cluster genomes. SGI1 is an integrative and mobilizable element found in several S. enterica serovars and other non-salmonellae and responsible for resistance to multiple antibiotics [53][54]. The structure of this island is known to be highly variable [44][55]. Four of five SGI1 encoding genomes encoded regions known for resistance to mercury. However, this regions is absent from in CVM 43756 (Fig 11). For all cluster 198.2 genomes excluding 915c, strB (streptomycin phosphotransferase) and strA were not detected.
Fig 11. BLASTN comparison of SGI1-K against the ST198 genomes (Top).
Blue squares indicate level of similarity, red squares indicate that homologs were not detected in the query genome sequence, orange squares = homolog was identified but at < 90% coverage. BLASTN comparison of a ca. 17 kb region inserted at the trmE-yidY locus in cluster 198.1 genomes (Bottom).
At the trmE-yidY insertion locus (AEX15_14100, AEX15_14180) all 198.1 genomes, excluding ATCC 9263, encoded a ca. 17 kb region with ORFs annotated as an integrase, transcriptional regulator, helicase, five ORFs annotated as hypothetical proteins, a single ORF annotated as 2-hydroxyacid dehydrogenase, ATP F0F1 synthase synthase, addiction module antitoxin, resolvase, and three restriction endonuclease ORFs. To-date, this region has only been identified in these six S. Kentucky ST198 genomes, S. Seftenberg ATCC 43845 and S. Agona strains 18.H.07, 557928, and 620239.
Based on the PlasmidFinder analysis, plasmids were not detected in any of the ST198 strains except for S. Kentucky CVM 51290 (Table 3). However, strains harboring plasmids carrying antibiotic resistance genes have been collected from human clinical cases [9] suggesting that, like ST152 and other S. enterica serovars, plasmid presence is variable in ST198. The IncI1 plasmid sequence identified in S. Kentucky CVM 51290 is similar to others currently in a NCBI plasmid database and shows 99% sequence similarity across 90% of the putative plasmid region with plasmid pSL476_91 from S. Heidelberg str. SL476, and 99% similarity across 88% of the plasmid region with S. Kentucky plasmid pCS0010A_95.
Few conclusions, based on genomic data, can be made about the ST198 group at this time due to the limited number of available S. Kentucky ST198 genome sequences deposited in public databases. However, it is clear that there is considerable genomic diversity among the genomes that have been sequenced to date. Sequencing of more ST198 genomes will allow for a more comprehensive understanding of the geophylogeny, global diversity, and presence of variable regions, such as SGI1, in these isolates.
It is important to note that ST198 has been isolated from avian and bovine sources in the United States [6] indicating that it may have the potential to become established in poultry flocks, dairy herds, other livestock, and/or wildlife in the United States. However, more work needs to be conducted to understand its apparent limited distribution in these animals as well as the presence of any potential wildlife reservoirs. Further, more work needs to be conducted to estimate the virulence potential of the ST198 that is endemic in the United States and abroad.
Differences between S. Kentucky ST152 and ST198 Isolates
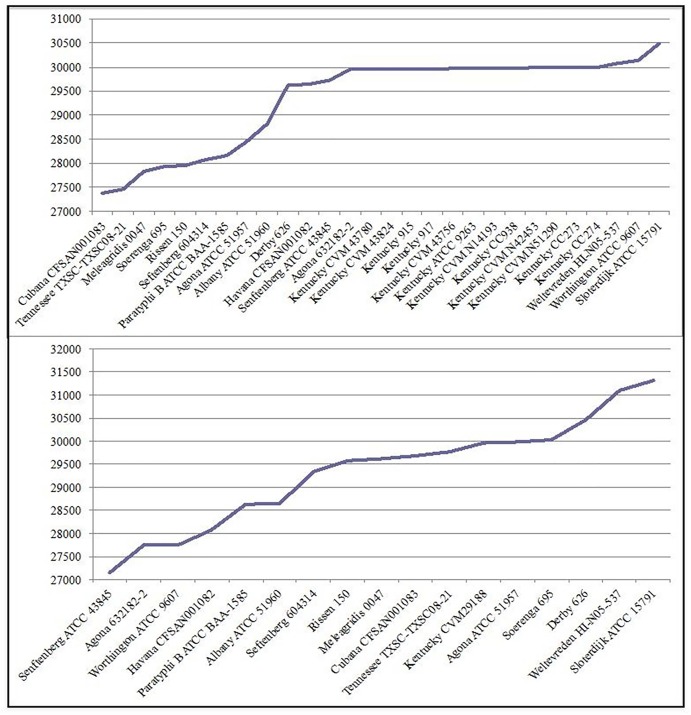
Although frequently considered to be similar groups of strains due to their serological-based nomenclature, S. Kentucky ST198 and S. Kentucky ST152 isolates demonstrate considerable differences in their genetic backgrounds that have not been adequately described. Using assembled genomes, with the chromosome of S. Kentucky CVM29188 as a reference, there were between 27,369 and 30,485 SNP differences between S. Kentucky CVM29188 and representatives of other subclade A1 serovars in a core-genome alignment of length 3,465,932 bp (ca. 72% coverage of the S. Kentucky CVM29188 chromosome) (Table 6, Fig 12). In this analysis there were between 29,952 and 29,990 SNPs between CVM29188 and ST198 genomes. Thirteen serovars had fewer SNP differences with S. Kentucky CVM29188 than did S. Kentucky ST198 with CVM29188 (Table 6, Fig 12), while, 11 serovars of subclade A1 demonstrated fewer SNP differences with ST198 S. Kentucky ATCC 9263 than did S. Kentucky CVM29188 with S. Kentucky ATCC 9263. These data indicate a significant difference in the nucleotide sequence content between ST152 and ST198 genomes and coupled with the core-genome phylogeny (Fig 1) indicate that S. Kentucky ST152 and ST198 are more similar to other serovars than they are to each other.
Table 6. Number of SNP differences between ST198/ST152 and subclade A1 serovars.
For this analysis representatives of subclade A1 were selected. Parsnp was used to identify core-genome SNPs among the sequenced isolates. The chromosome of S. Kentucky CVM29188 was used as a reference genome for the SNP analysis.
| Serovar/Strain | No. SNP differences with S. Kentucky CVM29188 | Serovar/Strain | No. SNP differences with S. Kentucky ATCC 9263 | |
|---|---|---|---|---|
| S. Cubana CFSAN001083 | 27369 | S. Senftenberg ATCC 43845 | 27140 | |
| S. Tennessee TXSC-TXSC08-21 | 27457 | S. Agona 632182–2 | 27743 | |
| S. Meleagridis 0047 | 27821 | S. Worthington ATCC 9607 | 27746 | |
| S. Soerenga 695 | 27927 | S. Havana CFSAN001082 | 28073 | |
| S. Rissen 150 | 27958 | S. Paratyphi B ATCC BAA-1585 | 28638 | |
| S. Seftenberg 604314 | 28066 | S. Albany ATCC 51960 | 28658 | |
| S. Paratyphi B ATCC BAA-1585 | 28156 | S. Seftenberg 604314 | 29329 | |
| S. Agona ATCC 51957 | 28445 | S. Rissen 150 | 29582 | |
| S. Albany ATCC 51960 | 28815 | S. Meleagridis 0047 | 29614 | |
| S. Derby 626 | 29621 | S. Cubana CFSAN001083 | 29685 | |
| S. Havana CFSAN001082 | 29635 | S. Tennessee TXSC-TXSC08-21 | 29768 | |
| S. Senftenberg ATCC 43845 | 29718 | S. Kentucky CVM29188 (ST152) | 29971 | |
| S. Agona 632182–2 | 29940 | S. Agona ATCC 51957 | 29974 | |
| S. Kentucky ATCC 9263 (ST198) | 29971 | S. Soerenga 695 | 30015 | |
| S. Weltevreden HI-N05-537 | 30078 | S. Derby 626 | 30451 | |
| S. Worthington ATCC 9607 | 30140 | S. Weltevreden HI-N05-537 | 31088 | |
| S. Sloterdijk ATCC 15791 | 30485 | S. Sloterdijk ATCC 15791 | 31309 |
Fig 12. Number of SNP differences between S. Kentucky CVM29188 (ST152) and other S. enterica subclade A1 genomes (Top).
Number of SNP differences between S. Kentucky ATCC 9263 (ST198) and other S. enterica subclade A1 genomes (bottom). X-axes show genomes used in this analysis and Y-axes show number of SNP differences.
In an analysis comparing only ST152 genomes to ST198 genomes 37,769 SNP differences were detected (S3 Table). These SNPs were annotated and results demonstrated a significant number of polymorphisms in the protein-coding regions of the genome (S4 Table). Of a total of 4452 annotated protein-coding genes, at least one SNP was detected in 3713 (83.5%) genes while no SNPs were detected in 737 (16.5%) genes. Three hundred seventeen of these genes had ≥ 20 conserved SNP differences with ST152. Of particular interest are several protein coding genes that are involved with interactions with the animal host environment, specifically in the colonization and infection processes by S. enterica. For example, 239 conserved SNP differences were identified in SeKA_A3913, a 16,683 bp ORF annotated as a conserved hypothetical protein, located within SPI-4, and involved in colonization of the bovine gastrointestinal tract [56]. It should be noted that there was an appreciable level of sequence divergence in this gene within ST152 isolates, particularly in isolate ABB1087-1, and this is most likely due to the likelihood of synonymous substitutions to accumulate across a large protein coding gene. SeKA_A0255 of ST152, an ORF homologous to the secreted virulence protein SlrP (STM0800) of S. Typhimurium LT2, had 84 conserved SNP differences with ST198 genomes. A large repetitive protein (SeKA_A2250) homologous to BapA (STM2689) involved in host-colonization, organ invasion, and biofilm formation [57] encoded 68 conserved SNP differences between the two STs. An invasion-like protein (SeKA_A1129), homologous to STM1669 (ZirT), involved in virulence modulation [58] encoded 51 conserved SNP differences between the two STs. ShdA (SeKA_A1896), an outer-membrane protein that is expressed in the intestine and is involved in long-term fecal shedding [59][60], had 48 conserved SNP differences between the two STs. Multiple effector proteins also demonstrated appreciable divergence between ST152 and ST198 (SeKA_A1084 = sseJ; SeKA_A1053 = sifB; SeKA_A1621 = sopA; SeKA_A0837 = sseC, SeKA_A0835 = sseB; SeKA_A2465 = sopD; SeKA_A2283 = pipB; SeKA_A2398 = sipD; SeKA_A0839 = sseE; SeKA_A2393 = sptP; SeKA_A0841 = sseF; SeKA_A2380 = avrA). These proteins interact with host-cells and are intimately involved in the infection process [61][62][63] and their sequence divergence may result in different interactions in the human, cow, and poultry hosts, with the potential for different outcomes for each host. However, to fully assess the biological consequences of these differences in vivo analyses targeting these regions would need to be conducted.
Regions identified in one ST but absent in the other, as well as regions present in both but flanked by unique ORFs were identified by conducting a whole-genome reciprocal BLASTP analysis (Table 7). Sixteen regions of contiguous protein-coding genes present in ST198 genomes but absent in ST152 genomes, or highly diverged and flanked by unique regions were identified. Of these, six were completely or partially identified as genomic islands. Of particular interest in the ST198 genomes is an eight ORF region (region 3 in Table 7) responsible for sialic acid transport, a key feature of mammalian pathogens that allows them to scavenge sialic acid and utilize it as a carbon source in the host intestine (AEX15_07505, AEX15_07550) [64][65][66]. This region is flanked on one side by a tRNA-Ser locus suggesting it is a putative transmissible genomic island. A BLASTP analysis demonstrates there is a unidirectional match at low to moderate amino acid similarity (23 to 70% similarity) when these ORFs are compared to ST152 genomes indicating they are highly diverged from ORFs with a similar functional annotations encoded in the ST152 backbone. This sialic acid transport region was identified in other S. enterica serotypes, many of which are known pathogens of humans and other mammals, such as S. Typhimurium, S. Paratyphi A, S. Enteritidis, and S. Dublin. Another region of interest within the ST198 genomes consists of 15 ORFs involved inositol catabolism (region 4) (AEX15_17525, AEX15_17625). This region was also identified in several other serovars and has been identified as a myo-Inositol utilization island by Kröger and Fuchs [67]. Chaudhuri et al. [68] demonstrated this region as being associated with proliferation in the host gut. Further, in vivo studies have shown that salmonellae with an inactive reiD (the orphan regulator of the myo-inositol utilization island) demonstrated reduced fitness following oral infections or chickens, calf, and pigs [68]. S. Kentucky ST152 isolates, however, lack this island and, based on their prevalence in this environment, are suitably adapted for persistence in the bovine gut. A third region of interest is a nine ORF region identified within SPI-6 and encoding genes involved in alpha-fimbriae expression, a transcriptional regulator and a mobile genetic element (region 5) (AEX15_23130, AEX15_23170). This region is homologous to the Typhi-colonization factor (tcf) operon (STY0345-STY0348) and should be further investigated as a possible mechanism by which ST198 isolates colonize and infect human and animal hosts.
Table 7. Regions identified in one S. Kentucky ST (ST152 or ST198) and absent or partially present in the other.
| ST | Region | Features | Size A | Flanking Loci in ST198 (CVM N51290 or ATCC 9263) B | Homologous Flanking Loci in ST152 (CVM29188) |
|---|---|---|---|---|---|
| 198 | 1 | histone acetyltransferase HPA2, hypothetical proteins | 4405 | AEX15_02835, AEX15_02865 | SeKA_A3272, SeKA_A3273 |
| 2 | putative superfamily I DNA helicases, putative restriction endonuclease, putative type II restriction enzyme methylase subunit, putative cytoplasmic protein, putative inner membrane protein | 24503 | SEEK9263_16827, SEEK9263_t17079 (tRNA-Leu) | SeKA_A4142 (tRNA-Leu), SeKA_A4161 | |
| 3 | sialic acid metabolism (transport) D | 9036 | AEX15_07510, AEX15_07550 (tRNA-Ser) | SeKA_A0529, SeKA_A0537 (tRNA-Ser) | |
| 4 | inositol catabolism | 23081 | AEX15_17525, AEX15_17625 | SeKA_A4089, SeKA_A4088 | |
| 5 | SPI-6 [Typhi colonization factor (tcf) operon] D | 8399 | AEX15_23130, AEX15_23170 | SeKA_A4630, SeKA_A4631 | |
| 6 | anaerobic dimethyl sulfoxide reductase chain A, B, C E | 13280 | AEX15_10355, AEX15_10415 C | SeKA_A1139, SeKA_A1140 C | |
| 7 | DNA-binding protein, phosphorylase, nucleotidyltransferas, exonuclease D | 9323 | AEX15_12395, AEX15_12455 (tRNA-Asn) | SeKA_A1557, SeKA_A1558 (tRNA-Asn) | |
| 8 | hypothetical protein (secretin domain/putative phage-related secreted protein) | 2459 | AEX15_09660, AEX15_09685 C | SeKA_A0993, SeKA_A1001 C | |
| 9 | carbon Starvation, D-gluconate and ketogluconates metabolism, Lactate utilization | 5674 | AEX15_10030, AEX15_10060 | SeKA_A1070, SeKA_A1073 | |
| 10 | SPI-13 with inserted iron reductase, FMN-dependent NADH-azoreductase, hypothetical proteins | 3314 | AEX15_04620 (tRNA-Phe), AEX15_04645 | SeKA_A2676 (tRNA-Phe), SeKA_A2689 | |
| 11 | hypothetical proteins, reverse transcriptase (adjacent to SPI-9) D | 4247 | AEX15_21645/AEX15_21650, AEX15_21675 C | SeKA_A2255 (tmRNA), SeKA_A2274 C | |
| 12 | glycine biosynthesis, glycine and serine utilization, serine-glyoxylate cycle, serine biosynthesis, glutamine, glutamate, aspartate and asparagine biosynthesis, threonine and homoserine biosynthesis | 3958 | AEX15_19945, AEX15_19970 | SeKA_A3752, SeKA_A3753 | |
| 13 | probable integrase remnant | 1050 | AEX15_01875, AEX15_01895 (tRNA-Ser) | SeKA_A0372, SeKA_A0399 (tRNA-Ser) | |
| 14 | type I restriction-modification system, putative inner membrane protein, hypothetical protein | 10325 | AEX15_20690, AEX15_20730 C | SeKA_A4188, SeKA_A4198 C | |
| 15 | hypothetical protein | 693 | AEX15_14050, AEX15_14060 | SeKA_A3461, SeKA_A3465 | |
| 16 | hypothetical proteins, phage tail protein, oxidoreductase, transglycosylase E | 25556 | AEX15_11375, AEX15_11530 | SeKA_A1348, SeKA_A1377 C | |
| ST | Region | Features | Size A | Flanking Loci in ST152 (CVM29188) | Homologous Flanking Loci in ST198 (CVM N51290 or ATCC 9263) |
| 152 | 1 | transcriptional regulator (LysR family) hypothetical protein, major facilitator superfamily MFS_1, 3-oxoacyl-[acyl-carrier-protein] reductase, transketolase, aldehyde-alcohol dehydrogenase 2 | 8040 | SeKA_A0275, SeKA_A0283 C | AEX15_01480, AEX15_01485 |
| 2 | Entner-Doudoroff pathway, glycolysis and gluconeogenesis, respiratory dehydrogenases 1, inositol catabolism, benzoate degradation D | 23239 | SeKA_A0372, SeKA_A0399 (tRNA-Ser) | AEX15_01875, AEX15_01895 (tRNA-Ser) | |
| 3 | hypothetical proteins D | 3951 | SeKA_A0529, SeKA_A0537 (tRNA-Ser) | AEX15_07510, AEX15_07550 (tRNA-Ser) | |
| 4 | hypothetical proteins, protein TolA | 4683 | SeKA_A0993, SeKA_A1001 C | AEX15_09685, AEX15_09660 C | |
| 5 | phage ORFs D | 33283 | SeKA_A1348, SeKA_A1377 C, F, G | AEX15_11530, AEX15_11375 C | |
| 6 | hypothetical proteins, protein YibA, protein RhsD, IS1-family insertion element D | 4847 | SeKA_A1701, SeKA_A1712 | AEX15_13120, AEX15_13125 | |
| 7 | hypothetical proteins, putative membrane protein, putative cytoplasmic proteins, transposases, HTH domain protein, integrase D | 11111 | SeKA_A2096, SeKA_A2114 C | AEX15_15380, AEX15_15440 C | |
| 8 | prophage integrase, hypothetical proteins, nuclease-related domain family protein, protein YpjI, DNA repair protein, toxin-antitoxin system (adjacent to SPI-9) D | 12402 | SeKA_A2255 (tmRNA), SeKA_A2274 C | AEX15_21645/AEX15_21650, AEX15_21675 C | |
| 9 | glycolysis and gluconeogenesis, mannitol Utilization | 5553 | SeKA_A2624, SeKA_A2631 | AEX15_04405, AEX15_04410 | |
| 10 | hypothetical protein, colicin-E7 immunity protein, bacteriophage integrase (fragments overlap with SPIs-8 and 13) D | 4943 | SeKA_A2676 (tRNA-Phe), SeKA_A2689 | AEX15_04620 (tRNA-Phe), AEX15_04645 | |
| 11 | hypothetical proteins, putative autotransporter | 5146 | SeKA_A3310, SeKA_A3314 | AEX15_02655, AEX15_02660 | |
| 12 | hypothetical proteins | 5073 | SeKA_A3672, SeKA_A3678 | AEX15_19610, AEX15_19615 | |
| 13 | DNA phosphorothioation, hypothetical proteins, integrase D | 17034 | SeKA_A4142 (tRNA-Leu), SeKA_A4161 C | SEEK9263_t17079 (tRNA-Leu), SEEK9263_16827 C | |
| 14 | type I restriction-modification system, putative inner membrane protein, hypothetical protein | 10240 | SeKA_A4188, SeKA_A4198 | AEX15_20690, AEX15_20730 | |
| 15 | oxidoreductase, inner membrane metabolite transport protein, sorbitol dehydrogenase, fructose-bisphosphate aldolase, kinase. | 9227 | SeKA_A0714, SeKA_A0725 C | AEX15_08380, AEX15_08375 C | |
| 16 | putative membrane transport protein, mandelate racemase/muconate lactonizing enzyme, putative transcriptional regulator | 3481 | SeKA_A3461, SeKA_A3465 | AEX15_14060, AEX15_14050 | |
| 17 | peroxisomal (S)-2-hydroxy-acid oxidase, putative glycolate oxidase | 1202 | SeKA_A1070, SeKA_A1073 | AEX15_10030, AEX15_10060 |
A = based on start/stop positions of proten encoding genes within region
B = flanking loci
C = encodes a string of homologous sequences but with unique flanking regions/may be unidirectional hits.
D = putative genomic island by as determined by IslandViewer3 [41].
E = part of this region predicted to be a genomic island by IslandViewer3 [41].
F = not detected in ARS-CC7487
G = truncated in CVM N45934
Seventeen contiguous regions of ST152 genomes were determined to be absent in ST198 genomes by a similar reciprocal BLASTP analysis. Eight of these regions were identified as putative genomic islands. Several of these regions were annotated as arrays of hypothetical proteins with no known function. Other islands encoded ORFs annotated as being involved in metabolic functions such as region 2 (ORFs involved in glycolysis and gluconeogenesis, inositol catabolism, benzoate degradation) (SeKA_A0372, SeKA_A0399), region 9 (ORFs involved in glycolysis and gluconeogenesis, mannitol utilization) (SeKA_A2624, SeKA_A2631), and region 15 (sorbitol dehydrogenase, fructose-bisphosphate aldolase) (SeKA_A0714, SeKA_A0725). The presence of these operons in strains frequently isolated from dairy cows and poultry suggests their roles in these environments should be further investigated.
In both STs ten homologous insertion loci flanking different protein encoding regions were identified, indicating that they may be hotspots for integration of laterally transferred DNA in S. enterica. Five insertion loci encoded tRNA sequences and one encoded a tmRNA sequence, which are known to be regions of genomic island insertion [69]. For the most part the ST-specific islands were conserved among all members of each ST indicating that they may play a significant role in the ecology of these strains and/or they are anchored in the genomes of either host ST.
Conclusions
Based on the analysis conducted herein, there is a phylogenetic difference between poultry-associated and bovine-associated North American S. Kentucky ST152 isolates which is discernible due to four core-genome SNPs. However, several clusters of bovine-associated S. Kentucky ST152 isolates are more closely related to some poultry-associated isolates than they are to other bovine-associated isolates. This divergence is also associated, in silico, with the presence of several plasmids, one of which (ColV) has been demonstrated to enhance the colonization capacity of these strains in the chicken gastrointestinal tract, the other being a high frequency IncX1 plasmid with no known biological role. The influence of the core-genome SNPs and this IncX1 plasmid on the ecology of S. Kentucky ST152 in the bovine and poultry hosts, or survival in food sources or the environment should be further evaluated in vivo.
A significant difference in gene content and core-genome nucleotide sequence divergence between S. Kentucky ST152 and ST198 isolates was also observed. Based on the methods used in this study, both sequence types are phylogenetically more closely related to other serovars than they are to each other and their shared nomenclature stems from the high level of similarity between their antigenic coding regions which may have transferred laterally between sequence types. Several genomic elements in ST198, such as a sialic acid transport region, inositol catabolism and a homolog of the Typhi colonization factor (tcf), and differences in amino acid sequence of virulence-associated proteins may result in different interactions in the human, bovine, and poultry gastrointestinal tracts, an area that requires further research.
Although the predominant S. Kentucky strains recovered from both dairy cows and poultry in the United States do not appear to cause considerable disease in the animal hosts, the apparent high prevalence in these food animals represents a food safety risk to consumers of beef, dairy, and poultry products. Although infrequent, human clinical salmonellosis caused by S. Kentucky has been reported in the United States, but there is not much available data on the sequence types of human-clinical S. Kentucky in this country. The recent establishment of CIPR S. Kentucky ST198 in poultry in France and Poland and other regions along with the detection of this ST in dairy cows, raw milk, ground beef, and ground turkey suggests the presence of this potential emerging pathogen should be monitored in food-producing animals.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We gratefully acknowledge Jakeitha Sonnier for laboratory assistance. This project was supported by internal USDA/ARS research funding. The mention of a trade name, proprietary product, or specific equipment does not constitute guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that might be suitable.
Data Availability
Accession numbers for genomic data in NCBI are available in the relevant Supporting Information file.
Funding Statement
This project was supported by internal USDA/ARS research funding.
References
- 1.Centers for Disease Control and Prevention (2011) Salmonella annual summary tables 2009 Atlanta, GA: US Department of Health and Human Services. [Google Scholar]
- 2.Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, Ricke SC (2011) Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Env. Micro 77: 4273–4279 10.1128/AEM.00598-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Kessel JS, Karns JS, Wolfgang DR, Hovingh E, Schukken YH (2012) Dynamics of Salmonella serotype shifts in an endemically infected dairy herd. Foodborne Pathog Dis. 9: 319–324. 10.1089/fpd.2011.1054 [DOI] [PubMed] [Google Scholar]
- 4.Parveen S, Taabodi M, Schwarz JG, Oscar TP, Harter-Dennis J, White DG. (2007) Prevalence and antimicrobial resistance of Salmonella recovered from processed poultry.J Food Prot. 70(11):2466–72. [DOI] [PubMed] [Google Scholar]
- 5.Van Kessel JS, Karns JS, Wolfgang DR, Hovingh E. (2013) Regional distribution of two dairy-associated Salmonella enterica serotypes. Foodborne Pathog Dis. 10(5):448–52 10.1089/fpd.2012.1380 [DOI] [PubMed] [Google Scholar]
- 6.University of Warwick MLST Database http://mlst.warwick.ac.uk/mlst/
- 7.Le Hello S, Hendriksen RS, Doublet B, Fisher I, Nielsen EM, Whichard JM, et al. (2011) International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J Infec Dis. 204: 675–684. 10.1093/infdis/jir409 [DOI] [PubMed] [Google Scholar]
- 8.Le Hello S, Bekhit A, Granier S, Barua H, Beutlich J, Zając M, et al. (2013) The global establishment of a highly-fluoroquinolone resistant Salmonella enterica serotype Kentucky ST198 strain. Frontiers in Microbiology 4 (2013). 10.3389/fmicb.2013.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Hello S, Harrois D, Bouchrif B, Sontag L, Elhani D, Guibert V, et al. (2013). Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: a microbiological study. Lancet Infect Dis. 13(8):672–9. 10.1016/S1473-3099(13)70124-5 [DOI] [PubMed] [Google Scholar]
- 10.Li X, Payne JB, Santos FB, Levine JF, Anderson KE, Sheldon BW. (2007) Salmonella populations and prevalence in layer feces from commercial high-rise houses and characterization of the Salmonella isolates by serotyping, antibiotic resistance analysis, and pulsed field gel electrophoresis. Poult Sci. 86(3):591–7. 10.1093/ps/86.3.591 [DOI] [PubMed] [Google Scholar]
- 11.Santos FB, Dsouza DH, Jaykus L, Ferket PR, Sheldon BW. (2007) Genotypes, serotypes, and antibiotic resistance profiles of Salmonella isolated from commercial North Carolina turkey farms. J Food Prot. 70(6):1328–33. [DOI] [PubMed] [Google Scholar]
- 12.Fricke WF, McDermott PF, Mammel MK, Zhao S, Johnson TJ, Rasko DA, et al. (2009) Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl Environ Microbiol. 75(18):5963–71. 10.1128/AEM.00786-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladely SR, Meinersmann RJ, Ball TA, Fedorka-Cray PJ. (2016) Antimicrobial Susceptibility and Plasmid Replicon Typing of Salmonella enterica Serovar Kentucky Isolates Recovered from Broilers. Foodborne Pathog Dis. 13(6):309–15. 10.1089/fpd.2015.2102 [DOI] [PubMed] [Google Scholar]
- 14.Edrington TS, Hume ME, Looper ML, Schultz CL, Fitzgerald AC, Callaway TR, et al. (2004) Variation in the faecal shedding of Salmonella and E. coli O157: H7 in lactating dairy cattle and examination of Salmonella genotypes using pulsed-field gel electrophoresis. Letters Appl. Micro 38: 366–372. 10.1111/j.1472-765X.2004.01495.x [DOI] [PubMed] [Google Scholar]
- 15.Van Kessel JS, Karns JS, Lombard JE, Kopral C (2011) Prevalence of Salmonella enterica, Listeria monocytogenes, and Escherichia coli virulence factors in bulk tank milk and in-line filters from U.S. dairies. J. Food Prot. 74: 759–68. 10.4315/0362-028X.JFP-10-423 [DOI] [PubMed] [Google Scholar]
- 16.Timme RE, Pettengill JB, Allard MW, Strain E, Barrangou R, et al. (2013) Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol 5: 2109–2123. 10.1093/gbe/evt159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng X, Desai PT, den Bakker HC, Mikoleit M, Tolar B, Trees E, et al. (2014). Genomic epidemiology of Salmonella enterica serotype Enteritidis based on population structure of prevalent lineages. Emerging Infectious Diseases. 20(9): 1481 10.3201/eid2009.131095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chun J., Grim CJ, Hasan NA, Lee JH, Choi SY, Haley B, Taviani E. et al. (2009) Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proceedings of the National Academy of Sciences 106(36): 15442–15447. 10.1073/pnas.0907787106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haley BJ, Allard M, Brown E, Hovingh E, Karns JS, Van Kessel JS. (2015) Molecular detection of the index case of a subclinical Salmonella Kentucky epidemic on a dairy farm. Epidemiology and Infection,143:682–686. 10.1017/S0950268814001289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera-Leon S, Ramiro R, Arroyo M, Diez R, Usera MA, Echeita MA (2007) Blind comparison of traditional serotyping with three multiplex PCRs for the identification of Salmonella serotypes. Res Microbiol. 158:122–127. 10.1016/j.resmic.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 21.Karns JS, Haley BJ, Van Kessel JA. (2015) Improvements to a PCR-Based Serogrouping Scheme for Salmonella enterica from Dairy Farm Samples. J Food Prot. 78(6):1182–5. 10.4315/0362-028X.JFP-14-475 [DOI] [PubMed] [Google Scholar]
- 22.Schmieder R, and Edwards R. (2011) Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 6:e17288 10.1371/journal.pone.0017288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. (2012) J Comput Biol. 19(5):455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albert MJ, Obaid K, Alfouzan W, Sheikh AR, Udo E, Izumiya H, Bulach DM, Seemann T.et al. (2014). Isolation of Salmonella enterica Serovar Kentucky Strain ST 198 and Its H2S-Negative Variant from a Patient: Implications for Diagnosis. Journal of Clinical Microbiology. 52(11): 4090–4093. 10.1128/JCM.01775-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.https://github.com/lskatz/lyve-SET
- 27.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL et al. (2013) Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio 4(4):e00398–13. 10.1128/mBio.00398-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.https://github.com/CFSAN-Biostatistics/snp-pipeline
- 29.https://peerj.com/articles/cs-20/
- 30.https://github.com/marbl/harvest
- 31.Treangen TJ, Ondov BD, Koren S, Phillippy AM. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15(524): 2 10.1186/PREACCEPT-2573980311437212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DMet al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 6(2):80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K., Stecher G., Peterson D., Filipski A., & Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular biology and evolution, 30(12), 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatakis A (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cummings MP, Neel MC, Shaw KL. (2008) A genealogical approach to quantifying lineage divergence. Evolution. 62(9):2411–22. 10.1111/j.1558-5646.2008.00442.x [DOI] [PubMed] [Google Scholar]
- 36.http://molevol.lysine.umiacs.umd.edu/software/phylogenetics/gsi
- 37.https://cge.cbs.dtu.dk/services/MLST/
- 38.Pritchard JK, Stephens M, Donnelly P. (2000) Inference of population structure using multilocus genotype data. Genetics. 155(2):945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenberg (2004) DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 4, 137–138. [Google Scholar]
- 40.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Aarestrup F, Hasman Het al. (2014) In in silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial agents and chemotherapy 5(7): 3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, Pereira SK, Waglechner N, McArthur AG, Langille MG, Brinkman FS et al. (2015) IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 43(W1):W104–8. 10.1093/nar/gkv401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wells SJ, Fedorka-Cray PJ, Dargatz DA, Ferris K, Green A. (2001) Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J Food Prot. 64(1):3–11. [DOI] [PubMed] [Google Scholar]
- 43.Blau DM, McCluskey BJ, Ladely SR, Dargatz DA, Fedorka-Cray PJ, Ferris KE, Headrick ML.et al. (2005) Salmonella in dairy operations in the United States: prevalence and antimicrobial drug susceptibility. J Food Prot. 68(4):696–702. [DOI] [PubMed] [Google Scholar]
- 44.Le Hello S, Weill FX, Guibert V, Praud K, Cloeckaert A, Doublet B. (2012) Early strains of multidrug-resistant Salmonella enterica serovar Kentucky sequence type 198 from Southeast Asia harbor Salmonella genomic island 1-J variants with a novel insertion sequence. Antimicrob Agents Chemother. 56(10):5096–102. 10.1128/AAC.00732-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baucheron S, Le Hello S, Doublet B, Giraud E, Weill FX, Cloeckaert A. (2013) ramR mutations affecting fluoroquinolone susceptibility in epidemic multidrug-resistant Salmonella enterica serovar Kentucky ST198. Front Microbiol. 4:213 10.3389/fmicb.2013.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uebeck A, Dougan J, Harrison LH, Brisse S, S. enterica MLST Study Group et al. (2012). Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathogens. 8, e1002776 10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniels MJ, Hutchings MR, Greig A. (2003) The risk of disease transmission to livestock posed by contamination of farm stored feed by wildlife excreta. Epidemiol Infect. 130(3):561–8. [PMC free article] [PubMed] [Google Scholar]
- 48.Callaway TR, Edrington TS, Nisbet DJ. (2014). Isolation of Escherichia coli O157:H7 and Salmonella from migratory brown-headed cowbirds (Molothrus ater), common grackles (Quiscalus quiscula), and cattle egrets (Bubulcus ibis). Foodborne Pathogens and Disease. 11:791–794. 10.1089/fpd.2014.1800 [DOI] [PubMed] [Google Scholar]
- 49.Carlson JC, Hyatt DR, Bentler K, Mangan AM, Russell M, Piaggio AJ, Linz GM et al. (2015) Molecular characterization of Salmonella enterica isolates associated with starling-livestock interactions. Vet Microbiol. 179(1–2):109–18. 10.1016/j.vetmic.2015.03.015 [DOI] [PubMed] [Google Scholar]
- 50.Chen YC, Chang MC, Chuang YC, Jeang CL. (2004). Characterization and virulence of hemolysin III from Vibrio vulnificus. Curr Microbiol. 49(3):175–9. 10.1007/s00284-004-4288-5 [DOI] [PubMed] [Google Scholar]
- 51.Baida GE, Kuzmin NP. (1996) Mechanism of action of hemolysin III from Bacillus cereus. Biochim Biophys Acta. 1284(2):122–4. 10.1016/s0005-2736(96)00168-x [DOI] [PubMed] [Google Scholar]
- 52.Johnson TJ, Thorsness JL, Anderson C, Lynne A, Foley S, Han J, Fricke WF, et al. (2010) Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS One 5(12): e15524 10.1371/journal.pone.0015524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levings RS, Lightfoot D, Partridge SR, Hall RM, Djordjevic SP. (2005) The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J Bacteriol. 187(13):4401–9. 10.1128/JB.187.13.4401-4409.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyd DA, Shi X, Hu QH, Ng LK, Doublet B, Cloeckaert A, Mulvey MR et al. (2008) Salmonella genomic island 1 (SGI1), variant SGI1-I, and new variant SGI1-O in Proteus mirabilis clinical and food isolates from China. Antimicrob Agents Chemother. 52(1):340–4. 10.1128/AAC.00902-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamidian M, Holt KE, Hall RM. (2015) The complete sequence of Salmonella genomic island SGI1-K. J Antimicrob Chemother. 70(1):305–6. 10.1093/jac/dku331 [DOI] [PubMed] [Google Scholar]
- 56.Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, Barrow PA, Maskell DJ, Wallis TS et al. (2004) Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol. 54(4):994–1010. 10.1111/j.1365-2958.2004.04323.x [DOI] [PubMed] [Google Scholar]
- 57.Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penadés JR, Lasa I et al. (2005) BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol.58(5):1322–39. 10.1111/j.1365-2958.2005.04907.x [DOI] [PubMed] [Google Scholar]
- 58.Gal-Mor O, Gibson DL, Baluta D, Vallance BA, Finlay BB. (2008) A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis. PLoS Pathog. 4(4):e1000036 10.1371/journal.ppat.1000036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kingsley RA, van Amsterdam K, Kramer N, Bäumler AJ. (2000) The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect Immun. 68(5):2720–7. 10.1128/iai.68.5.2720-2727.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kingsley RA, Santos RL, Keestra AM, Adams LG, Bäumler AJ. (2002) Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol Microbiol. 43(4):895–905. 10.1046/j.1365-2958.2002.02805.x [DOI] [PubMed] [Google Scholar]
- 61.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW et al. (1998) Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 30(1):163–74. 10.1046/j.1365-2958.1998.01047.x [DOI] [PubMed] [Google Scholar]
- 62.Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Bäumler AJ, Adams LG et al. (2002) The Salmonella enterica serotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect Immun. 70(7):3843–55. 10.1128/iai.70.7.3843-3855.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V (2009) Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12: 117–124 10.1016/j.mib.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Severi E, Hood DW, Thomas GH. (2007) Sialic acid utilization by bacterial pathogens. Microbiology. 153(Pt 9):2817–22. 10.1099/mic.0.2007/009480-0 [DOI] [PubMed] [Google Scholar]
- 65.Almagro-Moreno S, and Boyd EF. (2009). Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infection and immunity, 77(9), 3807–3816. 10.1128/IAI.00279-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Almagro-Moreno S, and Boyd EF. (2010). Bacterial catabolism of nonulosonic (sialic) acid and fitness in the gut. Gut Microbes, 1(1), 45–50. 10.4161/gmic.1.1.10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kröger C, and Fuchs TM. (2009). Characterization of the myo-inositol utilization island of Salmonella enterica serovar Typhimurium. Journal of bacteriologyBacteriology, 191(2), 545–554. 10.1128/JB.01253-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, et al. (2013) Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS 9(4): e1003456 10.1371/journal.pgen.1003456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobrindt U, Hochhut B, Hentschel U, Hacker J. (2004) Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2(5):414–24. 10.1038/nrmicro884 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
Accession numbers for genomic data in NCBI are available in the relevant Supporting Information file.