Abstract
Poor inhibitory processing of negative emotional content is central to many psychiatric disorders, including depression and anxiety. Moreover, increasing evidence suggests that core aspects of emotion-inhibitory processing are largely inherited and as such may represent a key intermediate or risk-related phenotype for common affective diseases (e.g., unipolar depressive, anxiety disorders). The current study employed a candidate-gene approach in order to most effectively examine this complex behavioral phenotype. We examined the novel interaction between BDNF (Val66Met) and TPH2 (rs4570625) polymorphisms and their influence on behavioral inhibition of negative emotion in two independent investigations of healthy adults. BDNF Met carriers consistently report greater symptoms of affective disease and display corresponding behavioral rigidity, while TPH2 T carriers display poor inhibitory processing. These genotypes are traditionally perceived as ‘risk’ genotypes when compared to their respective major Val and G homozygous genotypes, but evidence is mixed. Recent studies in humans and mutant mouse models suggest biological epistasis between BDNF and genes involved in serotonin regulation. Moreover, polymorphisms in the TPH2 gene may have greater influence on serotonergic function than other more commonly studied polymorphisms (e.g., 5-HTTLPR). We observed consistent evidence across two different emotion-inhibition paradigms, one with high internal validity (Study 1, n = 119) and one with high ecological validity (Study 2, n = 115) that the combination of Val/Val and G/G genotypes was clearly associated with impaired inhibition of negative emotional content. This was followed by individuals carrying the BDNF—Met allele (including Met/Val and Met/Met) when combined with the TPH2—T allele (including T/G and T/T combinations). The consistency of these results across tasks and studies suggests that these two groups may be particularly vulnerable to the most common psychiatric disorders and should be targets for future clinical investigation.
Introduction
Difficulty inhibiting negative emotional content has been consistently linked to specific and highly-impairing emotion-related disorders such as depression [1,2]. Further, impairments in inhibiting negative emotions are broadly associated with symptoms of common affective diseases (such as unipolar depressive, anxiety, and stress disorders) [3], including: patterns of negative self-referential thought [4–6]; enduring negative mood; inattention; impulsivity; agitation and arousal, as well as disruptions in sleep and eating [7–10]. Accordingly, poor inhibition of negative emotional content is now considered a key trans-diagnostic risk factor for affective disease. Here, we sought to evaluate a novel interaction between the BDNF Val66Met polymorphism and a less commonly studied serotonergic system polymorphism in the tryptophan hydroxylase 2 gene (TPH2) on negative emotion inhibition in two independent samples of healthy adults. Persistent risk-related differences in cognitive and emotional processes over a wide range of contexts may be influenced by genetic polymorphism interactions, such as those between BDNF and serotonergic systems [11–13]. Indeed, despite recent reduced enthusiasm for candidate-gene research, there is a clear need for detailed examination of complex behavioral phenotypes, work that is by necessity limited to candidate gene studies given the related methodological demands. Genome-wide association studies (GWAS) focused on affective disorders such as major depressive disorder are often limited to distal measurements given the immense sample sizes (tens of thousands) required [14,15]. In fact, despite considerable evidence implicating Val66Met in candidate gene research [16,17], multiple GWAS studies have thus far not implicated any BDNF or TPH2 polymorphisms in association with affective disorders [16–21]. Accordingly, here we focused on careful assessment of a novel yet potentially informative interaction between two polymorphisms in these systems in the prediction of a key trans-diagnostic risk factor: poor inhibition of negative emotional content.
TPH2 is the neuron-specific enzyme isoform catalyzing the rate-limiting step in serotonin synthesis [22–25]. Thus, polymorphisms in the TPH2 gene may have the potential for greater influence on serotonergic function than other identified and highly studied polymorphisms within this system (e.g., 5-HTTLPR). One common SNP within TPH2 is a T substitution for G in the 5′regulatory region of the gene (rs4570625; also noted as G-703T). The T allele of rs4570625 is hypothesized to be associated with reduced serotonin levels [26,27], and has been linked to deficits in executive attention and may be more common in human samples with ADHD and obsessive-compulsive disorder; two disorders characterized by poor inhibitory processes [28–30]. TPH2 variation has been linked to common affective disorders such as major depression in a recent meta-analysis [31] and is predictive of amygdala responses to emotional content [28,32–34]. Indeed, evidence points to the relevance of this polymorphism in affective processing as well as higher-order cognitive processes associated with inhibition [35]. Though some research has examined interactions between the TPH2 rs4570625 polymorphism and the 5-HTTLPR polymorphism [29,36–39], none have examined an interaction between TPH2 rs4570625 and BDNF Val66Met on cognitive function and never in relation to emotional content.
The broad influence of BDNF on neural plasticity and development has consistently suggested its key role in emotion-related learning and response patterns. The most widely studied BDNF polymorphism, the Val66Met SNP (rs6265) leads to a non-synonymous methionine (Met) substitution for valine (Val) at codon 66 in the prodomain of BDNF. This substitution results in reduced BDNF release under stimulated conditions [40]. The BDNF Val66Met Met allele has been associated with impaired fear extinction [41,42] and increased sensitivity to environment [43]. Further, the BDNF Val66Met polymorphism influences serotonergic system functioning in humans [44,45]. Animal studies further support reciprocal modulation occurring between serotonergic and BDNF systems [46–50].
Given these previous data suggesting individual contributions of the BDNF Val66Met Met allele or TPH2 G-703T T-allele to emotion regulation, we hypothesized these polymorphisms’ interactive influence would be compounded to result in more significant impairment of emotion-inhibitory processing. In particular, recent studies in humans and mutant mouse models suggest biological epistasis between BDNF and another gene involved in serotonin regulation (serotonin transporter, SLC6A4) [11,12,46,47,51] that may alter neural circuitry and thereby influence risk for psychiatric disease. For instance, reductions in brain BDNF levels through genetic knockout exacerbate serotonin level deficiencies already observed in serotonin transporter (5-HTT) knockout mice. Reduced BDNF also increases avoidance behavior and neuroendocrine stress responses in these double mutant mice [46,47,51]. Further, TPH2 knockout mice, but not 5-HTT knockouts, exhibit increases in hippocampal BDNF [50], together suggesting a reciprocal feedback system by which BDNF promotes serotonergic activity, while serotonin reduces BDNF activity. These effects are supported by earlier literature demonstrating BDNF-serotonergic interactions in vitro [52–56]. Most notable is work by Weinberger and colleagues, [11] in which the Met allele of the BDNF Val66Met polymorphism mitigated the 5-HTTLPR S allele-induced reductions in prefrontal cortex gray matter volume and amygdala connectivity. Although counterintuitive, these data suggest that levels of BDNF may protect against the potentially adverse neurodevelopmental effects of increased serotonergic signaling. Indeed, much of the literature recognizes that optimal serotonin function is essential for proper neurodevelopment, and sub- or supra-optimal functioning can shift cortical and limbic circuitry development resulting in lifelong emotional consequences [57–62].
Current study
In this investigation, we used a candidate gene approach in combination with behavioral measures to examine the interaction between BDNF Val66Met and TPH2 G-703T polymorphisms on inhibition of negative emotion. Although candidate gene approaches have been recently criticized because of non-replication issues and an inability to account for polygenic influences on phenotypes, this approach is appropriately suited for the highly detailed behavioral data collected in the present study. Moreover, by testing the predicted relationships across two distinct samples and lab paradigms, we begin to address issues related to reliability and replication.
We focused exclusively on testing the association between patterns of emotion-inhibitory processing and allelic combinations in healthy adults. Poor emotion-inhibitory processing is well established in clinical samples with common affective disorders. Individual differences in emotion-inhibition can predict responsivity to treatment or even identify individuals whose symptoms are particularly intractable [63–65]. However, linking specific allelic combinations to similar risk-related patterns in healthy adults might be a first step to understanding how genetic variation contributes to the onset of disease. Indeed, most models of emotion-related psychiatric disease involve complex interactions between inherited and learned/environmental factors. Characteristics influencing vulnerability to common affective disorders vary in heritability, from 20–30% for emotion regulation [66,67], 56% for temperament [68], and up to 99% for executive function [69]. Accordingly, though genes substantially contribute to multiple core aspects of emotion-inhibitory processing, identification of clear and unique gene-disorder relationships is unlikely [70]. In contrast, linking genetic variation to underlying and more proximal intermediate phenotypes shows considerable promise [71].
We conducted two studies involving distinct samples of adult participants to determine the combined contributions of these polymorphisms to variability in emotion-inhibitory processing and psychiatric risk. In Study 1 we examined genetic influences on performance during an Emotion-word Stroop task, a well-validated emotion-inhibition paradigm. In Study 2 we evaluated genetic influences on spontaneous and more naturalistic inhibition of negative emotion by indexing real-time down-regulation of negative emotion during emotionally-evocative films [72,73]. Naturalistic emotion-response paradigms are recognized for their ability to capture salient patterns of emotion that more closely reflect real-world responding [74] and are more highly predictive of salient clinical phenomena [65,75,76].
Methods and Materials
Ethics statement
The study was approved by the Kent State institutional review board governing human subjects research and has been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki (World Medical Association, 2013). The study also complies with the APA ethical standards for treatment of human subjects. All participants provided written informed consent before participating in the study.
Subjects
Study 1 and Study 2 were comprised of two independent samples. Study 1: Participants were 119 undergraduate students (62% female; 77% Caucasian, 11% African American, 3% Asian; 95% Non-Hispanic). Mean age was M = 20.14, SD = 4.15. Mean rated depression on the Center of Epidemiological Studies Depression Scale (CES-D) [77] was typical for community samples M = 11.74, SD = 8.37 (above 16 denotes clinically significant depression; n = 31 scored above 16; range was 0–50). Study 2: Participants were 115 undergraduate students (64% female; 79% Caucasian, 12% African American, 3% Asian, 6% Other; 91% Non-Hispanic). Mean age was M = 20.80, SD = 6.48. Mean rated depression on the CES-D was M = 11.54, SD = 7.43 (scores above 16 reflect clinically significant depression symptoms; n = 23 scored above 16; range was 1–39).
Genotyping and RFLP
2 mL of saliva was collected from each participant and was stored at -20°C [78]. DNA extraction was accomplished using prepIT-L2P and following the manufacturer’s recommendations (DNA Genotek inc., Ottawa, Canada). DNA was then purified using Genomic DNA Clean & Concentrator kit (Zymo Research, Irvine, CA) and quantified using SYBR Green I dye (Lonza, Walkersville, MD). Genomic DNA was diluted to 5 ng/μL prior to polymerase chain reaction (PCR). In a final volume of 20 μl, 1 μl DNA was amplified, and each reaction consisted of 0.5 μM forward primer, 0.5 μM reverse primer, 0.5 μl Taq polymerase solution, 5 mM MgCl2, 2 mM dNTPs, and 10% glycerol. Both genes were run on an Eppendorf PCR Mastercycler pro (model no. 6321, Hamburg,Germany) using a touchdown PCR cycle protocol adopted from Anchordoquy et al., [79] and modified to have a 65°C annealing temperature for 10 cycles, followed by a 55°C annealing temperature for 35 cycles.
rs4570625 (G-703T)
Primers used to amplify rs4570625 within the TPH2 gene were adopted from Mössner et al., [80] (Forward: 5'-TTT TAT GAA AGC CAT TAC ACA T; Reverse: 5'-TTC CAC TCT TCC AGT TAT TTT A). Following amplification, a restriction digest was performed to discern the presence or absence of the T to G substitution. 10 μl of the PCR product was used in the total reaction with 1X Cutsmart buffer and 4 U/reaction PsiI enzyme (New England BioLabs, Ipswich, MA) in a total volume of 20 μl and was incubated for 3 hours at 37°C. Restriction fragments were visualized using a 2% agarose gel. In the presence of the T substitution, the product yields 149 and 55 bp lengths. In the absence of the substitution (i.e., G allele), the product was undigested, and a 204 bp fragment was visualized. Ten percent of sample genotypes were separately reconfirmed with 100% concordance.
BDNF Val66Met
Primers used to amplify Val66Met polymorphism in the BDNF gene were adopted from Hünnerkopf et al. [81] (Forward: 5’-AAA GAA GCA AAC ATC CGA GGA CAA G; Reverse: 5’- ATT CCT CCA GCA GAA AGA GAA GAG G). Following amplification, a digest similar to above was performed, using NlaIII enzyme (New England BioLabs, Ipswich, MA) incubated at 37°C for 1 hour. Restriction fragments were visualized on a 3% agarose gel. In the presence of methionine substitution (A allele) yielded product lengths of 140, 77, and 57 bp. In the absence of the substitution, (G allele) the PCR product was undigested and a 274 bp product was visualized. Ten percent of sample genotypes were separately reconfirmed with 100% concordance.
Procedures
In each study, after providing written informed consent, participants provided demographic information, reported current depressive symptoms using the CES-D [82], and provided saliva samples (Oragene-DISCOVER OGR-500 kits). They then proceeded to complete the emotion-inhibitory processing tasks described below.
Emotion-inhibitory processing assessment.
Study 1, Emotion-word Stroop
Participants completed an Emotion-word Stroop [83] task via computer (ePrime, 2.0;Psychology Software Tools, Inc., Pittsburgh, PA). The task consisted of 180 trials of negative, positive, or neutral words (60 of each type) and participants were instructed to label the text color (red, green, yellow, or blue) by button press as quickly as possible and to ignore word meaning. Emotion-word Stroops are established indicators of emotion-inhibitory processing as individuals must inhibit the emotion-word content in order to attend to the text color [84,85]. Participants were seated comfortably in front of a computer monitor by themselves and were instructed to “work as quickly as possible while avoiding mistakes”. Participants completed practice trials and then began the task. Words were presented as one set, in random order, and randomized by valence. Before each trial, participants saw a white fixation point on a black screen for 500 ms, followed by the stimulus (affective or neutral word). Stimuli remained on the screen until the participant indicated color of the word. As soon as the participant responded to the stimuli, the fixation point re-appeared on the screen.
Data were cleaned following standard conventions [86]. Overall error rates were low and there were no differences in errors by word type (neutral % error M = 4.38, SD = 0.05; negative % error M = 4.31, SD = 0.05; positive % error M = 4.40, SD = 0.05). One individual was dropped from the analysis because of reaction times greater than 2 SDs from the mean (final N = 119). Only reaction times from correct responses were used in the analyses.
Study 2, Naturalistic emotion-inhibition
Participants were asked to “emotionally engage” with four previously validated highly emotionally evocative films [72,73], each approximately five minutes in duration, with a brief break between. As is customary when assessing emotion during naturalistic tasks, assessments were repetitive and multi-dimensional (emotion responses were based on both self-report and objective coding of facial emotion) so as to increase validity [65,72]. Films were presented in a sequence to maximize the intensity of elicited responses and to place greater demands on participants to naturally inhibit their negative emotion responses from a negative film to a positive film. Intense negative emotions were elicited in the first film clip (anger and disgust: Road to Guantanamo, Revolution Films, 2006) and third film clip (sadness: The Champ, Metro-Goldwyn Mayer, 1979). Each negative film was followed by an explicitly positive film, as such the second film clip (happiness: Alive, Paramount Pictures, 1993) and fourth film clip (amusement: Between two Ferns, www.comedyordie.com, 2010) depicted highly positive scenes. Response to the two positive films were aggregated. Given that the film sequence consistently shifted from negative to explicitly positive, inhibition or down-regulation of negative emotion should occur in response to each positive film.
To index self-reported emotion, participants used a 1–7 Likert scale to rate their negative (fear, sadness, disgust, guilt, distress, anger) and positive (happiness, enjoyment, amusement, affection, relief) emotion immediately following each film. To objectively index emotional facial behavior, participant expressions were coded by five research assistants naïve to study details. Coders rated global indicators of negative and positive emotional expressions to each film [87]. Coders were sufficiently reliable (average ICC = .80, range .74-.90) and ratings were averaged across coders by participant to increase reliability. Reported and coded emotion scores were standardized to z-scores and combined yielding one score for negative emotion during the positive films. Participants also had scores for positive emotion during both films.
Data analytic strategy
In both studies, the analyses specifically targeted the inhibition of emotional content in order to examine group differences based on genetic variation and interaction. In all analyses we controlled for reported depression because of considerable prior evidence demonstrating a strong link between depression and emotion-inhibitory processing [88] and our interest was in isolating genotypic influences on these processes. In each of Study 1 and Study 2, we performed omnibus ANCOVA tests. In Study 1, we isolated reaction times for emotion words (negative or positive) by controlling reaction times to neutral words as in previous research in the Emotion-word Stroop task [89,90]. In Study 2, we isolated negative emotion responses that lingered during explicitly positive films that followed negative films in a naturalistic emotion task. In this case, by controlling for negative emotions that were generated in response to the preceding negative films, we were able to isolate the degree to which participants could naturalistically inhibit or down-regulate negative emotion. Analyses of these polymorphisms followed literature convention, [91–95] by comparing Met carriers (Val/Met and Met/Met genotypes) with Val/Val homozygotes for BDNF Val66Met, and by comparing T carriers (T/G and T/T genotypes) with G/G homozygotes for TPH2 rs4570625. Consequently, this resulted in 4 possible genotype combinations for combinatorial analyses: 1) Val/Val—G/G; 2) Val/Val—T carrier; 3) Met carrier—G/G; 4) Met carrier—T carrier. Significance was set a priori at p<0.05. All data are graphed as mean ± S.E.M.
Results
All data underlying the findings described in the current manuscript are fully available to the scientific community. Full access to data can obtain by contacting the corresponding author through email.
Frequency of rs4570625 and BDNF Val66Met genotypes
As shown in Table 1the frequency of the rs4570625 alleles (T/T = 17 (7.26%); T/G = 81 (34.62%); G/G = 136 (58.12%) did not differ from Hardy-Weinberg equilibrium, χ2 = 1.03, p = 0.31. The frequency of BDNF Val66Met alleles (Val/Val = 156 (66.67%); Val/Met = 71 (30.34%); Met/Met = 7 (2.99%) also did not differ from Hardy-Weinberg equilibrium, χ2 = 0.10, p = 0.75. N’s for each genotype interaction are also shown in Table 1for Study 1 and Study 2. Combined analysis of the TPH2 and BDNF polymorphisms do not appear to be in linkage disequilibrium (D′ = -0.21, r2 = 0.01) [96].
Table 1.
| Study 1 | ||||||
| TPH2 rs4570625 Genotype | ||||||
| G/G | G/T | T/T | Row Total | Table Total | ||
| BDNF rs6265 Genotype | ValVal | 48 | 30 | 6 | 84 | |
| ValMet | 22 | 8 | 2 | 32 | ||
| MetMet | 2 | 1 | 0 | 3 | ||
| Column Total | 72 | 39 | 8 | 119 | ||
| Table Total | ||||||
| Study 2 | ||||||
| TPH2 rs4570625 Genotype | ||||||
| G/G | G/T | T/T | Row Total | Table Total | ||
| BDNF rs6265 Genotype | ValVal | 35 | 32 | 5 | 72 | |
| ValMet | 28 | 8 | 3 | 39 | ||
| MetMet | 1 | 2 | 1 | 4 | ||
| Column Total | 64 | 42 | 9 | 115 | ||
| Table Total | ||||||
Study 1: Emotion-word Stroop task
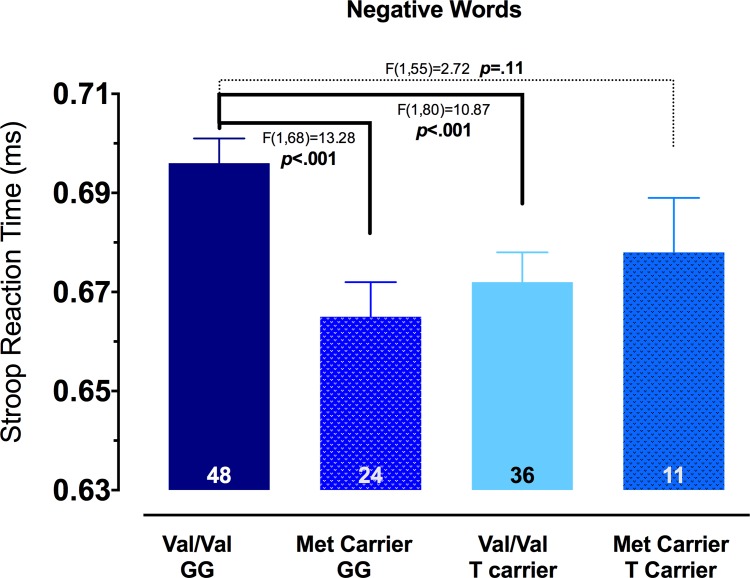
We first performed omnibus tests for group differences (BDNF Val/Val vs. BDNF Val/Met or Met/Met; TPH2 G/G vs. TPH2 T/G or T/T; and their combinations) in inhibitory responses to negative or positive emotion words using an ANCOVA, controlling for responses to the neutral words and reported depression. As predicted, we found a significant interaction, F(1,113) = 6.02, p = 0.016, for negative words. Follow-up tests revealed significant differences by allelic combination. Surprisingly, individuals with the Val/Val–G/G combination showed the weakest inhibition of negative content (i.e., slowest response time for negative words), relative to Val/Val–T carriers, F(1,80) = 10.87, p < .001, and Met carrier-G/Gs, F(1,68) = 13.28, p < .001, but were not significantly different from Met Carrier–T’s, F (1,55) = 2.72, p = 0.11. (See Fig 1 for all group comparisons). There were no individual main effects for genotype for either the BDNF polymorphism nor the TPH2 polymorphism for either negative or positive words, and no interaction for positive words.
Fig 1. Reaction time to negative words in the Emotion-word Stroop task.
Subjects from Study 1 that were Val/Val–G/G had significantly increased reaction times to negative words compared to Val/Val–T Carriers (F(1,80) = 10.87, p<0.001) and Met Carrier–G/Gs (F(1,68) = 13.28, p<0.001). There was also a trend for an increased reaction time of Val/Val–G/Gs compared to Met Carrier–T Carriers (F(1,55) = 2.72, p = 0.11). Met Carrier–G/Gs were not significantly different from Val/Val–T Carriers (F(1,56) = 0.11, p = 0.74) nor from Met Carrier–T Carriers (F(1,31) = 0.70, p = 0.41). Val/Val–T Carriers were also not significantly different from Met Carrier–T Carriers (F(1,43) = 0.22, p = 0.64). All data are presented as mean ± SEM.
Study 2: Naturalistic emotion-inhibition
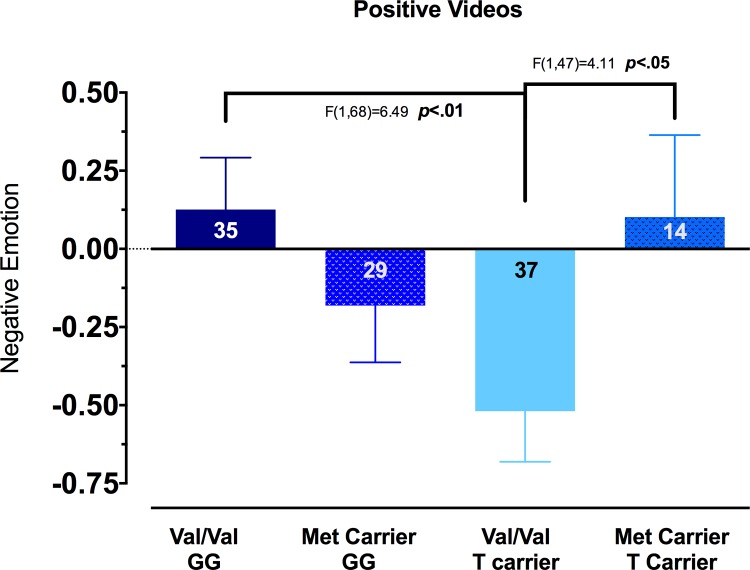
As in Study 1, we first analyzed for group differences (BDNF Val/Val vs. BDNF Val/Met or Met/Met; TPH2 G/G vs. TPH2 T/G or T/T; and their interaction) in inhibitory processing of negative emotion using an omnibus ANCOVA test, controlling for reported depression as well as emotion responses to the previous films. Specifically, we tested to see if there were group differences by genotype and interaction in negative emotion responses during the positive films. Because of the specific parameters of the paradigm, all negative films (eliciting intense negative emotions) were followed by explicitly positive films, placing demands on participants to inhibit negative emotions during the positive films. As such, negative emotions present during positive films would be indicative of poor inhibition of negative emotion. The results of the analysis of negative emotion were highly similar to the results from Study 1. Consistent with our predictions, there was a significant interaction between the two polymorphisms, F(1,109) = 5.50, p = .02, and no main effects for genotype for either polymorphism. As with Study 1, individuals with the Val/Val–G/G combination showed the weakest inhibition of negative content, demonstrating the highest negative emotion responses during positive films. As before, differences were most clear when compared to Val/Val–T carriers, F(1,68) = 6.49, p < .01 and also consistent with Study 1, there were no differences with Met Carrier-Ts. (See Fig 2 for all group comparisons).
Fig 2. Negative emotions expressed during positive contexts.
Reported and coded emotion scores from subjects in Study 2 were standardized to z-scores and combined yielding one score for negative emotion during the positive films. Negative emotion of Val/Val–T Carriers was significantly lower than those exhibited by Val/Val–G/Gs (F(1,68) = 6.49, p<0.01) and by Met Carrier–T Carriers (F(1,47) = 4.11, p<0.05). Negative emotion of Val/Val–T Carriers was not significantly different from Met Carrier–G/Gs (F(1,62) = 1.75, p = 0.19). Val/Val–G/Gs were not significantly different from Met Carrier–G/Gs (F(1,60) = 1.65, p = 0.20) nor from Met Carrier–T Carriers (F(1,45) = 0.02, p = 0.90). Met Carrier–G/Gs were also not significantly different from Met Carrier–T Carriers (F(1,39) = 0.96, p = 0.33. All data are presented as mean ± SEM.
Discussion
This is the first report of the potential risk-related consequences of a novel interaction between the BDNF Val66Met and TPH2 rs4570625 G-703T polymorphisms. Specifically, in this investigation we were able to detect the influence of a significant interaction between the TPH2 rs4570625 and BDNF Val66Met polymorphisms on negative emotion inhibitory processing. We observed consistent evidence across two different emotion-inhibition paradigms, one with high internal validity (Study 1) and one with high ecological validity (Study 2) that the combination of Val/Val and G/G genotypes was associated with impaired inhibition of negative emotional content. This was initially surprising given our original hypothesis that Met Carrier-Ts would exhibit the most pronounced impairments in negative emotion inhibition. Because in neither study did Met carrier-Ts differ from the Val/Val-G/Gs, it remains plausible that the former individuals are also at increased risk. There is growing consensus that inhibitory processes (regulating attention, behavior, and memory) are an essential component of emotional systems that influence nearly all psychological processes [97–103]. The ability to inhibit emotional responses or emotional information is well-recognized as a significant predictor and characteristic of common affective disease, including depression [88], post-traumatic stress disorder [104,105] and anxiety disorders [106]. Moreover, current evidence suggests that poor inhibition of negative emotion can reliably predict the onset of pathology following a stressful life event [87] as well as its persistence over time [75].
The current results indicate that the interaction of two polymorphisms–BDNF (Val66Met) and TPH2 (rs4570625; G-703T)–yields two at-risk groups, both characterized by significantly greater difficulty inhibiting negative emotional content. This was most clearly evident when the BDNF–Val/Val was combined with the TPH2 –G/G, but also BDNF–Met was combined with TPH2-T. This pattern is most evident in Study 1, but is effectively replicated in Study 2 (in a more naturalistic task), providing further evidence that these two genotype combinations contribute to high-risk patterns of emotion-inhibitory processing across measurement or methodological contexts.
When studied in isolation, the BDNF Val66Met Met allele has been associated with reduced hippocampal volume, poor performance on hippocampal-dependent tasks in humans [40,93,107,108] and thus has been considered a risk allele. However, it is also associated with some cognitive benefits, including improved performance on executive tasks with less hippocampal involvement [109–111], also consistent with our findings here. Collectively, these data suggest that mixed evidence of performance deficits associated with unique alleles may be task-specific and substantial epistatic relationships may exist between BDNF and genes involved in serotonin regulation. This could result in complex effects on emotion inhibitory processing and risk for psychiatric disease–findings that would not be predicted or detected if each gene was studied in isolation.
Although only limited prior data are available, the functional effect of the TPH2 rs4570625 polymorphism may be to alter DNA-protein interactions, ultimately affecting transcription of the TPH2 gene, as the presence of the T-allele is associated with reduced TPH2 promoter activity in vitro [26,27]. This is hypothesized to result in reduced serotonin levels, although this has yet to be confirmed in vivo. In two studies, the rare T/T genotype was associated with poor impulse control and decreased executive control [29,111]. Other studies have found that the T allele is predictive of amygdala and cortical responses to emotional content [28,32–34,112] and has been significantly associated with major depressive disorder [113]. Yet, similar to the BDNF Val66Met Met allele, the TPH2 T/T genotype has also been associated with significantly lower measures of trait anxiety [114,115] suggesting a trade-off between negative emotionality and cognitive functions. In the present study, however, only when the G/G genotype of rs4570625 was combined with Val/Val of BDNF Val66Met did we find impaired inhibition of negative emotional content, indicating a more intricate interaction between BDNF and TPH2 genes. Taken together, these findings suggest that the poor emotion-inhibitory responses exhibited by Val/Val-G/Gs and Met Carrier-Ts may leave them particularly vulnerable for the most common emotion-related psychiatric disorders.
Recent studies in humans and mutant mouse models provide supporting evidence for biological epistasis between BDNF and genes involved in serotonin regulation [11,12,46,47,51] that may alter neural circuitry and thereby influence risk for psychiatric disease. For instance, the BDNF Val66Met Met allele has been found to ameliorate differences in limbic circuitry associated with the 5-HTTLPR S allele [11]. Thus, reductions in BDNF may minimize the increased serotonergic tone hypothesized to be associated with reduced serotonin transporter function. Indeed, researchers are continuing to uncover the lifelong emotional consequences of dysregulated serotonergic signaling (particularly during neurodevelopment) [57–62]. For example, constitutive genetic or transient early life pharmacologic increases in serotonin signaling in rodents have been associated with increased anxiety- and depressive-like behaviors [116,117] as well as social impairments and stereotypy [118,119]. Though the physiological characterization of TPH2 rs4570625 is limited [26,27], such research supports our findings here, indicating that homozygosity for both the higher producing BDNF allele (Val) and the higher producing TPH2 allele (G) might contribute to a serotonergic tone that falls outside an optimal range. These findings are also supported by evidence across the two studies that presence of both the lower producing BDNF allele (Met) and the lower producing TPH2 allele (T) was linked to impairments in negative emotion inhibition that did not significantly differ from those of the at-risk Val/Val-G/G group. Indeed, it is the genotype groups with a combination of one lower producing allele and one higher producing allele that did not exhibit such pronounced negative emotion inhibition deficits. These particular genetic combinations (G/G and Met carrier, or T carrier and Val/Val) may, in combination, help maintain serotonergic tone within an optimal range, possibly during neurodevelopment.
There are some limitations to our study. The first of these being sample sizes that were constrained by the labor-intensive nature of assessing this complex behavioral phenotype. However, the benefit of this approach, which includes more objective and valid indicators of emotion-inhibitory processing, outweigh the limitations of small sample sizes. Our subject pool consisted of an overwhelming majority of Caucasian individuals, preventing evaluation of race- or ethnicity-specific stratification of effects. This shortcoming could be addressed by replication of these methods in regions without a predominant Caucasian presence. Additionally, the Met Carrier-Ts group itself was small in both studies, an inherent limitation due both to the overall sample sizes as well as the much lower frequency of these Met and T alleles. In addition, we focused our study on negative emotion inhibitory processing elicited in-lab. Future studies would benefit from including environmental and life-history data given the importance of gene × environment interactions on risk for psychiatric disease. Though our analyses indicate that the TPH2 and BDNF polymorphisms are not in linkage disequilibrium, there remains the possibility that we are unintentionally measuring the effects of an unidentified gene or polymorphism that is in linkage disequilibrium with one or both of these polymorphisms.
Conclusion
This investigation makes an important contribution to the understanding of the genetic basis of vulnerability to the most common emotion-related psychiatric diseases. In particular, ours is the first study to suggest an epistatic relationship between BDNF Val66Met and TPH2 rs4570625 polymorphisms for impaired regulation of negative emotional content, which has been consistently linked to common affective disorders. These data also emphasize the importance of considering multiple genes and their relationships when investigating their genetic influence on complex behavioral outcomes. Integrating multiple genetic influences on prominent underlying risk factors for emotional psychiatric diseases, such as poor inhibition of negative emotion, rather than associations with broader disease states, will help build more comprehensive models and identify groups at risk for psychiatric disease.
Data Availability
The full data includes video recordings of participants and those videos must remain confidential due to Kent State University Institutional Review Board (IRB) restrictions. Anyone wishing to have access to the data can contact the corresponding author (Aaron Jasnow-ajasnow@kent.edu) and all appropriate available data will be provided. Contact information for the Kent State University IRB: Office of Compliance, Division of Research and Sponsored Programs Room 222, Cartwright Hall, (330) 672-2384.
Funding Statement
The research described in this manuscript was funded, in part, by the Institute for Clinical and Translational Research, Kent State University; and The Applied Psychology Center, Department of Psychological Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Foland-Ross LC, Hamilton JP, Joormann J, Berman MG, Jonides J, et al. (2013) The neural basis of difficulties disengaging from negative irrelevant material in major depression. Psychological science 24: 334–344. 10.1177/0956797612457380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joormann J, Gotlib IH (2010) Emotion regulation in depression: relation to cognitive inhibition. Cognition and Emotion 24: 281–298. 10.1080/02699930903407948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuellar AK, Johnson SL, Winters R (2005) Distinctions between bipolar and unipolar depression. Clinical psychology review 25: 307–339. 10.1016/j.cpr.2004.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandell D, Siegle GJ, Shutt L, Feldmiller J, Thase ME (2014) Neural substrates of trait ruminations in depression. Journal of abnormal psychology 123: 35 10.1037/a0035834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mennin DS, Fresco DM (2013) What, Me Worry and Ruminate About DSM‐5 and RDoC? The Importance of Targeting Negative Self‐Referential Processing. Clinical Psychology: Science and Practice 20: 258–267. 10.1111/cpsp.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitmer AJ, Gotlib IH (2012) Switching and backward inhibition in major depressive disorder: The role of rumination. Journal of abnormal psychology 121: 570 10.1037/a0027474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barlow DH, Sauer-Zavala S, Carl JR, Bullis JR, Ellard KK (2014) The nature, diagnosis, and treatment of neuroticism back to the future. Clinical Psychological Science 2: 344–365. 10.1177/2167702613505532 [DOI] [Google Scholar]
- 8.Fisher AJ, Newman MG (2013) Heart rate and autonomic response to stress after experimental induction of worry versus relaxation in healthy, high-worry, and generalized anxiety disorder individuals. Biological psychology 93: 65–74. 10.1016/j.biopsycho.2013.01.012 [DOI] [PubMed] [Google Scholar]
- 9.Hofmann W, Friese M, Roefs A (2009) Three ways to resist temptation: The independent contributions of executive attention, inhibitory control, and affect regulation to the impulse control of eating behavior. Journal of Experimental Social Psychology 45: 431–435. 10.1016/j.jesp.2008.09.013 [DOI] [Google Scholar]
- 10.Jansson M, Linton SJ (2007) Psychological mechanisms in the maintenance of insomnia: arousal, distress, and sleep-related beliefs. Behaviour research and therapy 45: 511–521. 10.1016/j.brat.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 11.Pezawas L, Meyer-Lindenberg A, Goldman A, Verchinski B, Chen G, et al. (2008) Evidence of biologic epistasis between BDNF and SLC6A4 and implications for depression. Molecular psychiatry 13: 709–716. 10.1038/mp.2008.32 [DOI] [PubMed] [Google Scholar]
- 12.Dougherty LR, Klein DN, Congdon E, Canli T, Hayden EP (2010) Interaction between 5-HTTLPR and BDNF Val66Met polymorphisms on HPA axis reactivity in preschoolers. Biological psychology 83: 93–100. 10.1016/j.biopsycho.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anttila S, Huuhka K, Huuhka M, Rontu R, Hurme M, et al. (2007) Interaction between 5-HT1A and BDNF genotypes increases the risk of treatment-resistant depression. Journal of neural transmission 114: 1065–1068. 10.1007/s00702-007-0705-9 [DOI] [PubMed] [Google Scholar]
- 14.Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, et al. (2012) Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry 17: 36–48. 10.1038/mp.2010.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohoff FW (2010) Overview of the genetics of major depressive disorder. Curr Psychiatry Rep 12: 539–546. 10.1007/s11920-010-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Li J, He Z, Lu Q, Witte JS, et al. (2016) Testing Allele Transmission of an SNP Set Using a Family-Based Generalized Genetic Random Field Method. Genet Epidemiol 40: 341–351. 10.1002/gepi.21970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyekis JP, Yu W, Dong S, Wang H, Qian J, et al. (2013) No association of genetic variants in BDNF with major depression: a meta- and gene-based analysis. Am J Med Genet B Neuropsychiatr Genet 162B: 61–70. 10.1002/ajmg.b.32122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, et al. (2010) Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry 15: 589–601. 10.1038/mp.2008.131 [DOI] [PubMed] [Google Scholar]
- 19.Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, et al. (2011) Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry 16: 516–532. 10.1038/mp.2010.38 [DOI] [PubMed] [Google Scholar]
- 20.Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, et al. (2011) Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry 16: 193–201. 10.1038/mp.2009.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, et al. (2010) Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry 167: 949–957. 10.1176/appi.ajp.2010.09091380 [DOI] [PubMed] [Google Scholar]
- 22.Gutknecht L, Kriegebaum C, Waider J, Schmitt A, Lesch K- P (2009) Spatio-temporal expression of tryptophan hydroxylase isoforms in murine and human brain: convergent data from Tph2 knockout mice. European Neuropsychopharmacology 19: 266–282. 10.1016/j.euroneuro.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 23.Udenfriend S, Titus E, Weissbach H, Peterson RE (1956) Biogenesis and metabolism of 5-hydroxyindole compounds. Journal of Biological Chemistry 219: 335–344. [PubMed] [Google Scholar]
- 24.Walther DJ, Bader M (2003) A unique central tryptophan hydroxylase isoform. Biochemical pharmacology 66: 1673–1680. 10.1016/s0006-2952(03)00556-2 [DOI] [PubMed] [Google Scholar]
- 25.Walther DJ, Peter J-U, Bashammakh S, Hörtnagl H, Voits M, et al. (2003) Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 299: 76–76. 10.1126/science.1078197 [DOI] [PubMed] [Google Scholar]
- 26.Chen G- L, Vallender EJ, Miller GM (2008) Functional characterization of the human TPH2 5' regulatory region: untranslated region and polymorphisms modulate gene expression in vitro. Human genetics 122: 645–657. 10.1007/s00439-007-0443-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheuch K, Lautenschlager M, Grohmann M, Stahlberg S, Kirchheiner J, et al. (2007) Characterization of a functional promoter polymorphism of the human tryptophan hydroxylase 2 gene in serotonergic raphe neurons. Biological psychiatry 62: 1288–1294. 10.1016/j.biopsych.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 28.Canli T, Congdon E, Gutknecht L, Constable R, Lesch K (2005) Amygdala responsiveness is modulated by tryptophan hydroxylase-2 gene variation. Journal of neural transmission 112: 1479–1485. 10.1007/s00702-005-0391-4 [DOI] [PubMed] [Google Scholar]
- 29.Strobel A, Dreisbach G, Müller J, Goschke T, Brocke B, et al. (2007) Genetic variation of serotonin function and cognitive control. Journal of Cognitive Neuroscience 19: 1923–1931. 10.1162/jocn.2007.19.12.1923 [DOI] [PubMed] [Google Scholar]
- 30.Waider J, Araragi N, Gutknecht L, Lesch K-P (2011) Tryptophan hydroxylase-2 (TPH2) in disorders of cognitive control and emotion regulation: a perspective. Psychoneuroendocrinology 36: 393–405. 10.1016/j.psyneuen.2010.12.012 [DOI] [PubMed] [Google Scholar]
- 31.Gao J, Pan Z, Jiao Z, Li F, Zhao G, et al. (2012) TPH2 gene polymorphisms and major depression—a meta-analysis. PloS one 7: e36721 10.1371/journal.pone.0036721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown S, Peet E, Manuck S, Williamson D, Dahl R, et al. (2005) A regulatory variant of the human tryptophan hydroxylase-2 gene biases amygdala reactivity. Molecular psychiatry 10: 884–888. 10.1038/sj.mp.4001716 [DOI] [PubMed] [Google Scholar]
- 33.Harvey M, Shink E, Tremblay M, Gagne B, Raymond C, et al. (2004) Support for the involvement of TPH2 gene in affective disorders. Molecular psychiatry 9: 980–981. 10.1038/sj.mp.4001557 [DOI] [PubMed] [Google Scholar]
- 34.Zill P, Baghai T, Zwanzger P, Schüle C, Eser D, et al. (2004) SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Molecular psychiatry 9: 1030–1036. 10.1038/sj.mp.4001525 [DOI] [PubMed] [Google Scholar]
- 35.Osinsky R, Schmitz A, Alexander N, Kuepper Y, Kozyra E, et al. (2009) TPH2 gene variation and conflict processing in a cognitive and an emotional Stroop task. Behavioural brain research 198: 404–410. 10.1016/j.bbr.2008.11.022 [DOI] [PubMed] [Google Scholar]
- 36.Drago A, Liappas I, Petio C, Albani D, Forloni G, et al. (2009) Epistasis between IL1A, IL1B, TNF, HTR2A, 5-HTTLPR and TPH2 variations does not impact alcohol dependence disorder features. International journal of environmental research and public health 6: 1980–1990. 10.3390/ijerph6071980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rotberg B, Kronenberg S, Carmel M, Frisch A, Brent D, et al. (2013) Additive effects of 5-HTTLPR (serotonin transporter) and tryptophan hydroxylase 2 G-703T gene polymorphisms on the clinical response to citalopram among children and adolescents with depression and anxiety disorders. Journal of child and adolescent psychopharmacology 23: 117–122. 10.1089/cap.2012.0020 [DOI] [PubMed] [Google Scholar]
- 38.Lehto K, Vaht M, Mäestu J, Veidebaum T, Harro J (2015) Effect of tryptophan hydroxylase-2 gene polymorphism G-703 T on personality in a population representative sample. Progress in Neuro-Psychopharmacology and Biological Psychiatry 57: 31–35. 10.1016/j.pnpbp.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 39.Havranek M, Vonmoos M, Müller C, Büetiger J, Tasiudi E, et al. (2015) Serotonin Transporter and Tryptophan Hydroxylase Gene Variations Mediate Working Memory Deficits of Cocaine Users. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, et al. (2003) The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112: 257–269. 10.1016/s0092-8674(03)00035-7 [DOI] [PubMed] [Google Scholar]
- 41.Johnson D, Casey B (2015) Easy to remember, difficult to forget: The development of fear regulation. Developmental cognitive neuroscience 11: 42–55. 10.1016/j.dcn.2014.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galvin C, Lee FS, Ninan I (2015) Alteration of the Centromedial Amygdala Glutamatergic Synapses by the BDNF Val66Met Polymorphism. Neuropsychopharmacology. 10.1038/npp.2015.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayden EP, Klein DN, Dougherty LR, Olino TM, Dyson MW, et al. (2010) The role of brain-derived neurotrophic factor genotype, parental depression, and relationship discord in predicting early-emerging negative emotionality. Psychological science 21: 1678–1685. 10.1177/0956797610385357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan MJ, Ogden RT, Huang Y-y, Oquendo MA, Sullivan GM, et al. (2014) Genetic variation in brain-derived neurotrophic factor val66met allele is associated with altered serotonin-1A receptor binding in human brain. NeuroImage 94: 33–39. 10.1016/j.neuroimage.2014.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henningsson S, Borg J, Lundberg J, Bah J, Lindström M, et al. (2009) Genetic variation in brain-derived neurotrophic factor is associated with serotonin transporter but not serotonin-1A receptor availability in men. Biological psychiatry 66: 477–485. 10.1016/j.biopsych.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 46.Ren-Patterson RF, Cochran LW, Holmes A, Lesch K- P, Lu B, et al. (2006) Gender-dependent modulation of brain monoamines and anxiety-like behaviors in mice with genetic serotonin transporter and BDNF deficiencies. Cellular and molecular neurobiology 26: 753–778. 10.1007/s10571-006-9048-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren‐Patterson RF, Cochran LW, Holmes A, Sherrill S, Huang SJ, et al. (2005) Loss of brain‐derived neurotrophic factor gene allele exacerbates brain monoamine deficiencies and increases stress abnormalities of serotonin transporter knockout mice. Journal of neuroscience research 79: 756–771. [DOI] [PubMed] [Google Scholar]
- 48.Migliarini S, Pacini G, Pelosi B, Lunardi G, Pasqualetti M (2013) Lack of brain serotonin affects postnatal development and serotonergic neuronal circuitry formation. Molecular psychiatry 18: 1106–1118. 10.1038/mp.2012.128 [DOI] [PubMed] [Google Scholar]
- 49.Naumenko V, Kondaurova E, Bazovkina D, Tsybko A, Tikhonova M, et al. (2012) Effect of brain-derived neurotrophic factor on behavior and key members of the brain serotonin system in genetically predisposed to behavioral disorders mouse strains. Neuroscience 214: 59–67. 10.1016/j.neuroscience.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 50.Kronenberg G, Mosienko V, Gertz K, Alenina N, Hellweg R, et al. (2015) Increased brain-derived neurotrophic factor (BDNF) protein concentrations in mice lacking brain serotonin. European archives of psychiatry and clinical neuroscience: 1–4. [DOI] [PubMed] [Google Scholar]
- 51.Murphy D, Uhl G, Holmes A, Ren‐Patterson R, Hall F, et al. (2003) Experimental gene interaction studies with SERT mutant mice as models for human polygenic and epistatic traits and disorders. Genes, Brain and Behavior 2: 350–364. 10.1046/j.1601-1848.2003.00049.x [DOI] [PubMed] [Google Scholar]
- 52.Eaton MJ, Staley JK, Globus MY-T, Whittemore SR (1995) Developmental regulation of early serotonergic neuronal differentiation: the role of brain-derived neurotrophic factor and membrane depolarization. Developmental biology 170: 169–182. 10.1006/dbio.1995.1205 [DOI] [PubMed] [Google Scholar]
- 53.Galter D, Unsicker K (2000) Brain‐derived neurotrophic factor and trkB are essential for cAMP‐mediated induction of the serotonergic neuronal phenotype. Journal of neuroscience research 61: 295–301. [DOI] [PubMed] [Google Scholar]
- 54.Mamounas LA, Blue ME, Siuciak JA, Altar CA (1995) Brain-derived neurotrophic factor promotes the survival and sprouting of serotonergic axons in rat brain. The Journal of neuroscience 15: 7929–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pelleymounter MA, Cullen MJ, Wellman CL (1995) Characteristics of BDNF-induced weight loss. Experimental neurology 131: 229–238. 10.1016/0014-4886(95)90045-4 [DOI] [PubMed] [Google Scholar]
- 56.Siuciak JA, Boylan C, Fritsche M, Altar CA, Lindsay RM (1996) BDNF increases monoaminergic activity in rat brain following intracerebroventricular or intraparenchymal administration. Brain research 710: 11–20. 10.1016/0006-8993(95)01289-3 [DOI] [PubMed] [Google Scholar]
- 57.Daws LC, Gould GG (2011) Ontogeny and regulation of the serotonin transporter: providing insights into human disorders. Pharmacology & therapeutics 131: 61–79. 10.1016/j.pharmthera.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez SP, Gaspar P (2012) Investigating anxiety and depressive-like phenotypes in genetic mouse models of serotonin depletion. Neuropharmacology 62: 144–154. 10.1016/j.neuropharm.2011.08.049 [DOI] [PubMed] [Google Scholar]
- 59.Gaspar P, Cases O, Maroteaux L (2003) The developmental role of serotonin: news from mouse molecular genetics. Nature Reviews Neuroscience 4: 1002–1012. 10.1038/nrn1256 [DOI] [PubMed] [Google Scholar]
- 60.Leonardo ED, Hen R (2006) Genetics of affective and anxiety disorders. Annual review of psychology 57: 117–137. 10.1146/annurev.psych.57.102904.190118 [DOI] [PubMed] [Google Scholar]
- 61.Leonardo ED, Hen R (2008) Anxiety as a developmental disorder. Neuropsychopharmacology 33: 134–140. 10.1038/sj.npp.1301569 [DOI] [PubMed] [Google Scholar]
- 62.Gross C, Zhuang X, Stark K, Ramboz S, Oosting R, et al. (2002) Serotonin1A receptor acts during development to establish normal anxiety-like behaviour in the adult. Nature 416: 396–400. 10.1038/416396a [DOI] [PubMed] [Google Scholar]
- 63.McLaughlin KA, Nolen-Hoeksema S (2011) Rumination as a transdiagnostic factor in depression and anxiety. Behaviour research and therapy 49: 186–193. 10.1016/j.brat.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nolen-Hoeksema S (1991) Responses to depression and their effects on the duration of depressive episodes. Journal of abnormal psychology 100: 569 10.1037//0021-843x.100.4.569 [DOI] [PubMed] [Google Scholar]
- 65.Rottenberg J, Kasch KL, Gross JJ, Gotlib IH (2002) Sadness and amusement reactivity differentially predict concurrent and prospective functioning in major depressive disorder. Emotion 2: 135 10.1037//1528-3542.2.2.135 [DOI] [PubMed] [Google Scholar]
- 66.Soussignan R, Boivin M, Girard A, Perusse D, Liu X, et al. (2009) Genetic and environmental etiology of emotional and social behaviors in 5-month-old infant twins: influence of the social context. Infant Behav Dev 32: 1–9. 10.1016/j.infbeh.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 67.Canli T, Ferri J, Duman EA (2009) Genetics of emotion regulation. NSC 164: 43–54. 10.1016/j.neuroscience.2009.06.049 [DOI] [PubMed] [Google Scholar]
- 68.Emde R, Plomin R, Robinson J, Corley R, DeFries J, et al. (1992) Temperment, emotion, and cognition at fourteen months: The MacArthur Logitdudinal Twin Study. Child development 63: 1437–1455. 10.2307/1131567 [DOI] [PubMed] [Google Scholar]
- 69.Friedman NP, Miyake A, Young SE, Defries JC, Corley RP, et al. (2008) Individual differences in executive functions are almost entirely genetic in origin. J Exp Psychol Gen 137: 201–225. 10.1037/0096-3445.137.2.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cross-Disorder Group of the Psychiatric Genomics C, Lee SH, Ripke S, Neale BM, Faraone SV, et al. (2013) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45: 984–994. 10.1038/ng.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gottesman II, Gould TD (2003) The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 160: 636–645. 10.1176/appi.ajp.160.4.636 [DOI] [PubMed] [Google Scholar]
- 72.Gilman TL, Latsko M, Matt L, Flynn J, Cabrera OdlC, et al. (2015) Variation of 5-HTTLPR and deficits in emotion regulation: A pathway to risk? Psychology & Neuroscience 8: 397. [Google Scholar]
- 73.Gross JJ, Levenson RW (1995) Emotion elicitation using films. Cognition & emotion 9: 87–108. 10.1080/02699939508408966 [DOI] [Google Scholar]
- 74.Gyurak A, Gross JJ, Etkin A (2011) Explicit and implicit emotion regulation: a dual-process framework. Cognition and Emotion 25: 400–412. 10.1080/02699931.2010.544160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Diminich E, Bonanno G (2014) Faces, feelings, words: Divergence across channels of emotional responding in complicated grief. Journal of abnormal psychology 123: 350 10.1037/a0036398 [DOI] [PubMed] [Google Scholar]
- 76.Rottenberg J, Gross JJ, Gotlib IH (2005) Emotion context insensitivity in major depressive disorder. Journal of abnormal psychology 114: 627 10.1037/0021-843x.114.4.627 [DOI] [PubMed] [Google Scholar]
- 77.Radloff LS (1977) The CES-D Scale: A Self-Report Depression Scale for Research in the General Population. Applied Psychological Measurement 1: 385–401. 10.1177/014662167700100306 [DOI] [Google Scholar]
- 78.Beevers CG, Marti CN, Lee H- J, Stote DL, Ferrell RE, et al. (2011) Associations between serotonin transporter gene promoter region (5-HTTLPR) polymorphism and gaze bias for emotional information. Journal of Abnormal Psychology 120: 187 10.1037/a0022125 [DOI] [PubMed] [Google Scholar]
- 79.Anchordoquy HC, McGeary C, Liu L, Krauter KS, Smolen A (2003) Genotyping of three candidate genes after whole-genome preamplification of DNA collected from buccal cells. Behavior genetics 33: 73–78. [DOI] [PubMed] [Google Scholar]
- 80.Mössner R, Walitza S, Geller F, Scherag A, Gutknecht L, et al. (2006) Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in children and adolescents with obsessive–compulsive disorder. The International Journal of Neuropsychopharmacology 9: 437–442. 10.1017/S1461145705005997 [DOI] [PubMed] [Google Scholar]
- 81.Hünnerkopf R, Strobel A, Gutknecht L, Brocke B, Lesch KP (2007) Interaction between BDNF Val66Met and dopamine transporter gene variation influences anxiety-related traits. Neuropsychopharmacology 32: 2552–2560. 10.1038/sj.npp.1301383 [DOI] [PubMed] [Google Scholar]
- 82.Radloff LS (1977) The CES-D scale a self-report depression scale for research in the general population. Applied psychological measurement 1: 385–401. 10.1177/014662167700100306 [DOI] [Google Scholar]
- 83.Mathews A, MacLeod C (1985) Selective processing of threat cues in anxiety states. Behaviour research and therapy 23: 563–569. 10.1016/0005-7967(85)90104-4 [DOI] [PubMed] [Google Scholar]
- 84.Frings C, Englert J, Wentura D, Bermeitinger C (2010) Decomposing the emotional Stroop effect. The quarterly journal of experimental psychology 63: 42–49. 10.1080/17470210903156594 [DOI] [PubMed] [Google Scholar]
- 85.Williams JMG, Mathews A, MacLeod C (1996) The emotional Stroop task and psychopathology. Psychological bulletin 120: 3 10.1037//0033-2909.120.1.3 [DOI] [PubMed] [Google Scholar]
- 86.Tukey JW (1977) Exploratory data analysis.
- 87.Coifman KG, Bonanno GA (2010) When distress does not become depression: emotion context sensitivity and adjustment to bereavement. Journal of Abnormal Psychology 119: 479 10.1037/a0020113 [DOI] [PubMed] [Google Scholar]
- 88.Gotlib IH, Joormann J (2010) Cognition and depression: current status and future directions. Annual review of clinical psychology 6: 285 10.1146/annurev.clinpsy.121208.131305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Janes AC, Pizzagalli DA, Richardt S, Chuzi S, Pachas G, et al. (2010) Brain reactivity to smoking cues prior to smoking cessation predicts ability to maintain tobacco abstinence. Biological psychiatry 67: 722–729. 10.1016/j.biopsych.2009.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bremner JD, Vermetten E, Vythilingam M, Afzal N, Schmahl C, et al. (2004) Neural correlates of the classic color and emotional stroop in women with abuse-related posttraumatic stress disorder. Biological psychiatry 55: 612–620. 10.1016/j.biopsych.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 91.Hünnerkopf R, Strobel A, Gutknecht L, Brocke B, Lesch K- P (2007) Interaction between BDNF Val66Met and Dopamine Transporter Gene Variation Influences Anxiety-Related Traits. Neuropsychopharmacology 32: 2552–2560. 10.1038/sj.npp.1301383 [DOI] [PubMed] [Google Scholar]
- 92.Dougherty LR, Klein DN, Congdon E, Canli T, Hayden EP (2010) Interaction between 5-HTTLPR and BDNF Val66Met polymorphisms on HPA axis reactivity in preschoolers. Biological Psychology 83: 93–100. 10.1016/j.biopsycho.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hariri A, Goldberg T, Mattay V, Kolachana B, Callicott J, et al. (2003) Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. The Journal of neuroscience: the official journal of the Society for Neuroscience 23: 6690–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soliman F, Glatt CE, Bath KG, Levita L, Jones RM, et al. (2010) A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science (New York, NY) 327: 863–866. 10.1126/science.1181886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pezawas L, Meyer-Lindenberg A, Goldman AL, Verchinski BA, Chen G, et al. (2008) Evidence of biologic epistasis between BDNF and SLC6A4 and implications for depression. Molecular psychiatry 13: 709–716. 10.1038/mp.2008.32 [DOI] [PubMed] [Google Scholar]
- 96.Gaunt TR, Rodríguez S, Day IN (2007) Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool'CubeX'. BMC bioinformatics 8: 428 10.1186/1471-2105-8-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bargh JA, Williams LE (2007) The nonconscious regulation of emotion. Handbook of emotion regulation 1: 429Á445. [Google Scholar]
- 98.McRae K, Misra S, Prasad AK, Pereira SC, Gross JJ (2012) Bottom-up and top-down emotion generation: implications for emotion regulation. Social cognitive and affective neuroscience 7: 253–262. 10.1093/scan/nsq103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gross JJ (2013) Emotion regulation: taking stock and moving forward. Emotion 13: 359 10.1037/a0032135 [DOI] [PubMed] [Google Scholar]
- 100.Kaplan S, Berman MG (2010) Directed attention as a common resource for executive functioning and self-regulation. Perspectives on Psychological Science 5: 43–57. 10.1177/1745691609356784 [DOI] [PubMed] [Google Scholar]
- 101.LeDoux J (2012) A Neuroscientist’s Perspective on Debates about the Nature of Emotion. Emotion Review 4: 375–379. 10.1177/1754073912445822 [DOI] [Google Scholar]
- 102.Payer DE, Baicy K, Lieberman MD, London ED (2012) Overlapping neural substrates between intentional and incidental down-regulation of negative emotions. Emotion 12: 229 10.1037/a0027421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anderson MC, Spellman BA (1995) On the status of inhibitory mechanisms in cognition: memory retrieval as a model case. Psychological review 102: 68 10.1037//0033-295x.102.1.68 [DOI] [PubMed] [Google Scholar]
- 104.Charney DS, Deutch AY, Krystal JH, Southwick SM, Davis M (1993) Psychobiologic mechanisms of posttraumatic stress disorder. Archives of General Psychiatry 50: 294–305. 10.1001/archpsyc.1993.01820160064008 [DOI] [PubMed] [Google Scholar]
- 105.Cohen JE, Shalev H, Admon R, Hefetz S, Gasho CJ, et al. (2013) Emotional brain rhythms and their impairment in post‐traumatic patients. Human brain mapping 34: 1344–1356. 10.1002/hbm.21516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mathews A, MacLeod C (2005) Cognitive vulnerability to emotional disorders. Annu Rev Clin Psychol 1: 167–195. [DOI] [PubMed] [Google Scholar]
- 107.Bueller JA, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, et al. (2006) BDNF Val66Met allele is associated with reduced hippocampal volume in healthy subjects. Biological psychiatry 59: 812–815. 10.1016/j.biopsych.2005.09.022 [DOI] [PubMed] [Google Scholar]
- 108.Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, et al. (2004) The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. The Journal of Neuroscience 24: 10099–10102. 10.1523/jneurosci.2680-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gajewski PD, Hengstler JG, Golka K, Falkenstein M, Beste C (2011) The Met-allele of the BDNF Val66Met polymorphism enhances task switching in elderly. Neurobiology of Aging 32: 2327 e2327-2327. e2319. 10.1016/j.neurobiolaging.2011.06.010 [DOI] [PubMed] [Google Scholar]
- 110.Alfimova M, Korovaitseva G, Lezheiko T, Golimbet V (2012) Effect of BDNF Val66Met polymorphism on normal variability of executive functions. Bulletin of experimental biology and medicine 152: 606–609. 10.1007/s10517-012-1587-x [DOI] [PubMed] [Google Scholar]
- 111.Reuter M, Ott U, Vaitl D, Hennig J (2007) Impaired executive control is associated with a variation in the promoter region of the tryptophan hydroxylase 2 gene. Journal of Cognitive Neuroscience 19: 401–408. 10.1162/jocn.2007.19.3.401 [DOI] [PubMed] [Google Scholar]
- 112.Herrmann MJ, Huter T, Müller F, Mühlberger A, Pauli P, et al. (2007) Additive effects of serotonin transporter and tryptophan hydroxylase-2 gene variation on emotional processing. Cerebral Cortex 17: 1160–1163. 10.1093/cercor/bhl026 [DOI] [PubMed] [Google Scholar]
- 113.Gao J, Pan Z, Jiao Z, Li F, Zhao G, et al. (2012) TPH2 gene polymorphisms and major depression–a meta-analysis. PloS one 7: e36721 10.1371/journal.pone.0036721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reuter M, Kuepper Y, Hennig J (2007) Association between a polymorphism in the promoter region of the TPH2 gene and the personality trait of harm avoidance. The International Journal of Neuropsychopharmacology 10: 401–404. 10.1017/S1461145706007073 [DOI] [PubMed] [Google Scholar]
- 115.Gutknecht L, Jacob C, Strobel A, Kriegebaum C, Müller J, et al. (2007) Tryptophan hydroxylase-2 gene variation influences personality traits and disorders related to emotional dysregulation. The International Journal of Neuropsychopharmacology 10: 309–320. 10.1017/S1461145706007437 [DOI] [PubMed] [Google Scholar]
- 116.Olivier JD, Van Der Hart MG, Van Swelm RP, Dederen PJ, Homberg JR, et al. (2008) A study in male and female 5-HT transporter knockout rats: an animal model for anxiety and depression disorders. Neuroscience 152: 573–584. 10.1016/j.neuroscience.2007.12.032 [DOI] [PubMed] [Google Scholar]
- 117.Freund N, Thompson BS, Denormandie J, Vaccarro K, Andersen SL (2013) Windows of vulnerability: maternal separation, age, and fluoxetine on adolescent depressive-like behavior in rats. Neuroscience 249: 88–97. 10.1016/j.neuroscience.2013.06.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Veenstra-VanderWeele J, Muller CL, Iwamoto H, Sauer JE, Owens WA, et al. (2012) Autism gene variant causes hyperserotonemia, serotonin receptor hypersensitivity, social impairment and repetitive behavior. Proc Natl Acad Sci U S A 109: 5469–5474. 10.1073/pnas.1112345109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kepser LJ, Homberg JR (2015) The neurodevelopmental effects of serotonin: a behavioural perspective. Behav Brain Res 277: 3–13. 10.1016/j.bbr.2014.05.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The full data includes video recordings of participants and those videos must remain confidential due to Kent State University Institutional Review Board (IRB) restrictions. Anyone wishing to have access to the data can contact the corresponding author (Aaron Jasnow-ajasnow@kent.edu) and all appropriate available data will be provided. Contact information for the Kent State University IRB: Office of Compliance, Division of Research and Sponsored Programs Room 222, Cartwright Hall, (330) 672-2384.