Abstract
Objectives
Minichromosome maintenance (MCM) proteins play important roles in DNA replication by interacting with other factors which participate in the regulation of DNA synthesis. Abnormal over-expression of MCMs was observed in numerous malignancies, such as colorectal cancer. However, the expression of MCMs in pancreatic cancer (PC) was less investigated so far. This study was designed to analyze the expression and prognostic roles of MCM1-10 in PC based on the data provided by The Cancer Genome Atlas (TCGA).
Methods
Pearson χ2 test was applied to evaluate the association of MCMs expression with clinicopathologic indicators, and biomarkers for tumor biological behaviors. Kaplan-Meier plots and log-rank tests were used to assess survival analysis, and univariate and multivariate Cox proportional hazard regression models were used to recognize independent prognostic factors.
Results
MCM1-10 were generally expressed in PC samples. The levels of some molecules were markedly correlated with that of biomarkers for S phase, proliferation, gemcitabine resistance. And part of these molecules over-expression was significantly associated with indicators of disease progression, such as depth of tumor invasion and lymph node metastasis. Furthermore, MCM2, 4, 6, 8, and 10 over-expression was remarkably associated with shorter disease free survival time, and MCM2, 4,8, and 10 over-expression was associated with shorter overall survival time. Further multivariate analysis suggested that MCM8 was an independent prognostic factor for PC.
Conclusion
MCMs abnormal over-expression was significantly associated with PC progression and prognosis. These molecules could be regarded as prognostic and therapeutic biomarkers for PC. The roles of MCMs may be vitally important and the underlying mechanisms need to be furtherinvestigated.
Introduction
Pancreatic cancer is a digestive malignancy with extremely aggressive behavior. It is the fifth leading cause of cancer-related deathsworldwide, and the five-year survival rate of such cancer is approximately8% on the basis of latest data provided by Siegel RL, et al [1]. Chemotherapy and radiotherapy are not effective for PC due to several reasons, such as complex genetic mutations, hypoxic tolerance, and excessive fibrosis [2]. At present, the only available treatment for PC is radical operation; however, the rate of surgical resection is less than 20% because of the absence of obvious symptoms at the early stage [3].Therefore, to find novel therapeutic strategies based on molecular biomarkers of PC is extremely urgent. Recently, an increasing number of molecular contribute to the transformation and progression of PC were identified, some of these molecular was regarded as prognostic and/or therapeutic markers, such as MUC4, LSD1, and FHL2 [4–6].
MCM family is composed of ten proteins which primarily promote the process of DNA replication of eukaryotes [7]. MCM1 is an important member of MADS box transcription factor family, this protein affects the process of cell cycle, apoptosis, growth, and differentiationthrough regulating many gene activation [8]. The MCM2-7 heterohexamer complex was first detected in the yeast Saccharomyces cerevisiae[9], and the functions of this complex were extensively studied nowdays.MCM2-7interact with each other to form a functional DNA helicases which trigger the initial step of DNA synthesis [10, 11]. MCM8 and MCM9 were generally considered as additional members of MCM2-7 family [7]. Like MCM2-7 complex, MCM8 and MCM9 were also crucial components of the pre-replication complex. MCM8 and MCM9 were also involved in drivingthe initiation of Sphase [12, 13]. MCM10 is another necessary molecule for initializing the DNA synthesis due to its interaction with MCM2-7 complex[14, 15]. Furthermore, it has been reported that some members ofMCMs were abnormally up-regulated in various malignancies, and over-expression of them could promote the progression of malignant cells and predict the survival times of suffered patients [16–18]. However, the roles of MCMs in PC was absolutely unknown.
Here we will assess the expression of MCMs in PC according to the data provided by TCGA, and analyze the association between MCMs expression and the progression, prognosis of PC.
Methods and Materials
Clinicopathologic Features and MCMs Expression
Clinicopathologic features and MCMs expression (level 3 data, log2(RSEM+1) transformed) for pancreatic cancer patients were downloaded from TCGA data portal (http://cancergenome.nih.gov/). The National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI) work with physicians who collect tissue for TCGA to gain approval with local Institutional Review Boards (IRBs). An IRB is a group of scientists, doctors, clergy and consumers who review and approve the research proposal for every research project that involves human subjects. 165 patients were finally included in our study, others were excluded due to the lack of critical information (such as overall survival time, age, gender, et al). Main clinicopathologic features for PC patients were shown in Table 1. All patients were divided into low and high expression group according to the median value of each gene expression. Gene was defined as high expression if this gene expression was more than or equal to median value, otherwise it was defined as low expression.
Table 1. Clinicopathologic features of the patients with PC.
| Pancreatic Cancer Patients (n = 165) | ||
|---|---|---|
| Age | Median age | 65 |
| Range | 35–88 | |
| Gender | Male | 90(54.55%) |
| Female | 75(45.45%) | |
| TMN stage | 0/I/IIA | 0(0.00%)/20(12.12%)/25(15.15%) |
| IIB/III/IV | 113(68.48%)/4(2.42%)/3(1.82%) |
Statistical Analysis
The different expression groups (high vs. low) was defined by the median value of MCMs expression. The association of MCMs expression with clinicopathologic indicators was accessed by Pearson χ2 test. The correction between MCMs expression and overall survival time was evaluated by Kaplan-Meier plots and log-rank tests, as well as disease free time. Independent prognostic factors were recognized byunivariate and multivariate Cox proportional hazard regression models were used to recognize. All statistical analyses were performed by SPSS 20.0 software. Differences between groups were considered significant at P<0.05.
Results
MCMs Expression and Association with Biomarkers for Tumor Biological Behaviors
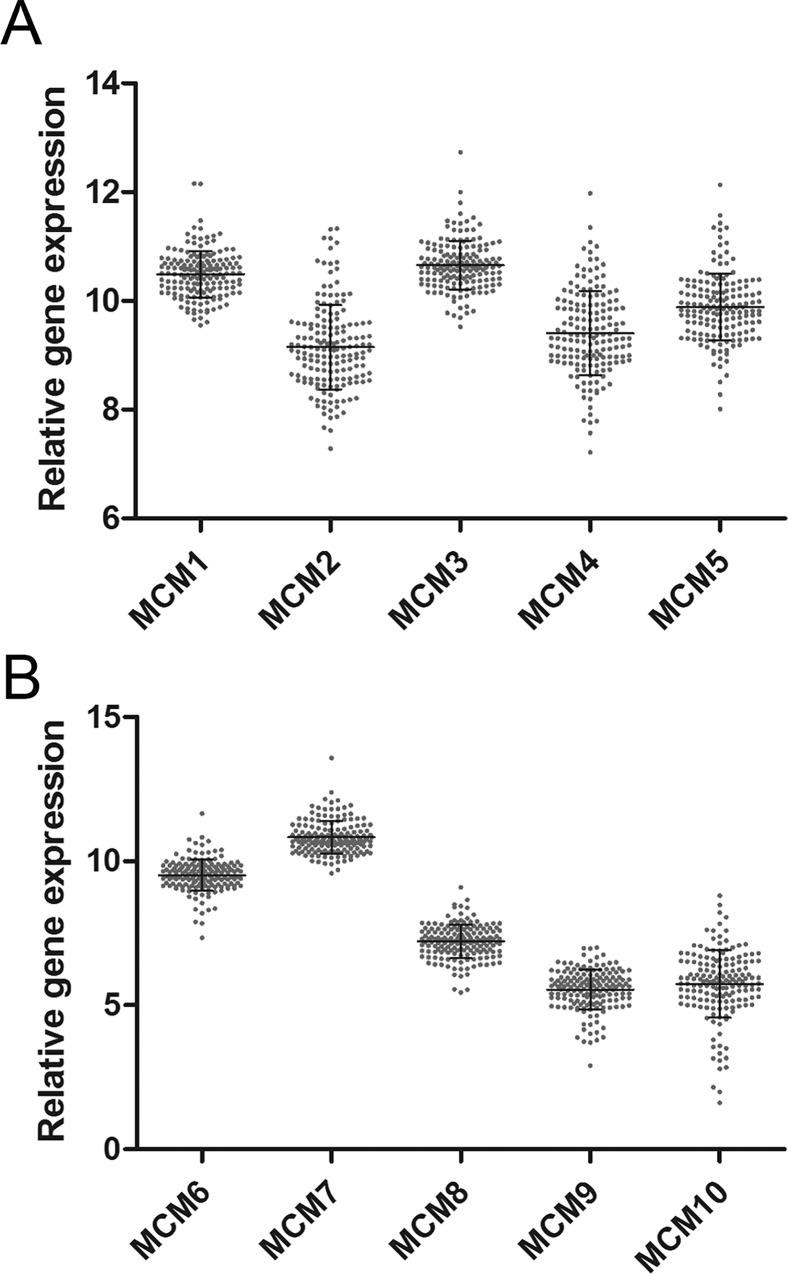
As shown in Fig 1, all MCMs were generally determined in PC samples.
Fig 1. The relative expression of MCM1-10 in 165 PC samples.
(A) The relative expression of MCM1-5 in 165 PC samples. (B) The relative expression of MCM6-10 in 165 PC samples.
As MCMs were reported to participate in the synthesis of DNA, we firstly analyzed the association between MCMs and markers for S phase and proliferation. MCM2-7 complex, MCM8, and MCM10 expression was positively associated with CDK1 and CCNB1 expression which worked as critical proteins in S phase. Almost all assessed molecules were positively corrected with the proliferative biomarker Ki-67 in addition to MCM1. We further assessed the functions of MCMs in cell invasion, migration, and EMT. A few molecules were weakly or moderately associated with MMP2, MMP7, and MMP9; little correlation between MCMs expression and EMT biomarkers expression was observed. Furthermore, we evaluated the roles of MCMs in gemcitabine resistance, and analytic data suggested that most of these molecules over-expression was associated with gemcitabine resistance biomarker RRM1 in different degrees. All results in this aspect were shown in Table 2. These results demonstrated that higher MCMs levels might be responsible for PC cell proliferation and gemcitabine resistance.
Table 2. Association between MCMs and biomarkers for tumor biological behaviors.
| CDK1 | CCNB1 | Ki67 | RRM1 | hENT | MMP2 | MMP7 | MMP9 | ECAD | VIM | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MCM1 | r | -0.162 | -0.276 | -0.104 | -0.088 | 0.211 | 0.357 | 0.062 | 0.112 | -0.291 | 0.36 |
| P | 0.038 | 0.000 | 0.184 | 0.264 | 0.006 | 0.000 | 0.428 | 0.152 | 0.000 | 0.000 | |
| MCM2 | r | 0.639 | 0.675 | 0.606 | 0.666 | -0.126 | 0.114 | 0.036 | 0.222 | 0.026 | 0.015 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.106 | 0.145 | 0.643 | 0.004 | 0.738 | 0.850 | |
| MCM3 | r | 0.460 | 0.439 | 0.485 | 0.408 | 0.114 | 0.009 | 0.102 | 0.129 | -0.055 | 0.028 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.146 | 0.912 | 0.194 | 0.097 | 0.480 | 0.720 | |
| MCM4 | r | 0.660 | 0.681 | 0.678 | 0.667 | -0.086 | 0.130 | 0.124 | 0.163 | 0.184 | -0.123 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.274 | 0.097 | 0.112 | 0.037 | 0.018 | 0.115 | |
| MCM5 | r | 0.419 | 0.382 | 0.409 | 0.341 | 0.024 | 0.106 | 0.082 | 0.401 | -0.197 | 0.200 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.760 | 0.175 | 0.298 | 0.000 | 0.011 | 0.010 | |
| MCM6 | r | 0.613 | 0.544 | 0.609 | 0.564 | -0.029 | 0.423 | 0.235 | 0.330 | -0.129 | 0.271 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.707 | 0.000 | 0.002 | 0.000 | 0.097 | 0.000 | |
| MCM7 | r | 0.453 | 0.574 | 0.390 | 0.429 | -0.124 | -0.148 | -0.141 | 0.060 | -0.006 | -0.181 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.112 | 0.057 | 0.071 | 0.446 | 0.942 | 0.020 | |
| MCM8 | r | 0.509 | 0.469 | 0.542 | 0.472 | -0.119 | 0.035 | -0.053 | 0.179 | -0.008 | -0.149 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.128 | 0.660 | 0.501 | 0.022 | 0.915 | 0.056 | |
| MCM9 | r | 0.067 | -0.152 | 0.190 | -0.081 | 0.054 | 0.135 | 0.287 | 0.113 | 0.044 | -0.066 |
| P | 0.394 | 0.052 | 0.015 | 0.302 | 0.489 | 0.083 | 0.000 | 0.149 | 0.579 | 0.406 | |
| MCM10 | r | 0.889 | 0.731 | 0.857 | 0.569 | -0.038 | 0.230 | 0.339 | 0.321 | 0.172 | -0.013 |
| P | 0.000 | 0.000 | 0.000 | 0.000 | 0.625 | 0.003 | 0.000 | 0.000 | 0.027 | 0.868 | |
Association between MCMs Expression and Clinicopathologic Variables
Analytic results suggested that MCM4 over-expression was detected in patients with aggressive T stage, and MCM9 was over-expressed in patients with lymph node metastasis (Table 3). Furthermore, no other association between these genes expression and clinicopathologic features were observed (Table 3). Results in this aspect suggested that MCM4 and MCM9 could be considered as biomarkers for PC progression.
Table 3. Association between MCMs expression and clinicopathologic variables.
| Histology stage | N stage | T stage | TNM stage | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Well or moderate | Poor | Absent | Present | T1 or T2 | T3 or T4 | I-IIA | IIB-IV | ||
| MCM1 | Low | 54 | 29 | 25 | 58 | 16 | 67 | 23 | 60 |
| High | 62 | 20 | 23 | 59 | 11 | 71 | 22 | 60 | |
| P | 0.138 | 0.770 | 0.309 | 0.899 | |||||
| MCM2 | Low | 64 | 19 | 23 | 60 | 16 | 67 | 23 | 60 |
| High | 52 | 30 | 25 | 57 | 11 | 71 | 22 | 60 | |
| P | 0.054 | 0.695 | 0.309 | 0.899 | |||||
| MCM3 | Low | 61 | 22 | 23 | 60 | 17 | 66 | 23 | 60 |
| High | 55 | 27 | 25 | 57 | 10 | 72 | 22 | 60 | |
| P | 0.367 | 0.695 | 0.150 | 0.899 | |||||
| MCM4 | Low | 63 | 20 | 25 | 58 | 19 | 64 | 24 | 59 |
| High | 53 | 29 | 23 | 59 | 8 | 74 | 21 | 61 | |
| P | 0.113 | 0.770 | 0.023 | 0.634 | |||||
| MCM5 | Low | 62 | 21 | 27 | 56 | 15 | 68 | 25 | 58 |
| High | 54 | 28 | 21 | 61 | 12 | 70 | 20 | 62 | |
| P | 0.214 | 0.328 | 0.551 | 0.409 | |||||
| MCM6 | Low | 59 | 24 | 27 | 56 | 16 | 67 | 26 | 57 |
| High | 57 | 25 | 21 | 61 | 11 | 71 | 19 | 63 | |
| P | 0.825 | 0.328 | 0.309 | 0.240 | |||||
| MCM7 | Low | 63 | 20 | 22 | 61 | 17 | 66 | 22 | 61 |
| High | 53 | 29 | 26 | 56 | 10 | 72 | 23 | 59 | |
| P | 0.113 | 0.462 | 0.150 | 0.824 | |||||
| MCM8 | Low | 63 | 20 | 25 | 58 | 15 | 68 | 23 | 60 |
| High | 53 | 29 | 23 | 59 | 12 | 70 | 22 | 60 | |
| P | 0.113 | 0.770 | 0.551 | 0.899 | |||||
| MCM9 | Low | 64 | 19 | 30 | 53 | 16 | 67 | 27 | 56 |
| High | 52 | 30 | 18 | 64 | 11 | 71 | 18 | 64 | |
| P | 0.054 | 0.045 | 0.309 | 0.127 | |||||
| MCM10 | Low | 61 | 22 | 22 | 60 | 17 | 66 | 22 | 61 |
| High | 55 | 27 | 25 | 57 | 10 | 72 | 23 | 59 | |
| P | 0.367 | 0.695 | 0.150 | 0.824 | |||||
Survival Outcomes and Multivariate Analysis
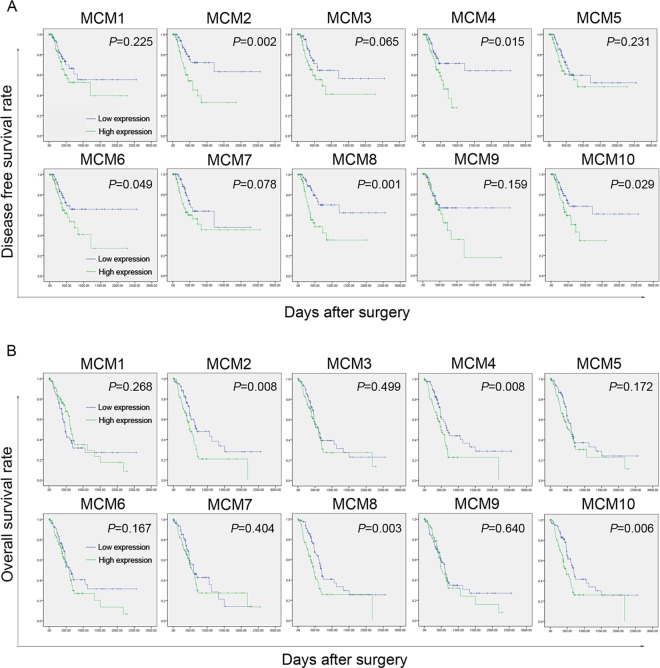
Firstly, the influence of MCMs on disease free survival time was evaluated. A total of 140 patients with disease free survival time related data were enrolled in this section. Analytic results suggested that MCM2, 4, 6, 8, and 10 expression was significantly associated with disease free survival time (Fig 2A). Specifically patients with lower MCM2, 4, 6, 8, 10 levels had longer disease free survival time.And then, the association between MCMs and overall survival time was also assessed. Similar to disease free survival time, lower MCM2, 4, 8, 10 expression was markedly correlated with better overall survival (Fig 2B). Other molecules did not show any correlation with overall survival. Finally, independent prognostic factors was investigated by using Cox proportional hazard regression models. Results of univariate analysis demonstrated that not only some MCM proteins but also some clinicopathologic indicators corrected with overall survival, including T stage, N stage, and TNM stage (Table 4). Multivariate analytic results suggested that MCM8 could independently predict the overall survival of PC (Table 4).
Fig 2. Survival analysis of PC patients correlated with MCM1-10 expression.
(A) The association between MCM1-10 expression and disease free survival time. The higher expression of MCM2, 4, 6, 8, and 10 was significantly associated with shorter disease free survival time. (B) The correlation between MCM1-10 expression and overall survival time. The higher levels of MCM2, 4, 8, and 10 were markedly corrected with poorer outcome.
Table 4. Survival outcomes.
| Univariable analysis | Multivariable analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| MCM1 | 0.784 | 0.509–1.207 | 0.270 | |||
| MCM2 | 1.786 | 1.154–2.734 | 0.009 | 1.322 | 0.700–2.498 | 0.389 |
| MCM3 | 1.16 | 0.754–1.783 | 0.500 | |||
| MCM4 | 1.805 | 1.159–2.809 | 0.009 | 1.240 | 0.684–2.248 | 0.478 |
| MCM5 | 1.349 | 0.876–2.076 | 0.174 | 1.126 | 0.687–1.845 | 0.639 |
| MCM6 | 1.353 | 0.880–2.079 | 0.169 | 0.565 | 0.309–1.035 | 0.065 |
| MCM7 | 1.201 | 0.780–1.850 | 0.405 | |||
| MCM8 | 1.895 | 1.227–2.928 | 0.004 | 1.707 | 1.047–2.863 | 0.032 |
| MCM9 | 1.108 | 0.720–1.707 | 0.640 | |||
| MCM10 | 1.818 | 1.176–2.811 | 0.007 | 1.588 | 0.881–2.863 | 0.124 |
| Age | 1.456 | 0.942–2.250 | 0.091 | 1.448 | 0.917–2.288 | 0.112 |
| Gender | 0.934 | 0.608–1.434 | 0.755 | |||
| HS | 1.556 | 0.990–2.445 | 0.055 | 1.172 | 0.725–1.895 | 0.517 |
| N stage | 2.001 | 1.193–3.358 | 0.009 | 1.169 | 0.358–2.816 | 0.796 |
| T stage | 2.396 | 1.186–4.838 | 0.015 | 1.383 | 0.621–3.080 | 0.427 |
| TMN stage | 2.323 | 1.333–4.046 | 0.003 | 2.052 | 0.557–7.554 | 0.280 |
CI, confidence interval; HR, hazard ratio; HS, Histology stage.
Discussion
PC is a digestive malignancy with extremely high mortality and less therapeutic options. At present the most urgent work for this disease is to investigate novel and efficient biomarkers for prognosis and therapy [19]. In this study, we explored the roles of MCMs in PC for the first time. MCMs abnormal over-expression was significantly associated with PC progression, aggressive PC cell behaviors, poorer disease free survival, and poorer overall survival.
It has been reported thatMCM2-7 complex could work together as DNA helicases to promote the initial stage of DNA replication through participating the formation of pre-replication complex [20–22]. Once binding with some crucial factors in cell cycle (such as Cdc6, Cdt1, and Dbf4/Cdc7), MCM2-7 complex was activated to further enhance DNA synthesis by triggering DNA unwind [23–25]. In other words, MCM2-7 complex could promote cell cycle from S phase to G2/M phase, and finally enhance cell proliferation. Our analytic results also revealed MCM2-7 complex was very important in S phase. The expression of all members of MCM2-7 complex was positively associated with that of S phase biomarkers (CDK1 and CCNB1), as well as cell proliferation biomarker (Ki-67). However, there was no connection between MCM2-7 complex and PC cell migration, invasion, and EMT. Furthermore, MCM2-7 complex members were abnormally up-regulated in various cancers, such as gastric cancer and colon cancer. And they could be regarded as indicators for certain cancer progression and prognosis [26, 27].Our results also suggested that some members of MCM2-7 complex could predict PC progression (MCM4 for T stage). In accordance with the data provided by several past studies in other malignancies, PC patients with higher levels of MCM2, MCM4 and MCM 6 had poorer outcomes (MCM2, 4, and 6 for disease free survival time and MCM2 and 2 for overall survival time). However, none of them was an independent predictor of worse outcome.
There were other four members belong to MCM family, including MCM1, 8, 9, and 10. All of these four molecules were identified as critical factors in the regulation of cell cycle through different mechanisms. MCM1 could activate many immediate-early genes to further perform its function by interacting with serum response element [28, 29].As reported, the interaction between MCM8and CDC6 couldeffectively accelerate pre-replication complex assembly [30], as well as MCM9 and Cdt1 interaction [31]. It has also been reported that MCM8 and MCM9 could form a complex to facilitate homologous recombination which was mediated by RAD51 recruitment at DNA damage sites [32]. MCM10 was reported to promote chromosome replication by enhancing the assembly of the Cdc45-Mcm2-7-GINS complex [33]. And MCM10-RECQ4 interaction was necessary for this process [34]. All reported results above revealed that these four MCMs could also promote cell proliferation by influencing cell cycle. Our analytic data were similar to the report of literatures that MCM8-10 were positively correlated with biomarkers for S phase and cell proliferation in different degrees. Furthermore, many articles focused on these MCMs in malignancies demonstrated that their over-expression was significantly associated with clinicopathologic features for disease progression [17, 35, 36]. However, we just found that MCM9 over-expression was markedly associated lymph node metastasis of PC. Moreover, the prognostic values of MCM8 and MCM10 in PC were similar to MCM2 and MCM4, and MCM8 could be regarded as an independent prognostic factor for PC patients.
Furthermore, latest studies revealed that suppression of the MCMs could sensitize cancer cells to chemotherapeutic agents by inhibiting replicative fork progression, for example, gemcitabine and 5-Fluorouracil [37]. Therefore, we assessed the association between MCMs and gemcitabine resistance biomarkers. Meaningly, MCM2-7 complex expression was significantly associated with gemcitabine resistance biomarker RRM1, especially MCM2 and MCM4 which has high association with RRM1. And then, MCM7 and MCM9 was also moderately associated with RRM1. According to these results, we proposed a hypothesis that gemcitabine resistance might be improved after some MCMs down-regulation.
Conclusion
In conclusion, abnormally up-regulated MCMs in PC were significantly associated with cancer cell proliferation, gemcitabine resistance, disease progression, and poorer outcomes. These results suggested that MCMs were potential biomarkers for PC progression and prognosis, and MCMs could also be considered as targets to improve the efficiency of gemcitabine-based therapy.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The results shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Data Availability
All relevant data applied to perform the analyses in our study was provided in Supporting Information files.
Funding Statement
Funded by grant numbers: 81170336 and 81272239, website: http://www.nsfc.gov.cn/, full name of the funding institutions: National Natural Science Foundation of China, authors who received the funding: Yi Miao. Dr. Yi Miao designed this study.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66(1):7–30. 10.3322/caac.21332 . [DOI] [PubMed] [Google Scholar]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371(22):2140–1. 10.1056/NEJMc1412266 . [DOI] [PubMed] [Google Scholar]
- 3.Tuveson DA, Neoptolemos JP. Understanding metastasis in pancreatic cancer: a call for new clinical approaches. Cell. 2012;148(1–2):21–3. 10.1016/j.cell.2011.12.021 . [DOI] [PubMed] [Google Scholar]
- 4.Qin Y, Zhu W, Xu W, Zhang B, Shi S, Ji S, et al. LSD1 sustains pancreatic cancer growth via maintaining HIF1alpha-dependent glycolytic process. Cancer Lett. 2014;347(2):225–32. 10.1016/j.canlet.2014.02.013 . [DOI] [PubMed] [Google Scholar]
- 5.Zhu Y, Zhang JJ, Xie KL, Tang J, Liang WB, Zhu R, et al. Specific-detection of clinical samples, systematic functional investigations, and transcriptome analysis reveals that splice variant MUC4/Y contributes to the malignant progression of pancreatic cancer by triggering malignancy-related positive feedback loops signaling. J Transl Med. 2014;12:309 10.1186/s12967-014-0309-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zienert E, Eke I, Aust D, Cordes N. LIM-only protein FHL2 critically determines survival and radioresistance of pancreatic cancer cells. Cancer Lett. 2015;364(1):17–24. 10.1016/j.canlet.2015.04.019 . [DOI] [PubMed] [Google Scholar]
- 7.Maiorano D, Lutzmann M, Mechali M. MCM proteins and DNA replication. Curr Opin Cell Biol. 2006;18(2):130–6. 10.1016/j.ceb.2006.02.006 . [DOI] [PubMed] [Google Scholar]
- 8.Pramila T, Miles S, GuhaThakurta D, Jemiolo D, Breeden LL. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002;16(23):3034–45. 10.1101/gad.1034302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen VQ, Co C, Irie K, Li JJ. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol. 2000;10(4):195–205. 10.1016/s0960-9822(00)00337-7 . [DOI] [PubMed] [Google Scholar]
- 10.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106(48):20240–5. 10.1073/pnas.0911500106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder M, Huang XY, Zhang JJ. The minichromosome maintenance proteins 2–7 (MCM2-7) are necessary for RNA polymerase II (Pol II)-mediated transcription. J Biol Chem. 2009;284(20):13466–72. 10.1074/jbc.M809471200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura K, Ishiai M, Horikawa K, Fukagawa T, Takata M, Takisawa H, et al. Mcm8 and Mcm9 form a complex that functions in homologous recombination repair induced by DNA interstrand crosslinks. Mol Cell. 2012;47(4):511–22. 10.1016/j.molcel.2012.05.047 . [DOI] [PubMed] [Google Scholar]
- 13.Traver S, Coulombe P, Peiffer I, Hutchins JR, Kitzmann M, Latreille D, et al. MCM9 Is Required for Mammalian DNA Mismatch Repair. Mol Cell. 2015;59(5):831–9. 10.1016/j.molcel.2015.07.010 . [DOI] [PubMed] [Google Scholar]
- 14.Quan Y, Xia Y, Liu L, Cui J, Li Z, Cao Q, et al. Cell-Cycle-Regulated Interaction between Mcm10 and Double Hexameric Mcm2-7 Is Required for Helicase Splitting and Activation during S Phase. Cell Rep. 2015;13(11):2576–86. 10.1016/j.celrep.2015.11.018 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000;14(8):913–26. [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi J, Kinoshita I, Shimizu Y, Kikuchi E, Takeda K, Aburatani H, et al. Minichromosome maintenance (MCM) protein 4 as a marker for proliferation and its clinical and clinicopathological significance in non-small cell lung cancer. Lung Cancer. 2011;72(2):229–37. 10.1016/j.lungcan.2010.08.020 . [DOI] [PubMed] [Google Scholar]
- 17.Kwok HF, Zhang SD, McCrudden CM, Yuen HF, Ting KP, Wen Q, et al. Prognostic significance of minichromosome maintenance proteins in breast cancer. Am J Cancer Res. 2015;5(1):52–71. [PMC free article] [PubMed] [Google Scholar]
- 18.Hua C, Zhao G, Li Y, Bie L. Minichromosome Maintenance (MCM) Family as potential diagnostic and prognostic tumor markers for human gliomas. BMC Cancer. 2014;14:526 10.1186/1471-2407-14-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain SP. Pancreatic Cancer: Current Progress and Future Challenges. Int J Biol Sci. 2016;12(3):270–2. 10.7150/ijbs.14950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieduszynski CA, Blow JJ, Donaldson AD. The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic Acids Res. 2005;33(8):2410–20. 10.1093/nar/gki539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C, Hong B, Choi JM, Kim Y, Watanabe S, Ishimi Y, et al. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature. 2004;430(7002):913–7. 10.1038/nature02813 . [DOI] [PubMed] [Google Scholar]
- 22.Saxena S, Yuan P, Dhar SK, Senga T, Takeda D, Robinson H, et al. A dimerized coiled-coil domain and an adjoining part of geminin interact with two sites on Cdt1 for replication inhibition. Mol Cell. 2004;15(2):245–58. 10.1016/j.molcel.2004.06.045 . [DOI] [PubMed] [Google Scholar]
- 23.Sun J, Evrin C, Samel SA, Fernandez-Cid A, Riera A, Kawakami H, et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20(8):944–51. 10.1038/nsmb.2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Cid A, Riera A, Tognetti S, Herrera MC, Samel S, Evrin C, et al. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell. 2013;50(4):577–88. 10.1016/j.molcel.2013.03.026 . [DOI] [PubMed] [Google Scholar]
- 25.Ramer MD, Suman ES, Richter H, Stanger K, Spranger M, Bieberstein N, et al. Dbf4 and Cdc7 proteins promote DNA replication through interactions with distinct Mcm2-7 protein subunits. J Biol Chem. 2013;288(21):14926–35. 10.1074/jbc.M112.392910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giaginis C, Georgiadou M, Dimakopoulou K, Tsourouflis G, Gatzidou E, Kouraklis G, et al. Clinical significance of MCM-2 and MCM-5 expression in colon cancer: association with clinicopathological parameters and tumor proliferative capacity. Dig Dis Sci. 2009;54(2):282–91. 10.1007/s10620-008-0305-z . [DOI] [PubMed] [Google Scholar]
- 27.Giaginis C, Giagini A, Tsourouflis G, Gatzidou E, Agapitos E, Kouraklis G, et al. MCM-2 and MCM-5 expression in gastric adenocarcinoma: clinical significance and comparison with Ki-67 proliferative marker. Dig Dis Sci. 2011;56(3):777–85. 10.1007/s10620-010-1348-5 . [DOI] [PubMed] [Google Scholar]
- 28.Abraham DS, Vershon AK. N-terminal arm of Mcm1 is required for transcription of a subset of genes involved in maintenance of the cell wall. Eukaryot Cell. 2005;4(11):1808–19. 10.1128/EC.4.11.1808-1819.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wynne J, Treisman R. SRF and MCM1 have related but distinct DNA binding specificities. Nucleic Acids Res. 1992;20(13):3297–303. 10.1093/nar/20.13.3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volkening M, Hoffmann I. Involvement of human MCM8 in prereplication complex assembly by recruiting hcdc6 to chromatin. Mol Cell Biol. 2005;25(4):1560–8. 10.1128/MCB.25.4.1560-1568.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutzmann M, Mechali M. MCM9 binds Cdt1 and is required for the assembly of prereplication complexes. Mol Cell. 2008;31(2):190–200. 10.1016/j.molcel.2008.07.001 . [DOI] [PubMed] [Google Scholar]
- 32.Park J, Long DT, Lee KY, Abbas T, Shibata E, Negishi M, et al. The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination. Mol Cell Biol. 2013;33(8):1632–44. 10.1128/MCB.01503-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci U S A. 2009;106(37):15628–32. 10.1073/pnas.0908039106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28(19):3005–14. 10.1038/emboj.2009.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das M, Prasad SB, Yadav SS, Govardhan HB, Pandey LK, Singh S, et al. Over expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesis. PLoS One. 2013;8(7):e69607 10.1371/journal.pone.0069607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padmanabhan V, Callas P, Philips G, Trainer TD, Beatty BG. DNA replication regulation protein Mcm7 as a marker of proliferation in prostate cancer. J Clin Pathol. 2004;57(10):1057–62. 10.1136/jcp.2004.016436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryant VL, Elias RM, McCarthy SM, Yeatman TJ, Alexandrow MG. Suppression of Reserve MCM Complexes Chemosensitizes to Gemcitabine and 5-Fluorouracil. Mol Cancer Res. 2015;13(9):1296–305. 10.1158/1541-7786.MCR-14-0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data applied to perform the analyses in our study was provided in Supporting Information files.