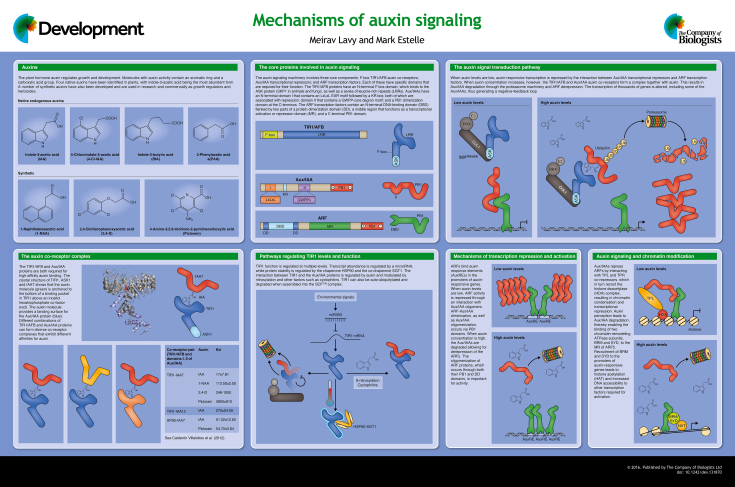
Abstract
The plant hormone auxin triggers complex growth and developmental processes. Its underlying molecular mechanism of action facilitates rapid switching between transcriptional repression and gene activation through the auxin-dependent degradation of transcriptional repressors. The nuclear auxin signaling pathway consists of a small number of core components. However, in most plants each component is represented by a large gene family. The modular construction of the pathway can thus produce diverse transcriptional outputs depending on the cellular and environmental context. Here, and in the accompanying poster, we outline the current model for TIR1/AFB-dependent auxin signaling with an emphasis on recent studies.
KEY WORDS: Auxin, Cellular response, Transcriptional response
Summary: This poster article provides an overview of the auxin signal transduction pathway, highlighting how it facilitates rapid switching between transcriptional repression and gene activation.
Introduction
Auxin is a plant hormone that has a role in most aspects of plant growth and development. Four native auxins have been identified in plants, with indole-3-acetic acid (IAA) being the most abundant form. Auxin distribution within plant tissues is regulated by biosynthesis, inactivation and transport pathways (Zazimalova et al., 2010; Zhao, 2010). Upon perception in the nucleus, auxin can trigger broad and specific transcriptional responses. The core components of the auxin signaling machinery belong to three protein families: the F-box TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX PROTEIN (TIR1/AFB) auxin co-receptors, the Auxin/INDOLE-3-ACETIC ACID (Aux/IAA) transcriptional repressors, and the AUXIN RESPONSE FACTOR (ARF) transcription factors. Auxin promotes an interaction between TIR1/AFB and Aux/IAA proteins, resulting in degradation of the Aux/IAAs and the release of ARF repression (Wang and Estelle, 2014; Salehin et al., 2015).
Gene expression associated with ARF activation has been implicated in diverse processes in land plants, including tropic responses and the establishment of polarity, as well as embryogenesis and organogenesis in flowering plants, and both gametophyte and sporophyte development in nonflowering plants (De Smet and Jurgens, 2007; Banks, 2009; Moller and Weijers, 2009; Prigge et al., 2010; Vernoux et al., 2010; Bennett et al., 2014; Flores-Sandoval et al., 2015; Kato et al., 2015). At the cellular level, auxin affects all aspects of cellular growth, including cell elongation, cell division and differentiation (Perrot-Rechenmann, 2010; Takatsuka and Umeda, 2014).
The repression of auxin-induced genes
Auxin functions by triggering genome-wide transcriptional responses via its effects on ARF activity. At low auxin levels, Aux/IAA transcriptional repressors interact with ARFs and repress their activity. However, in the presence of auxin the TIR1/AFB proteins bind to Aux/IAA transcriptional repressors and mediate their polyubiquitylation and subsequent proteasomal degradation. Another important aspect of the pathway is the rapid induction of auxin-responsive genes, including Aux/IAAs and the GH3 family of auxin homeostasis modulators, triggering negative-feedback loops (Benjamins and Scheres, 2008).
The Aux/IAA transcriptional repressors interact with proteins in the auxin signaling pathway through a number of protein domains. In general, the Aux/IAAs comprise three functional domains: a leucine repeat EAR motif within domain I (Kagale and Rozwadowski, 2011), an internal domain II that contains a GWPP-core degron motif, and a C-terminal region that forms a type I/II Phox and Bem1 (PB1) domain (Guilfoyle, 2015). The PB1 domain facilitates interactions with ARF proteins as well as self-dimerization (Vernoux et al., 2011), while the degron motif is required for interaction with TIR1/AFB proteins and therefore determines Aux/IAA stability. Domain I functions as a repression motif by recruiting transcriptional co-repressors. Although domain II is necessary for the interaction with TIR1/AFBs, other sequences outside of domain II, including an N-terminal lysine-arginine (KR) motif, contribute to TIR1/AFB binding and/or Aux/IAA degradation (Dreher et al., 2006; Calderon Villalobos et al., 2012; Moss et al., 2015).
Auxin perception
Polyubiquitylation of the Aux/IAA transcriptional repressors requires an E3 ubiquitin ligase SCFTIR/AFB complex. SCF complexes consist of an F-box protein that provides substrate recognition, an ARABIDOPSIS SKP1 HOMOLOG1 (ASK1) adaptor (SKP1 in animals and fungi), the scaffold protein CULLIN1 (CUL1), and RING-BOX PROTEIN1 (RBX1) that promotes transfer of ubiquitin molecules to the substrate. The SCFTIR/AFB complex binds the Aux/IAA substrate in an auxin-dependent manner through the TIR1 or AFB F-box protein (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Biochemical studies have revealed that both the TIR1/AFB and the Aux/IAA protein are required for high-affinity auxin binding, indicating that the two proteins can be referred to as auxin co-receptors (Calderon Villalobos et al., 2012). TIR1/AFB proteins are composed of an F-box motif and a leucine-rich repeat (LRR) domain. Crystallographic studies have demonstrated that an auxin molecule is anchored to the bottom of a hydrophobic binding pocket in TIR1 formed by the LRR domain, providing a binding surface for an Aux/IAA protein. In turn, the Aux/IAA protein binds to the upper part of the auxin-binding pocket through its degron motif. Thus, auxin functions as a molecular ‘glue’ that stabilizes the TIR1/AFB-Aux/IAA interaction (Tan et al., 2007). This interaction is also facilitated by other factors. For example, S-nitrosylation of conserved cysteine residues in the LRR of TIR1 can enhance the interaction between TIR1 and Aux/IAA proteins (Terrile et al., 2012). In addition, the cyclophilin LATERAL ROOTLESS2 (LRT2) was recently shown to facilitate TIR1/AFB-Aux/IAA interaction in rice by allowing for correct folding of the degron motif-containing domain (Jing et al., 2015).
Flowering plants possess several related TIR1/AFB auxin co-receptors, and the number of Aux/IAA family members is also large. In vitro auxin binding assays have revealed that different TIR1/AFB-Aux/IAA co-receptor pairs have different affinities for auxin. Whereas the Arabidopsis proteins TIR1 and IAA7 bind the natural auxin with high affinity, the TIR1-IAA12 complex has an IAA affinity that is one order of magnitude lower (Calderon Villalobos et al., 2012). In addition, in vivo experiments, as well as studies using a reconstituted auxin response pathway in yeast, have shown that different Aux/IAAs have very different degradation rates (Havens et al., 2012; Dreher et al., 2006). Notably, AFB5-IAA7 has a similar affinity for IAA as TIR1-IAA7, but exhibits a much higher affinity for the synthetic auxin picloram. These findings, together with results from earlier genetic studies, indicate that the AFB5 protein is a major target of picloram and structurally related herbicides (Walsh et al., 2006; Calderon Villalobos et al., 2012). The existence of different co-receptor complexes with different affinities for auxin in diverse tissues and developmental stages might contribute to context-specific auxin responses.
TIR1/AFB levels are regulated by multiple post-transcriptional and post-translational mechanisms, adding additional regulatory layers to auxin receptor function. For example, the microRNA miR393 targets TIR1, AFB2 and AFB3, and it can reduce their transcript levels in response to different environmental cues, including pathogens and abiotic stress (Windels and Vazquez, 2011; Iglesias et al., 2014). Recent studies show that post-translational changes in the stability of TIR1 are associated with its autocatalytic degradation as well as HSP90 activity. When assembled into the SCF, TIR1 is relatively unstable. Correspondingly, mutations in the region encoding the F-box domain of TIR1/AFB, as well as amino acid variations that untether TIR1/AFB from CUL1, result in increased stability of TIR1/AFBs (Yu et al., 2015). TIR1 is also a client of the HEAT SHOCK FACTOR 90 (HSP90)-SUPPRESSOR OF G2 ALLELE SKP1 (SGT1) co-chaperone complex. TIR1 interacts directly with both HSP90 and SGT1, which in turn act to increase TIR1 stability. This effect of the HSP90-SGT1 co-chaperone complex on TIR1 levels is implicated in the environmental response to an increase in ambient temperature (Wang et al., 2016).
Auxin-dependent regulation of transcription
Auxin perception by the auxin receptor complex and the subsequent degradation of the Aux/IAAs allow ARF-mediated transcriptional responses. ARF transcription factors bind to the promoters of auxin-responsive genes through cis-regulatory auxin response elements (AuxREs). The TGTCTC sequence was first identified in the promoter of the soybean GH3 gene as a functional AuxRE and has been commonly used in auxin-responsive reporters (Ulmasov et al., 1995). However, whereas the TGTCTC canonical motif is not present in every promoter of auxin-responsive genes, the core element TGTC appears to be required for the recruitment of ARFs. In addition, recent structural analyses have revealed that Arabidopsis ARF1 and ARF5 preferentially interact with a TGTCCG motif (Boer et al., 2014), which led to the development of a new set of genetic tools (Liao et al., 2015).
ARFs contain an N-terminal DNA-binding domain (DBD), a variable middle region (MR) and, similar to Aux/IAAs, a C-terminal type I/II PB1 dimerization domain. Based on bioinformatic analyses of their MR domains and their behavior in transient protoplast assays, ARFs can be characterized as transcriptional activators or repressors (Ulmasov et al., 1999; Tiwari et al., 2003). ARFs can dimerize through their PB1 domain, as well as through an N-terminal motif formed by the flanking regions of their designated DBDs, as was recently shown (Boer et al., 2014). The same study further demonstrated that different ARF monomers bind to similar elements, but that as homodimers, as represented by ARF1 and ARF5, their preferred spacing between AuxREs varies. This variability of ARF dimers forming on different promoters can contribute to complex transcriptional responses.
Additional structural studies have indicated that the C-terminal regions of both ARF and Aux/IAAs adopt a type I/II PB1 domain (Korasick et al., 2015), which is characterized by two opposing electrostatic faces that allow for directional protein interactions and the formation of high-order oligomers. Indeed, genetic studies verify that the oligomerization of Aux/IAAs is important for the efficient repression of ARF activity (Korasick et al., 2014). The PB1 domain facilitates both ARF-ARF and Aux-Aux dimerization, as well as the heterodimerization of ARF-Aux/IAA. Structural studies of ARF5 and IAA17 demonstrate that their heterodimerization is stronger than self-interactions (Han et al., 2014). The formation of favored ARF-Aux/IAA dimers may destabilize ARF interactions with DNA and contribute to ARF repression in the absence of auxin. Furthermore, the level of transcriptional repression could be tuned by the composition of oligomers formed on promoters of auxin-responsive genes. Interestingly, some ARFs and Aux/IAAs were shown to interact with other transcriptional regulators, suggesting additional layers of transcriptional regulation (Wang and Estelle, 2014).
Auxin-regulated chromatin switches
The recruitment of Aux/IAAs to the promoters of auxin-responsive genes by activating ARFs results in gene repression. This repression mechanism involves chromatin modifications that result in decreased accessibility of target genes. Aux/IAA proteins can interact with the TOPLESS (TPL) and TPL-related (TPR) co-repressor proteins through their EAR motif. In turn, TPL and TPRs interact with histone deacetylases that catalyze the removal of acetyl groups from histone proteins, leading to DNA condensation and transcriptional repression (Szemenyei et al., 2008; Kagale and Rozwadowski, 2011).
A new paradigm of an auxin-mediated chromatin switch has also emerged. It was recently shown that, in flower primordia, ARF5 interacts with BRAHMA (BRM) and SPLAYED (SYD) (Wu et al., 2015), both of which are chromatin-remodeling ATPase subunits of the SWI/SNF complex (Wagner and Meyerowitz, 2002; Farrona et al., 2004). Through their interaction with ARF5, BRM and SYD are recruited to promoters of auxin-responsive genes involved in flower formation. As a result, the accessibility of DNA to additional transcription factors is increased, leading to induction of the corresponding target genes. This study further showed that Aux/IAAs prevent the association of BRM and SYD with gene promoters. Thus, the switching between gene repression and gene activation is enabled by auxin and Aux/IAA degradation. Although the involvement of the SWI/SNF complex in other auxin-regulated developmental processes has not yet been shown, it is possible that ARF5, as well as other ARFs, recruits BRM and SYD to additional target genes.
Perspectives
The elucidation of a complete nuclear auxin signaling pathway has provided the link between auxin perception and a transcriptional response. However, how the hormonal signal is differentially interpreted, giving rise to complex and context-dependent responses, is still not clear. Recent studies are beginning to bridge the gap between the seemingly simple auxin signaling pathway and the resulting complex responses. The answers appear to lie, at least in part, in the size of the protein families that function in auxin signaling, as well as in their ability to oligomerize. Combinatorial interactions between members of the TIR1/AFB and Aux/IAA co-receptor families may thus result in a broad range of auxin sensitivity. Similarly, levels of gene activation and repression can be tuned by the action of different Aux/IAA and ARF oligomers. Additional recent advances include the identification of new players affecting auxin signaling, including chaperones and chromatin modifiers, placing the auxin signal transduction pathway in a broader context of cellular events. New structural and biochemical insights into the complex interactions between core auxin signaling components have also provided a framework for future studies of their biological relevance. Such studies are likely to include a combination of genetic and high-throughput approaches to facilitate correlations between individual signaling complexes and the corresponding specific outputs.
Auxin has been implicated in developmental switches as well as in transient and dynamic cellular responses. Whether these two distinctive roles differ mechanistically is unclear. The studies described here imply that gene expression is regulated by chromatin-remodeling factors, as well as by differential interactions between transcription factors. Therefore, a better understanding of how auxin-regulated transcriptional changes reflect the chromatin state is a further interesting research goal. Finally, the importance of additional factors and other pathways that interact with the auxin signaling pathway is apparent. Gaining a better understanding of how these interactions are regulated and how they are affected by developmental and environmental signals will be required. An increased understanding of the principles of auxin signaling will clearly improve our ability to dissect complex auxin response networks in different developmental contexts as well as in different model and crop plants.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Research in the authors' laboratory is supported by a grant from the National Institutes of Health [GM43644 to M.E.] and grants from the Gordon and Betty Moore Foundation (to M.E.) and the Howard Hughes Medical Institute (to M.E.). Deposited in PMC for release after 12 months.
Development at a Glance
A high-resolution version of the poster is available for downloading in the online version of this article at http://dev.biologists.org/content/143/18/3226/F1.poster.jpg
References
- Banks J. A. (2009). Selaginella and 400 million years of separation. Annu. Rev. Plant Biol. 60, 223-238. 10.1146/annurev.arplant.59.032607.092851 [DOI] [PubMed] [Google Scholar]
- Benjamins R. and Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59, 443-465. 10.1146/annurev.arplant.58.032806.103805 [DOI] [PubMed] [Google Scholar]
- Bennett T. A., Liu M. M., Aoyama T., Bierfreund N. M., Braun M., Coudert Y., Dennis R. J., O'Connor D., Wang X. Y., White C. D. et al. (2014). Plasma membrane-targeted PIN proteins drive shoot development in a moss. Curr. Biol. 24, 2776-2785. 10.1016/j.cub.2014.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer D. R., Freire-Rios A., van den Berg W. A. M., Saaki T., Manfield I. W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J. M., de Vries S. C., Solano R. et al. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577-589. 10.1016/j.cell.2013.12.027 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos L. I. A., Lee S., De Oliveira C., Ivetac A., Brandt W., Armitage L., Sheard L. B., Tan X., Parry G., Mao H. et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477-485. 10.1038/nchembio.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I. and Jürgens G. (2007). Patterning the axis in plants--auxin in control. Curr. Opin. Genet. Dev. 17, 337-343. 10.1016/j.gde.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S. and Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441-445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dreher K. A., Brown J., Saw R. E. and Callis J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18, 699-714. 10.1105/tpc.105.039172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S., Hurtado L., Bowman J. L. and Reyes J. C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131, 4965-4975. 10.1242/dev.01363 [DOI] [PubMed] [Google Scholar]
- Flores-Sandoval E., Eklund D. M. and Bowman J. L. (2015). A simple auxin transcriptional response system regulates multiple morphogenetic processes in the liverwort marchantia polymorpha. PLoS Genet. 11, e1005207 10.1371/journal.pgen.1005207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T. J. (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27, 33-43. 10.1105/tpc.114.132753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Park Y., Kim I., Kim E.-H., Yu T.-K., Rhee S. and Suh J.-Y. (2014). Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc. Natl. Acad. Sci. USA 111, 18613-18618. 10.1073/pnas.1419525112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens K. A., Guseman J. M., Jang S. S., Pierre-Jerome E., Bolten N., Klavins E. and Nemhauser J. L. (2012). A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 160, 135-142. 10.1104/pp.112.202184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M. J., Terrile M. C., Windels D., Lombardo M. C., Bartoli C. G., Vazquez F., Estelle M. and Casalongué C. A. (2014). MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS ONE 9, e107678 10.1371/journal.pone.0107678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H., Yang X., Zhang J., Liu X., Zheng H., Dong G., Nian J., Feng J., Xia B., Qian Q. et al. (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 6, 7395 10.1038/ncomms8395 [DOI] [PubMed] [Google Scholar]
- Kagale S. and Rozwadowski K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141-146. 10.4161/epi.6.2.13627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Ishizaki K., Kouno M., Shirakawa M., Bowman J. L., Nishihama R. and Kohchi T. (2015). Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in marchantia polymorpha. PLoS Genet. 11, e1005084 10.1371/journal.pgen.1005084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S. and Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446-451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Korasick D. A., Westfall C. S., Lee S. G., Nanao M. H., Dumas R., Hagen G., Guilfoyle T. J., Jez J. M. and Strader L. C. (2014). Molecular basis for auxin response factor protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 111, 5427-5432. 10.1073/pnas.1400074111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D. A., Jez J. M. and Strader L. C. (2015). Refining the nuclear auxin response pathway through structural biology. Curr. Opin. Plant Biol. 27, 22-28. 10.1016/j.pbi.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C. Y., Smet W., Brunoud G., Yoshida S., Vernoux T. and Weijers D. (2015). Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207-210. 10.1038/nmeth.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller B. and Weijers D. (2009). Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 1, a001545 10.1101/cshperspect.a001545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. L., Mao H., Guseman J. M., Hinds T. R., Hellmuth A., Kovenock M., Noorassa A., Lanctot A., Villalobos L. I. A. C., Zheng N. et al. (2015). Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol. 169, 803-813. 10.1104/pp.15.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. (2010). Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2, a001446 10.1101/cshperspect.a001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M. J., Lavy M., Ashton N. W. and Estelle M. (2010). Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr. Biol. 20, 1907-1912. 10.1016/j.cub.2010.08.050 [DOI] [PubMed] [Google Scholar]
- Salehin M., Bagchi R. and Estelle M. (2015). SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27, 9-19. 10.1105/tpc.114.133744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M. and Long J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384-1386. 10.1126/science.1151461 [DOI] [PubMed] [Google Scholar]
- Takatsuka H. and Umeda M. (2014). Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 65, 2633-2643. 10.1093/jxb/ert485 [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L. I. A., Sharon M., Zheng C., Robinson C. V., Estelle M. and Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640-645. 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Terrile M. C., París R., Calderón-Villalobos L. I. A., Iglesias M. J., Lamattina L., Estelle M. and Casalongué C. A. (2012). Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis transport inhibitor response 1 auxin receptor. Plant J. 70, 492-500. 10.1111/j.1365-313X.2011.04885.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S. B., Hagen G. and Guilfoyle T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533-543. 10.1105/tpc.008417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Liu Z. B., Hagen G. and Guilfoyle T. J. (1995). Composite structure of auxin response elements. Plant Cell 7, 1611-1623. 10.1105/tpc.7.10.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G. and Guilfoyle T. J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844-5849. 10.1073/pnas.96.10.5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., Besnard F. and Traas J. (2010). Auxin at the shoot apical meristem. Cold Spring Harb. Perspect. Biol. 2, a001487 10.1101/cshperspect.a001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., Brunoud G., Farcot E., Morin V., Van den Daele H., Legrand J., Oliva M., Das P., Larrieu A., Wells D. et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7, 508 10.1038/msb.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. and Meyerowitz E. M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12, 85-94. 10.1016/S0960-9822(01)00651-0 [DOI] [PubMed] [Google Scholar]
- Walsh T. A., Neal R., Merlo A. O., Honma M., Hicks G. R., Wolff K., Matsumura W. and Davies J. P. (2006). Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 142, 542-552. 10.1104/pp.106.085969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. and Estelle M. (2014). Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 21, 51-58. 10.1016/j.pbi.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang Y., Kieffer M., Yu H., Kepinski S. and Estelle M. (2016). HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 7, 10269 10.1038/ncomms10269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels D. and Vazquez F. (2011). miR393: integrator of environmental cues in auxin signaling? Plant Signal. Behav. 6, 1672-1675. 10.4161/psb.6.11.17900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. F., Yamaguchi N., Xiao J., Bargmann B., Estelle M., Sang Y. and Wagner D. (2015). Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4, e09269 10.7554/elife.09269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Zhang Y., Moss B. L., Bargmann B. O., Wang R., Prigge M., Nemhauser J. L. and Estelle M. (2015). Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat. Plants 1, 14030 10.1038/nplants.2014.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazimalova E., Murphy A. S., Yang H., Hoyerova K. and Hosek P. (2010). Auxin transporters--why so many? Cold Spring Harb. Perspect. Biol. 2, a001552 10.1101/cshperspect.a001552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49-64. 10.1146/annurev-arplant-042809-112308 [DOI] [PMC free article] [PubMed] [Google Scholar]