ABSTRACT
After being severed from the cell body, axons initiate an active degeneration program known as Wallerian degeneration. Although dendrites also seem to have an active injury-induced degeneration program, no endogenous regulators of this process are known. Because microtubule disassembly has been proposed to play a role in both pruning and injury-induced degeneration, we used a Drosophila model to identify microtubule regulators involved in dendrite degeneration. We found that, when levels of fidgetin were reduced using mutant or RNA interference (RNAi) strategies, dendrite degeneration was delayed, but axon degeneration and dendrite pruning proceeded with normal timing. We explored two possible ways in which fidgetin could promote dendrite degeneration: (1) by acting constitutively to moderate microtubule stability in dendrites, or (2) by acting specifically after injury to disassemble microtubules. When comparing microtubule dynamics and stability in uninjured neurons with and without fidgetin, we could not find evidence that fidgetin regulated microtubule stability constitutively. However, we identified a fidgetin-dependent increase in microtubule dynamics in severed dendrites. We conclude that fidgetin acts after injury to promote disassembly of microtubules in dendrites severed from the cell body.
KEY WORDS: Wallerian degeneration, Dendrite degeneration, Dendrite pruning, Microtubule severing
Highlighted Article: Dendrite degeneration is analogous to axon degeneration, but no regulators of this process had previously been identified. We find that fidgetin helps take dendrites apart by severing microtubules.
INTRODUCTION
During development axons and dendrites grow extensively, and are often later trimmed back to generate specific mature connections (Luo and O'Leary, 2005; Schuldiner and Yaron, 2015). A large-scale version of this pruning process occurs during metamorphosis in some insects as larval body parts are removed and adult ones are innervated (Luo and O'Leary, 2005; Yu and Schuldiner, 2014). Removal of regions of axons and dendrites is also important after neuronal damage. For example, after irreparable axonal damage, distal regions of the axon are cleared by a process known as Wallerian degeneration (Coleman and Freeman, 2010; Conforti et al., 2014; Gerdts et al., 2016).
Developmental pruning and injury-induced degeneration proceed through morphologically similar steps. For example, formation of filopodia-like structures occurs relatively early during dendrite pruning (Williams and Truman, 2005a) and dendrite degeneration (Tao and Rolls, 2011) in Drosophila. Membrane internalization is also observed during injury-induced degeneration and pruning (Tao and Rolls, 2011). Microtubule disassembly seems to be a relatively early step in dendrite pruning (Williams and Truman, 2005a) and axon degeneration (MacDonald et al., 2006). These early rearrangements are followed by beading and clearance of debris by neighboring cells in all cases (MacDonald et al., 2006; Tao and Rolls, 2011; Williams and Truman, 2005a).
As developmental pruning and injury-induced degeneration proceed through a defined series of morphological changes, they are thought to operate as programmed cellular pathways analogous to apoptosis. If they are controlled processes like apoptosis, then specific regulators should be involved. Indeed, regulators of dendrite pruning have been identified in Drosophila. These include caspases (Kuo et al., 2006; Williams et al., 2006), Sox14 and Mical (Kirilly et al., 2009), IK2 and katanin p60-like 1 (Lee et al., 2009), and headcase (Loncle and Williams, 2012). Regulators of axon degeneration have also been identified, and in many cases have been shown to function in both Drosophila and mammals. Endogenous factors that control Wallerian axon degeneration include MORN4 (Bhattacharya et al., 2012), Sarm1 (Osterloh et al., 2012) and DLK (also known as wallenda, wnd) (Miller et al., 2009). Axon degeneration after growth factor withdrawal seems to involve different regulators, including caspases (Nikolaev et al., 2009; Schoenmann et al., 2010), and thus might be mechanistically more similar to dendrite pruning during insect metamorphosis than injury-induced axon degeneration.
In contrast to dendrite pruning and axon degeneration, no factors required for dendrite degeneration are known. In addition, it is unclear whether the three processes, pruning, axon degeneration and dendrite degeneration, are controlled by completely independent sets of machinery, or whether a core set of disassembly factors might be shared.
In order to identify regulators of dendrite disassembly after injury, and to gain insight into whether a core set of downstream components might act during pruning and degeneration, we focused on microtubule destabilization. Microtubule loss has been observed as an early event during both injury-induced axon degeneration (MacDonald et al., 2006) and dendrite pruning (Williams and Truman, 2005b) in Drosophila, and has also been reported to be important for axon degeneration induced by injury and growth factor withdrawal in mammals (Maor-Nof et al., 2013). Microtubule disassembly is therefore a good candidate for a convergence point of the pathways that regulate the different types of degeneration. To identify microtubule regulators involved in degeneration or pruning, we used a Drosophila model system.
Drosophila dendritic arborization (da) neurons are sensory neurons present in the body wall. They are grouped into different classes based on dendrite branching patterns (Grueber et al., 2002), and the most complex class, class IV, is responsible for nociception (Hwang et al., 2007). During pupariation, the dendrites of class IV neurons are pruned completely while the cell body and axons remain in place (Williams and Truman, 2005a). At the end of the pupal stage, dendrites regrow into the adult body wall (Shimono et al., 2009; Yasunaga et al., 2010). These cells are thus a useful model for studying large-scale developmental dendrite pruning. We have also shown that these cells initiate stereotyped degeneration after axon and dendrite injury (Tao and Rolls, 2011). In the same cell type, we can thus compare injury-induced axon and dendrite degeneration as well as developmental dendrite pruning. We used this system to screen known regulators of microtubule stability for roles in any of the three processes. We identified fidgetin (CG3326 in Drosophila) as a specific regulator of dendrite degeneration, and show that it acts to increase microtubule dynamics after dendrites are severed from the cell body.
RESULTS
Identification of microtubule regulators involved in degeneration
To identify microtubule regulators involved in pruning or injury-induced degeneration, we conducted a three-pronged candidate screen. The screen was designed to identify cell-autonomous regulators of degeneration that act in neurons; RNA hairpins targeting each candidate were expressed only in the neurons of interest. Assays for injury-induced axon degeneration, injury-induced dendrite degeneration and dendrite pruning were conducted in the class IV da neuron ddaC. Neurons were visualized with a membrane-localized GFP (mCD8–GFP) expressed in class IV neurons with ppk-Gal4. Precise severing of axons or dendrites was accomplished with a pulsed UV laser as in previous studies (Stone et al., 2014, 2010; Tao and Rolls, 2011). Complete absence of all dendrite remnants distal to the cut site was scored 18 h after injury (Fig. 1B). Because clearance of axon remnants was more variable than clearance of dendrite remnants, we scored axon beading at 12 h after injury (Fig. 1A), rather than complete absence of debris. Dendrite pruning was assayed by scoring dendrite remnants 18 h after the onset of pupariation (Fig. 1C). The timing of events during these three processes has been described in more detail previously (Tao and Rolls, 2011).
Fig. 1.
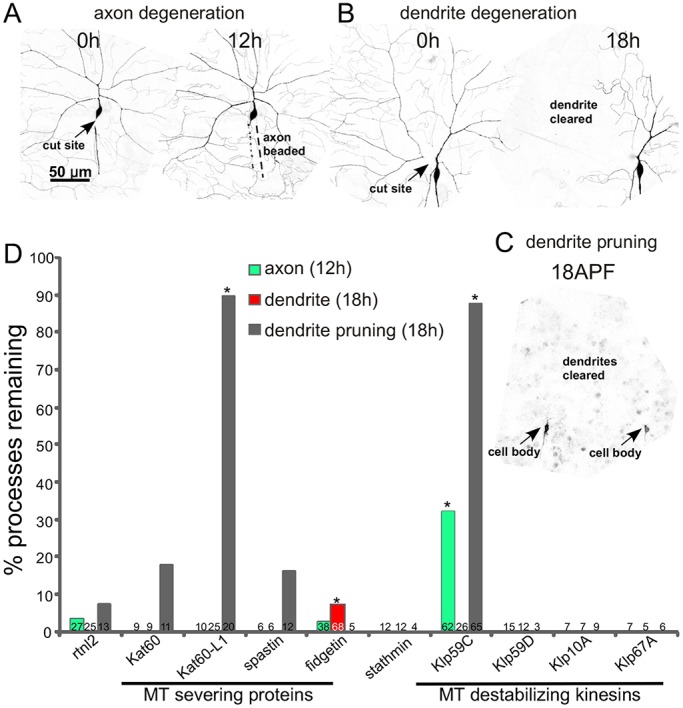
Identification of microtubule regulators required for degeneration and/or pruning. Class IV ddaC neurons were labeled with mCD8–GFP under the control of ppk-Gal4. (A) The assay for injury-induced axon degeneration is shown. 12 h after axon severing with a pulsed UV laser, axons were scored as beaded (normal degeneration) or continuous. (B) The assay for dendrite degeneration is shown. A ddaC dendrite was severed near its base with a pulsed UV laser at 0 h. The neuron was scored for complete clearance of the dendrite 18 h later. (C) Dendrite pruning was assayed 18 h after puparium formation (APF). An example image from a pupa is shown. Two adjacent cell bodies can be seen (arrows). Complete clearance of dendrites was scored as normal degeneration. (D) Tester animals (UAS-dicer2; ppk-Gal4, UAS-mCD8-GFP) were crossed with UAS-RNAi lines to target the microtubule regulators indicated. We used an RNAi line targeting rtnl2 as a control as it has never generated a phenotype in any assay we have performed. Kat60-L1 dendrite data was previously reported (Tao and Rolls, 2011), but is shown here for completeness. *P<0.05 compared to the appropriate rtnl2 control, all unmarked columns were not significantly different from controls (two-tailed Fisher's exact test). Numbers on each column are the number of animals tested for that condition.
To reduce levels of candidate microtubule regulators, a tester fly line (UAS-dicer2; ppk-Gal4, UAS-mCD8-GFP) was crossed with lines that contain transgenes that encode large hairpin RNAs under UAS control (Dietzl et al., 2007). These large hairpin RNAs are expressed with the Gal4-UAS system so that they can be controlled with cell-type-specific promoters. We used Gal4 drivers that result in expression of transgenes specifically in class IV neurons after these cells are specified. This means that most of the animal is normal and neuronal functions of essential genes can be studied. For example, many microtubule regulators are required for mitosis and so can be difficult to study in neurons. This RNA interference (RNAi) strategy allows the function of these genes to be examined in neurons as the hairpins are expressed only once the cells begin to differentiate. Candidates tested in the initial screen included microtubule depolymerases and severing proteins. Kinesin-13 proteins are depolymerases that function to increase subunit loss from microtubule ends (Ems-McClung and Walczak, 2010), and the AAA ATPases spastin, fidgetin and katanin can cut microtubules (Roll-Mecak and McNally, 2010; Sharp and Ross, 2012). Two candidates emerged from this screen: Klp59C (a kinesin-13 depolymerase) and fidgetin (CG3326) (Fig. 1D). However, we subsequently found that the Klp59C RNAi line used in this screen is not phenocopied by other Klp59C RNAi lines or mutants (data not shown). We therefore focused on fidgetin.
Drosophila fidgetin is an AAA ATPase protein proposed to be the single member of the fidgetin family present in Drosophila (Zhang et al., 2007). Fidgetins are part of a family of microtubule-severing proteins that includes spastin and katanin. Like spastin and katanin, overexpression of Drosophila fidgetin completely disrupted microtubules in cultured Drosophila cells (Zhang et al., 2007). This phenotype is consistent with all three proteins having microtubule-severing activity. During mitosis, fidgetin seems to have a similar role to spastin, controlling microtubule numbers and the turnover of tubulin at the ends of microtubules (Zhang et al., 2007). We did not, however, find similar phenotypes for spastin in dendrite degeneration (Fig. 1D), although axon regeneration is sensitive to spastin RNAi (Stone et al., 2012). Katanin p60-Like1 (Kat-60L1) is involved in dendrite pruning (Lee et al., 2009 and Fig. 1D); however, we did not find any delay in axon or dendrite degeneration in Kat-60L1 RNAi flies (Fig. 1D). Based on this candidate screen, we hypothesized that fidgetin is required for normal timing of dendrite degeneration, but not dendrite pruning or axon degeneration.
Fidgetin functions specifically in injury-induced dendrite degeneration
In the candidate screen, fidgetin RNAi resulted in a small, but significant, reduction in complete dendrite clearance 18 h after severing. To probe this phenotype further, we took two approaches. First, we investigated dendrite degeneration at an earlier time point, and second, we used mutants in addition to RNAi. We determined that assaying continuity of the severed dendrite at 9 h after injury provided a more sensitive assay for dendrite degeneration than assaying for complete absence of dendrite remnants at 18 h (Fig. 2). This 9-h time point was used in the rest of the experiments. Three different Deficiency (Df) lines that remove the fidgetin gene [Df(2L)Exel8008, Df(2L)BSC163, and Df(2L)BSC164] and one P element insertion allele (CG3326SH1400) were used to lower fidgetin levels.
Fig. 2.
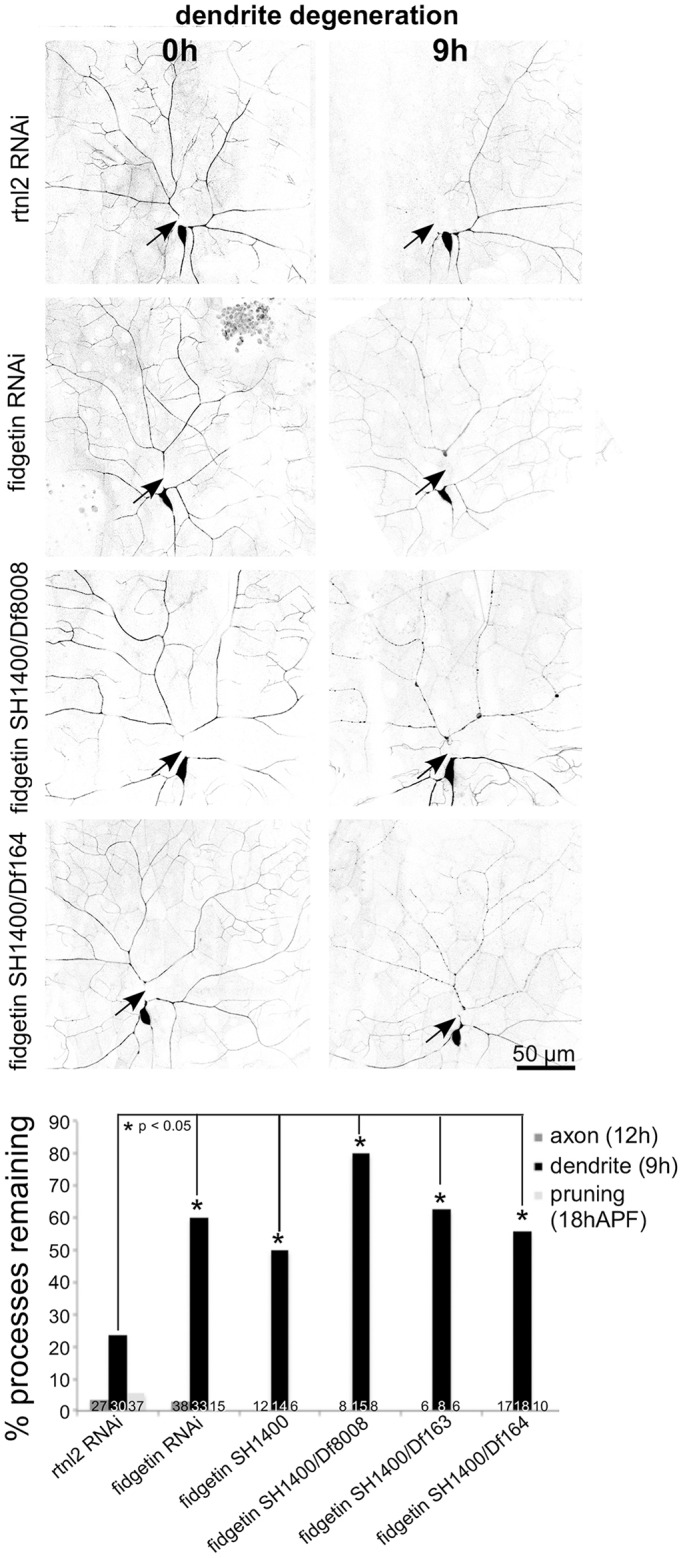
Fidgetin is required for timely dendrite, but not axon, degeneration. RNAi and different mutant alleles of fidgetin were used to reduce fidgetin levels in neurons (RNAi) or globally (mutants). Axon degeneration and pruning were assayed as in the initial screen. Dendrite degeneration was scored by assaying beading at 9 h after cutting. *P<0.05 (two-tailed Fisher's exact test). Numbers on each bar indicate the number of animals assayed.
All trans-heterozygous combinations of the SH1400 insertion allele and Df lines, or homozygous SH1400 animals, had more dendrites that were continuous 9 h after severing than controls (Fig. 2). The phenotype of SH1400/Df was not more severe than SH1400 homozygotes, indicating that SH1400 is a strong loss-of-function allele. The phenotype of the mutant combinations was also not significantly different from that with fidgetin RNAi, indicating the RNAi also induces a strong loss-of-function for fidgetin. All mutant combinations were tested for dendrite pruning and axon degeneration, but as in the initial screen, neither was affected by loss of fidgetin (Fig. 2). We conclude that fidgetin is required for normal timing of dendrite beading after injury, but not for axon degeneration or dendrite pruning.
Fidgetin reduction does not alter microtubule behavior in uninjured neurons
Loss of fidgetin could delay dendrite degeneration in two different ways. First, if fidgetin severs microtubules in dendrites constitutively, then reduction of fidgetin levels might result in more stable dendritic microtubules in uninjured neurons. These more stable microtubules might resist disassembly after injury and slow degeneration. A second possibility is that fidgetin is specifically activated after dendrite injury to destabilize microtubules and promote degeneration. To distinguish between these two alternatives, we began by examining uninjured neurons.
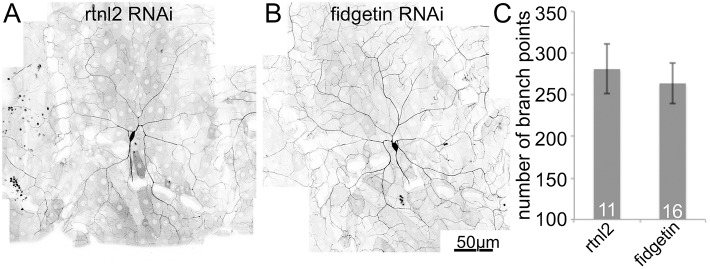
Reduction of any of the other three severing proteins, spastin, Kat60 and Kat60-L1, in ddaC neurons, alters dendrite morphology and this is thought to reflect a role in microtubule regulation in these neurons (Jinushi-Nakao et al., 2007; Mao et al., 2014; Stewart et al., 2012). We therefore examined dendrite morphology in uninjured ddaC neurons with normal and reduced levels of fidgetin. Overall neuron shape was indistinguishable in the two genotypes, and the number of branch points was the same (Fig. 3). This result does not support the idea that fidgetin acts constitutively in dendrites.
Fig. 3.
Dendrite shape is normal in fidgetin RNAi neurons. Images of dendrite arbors of ddaC neurons expressing mCD8–GFP and either control (A, rtnl2) or fidgetin (B) RNAi were acquired. The number of dendrite branch points was counted for each genotype (C). Results are mean±s.d. Numbers on the bars in the graph indicate the number of neurons analyzed for each genotype.
To more directly assay microtubule behavior in dendrites with and without fidgetin, we expressed EB1–GFP in both backgrounds. EB1 binds to the plus ends of microtubules when they are growing and falls off when they shrink (Akhmanova and Steinmetz, 2015). When tagged with GFP, it can be used to assay microtubule polarity in neurons (Stepanova et al., 2003) as well as the number of growing microtubules (Stone et al., 2010). We did not observe any changes in the behavior of growing ends of microtubules in fidgetin RNAi neurons, including the speed of EB1–GFP comet movement, microtubule polarity or number of growing microtubules (Fig. 4).
Fig. 4.
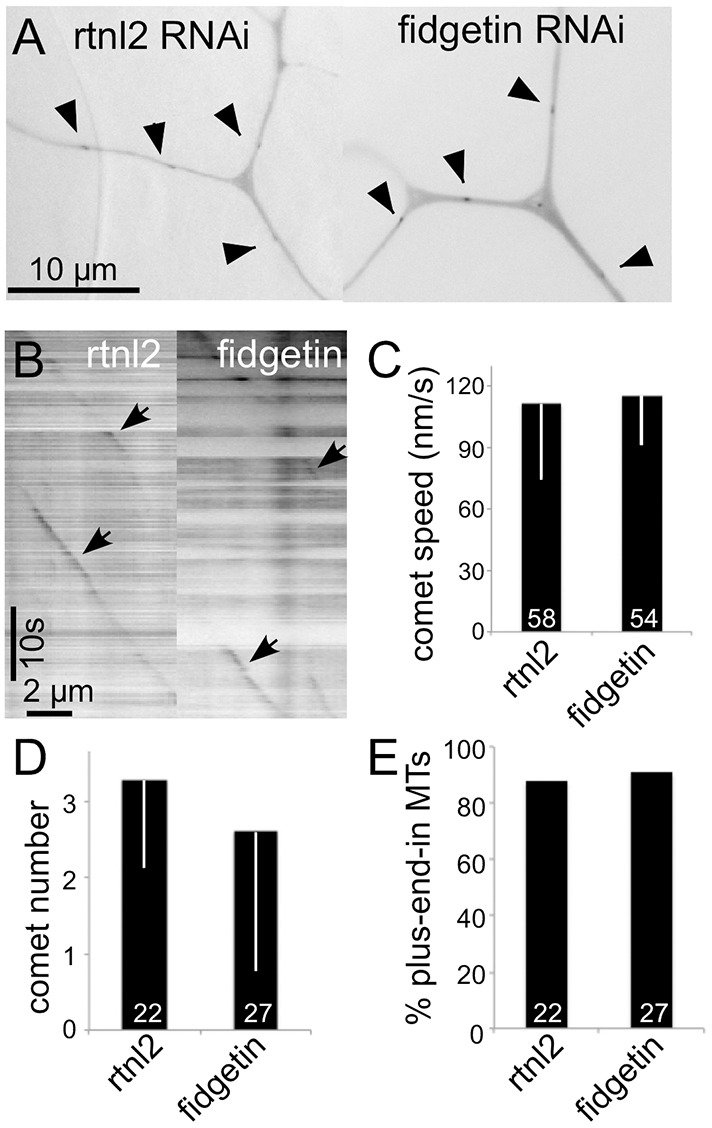
Microtubule growth parameters are similar in control and fidgetin RNAi neurons. Movies of EB1–GFP dynamics were acquired in control (rtnl2) or fidgetin RNAi ddaC neurons. (A) Examples of single frames. (B) Kymographs generated from a dendrite. (C–E) Measurements of EB1–GFP comet speed (C), number of comets in a 10-μm length of dendrite through the entire 200-s movie (D) and direction of movement (E) as assessed from the movies. Results in C and D are mean±s.d. Numbers on the bars are numbers of microtubules analyzed.
To further test whether microtubules were more stable in neurons with reduced levels of fidgetin, we immunostained larval body walls for acetylated tubulin, a modified form of tubulin associated with microtubule stability (Hammond et al., 2008). We expressed RNA hairpins and GFP in class IV neurons. Other neurons, including the easily identifiable ddaE neuron, had normal levels of fidgetin in this experiment. We then calculated the ratio of acetylated tubulin fluorescence in ddaC (class IV) and ddaE (class I) to normalize for antibody penetration and other variations in fluorescence signal. Reduction of fidgetin did not result in an increased amount of acetylated tubulin in the ddaC cell body or dendrites compared to ddaE (Fig. 5).
Fig. 5.
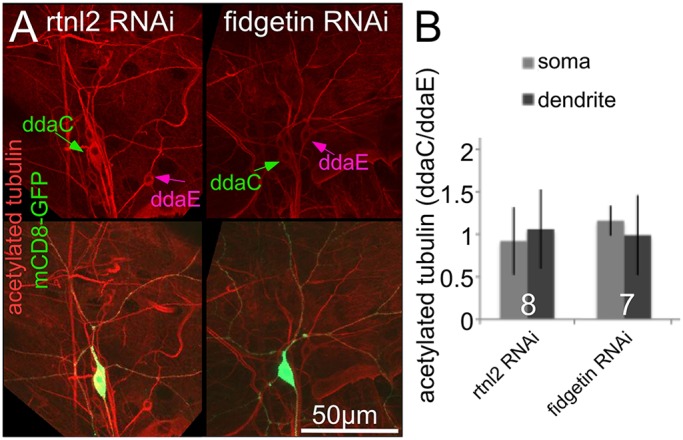
Reduction of fidgetin does not increase microtubule acetylation in uninjured neurons. Larvae expressing mCD8–GFP and dicer2 under the control of the ppk-Gal4 together with (rtnl2) RNAi, or fidgetin RNAi were fixed and stained for acetylated tubulin. (A) Example images are shown. The top row shows the acetylated tubulin channel alone, and the bottom row also shows GFP, which marks the ddaC neurons. Cell bodies of ddaE and ddaC neurons are indicated with arrows. (B) The intensity of acetylated tubulin staining was measured in cell bodies and dendrites of ddaE and ddaC neurons. The ddaE neuron is a control that does not express the RNAi hairpin and was used to normalize the ddaC staining. The ratios of fluorescence intensity in the ddaC to ddaE neuron are shown in the graph. No significant differences between the intensities in control and fidgetin RNAi neurons were found with an unpaired t-test. Results are mean±s.d. The numbers on the bars represent the number of animals tested.
In case either of the assays described above was not sensitive enough to detect subtle changes in microtubule stability, we also developed an assay to probe microtubule turnover. A tandem dimer of photocovertible Eos (tdEos) fused to α-tubulin has previously been used to analyze microtubule sliding in Drosophila S2 cells (Barlan et al., 2013) and cultured Drosophila neurons (Lu et al., 2013). As neurons mature, sliding is downregulated (Lu et al., 2013; Lu et al., 2015). We therefore reasoned that if we photoconverted tdEos–α-tubulin in mature neurons, any converted tubulin incorporated into a microtubule would only be able to move away after the microtubule depolymerized, and thus by tracking signal from photoconverted tubulin in a segment of a dendrite, we would be able to monitor microtubule turnover. Unconverted tdEos–α-tubulin emits a strong green fluorescence. After illuminating a 7-μm segment of the proximal dendrite with 405-nm light, tdEos–α-tubulin emitted red fluorescence (Fig. 6). The red mark was still visible 30 min after conversion, and gradually decayed to baseline by ∼2 h. The rate of red tubulin disappearance from the converted region was similar in control and fidgetin RNAi dendrites suggesting microtubule turnover is similar in these two genotypes. We conclude that it is unlikely that loss of fidgetin acts to delay dendrite degeneration by stabilizing dendritic microtubules before injury.
Fig. 6.
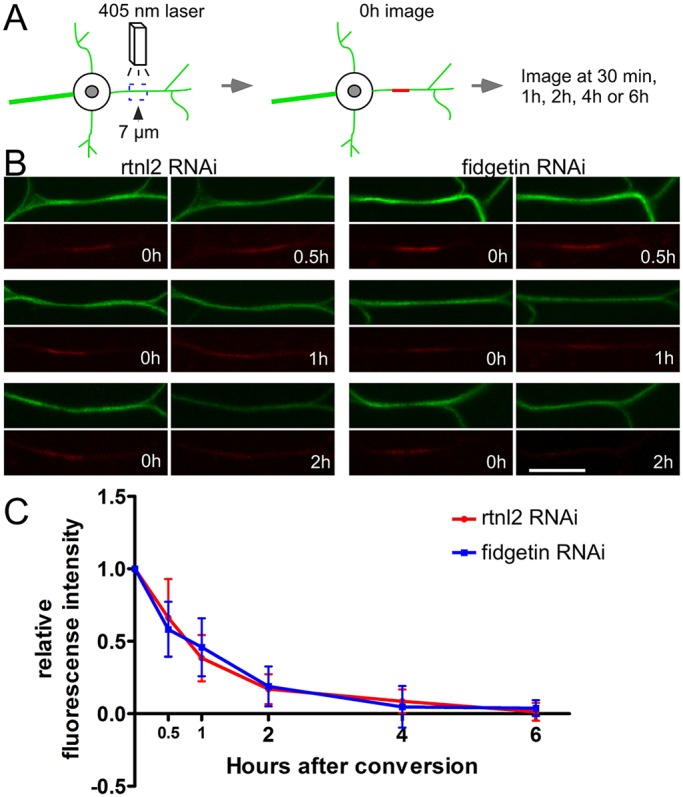
Microtubule turnover is similar in control and fidgetin RNAi neurons. A schematic of the assay for dendritic microtubule turnover using tdEOS-labeled α-Tubulin is shown in A. A 7-μm region in the proximal dendrite was photoconverted by using a 405-nm laser. After 0.5 h, 1 h, 2 h, 4 h or 6 h, the converted dendrite was checked for the remaining red tdEOS. Examples of photoconversion are shown in B. Scale bar: 10 μm. Photoconverted segments were imaged at different time points and the red fluorescence was measured both in the conversion region and outside it. The difference between the signal in the conversion region and neighboring regions was normalized to the same measurement at 0 h. Data points are mean±s.d. The n-values for each genotype and time point were as follows: control, 0.5 h, n=16; 1 h, n=21; 2 h, n=15; 4 h, n=18; 6 h, n=16; fidgetin RNAi, 0.5 h, n=20; 1 h, n=18; 2 h, n=16; 4 h, n=16; 6 h, n=15.
Fidgetin is required to increase the number of microtubule ends after dendrite severing
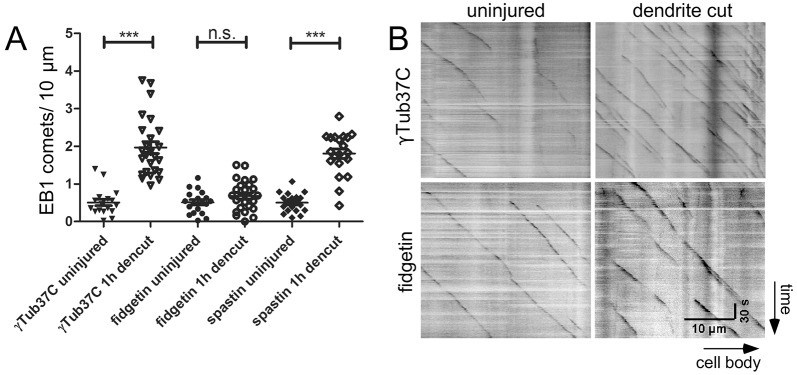
If microtubule stabilization before injury does not account for the degeneration phenotype in fidgetin RNAi neurons, perhaps this protein acts to disassemble microtubules after neuronal injury. To test this idea, we examined the behavior of microtubules in severed dendrites. If fidgetin acts to sever microtubules after dendrite injury, we might expect to see an increase in the number of microtubule plus-ends after the dendrite is removed as each long microtubule is cut into smaller pieces. As a readout of the number of microtubule ends, we expressed EB1–GFP in ddaC neurons. Dendrites were severed from the cell body, and movies of EB1–GFP were acquired 1 h later in the severed piece of the dendrite. The number of EB1–GFP comets increased by, on average, fourfold after dendrite injury in neurons expressing a control RNAi (Fig. 7; Movies 1 and 2). γ-tubulin37C was selected as a control to target with RNAi as it is a γ-tubulin expressed maternally that is not present in somatic tissues including neurons (Wiese, 2008); γ-tubulin23C is the somatic replacement that nucleates microtubules in neurons (Nguyen et al., 2014).
Fig. 7.
The number of growing microtubules increases in severed dendrites in a fidgetin-dependent manner. Movies of EB1–GFP dynamics were acquired in ddaC neurons expressing control (γTub37C RNAi), fidgetin or spastin RNA hairpins. EB1–GFP dots were counted in the cut-off part of dendrite arbor in injured animals or the whole dendrite arbor in uninjured ones, and the length of in-focus dendrites was measured. Total comet number was divided by length to give the average number of comets in a 10-μm stretch. (A) Comet number per 10-μm length is shown in uninjured or severed dendrites 1 h after laser surgery. ***P<0.001; n.s., not significant (unpaired t-test). (B) Example kymographs of EB1–GFP movement are shown.
To test whether fidgetin is responsible for the increase in microtubule ends after dendrite injury, we performed the EB1–GFP assay in a fidgetin RNAi background. The injury-induced increase in microtubule dynamics was completely abrogated in this background (Fig. 7; Movies 3 and 4). To test for specificity, we also performed the assay in spastin RNAi neurons. As in control cells, the number of microtubule plus ends increased dramatically in the severed dendrite 1 h after injury (Fig. 7).
All the experiments described so far were performed in the class IV nociceptive neuron ddaC. To test whether fidgetin functions to increase microtubule ends in other neuron types after dendrite removal, we performed parallel experiments in the ddaE neuron. ddaE is a class 1 dendritic arborization neuron, meaning it has the simplest dendrite arbor of these branched body wall neurons (Grueber et al., 2002). Rather than nociception, these neurons are involved in proprioception and help coordinate movement (Hughes and Thomas, 2007). In ddaE neurons expressing a control (γtub37C) RNA hairpin, the number of growing microtubule plus ends labeled with EB1–GFP increased in the severed region of the dendrite 1 h after injury (Fig. S1, Movies 5 and 6) as in ddaC neurons. Expression of fidgetin, but not spastin, RNA hairpins eliminated the injury-induced increase in microtubule ends (Fig. S1, Movies 7 and 8). Fidgetin therefore functions to increase microtubule dynamics after dendrite injury in at least two different neuron types.
DISCUSSION
In order to understand how axons and/or dendrites fragment after injury and during developmental clearance, we focused on microtubule breakdown. The microtubule-severing protein kat-60L1 was previously identified as a regulator of dendrite pruning (Lee et al., 2009), but does not play a role in injury-induced dendrite (Tao and Rolls, 2011) or axon (Fig. 1) degeneration. In mouse neurons, a microtubule depolymerase is required for normal timing of axon degeneration induced by trophic factor withdrawal (Maor-Nof et al., 2013). It therefore seemed possible that other microtubule destabilizers might be involved in pruning or degeneration. By using assays for injury-induced axon or dendrite degeneration and developmental pruning in the same cell type, we were able to compare the molecular requirements for the three processes (Fig. 1). We found that fidgetin is required for normal timing of dendrite degeneration (Figs 1 and 2), but in the same cells does not delay axon degeneration or dendrite pruning (Figs 1 and 2).
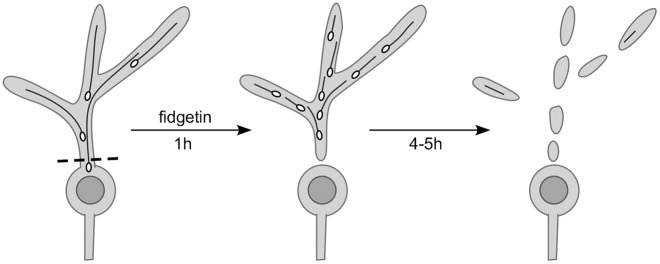
One challenge in studies on the role of microtubule regulators in pruning and axon degeneration is to demonstrate whether they act to destabilize microtubules constitutively or act specifically during the neurite deconstruction process. We therefore focused on determining whether fidgetin acts in uninjured neurons to control the balance of microtubule stability, or is activated to disassemble microtubules only after injury. In this study, we were able to visualize a dramatic increase in the number of dynamic microtubule plus-ends in dendrites after injury, and to demonstrate that fidgetin is responsible for this injury-induced increase (Fig. 8). This means that fidgetin is likely to be activated specifically in the severed piece of dendrite. The protein could either be present in the dendrite before injury, but inactive, or could be locally translated in dendrites after injury. Ribosomes are present in dendrites of dendritic arborization neurons and concentrate at dendrite branch points (Hill et al., 2012) so either mechanism is possible. In future studies, it will be extremely interesting to determine how injury controls fidgetin levels or activity.
Fig. 8.
Model to show how increase in microtubule ends relates to microtubule severing. Before dendrite injury relatively few long microtubules are present in the dendrites, so few EB1 comets (ovals) are observed. Activation of fidgetin in the injured dendrite could cut the long microtubules into shorter ones, with each piece now having a dynamic plus end. This increase in number of growing plus ends precedes beading of the entire dendrite (right panel) by 4–5 h.
Although microtubule dynamics are initially upregulated during dendrite degeneration (Fig. 7), microtubule comets eventually disappear completely at between 4 and 6 h after dendrite injury (Tao and Rolls, 2011), just prior to membrane beading. Interestingly, a spike in microtubule dynamics followed by a subsequent decrease has also been observed in the severed region of mouse axons (Kleele et al., 2014), hence, it is possible that a microtubule-severing protein also acts in this context. We hypothesize that increasing the number of microtubule ends soon after dendrite or axon injury facilitates subsequent microtubule disassembly. It is possible that a microtubule depolymerase acts on these microtubule ends to further eliminate microtubules. Although targeting Klp59c, which is in the same kinesin-13 family as the depolymerase involved in axon degeneration in mice (Maor-Nof et al., 2013), delayed degeneration in our initial screen, we could not confirm a role for this enzyme with other RNAi or mutant alleles. It therefore remains to be seen whether microtubule dynamics in dendrites is terminated during degeneration by an active depolymerase or by eventual exhaustion of energy, which is required for microtubule growth.
Reduction of fidgetin delays dendrite degeneration by several hours, but degeneration eventually proceeds. This is consistent with a model in which several parallel processes are activated by injury to induce disassembly of severed dendrites. Active disassembly of microtubules seems to be one aspect of degeneration that likely makes it easier for the axon or dendrite to bead. However, microtubules are dynamic polymers that constantly grow and shrink and require energy to do this. During axon degeneration at least, energy levels eventually fall (Gerdts et al., 2016), and this could allow passive disassembly of microtubules if the active mechanism is impaired.
Interestingly, alterations in microtubule dynamics seem broadly associated with neuronal injury and stress responses. A dramatic increase in the number of microtubule plus ends is also observed after axon injury in Drosophila, but on the cell body side of the cut, rather than in the severed piece (Stone et al., 2010). This longer term (6–48-h time range) global increase in microtubule dynamics after axon injury is also observed in mouse neurons (Kleele et al., 2014). We have been able to show in Drosophila that these global increases in microtubule dynamics on the cell body side of the axon injury are mediated by microtubule nucleation and not severing (Chen et al., 2012). In addition to being mechanistically distinct from the increase in dynamics associated with degeneration here, this nucleation-induced increase in dynamics acts to stabilize the injured neuron (Chen et al., 2012) rather than as part of the disassembly process. Thus, increases in the number of microtubule plus ends after injury can be part of either stabilizing or destabilizing pathways, depending on the mechanism in which they are induced and probably also on the other microtubule regulators, for example depolymerases, active at the same time.
MATERIALS AND METHODS
Drosophila stocks
The tester fly lines for RNAi experiments included the following: UAS-dicer2; ppk-Gal4, UAS-mCD8-GFP and 477-Gal4, UAS-EB1-GFP/CyO; UAS-dicer2 and 477-Gal4, UAS-tdEOS-α-tubulin/CyO-actinGFP; UAS-dicer2. All the RNAi experiments included dicer2 to increase the knockdown efficiency of the neuronal RNAi (Dietzl et al., 2007). The following RNAi lines were obtained from the Vienna Drosophila RNAi center (VDRC, Vienna, Austria): UAS-rtnl2-RNAi (33320), UAS-fidgetin-RNAi (24746), UAS-katanin-60-RNAi (38368), UAS-katanin-p60L1-RNAi (31598), UAS-spastin-RNAi (33110), UAS-stathmin-RNAi (32370), CG10793 (31351), UAS-Klp59C-RNAi (48576), UAS-Klp59D-RNAi (100530), UAS-Klp10A-RNAi (41534), UAS-Klp67A- RNAi (52105).
The following mutant and deficiency lines were provided by the Bloomington Drosophila Stock Center: fidgetinSH1400/CyO, fidgetin Df(2L)Exel8008/CyO and fidgetin Df(2L)BSC164/CyO. We rebalanced all the mutant and deficiency lines with CyO-actinGFP in our laboratory. For homozygous fidgetin mutants or fidgetin mutant over-deficiency genotypes, lines were generated that contained the fidgetinSH1400 mutant on the second chromosome balanced with CyO-actinGFP, and the ppk-Gal4 and mCD8–GFP transgenes on the third chromosome balanced with TM6 (Tb). These lines were crossed to fidgetin mutant or deficiency lines balanced with CyO-actinGFP. Then, non-Tb progeny without actin–GFP were analyzed.
Axon and dendrite injury experiments
The injury assays in this paper were performed in ddaC neurons in Drosophila larvae, with the exception of those in Fig. S1. mCD8–GFP driven by ppk-Gal4 was used to visualize the ddaC neurons. Embryos were collected overnight and then aged at 25°C for 3 days before imaging. A whole larva was mounted on a slide with a dry agarose cushion and covered with a coverslip that was held in place with tape. Live imaging was performed on an FV1000 confocal microscope (Olympus), an LSM510 confocal microscope (Zeiss) or, for microtubule dynamics, a Zeiss AxioImager with a Colibri LED illuminator. ImageJ software was used to generate maximum-intensity projections and perform image analysis (http://rsb.info.nih.gov/ij/).
To injure neurons, we used a MicroPoint pulsed UV laser (Andor Technology) to sever a single axon or dendrite close to the cell body. Images were acquired immediately after severing to make sure that axons or dendrites were completely cut. The larvae were then returned to normal Drosophila medium to recover at 25°C. The same larvae were later remounted on the microscope to examine the severed axons or dendrites. Dendrites that remained continuous without signs of fragmentation were scored at 18 h or 9 h after injury. Axons without beading 12 h after severing were used as the readout of axon integrity.
Dendrite pruning
Pruning was studied in ddaC neurons. White pre-pupae were collected and maintained at 25°C. At 18 h after puparium formation, the pupal cases were first removed as described previously (Williams et al., 2006) and then imaged on a confocal microscope. The presence of dendrites at this time point was scored as a pruning defect.
Microtubule behavior in uninjured neurons
To assay microtubules in uninjured neurons (Fig. 4), females from the 477Gal4, UAS-EB1-GFP; Dicer2 tester line were crossed to control or fidgetin RNAi flies. Three-day-old larvae were mounted on slides and imaged with a Zeiss Imager.M2 equipped with Colibri LED illumination. Movies were taken at a rate of one frame per second and 200 frames were acquired. To quantify the EB1–GFP dynamics, a 10-μm-long dendrite segment close to the cell body in the ddaC neuron was chosen. EB1 comets that went through the segment were counted during the whole movie and the total number of comets is shown in Fig. 4E. These comets were also scored based on direction of movement towards or away from the cell body. Velocity of comets was also measured from these movies.
Microtubule dynamics after dendrite injury
To examine the persistence of EB1–GFP comets after dendrite severing, flies with UAS-RNAi transgenes were crossed to the 477-Gal4, UAS-EB1-GFP; dUAS-dicer2 line for ddaC neurons or UAS-dicer2; 221-Gal4, UAS-dicer2 for ddaE neurons. Standard dendrite severing was performed on a 3-day-old larva. After 1 h, a time series was taken of the transected dendrite using a Zeiss Imager.M2 equipped with Colibri LED illumination. Movies were taken a rate of one frame per second and 300 frames were acquired. To quantify the EB1–GFP dynamics, comets in six in-focus frames (usually frame 1, 51, 101, 151, 201 and 251) were counted. The number of EB1–GFP comets was counted in the severed part of the dendrite arbor in injured neurons or the whole dendrite arbor in uninjured ones, and the length of in-focus dendrites was measured. Only mobile comets appearing in three consecutive frames were counted. EB1–GFP comet number was normalized to the length of in-focus dendrite for every neuron.
Immunostaining
For immunostaining of ddaC neurons, UAS-RNAi transgenic flies were crossed to UAS-dicer2; ppk-gal4, UAS-mCD8 GFP flies. Third-instar larval progeny were dissected in Schneider's medium and fixed in 4% paraformaldehyde for 30 min. After fixation, the larvae were washed four times in blocking solution (PBS, 1% BSA, 0.2% Triton X-100 and 10 mM glycine) and each wash lasted at least 10 min. Larvae were then incubated overnight at 4°C with primary antibody against acetylated-α-tubulin (1:1000, mouse monoclonal antibody T7451, Sigma) in the blocking solution. The next day, they were washed in the blocking solution several times for a few hours to remove the primary antibody. Then a Rhodamine-Red-X-coupled secondary antibody (Jackson ImmunoResearch) was applied to the larval preparations for 2 h at room temperature. The secondary antibody was removed with a final four washes with the blocking solution, and larval preparations were stored at 4°C in 85% glycerol and 15% 50 mM Tris-HCl, pH 8 until imaging. Images were obtained on a Zeiss LSM 510 confocal microscope.
Tubulin photoconversion assay
477-Gal4 was used to drive the expression of UAS-tdEos-α-tubulin, UAS-Dicer2 and control or fidgetin hairpin RNAs in class IV neurons. In one ddaC neuron per animal, a segment of ∼7-μm (50 pixels on an Olympus FV1000 microscope, 60× lens, 3× zoom) was chosen near the soma and illuminated with a 405-nm laser to photoconvert the tdEos–α-tubulin from green to red. Converted neurons were imaged immediately after conversion (0 h) and re-checked for red fluorescence at 30 min, 1 h, 2 h, 4 h or 6 h later using a 543-nm laser. Counter-images of the unconverted tdEos–α-tubulin were acquired with a 488-nm laser. Different sets of cells were used for each time point rather than assaying the same cells at multiple time points as the red signal bleached easily. Fluorescence intensity was measured within the 7-μm segment using ImageJ. Red fluorescence intensity was measured in converted segment and in the neighboring non-converted region using ImageJ. Remaining fluorescence intensity (FI) was defined as (FIconverted−FIneighboring)timecourse/(FIconverted−FIneighboring)0h.
Acknowledgements
We are very grateful to both the Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center for maintaining and providing valuable reagents.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
J.T. and C.F. designed and performed experiments and analyzed data. M.M.R. supervised experiments and wrote the manuscript.
Funding
Funding was provided by the National Institutes of Health [grant number R01GM085115] and the Pew Charitable Trusts; M.M.R. was a Pew Scholar in the Biomedical Sciences. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.188540.supplemental
References
- Akhmanova A. and Steinmetz M. O. (2015). Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711-726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- Barlan K., Lu W. and Gelfand V. I. (2013). The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol. 23, 317-322. 10.1016/j.cub.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M. R. C., Gerdts J., Naylor S. A., Royse E. X., Ebstein S. Y., Sasaki Y., Milbrandt J. and DiAntonio A. (2012). A model of toxic neuropathy in Drosophila reveals a role for MORN4 in promoting axonal degeneration. J. Neurosci. 32, 5054-5061. 10.1523/JNEUROSCI.4951-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Stone M. C., Tao J. and Rolls M. M. (2012). Axon injury and stress trigger a microtubule-based neuroprotective pathway. Proc. Natl. Acad. Sci. USA 109, 11842-11847. 10.1073/pnas.1121180109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. P. and Freeman M. R. (2010). Wallerian degeneration, wld(s), and nmnat. Annu. Rev. Neurosci. 33, 245-267. 10.1146/annurev-neuro-060909-153248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti L., Gilley J. and Coleman M. P. (2014). Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci. 15, 394-409. 10.1038/nrn3680 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Ems-McClung S. C. and Walczak C. E. (2010). Kinesin-13s in mitosis: key players in the spatial and temporal organization of spindle microtubules. Semin. Cell Dev. Biol. 21, 276-282. 10.1016/j.semcdb.2010.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J., Summers D. W., Milbrandt J. and DiAntonio A. (2016). Axon Self-destruction: new links among SARM1, MAPKs, and NAD+ Metabolism. Neuron 89, 449-460. 10.1016/j.neuron.2015.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber W. B., Jan L. Y. and Jan Y. N. (2002). Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129, 2867-2878. [DOI] [PubMed] [Google Scholar]
- Hammond J. W., Cai D. and Verhey K. J. (2008). Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20, 71-76. 10.1016/j.ceb.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. E., Parmar M., Gheres K. W., Guignet M. A., Huang Y., Jackson F. R. and Rolls M. M. (2012). Development of dendrite polarity in Drosophila neurons. Neural Dev. 7, 34 10.1186/1749-8104-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C. L. and Thomas J. B. (2007). A sensory feedback circuit coordinates muscle activity in Drosophila. Mol. Cell. Neurosci. 35, 383-396. 10.1016/j.mcn.2007.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang R. Y., Zhong L., Xu Y., Johnson T., Zhang F., Deisseroth K. and Tracey W. D. (2007). Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 17, 2105-2116. 10.1016/j.cub.2007.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinushi-Nakao S., Arvind R., Amikura R., Kinameri E., Liu A. W. and Moore A. W. (2007). Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron 56, 963-978. 10.1016/j.neuron.2007.10.031 [DOI] [PubMed] [Google Scholar]
- Kirilly D., Gu Y., Huang Y., Wu Z., Bashirullah A., Low B. C., Kolodkin A. L., Wang H. and Yu F. (2009). A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat. Neurosci. 12, 1497-1505. 10.1038/nn.2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleele T., Marinkovic P., Williams P. R., Stern S., Weigand E. E., Engerer P., Naumann R., Hartmann J., Karl R. M., Bradke F. et al. (2014). An assay to image neuronal microtubule dynamics in mice. Nat. Commun. 5, 4827 10.1038/ncomms5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. T., Zhu S., Younger S., Jan L. Y. and Jan Y. N. (2006). Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51, 283-290. 10.1016/j.neuron.2006.07.014 [DOI] [PubMed] [Google Scholar]
- Lee H.-H., Jan L. Y. and Jan Y.-N. (2009). Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc. Natl. Acad. Sci. USA 106, 6363-6368. 10.1073/pnas.0902051106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncle N. and Williams D. W. (2012). An interaction screen identifies headcase as a regulator of large-scale pruning. J. Neurosci. 32, 17086-17096. 10.1523/JNEUROSCI.1391-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Fox P., Lakonishok M., Davidson M. W. and Gelfand V. I. (2013). Initial neurite outgrowth in Drosophila neurons is driven by Kinesin-powered microtubule sliding. Curr. Biol. 23, 1018-1023. 10.1016/j.cub.2013.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Lakonishok M. and Gelfand V. I. (2015). Kinesin-1-powered microtubule sliding initiates axonal regeneration in Drosophila cultured neurons. Mol. Biol. Cell 26, 1296-1307. 10.1091/mbc.E14-10-1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. and O'Leary D. D. M. (2005). Axon retraction and degeneration in development and disease. Annu. Rev. Neurosci. 28, 127-156. 10.1146/annurev.neuro.28.061604.135632 [DOI] [PubMed] [Google Scholar]
- MacDonald J. M., Beach M. G., Porpiglia E., Sheehan A. E., Watts R. J. and Freeman M. R. (2006). The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron 50, 869-881. 10.1016/j.neuron.2006.04.028 [DOI] [PubMed] [Google Scholar]
- Mao C.-X., Xiong Y., Xiong Z., Wang Q., Zhang Y. Q. and Jin S. (2014). Microtubule-severing protein Katanin regulates neuromuscular junction development and dendritic elaboration in Drosophila. Development 141, 1064-1074. 10.1242/dev.097774 [DOI] [PubMed] [Google Scholar]
- Maor-Nof M., Homma N., Raanan C., Nof A., Hirokawa N. and Yaron A. (2013). Axonal pruning is actively regulated by the microtubule-destabilizing protein kinesin superfamily protein 2A. Cell Rep. 3, 971-977. 10.1016/j.celrep.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Miller B. R., Press C., Daniels R. W., Sasaki Y., Milbrandt J. and DiAntonio A. (2009). A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat. Neurosci. 12, 387-389. 10.1038/nn.2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. M., McCracken C. J., Milner E. S., Goetschius D. J., Weiner A. T., Long M. K., Michael N. L., Munro S. and Rolls M. M. (2014). Gamma-tubulin controls neuronal microtubule polarity independently of Golgi outposts. Mol. Biol. Cell 25, 2039-2050. 10.1091/mbc.E13-09-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev A., McLaughlin T., O'Leary D. D. M. and Tessier-Lavigne M. (2009). APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457, 981-989. 10.1038/nature07767 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Osterloh J. M., Yang J., Rooney T. M., Fox A. N., Adalbert R., Powell E. H., Sheehan A. E., Avery M. A., Hackett R., Logan M. A. et al. (2012). dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481-484. 10.1126/science.1223899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A. and McNally F. J. (2010). Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22, 96-103. 10.1016/j.ceb.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmann Z., Assa-Kunik E., Tiomny S., Minis A., Haklai-Topper L., Arama E. and Yaron A. (2010). Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. J. Neurosci. 30, 6375-6386. 10.1523/JNEUROSCI.0922-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O. and Yaron A. (2015). Mechanisms of developmental neurite pruning. Cell. Mol. Life Sci. 72, 101-119. 10.1007/s00018-014-1729-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J. and Ross J. L. (2012). Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125, 2561-2569. 10.1242/jcs.101139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono K., Fujimoto A., Tsuyama T., Yamamoto-Kochi M., Sato M., Hattori Y., Sugimura K., Usui T., Kimura K.-I. and Uemura T. (2009). Multidendritic sensory neurons in the adult Drosophila abdomen: origins, dendritic morphology, and segment- and age-dependent programmed cell death. Neural Dev. 4, 37 10.1186/1749-8104-4-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T., Slemmer J., Hoogenraad C. C., Lansbergen G., Dortland B., De Zeeuw C. I., Grosveld F., van Cappellen G., Akhmanova A. and Galjart N. (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23, 2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A., Tsubouchi A., Rolls M. M., Tracey W. D. and Sherwood N. T. (2012). Katanin p60-like1 promotes microtubule growth and terminal dendrite stability in the larval class IV sensory neurons of Drosophila. J. Neurosci. 32, 11631-11642. 10.1523/JNEUROSCI.0729-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M. C., Nguyen M. M., Tao J., Allender D. L. and Rolls M. M. (2010). Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol. Biol. Cell 21, 767-777. 10.1091/mbc.E09-11-0967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M. C., Rao K., Gheres K. W., Kim S., Tao J., La Rochelle C., Folker C. T., Sherwood N. T. and Rolls M. M. (2012). Normal Spastin gene dosage is specifically required for axon regeneration. Cell Rep. 2, 1340-1350. 10.1016/j.celrep.2012.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M. C., Albertson R. M., Chen L. and Rolls M. M. (2014). Dendrite injury triggers DLK-independent regeneration. Cell Rep. 6, 247-253. 10.1016/j.celrep.2013.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J. and Rolls M. M. (2011). Dendrites have a rapid program of injury-induced degeneration that is molecularly distinct from developmental pruning. J. Neurosci. 31, 5398-5405. 10.1523/JNEUROSCI.3826-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C. (2008). Distinct Dgrip84 isoforms correlate with distinct gamma-tubulins in Drosophila. Mol. Biol. Cell 19, 368-377. 10.1091/mbc.E07-08-0801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. W. and Truman J. W. (2005a). Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development 132, 3631-3642. 10.1242/dev.01928 [DOI] [PubMed] [Google Scholar]
- Williams D. W. and Truman J. W. (2005b). Remodeling dendrites during insect metamorphosis. J. Neurobiol. 64, 24-33. 10.1002/neu.20151 [DOI] [PubMed] [Google Scholar]
- Williams D. W., Kondo S., Krzyzanowska A., Hiromi Y. and Truman J. W. (2006). Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 9, 1234-1236. 10.1038/nn1774 [DOI] [PubMed] [Google Scholar]
- Yasunaga K.-I., Kanamori T., Morikawa R., Suzuki E. and Emoto K. (2010). Dendrite reshaping of adult Drosophila sensory neurons requires matrix metalloproteinase-mediated modification of the basement membranes. Dev. Cell 18, 621-632. 10.1016/j.devcel.2010.02.010 [DOI] [PubMed] [Google Scholar]
- Yu F. and Schuldiner O. (2014). Axon and dendrite pruning in Drosophila. Curr. Opin. Neurobiol. 27, 192-198. 10.1016/j.conb.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Rogers G. C., Buster D. W. and Sharp D. J. (2007). Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177, 231-242. 10.1083/jcb.200612011 [DOI] [PMC free article] [PubMed] [Google Scholar]