Abstract
Lactobacilli convert linoleic acid to the antifungal compound 10-hydroxy-12-octadecenoic acid (10-HOE) by linoleate 10-hydratase (10-LAH). However, the effect of this conversion on cellular membrane physiology and properties of the cell surface have not been demonstrated. Moreover, Lactobacillus plantarum produces 13-hydroxy-9-octadecenoic acid (13-HOE) in addition to 10-HOE, but the antifungal activity of 13-HOE was unknown. Phylogenetic analyses conducted in this study did not differentiate between 10-LAH and linoleate 13-hydratase (13-LAH). Thus, linoleate hydratases (LAHs) must be characterized through their differences in their activities of linoleate conversion. Four genes encoding putative LAHs from lactobacilli were cloned, heterologous expressed, purified and identified as FAD-dependent 10-LAH. The unsaturated fatty acid substrates stimulated the growth of lactobacilli. We also investigated the role of 10-LAH in ethanol tolerance, membrane fluidity and hydrophobicity of cell surfaces in lactobacilli by disruption of lah. Compared with the L. plantarum lah deficient strain, 10-LAH in wild-type strain did not exert effect on cell survival and membrane fluidity under ethanol stress, but influenced the cell surface hydrophobicity. Moreover, deletion of 10-LAH in L. plantarum facilitated purification of 13-HOE and demonstration of its antifungal activity against Penicillium roqueforti and Aspergillus niger.
Keywords: linoleate 10-hydratase, cell membrane fluidity, cell surface hydrophobicity, 13-hydroxy-9-octadecenoic acid, antifungal activity
Introduction
Antifungal metabolites of lactic acid bacteria have potential for applications as antifungal preservatives in cereal products, and in silage (Magnusson et al., 2003; Oliveira et al., 2014). Several hydroxy fatty acids have antifungal activity (Hou and Forman Iii, 2000; Hou, 2008) and antifungal 3-hydroxy fatty acids of C10 to C14 chain lengths are formed by Lactobacillus plantarum MiLAB 14 (Sjögren et al., 2003). Lactobacillus hammesii accumulates 10-HOE, an antifungal compound that increased the mold-free shelf life of bread (Black et al., 2013a,b). The biosynthesis of antifungal hydroxy fatty acids and the application of hydroxy fatty acids in food are dependent on knowledge of enzymes involved in microbial fatty acid-hydroxylation (Kim and Oh, 2013).
Linoleate hydratase (LAH) activity was first characterized in Streptococcus pyogenes; the hydratase was previously described as myosin cross-reactive antigen (Kil et al., 1994). The FAD containing LAH in S. pyogenes hydrates the cis-9 and cis-12 double bonds of C16 and C18 fatty acids to produce 10-hydroxy and 10,13-dihydroxy fatty acids (Volkov et al., 2010). Remarkably, the LAHs of bifidobacteria, lactobacilli and Nocardia spp. exclusively hydrate cis-9 double bond (Koritala and Bagby, 1992; Rosberg-Cody et al., 2011; Yang et al., 2013). The crystal structure of the LAH from Lactobacillus acidophilus provided the structural basis for the selective substrate recognition of linoleate 10-hydratase (Volkov et al., 2013). A second LAH in L. acidophilus hydrates the cis-12 double bond to produce 13-hydroxy fatty acid (Kim et al., 2015; Park et al., 2015).
The physiological and ecological function of LAHs -have been investigated. The LAH/myosin-cross reactive antigen of S. pyogenes mediated adherence to human keratinocytes; LAH was also suggested to detoxify linoleic acid by conversion to a hydroxyl-product with lower antibacterial activity (Volkov et al., 2010). Similarly, a L. acidophilus mutant with truncated LAH exhibited a decreased adherence to intestinal epithelial cells, and was more sensitive to stresses (O’Flaherty and Klaenhammer, 2010). Heterologous expression of a LAH of Bifidobacterium breve in Lactococcus lactis increased resistance to heat and solvent stresses (Rosberg-Cody et al., 2011). Linoleate 10-hydratase is predicted to be a membrane-associated protein with one trans-membrane helix (Kil et al., 1994; Rosberg-Cody et al., 2011), and hydration of linoleic acid occurs in the cell periphery (Kishino et al., 2011). However, the effect of linoleic acid hydratase on properties of cell membranes and the cell surface have not been demonstrated. Moreover, 10-HOE has antifungal activity (Black et al., 2013b) but a corresponding activity of alternative hydration products of fatty acids remains unknown.
This study aimed to characterize linoleic acid hydratases in L. plantarum, Lactobacillus reuteri, L. hammesii, and Lactobacillus spicheri. Strain selection included strains which produce only 10-HOE and strains that produce 10-HOE and 13-HOE (Black et al., 2013b). Four enzymes were characterized after heterologous expression in Escherichia coli. The physiological function of the linoleate 10-hydratase of L. plantarum was studied in more detail by comparison of the ethanol resistance and cell surface properties of L. plantarum TMW1.460 and its linoleate 10-hydratase deficient mutant L. plantarum TMA1.460Δlah. The antifungal activity of 13-HOE was compared to the activities of other hydroxy fatty acids.
Materials and Methods
Bacterial Strains and Fermentation
Lactobacillus reuteri LTH2584, L. plantarum TMW1.460, L. hammesii DSM16381, L. spicheri Lp38 and Lactobacillus sanfranciscensis ATCC 27651 were anaerobically cultivated at 37°C (L. reuteri) or 30°C (all other strains) in modified De Man Rogosa Sharpe (mMRS) medium containing 0.1% Tween 80 (mMRS-Tween 80; Zhang et al., 2010). mMRS-Tween 20 was prepared by replacing Tween 80- with an equal weight of Tween 20. E. coli DH5α (New England Biolabs) served as a host for plasmids in the cloning procedures, and E. coli BL21 Star (DE3) (ThermoFisher Scientific) was used for protein overexpression. E. coli strains were cultivated in Luria-Bertani (LB) medium (BD, Mississauga, CA, USA) with agitation at 200 rpm and 37°C. Antibiotic-resistant E. coli carrying plasmid pET-28a(+), pUC19 or pJRS233 were cultured in media containing 50 mg/L kanamycin, 50 mg/L ampicillin, or 500 mg/L erythromycin, respectively. Erythromycin-resistant L. plantarum was grown in presence of 5 mg/L erythromycin. Aspergillus niger FUA5001 and Penicillium roqueforti FUA5005 were grown at 25°C for 72 h on malt extract agar.
Linoleic acid metabolism of lactobacilli was analyzed after incubation in mMRS-Tween80 broth supplied with 5% inoculum and 4 g/L linoleic acid anaerobically for 4 days. Lipids were isolated by addition of one volume of 85:15 (vol/vol) chloroform-methanol to cultures prior to incubation 4°C overnight; cultures were then extracted twice with additional two volumes of chloroform–methanol (85:15, vol/vol). The organic solvent was evaporated under reduced pressure and the dry residue stored at -20°C under nitrogen.
DNA Manipulations
Genomic DNA was isolated using the Blood & Tissue Kit (Qiagen, Hilden, Germany). Plasmid DNA from E. coli was extracted with a QIAprep Spin Miniprep kit (Qiagen). PCR primers used in this study were synthesized by Integrated DNA Technologies (San Diego, CA, USA). The Taq DNA polymerase was purchased from TaKaRa Bio (Shiga, Japan). T4 DNA ligase and restriction enzymes were obtained from Thermo Scientific (Mississauga, CA, USA). PCR products were purified by using the DNA gel extraction kit (Qiagen). DNA sequencing was performed by Macrogen (Rockville, MD, USA).
Sequence and Phylogenetic Analysis of Linoleate Hydratases in Lactobacilli
Genes of putative LAHs in L. spicheri Lp38, L. reuteri LTH2584 and L. plantarum TMW1.460 were amplified with primers that are specific to LAHs in genome-sequenced strains of these species (Table 1). To identify the LAH in L. hammesii, lah sequences from the closely related L. spicheri and L. brevis were aligned and specific primers targeting conserved sequences upstream and downstream of lah coding sequences were designed (Table 1). The four lah genes were sequenced by service of Macrogen (Rockville, MD, USA).
Table 1.
Primers used in this study.
| Primers [forward (F); reverse (R)] | Sequence (5′–3′) | Restriction sitea |
|---|---|---|
| DSM16381 sequencing, F1 | 5′-TACGGAGGTGTTTTTTGATGGT-3′ | — |
| DSM16381 sequencing, R1 | 5′-CGTAAATTCATAAATCATTTGGTGCATGTA-3′ | — |
| DSM16381 sequencing, F2 | 5′-TACATGCACCAAATGATTTATGAATTTACG-3′ | — |
| DSM16381 sequencing, R2 | 5′-TACTTCGTCTTAGGTGACCA-3′ | — |
| LTH2584 cloning, F | 5′-CGCCATATGTACTATTCAAACGGAAATTATG-3′ | NdeI |
| LTH2584 cloning, R | 5′-ATTTGCGGCCGCTTAAAGTAAATGTTGTTCTTCCATT-3′ | NotI |
| TMW1.460 cloning, F | 5′-CCGGAATTCATGGTTAAAAGTAAAGCAATTATGA-3′ | EcoRI |
| TMW1.460 cloning, R | 5′-ATTTGCGGCCGCTTAATCAAACATCTTCTTAGTTGC-3′ | NotI |
| DSM16381 cloning, F | 5′-CGCGGATCCATGGTTAAAACAAAAGCAGTAATG-3′ | BamHI |
| DSM16381 cloning, R | 5′-CCCAAGCTTTTAGCTAAACATCCGCTTCGTTGC-3′ | HindIII |
| Lp38 cloning, F | 5′-CCGGAATTCATGGTTAAGACAAAAGCTGTAATG-3′ | EcoRI |
| Lp38 cloning, R | 5′-ATAGTTTAGCGGCCGCGTTAACTAAACATTTTCTTCGTTGCC-3′ | NotI |
| Lah-upstream A, F | 5′-AACTGCAGGCCTAAAACGAGCTAAACGAC-3′ | PstI |
| Lah-upstream A, R | 5′-ACATGCATGCCCCGGCACCAATCATAATTGCTTTAC-3′ | SphI |
| Lah-downstream B, F | 5′-ACATGCATGCAAGAAGATGTTTGATTAATTAAA-3′ | SphI |
| Lah-downstream B, R | 5′-CCCAAGCTTATGAAAAAATTAACATCAGTCG-3′ | HindIII |
aRestriction sites are underlined.
The phylogenetic analysis of LAHs included the type strains of the 24 groups of the -genus Lactobacillus sensu lato (Zheng et al., 2015) and the genera Weissella, Leuconostoc, and Oenococcus. Sequences of biochemically characterized linoleate 10-hydratase and linoleate 13-hydratase from Lactobacillus spp., Bifidobacterium spp., and Streptococcus spp. were included in the phylogenetic analysis (Volkov et al., 2010; Rosberg-Cody et al., 2011; Yang et al., 2013; Kim et al., 2015). Protein sequences of LAH were retrieved from GenBank1, using NCBI BLAST analysis with organism specific search. Protein phylogenetic tree was built in MEGA7.
Cloning and Heterologous Expression of Linoleate Hydratases of Lactobacilli
Coding regions of the four lah were amplified from genomic DNA of the respective strains with primers listed in Table 1. Amplicons were purified and cloned into pGEM-T Easy vector (Promega, Madison, WI, USA). The lah fragments in recombinant pGEM-T vectors were digested with restriction endonucleases (Table 1), purified and ligated into expression vector pET-28a(+) (Novagen, Toronto, ON, Canada), yielding the respective constructs pET28a/LAH for each strain. Recombinant plasmids were introduced into chemically competent E. coli BL21 Star (DE3) (Thermo Fisher Scientific) and transformants were plated on LB agar containing 50 mg/L kanamycin. The gene cloning was verified by PCR amplification and sequencing.
Four recombinant E. coli BL21 (DE3) strains were grown to an optical density (OD) at 600 nm of 0.6. Protein expression was induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1.0 mM, followed by incubation for 4 h and harvesting of cells by centrifugation at 4°C.
Purification of Linoleate Hydratases
Overexpressed LAHs were present mainly as inclusion bodies. Solubilization and refolding of inclusion bodies were carried out with the protein refolding kit (Novagen) according to the manufacturer’s instructions. The refolded proteins were concentrated by using a 10 KDa Amicon Ultra-15 centrifugal filter Unit (Millipore, Germany).
After concentration, the His-tagged LAHs were purified by Ni-NTA spin columns (Qiagen). The purified LAHs were finally dialyzed against 50 mM 4-morpholineethanesulfonic acid (MES) buffer (pH 6.1) (Sigma-Aldrich) overnight at 4°C. The purified enzymes were assessed by SDS-PAGE and staining with Coomassie blue. FAD was added to the purified enzymes at a final concentration of 0.2 mM and incubated at 4°C for 24 h (Joo et al., 2012).
Enzymatic Activity Assay and Fatty Acid Analysis
To determine the enzymatic activity, 4.5 mg linoleic acid and 25 μg purified LAH were incubated in 1 ml of 50 mM MES buffer (pH 6.1) containing 50 mM NaCl, 2% ethanol and 10% glycerol at 25°C for 3 h. Fatty acids were extracted following the procedure described above. The organic phase of the extracts was collected and analyzed with LC-APPI-MS as described (Black et al., 2013a,b).
Construction of L. plantarum TMW1.460Δlah by Double-Crossover Mutagenesis
Gene disruption of lah in L. plantarum TMW1.460 was achieved by an in-frame, unmarked deletion (Su et al., 2011). The approximately 900 bp 5′-flanking regions (fragment A) and 1000 bp 3′-flanking regions (fragment B) of lah were amplified by PCR with primers listed in Table 1. The fragment A was digested with PstI, SphI, and fragment B was digested with SphI, HindIII. The resulting fragments were purified and sequentially ligated into vector pUC19 to generate pUC19/AB. The AB fragment in pUC19/AB was confirmed by sequencing, and sub-cloned into the PstI and HindIII restriction sites of pJRS233 to create pJRS233/Δlah. Recombinant plasmids were transformed into electrocompetent L. plantarum TMW1.460 at 12.5 kV/cm, 25 μF, and 200 Ω. The cells were grown in mMRS-Tween80 broth containing 5 mg/l erythromycin at 42–44°C for 80 generations to select for single-crossover mutants. Several colonies were isolated, cultured in mMRS-Tween80 broth for approximately 100 generations, and plated on mMRS-Tween80 agar at 30°C. The colonies were replica plated on mMRS-Tween80-erythromycin agar to identify erythromycin-sensitive double-crossover mutants. Gene replacement was confirmed by PCR amplification and sequencing. The phenotype was determined by LC-APPI-MS analysis of culture supernatant of L. plantarum TMW1.460Δlah grown in mMRS-Tween80 supplemented with 4 g/L linoleic acid.
Determination of Ethanol Resistance
Ethanol tolerance of L. plantarum TMW1.460 and TMW1.460Δlah was carried out with strains grown in mMRS-Tween 80, or mMRS-Tween 20. Ethanol was added as indicated and media were sterilized by filtration after addition of ethanol. Stationary phase cells were harvested by centrifugation, and resuspended in the same volume of mMRS-Tween 80 or -Tween 20 containing 20% ethanol. A 100 μl aliquot of cell suspension was analyzed as untreated control. The samples were incubated in 30°C and aliquots were removed in 1.5 h intervals and serially diluted in 0.85% NaCl. The appropriate dilutions were surface plated in duplicate on respective mMRS-Tween80 or -Tween20 agar and incubated at 30°C for 24 h. Ethanol resistance was determined in three independent experiments.
Determination of the Membrane Fluidity under Ethanol Stress
To investigate the membrane fluidity, LAURDAN (6-dodecanoyl-2-dimethylaminonaphthalene) (Thermo Fisher Scientific) was employed to measure generalized polarization (GP) value. L. plantarum TMW1.460 and TMW1.460Δlah were cultivated at 30°C for 20 h in mMRS-Tween80 or mMRS-Tween 20. The membrane fluidity of the cells influenced by ethanol at the concentration ranging from 0 to 16% was assessed as described (Molina-Höppner et al., 2004). The effect was determined in three independent experiments.
Physicochemical Properties of Cells Surface
Cell surface hydrophobicity was assessed by quantification of microbial adhesion to solvents (MATS; Kankaanpää et al., 2004). Solvents used were: chloroform (polar and electron acceptor) and tetradecane (non-polar), ethyl acetate (polar and electron donor) and octane (non-polar). The MATS is based on the comparison between microbial cell affinity to a polar and non-polar solvent within a couple that pose similar van der Waals surface tension components.
Lactobacillus plantarum TMW1.460 and TMW1.460Δlah were grown in mMRS-Tween 20, or mMRS without Tween but supplemented with 1 g/L oleic acid or linoleic acid. Cells were harvested by centrifugation, washed twice and resuspended in 150 mM NaCl to a final cell concentration of 108 CFU/ml. A 1 ml aliquot was removed as untreated control (A0). A 2.4 ml aliquot of cell suspension was mixed with 0.4 ml of solvent by vortexing for 60 s, respectively. The emulsified mixture was allowed to stand for 20 min to ensure the complete separation of the two phases. A sample (1 ml) was carefully taken from the aqueous phase (A). The optical cell density of sample A0 and A was measured at 600 nm. The microbial adhesion percentage to each solvent was calculated with the equation: percent affinity = 100 × [1-(A/A0)]. Each measurement was determined in three independent experiments.
Extraction and Purification of 13-HOE and 10-HOE
Extraction and purification of 13-HOE and 10-HOE was based on a protocol developed for 10-HOE (Black et al., 2013b). Fermentation of L. hammesii or L. plantarum TMW1.460Δlah in mMRS-Tween 80 with 4 g/L linoleic acid was conducted for production of 10-HOE or 13-HOE, respectively. The crude extraction of cellular lipids was performed as described above. Subsequently, the purification of 10-HOE or 13-HOE was based on a protocol described previously (Black et al., 2013b). The fermentation of L. hammesii or L. plantarum TMW1.460Δlah in mMRS-Tween 80 with 4 g/L linoleic acid and the crude extraction of fatty acid mixtures were performed as described above. For further purification, 25 mg of sample dissolved in chloroform was applied to a 500-mg Sep-Pak silica cartridge (Waters, Ltd, Mississauga, ON, Canada) previously equilibrated with 6 ml chloroform. The cartridge was successively washed with the following gradient of isopropanol in chloroform: 35 ml of chloroform, followed by 18 ml of 1, 5, 10, and 50% (vol/vol) isopropanol in chloroform. Eluates were collected and concentrated to dryness under nitrogen. The dry residues were dissolved in chloroform for analysis by LC-APPI-MS to identity of the product, and the removal of contaminating lipids.
Determination of the Antimicrobial Activity of Fatty Acids
The minimum inhibitory concentration (MIC) was determined to assess the toxicity of linoleic acid to lactobacilli, and to determine the antifungal activity of 10-HOE, 13-HOE, coriolic acid, ricinoleic acid, and linoleic acid. Lipids dissolved in ethanol were serially diluted twofold in mMRS-Tween80 using 96-well microtiter plates. Ethanol in the samples was evaporated under a sterile laminar flow hood before inoculation with indicator strains. Stationary phase cells of lactobacilli were harvested by centrifugation, washed twice in mMRS-Tween80 and diluted to approximately 107 CFU/ml in the same medium. The plates were incubated at 30°C.
For evaluation of the antifungal activity, lipids diluted with serial twofold dilutions. Conidiospores of A. niger and P. roqueforti were prepared as reported (Black et al., 2013b). The plate was inoculated with A. niger as an indicator organism and incubated at 25°C for 2 days, while the plate inoculated with P. roqueforti was incubated for 3 days. Fungal growth without addition of lipids served as the positive control and media alone as the negative control. The MIC was defined as the lowest concentration of lipids that inhibited the growth of fungi when growth was visible in the positive control. MIC values were determined by six independent experiments.
Statistical Analysis
Data analysis was conducted with R 3.1.2 (R Core Team, 2014). Significant differences in the assessment of cell survival and membrane fluidity under ethanol stress were determined by one-way analysis (ANOVA). Significance was assessed at a 5% probability of error (P < 0.05).
Accession Numbers
The sequences of linoleate 10-hydratase in L. reuteri, L. plantarum, L. hammesii, and L. spicheri were deposited in GenBank with accession numbers KX827285, KX827286, KX827287, KX827288, respectively.
Results
Identification of the Products of Linoleate Conversion by Lactobacilli
The products of linoleic acid conversion by five lactobacilli were analyzed by negative ion LC/APPI-MS/MS (Table 2). MS/MS spectra of the products confirmed the position of hydroxyl groups (Figure 1; Black et al., 2013a). The strains of lactobacilli differed in with respect to their conversion of linoleic acid to hydroxy fatty acids (Table 2). L. reuteri, L. hammesii, and L. spicheri produced 10-HOE only, while L. plantarum produced 10-HOE, 13-HOE and 10,13-dihydroxy octadecanoic acid. L. plantarum TMW1.460Δlah produced 13-HOE but not 10-HOE or 10,13-dihydroxy octadecanoic acid, demonstrating that their formation by L. plantarum TMW1.460 is attributable to a dedicated linoleate 13-hydratase acting on linoleic acid and 10-HOE, respectively. L. sanfranciscensis did not convert linoleic acid.
Table 2.
Comparison of products obtained from strain fermentation and enzymatic reaction with linoleic acid as substrate.
| Strain | Products of fermentation | Products of LAH | Fragmentation ions (m/z) |
|---|---|---|---|
| L. reuteri LTH2584 | 10-HOEa | 10-HOE | 297[M-H]-, 185 |
| L. hammesii DSM16381 | 10-HOE | 10-HOE | 297[M-H]-, 185 |
| L. spicheri LS38 | 10-HOE | 10-HOE | 297 [M-H]-, 185 |
| L. plantarum TMW1.460 | 10-HOE, 13-HOEa and 10,13-HOAa |
10-HOE | 297 [M-H]-, 185 297 [M-H]-, 99, 197 315 [M-H]-, 99, 127, 185, 243 |
| L. plantarum TMW1.460 Δlah | 13-HOE | — | 297 [M-H]-, 99, 197 |
| L. sanfranciscensis ATCC 27651 | No products | No products | — |
a10-HOE, 10-hydroxy-12-octadecenoic acid; 13-HOE, 13-hydroxy-9-octade cenoic acid; 10,13-HOA, 10,13-dihydroxy octadecanoic acid.
FIGURE 1.
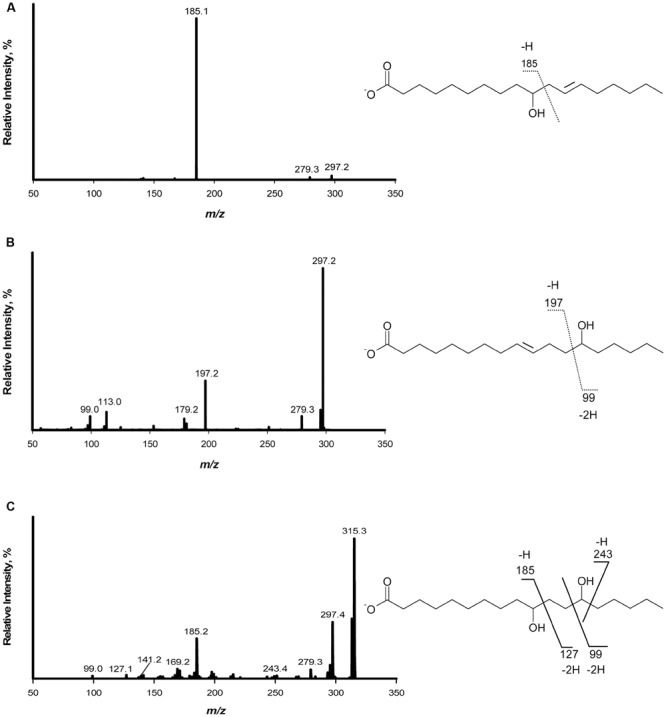
Fragmentation pattern and APPI-MS/MS spectra of hydroxy fatty acids produced when linoleic acid was used as substrate. (A) Mass spectrum of 10-hydroxy-12- octadecenoic acid; (B) Mass spectrum of 13-hydroxy-9-octadecenoic acid; (C) Mass spectrum of 10,13-dihydroxy octadecanoic acid.
Phylogenetic Analysis of Linoleate Hydratase
Phylogenetic relationships of putative LAHs from lactobacilli were compared to the corresponding enzymes of the 24 type strains in the genus Lactobacillus, and type strains from the genera Pediococcus, Weissella, Leuconostoc, and Oenococcus (Figure 2). All four hydratases from lactobacilli that were investigated in this study belonged to myosin cross reactive antigen family. L. sanfranciscensis harbored no LAH. The topology of the protein tree did not conform to the evolutionary relationship of the organisms (Zheng et al., 2015). The tree displayed two clusters but the two clusters do not differentiate between linoleate 10-hydratases and linoleate 13-hydratases (Figure 2).
FIGURE 2.
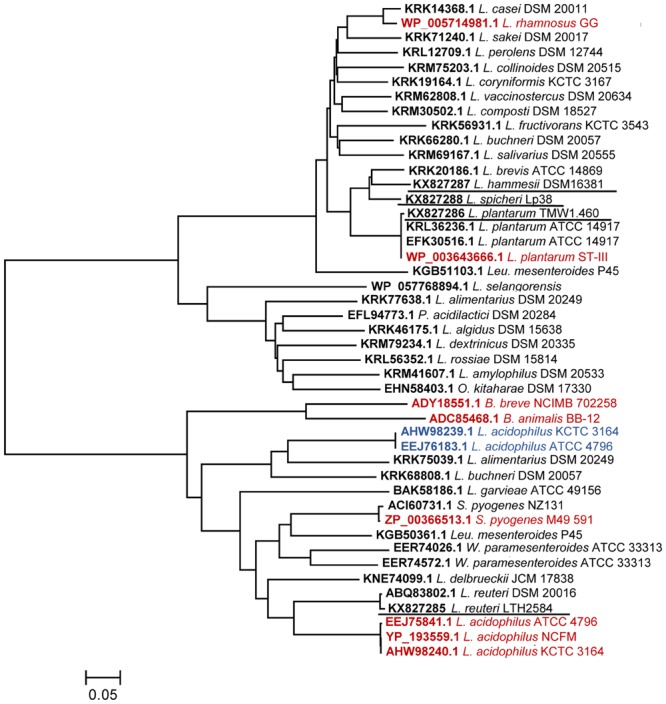
Phylogenetic tree of linoleate hydratases (LAHs). The evolutionary relationships are shown with scale bar line which represents an evolutionary distance of 0.05. Linoleate 10-hydratases reported in the literature are highlighted in red, linoleate 13-hydratases are highlighted in blue, linoleate 10-hydratases that were characterized in this study are underlined.
Characterization of Linoleate 10-Hydratase
Sequence analysis did not distinguish 10-hydratases from 13-hydratases, therefore, LAHs from four strains of lactobacilli were characterized biochemically after heterologous expression in E. coli and purification by affinity chromatography. A single band was observed by SDS-PAGE analysis after purification of the four enzymes (Figure 3); this band was absent in crude cellular extracts of E. coli strains prior to induction (data not shown). The genes from L. reuteri, L. plantarum, L. hammesii, and L. spicheri encoded a protein of 590, 564, 564, 564 amino acids, respectively, matching the size of the major band observed by SDS-PAGE analysis (Figure 3). The addition of cofactor FAD to apoenzyme was essential for activity. LC-APPI-MS analysis revealed that all four recombinant proteins produced 10-HOE from linoleic acid, as demonstrated by the fragment ion at m/z 185.1 in the MS/MS spectra (Table 2). Hence, all four recombinant proteins were shown to be 10-hydratases. The substrate specificity of the linoleate 10-hydratase of L. plantarum TMW1.460 confirmed that formation of 13-HOE and 10,13-dihydroxy octadecanoic acid by this strain is likely attributable to a second and dedicated linoleate 13-hydratase.
FIGURE 3.
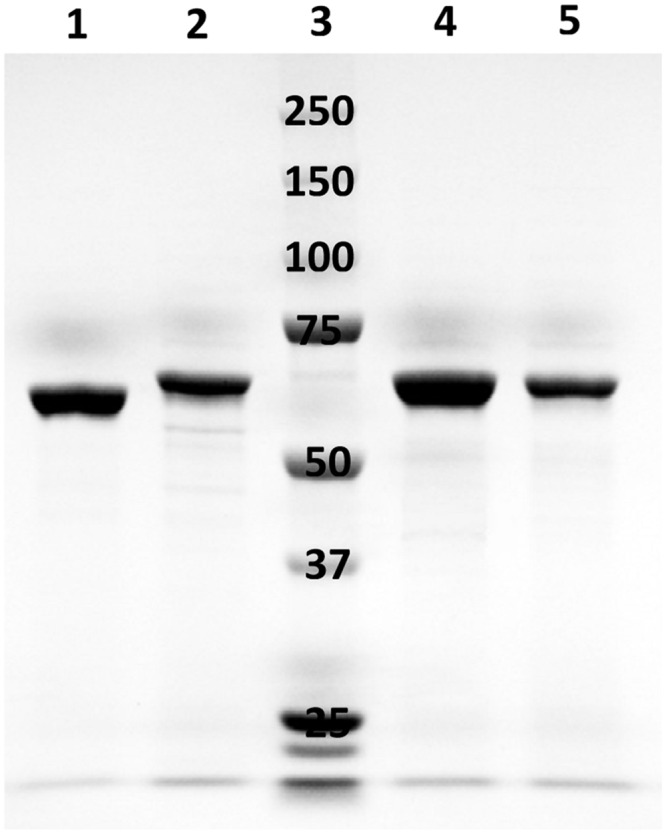
SDS-PAGE analysis of purified LAHs expressed from respective recombinant Escherichia coli BL21 (DE3) cells. Lane 1, LAH of Lactobacillus reuteri; lane 2, LAH of Lactobacillus plantarum; lane 3, molecular mass marker proteins (250, 150, 100, 75, 50, 37, and 25 kDa); lane 4, LAH of Lactobacillus hammesii; lane 5, LAH of Lactobacillus spicheri.
Most Lactobacilli Require Oleic Acid or Linoleic Acid for Growth
Tween 80 is a derivative of oleate and a component of mMRS (Johnsson et al., 1995; Gänzle et al., 2000). mMRS-Tween20 did not support the growth of L. reuteri, L. hammesii, and L. sanfranciscensis but did support the growth of L. plantarum and its Δlah mutant (data not shown). However, unsaturated fatty acids (UFAs) may also exhibit antibacterial activity (Desbois and Smith, 2010). Therefore, the MICs of oleic and linoleic acid to lactobacilli including L. plantarum TMW1.460Δlah were assessed. The MICs of oleic and linoleic acids were >8 g/L for all strains, which indicates a high tolerance toward oleic and linoleic acid. L. sanfranciscensis showed less tolerance and its growth was inhibited by 1 g/L oleic acid and 0.5 g/L linoleic acid.
The Effect of lah on Stress Tolerance in L. plantarum
To investigate the effect of hydroxy fatty acids or 10-LAH itself on ethanol resistance, the survival of L. plantarum TMW1.460 and TMW1.460Δlah was assessed in mMRS-Tween80 and mMRS-Tween20 containing 20% ethanol. The ethanol tolerance of L. plantarum TMW1.460 and TMW1.460Δlah did not differ (Figure 4). However, the presence of Tween 80 in the growth medium enhanced bacterial survival, especially for L. plantarum TMW1.460 (Figure 4).
FIGURE 4.
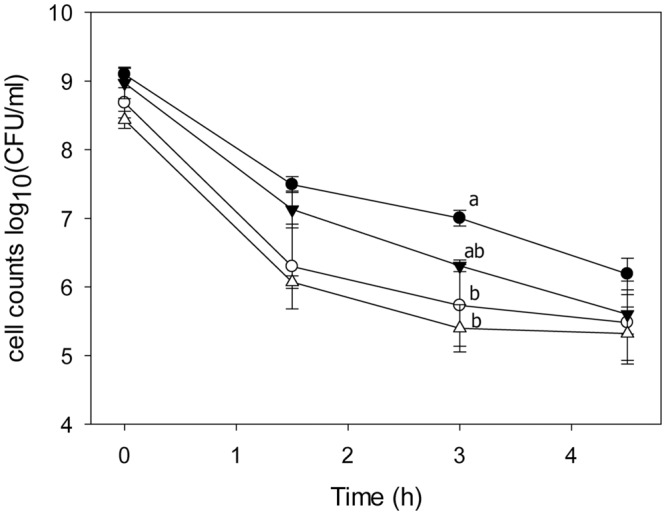
Survival of L. plantarum and its lah deficient derivative under 20% ethanol treatment. L. plantarum TMW1.460 was incubated in mMRS-Tween80 (closed circles) or mMRS-Tween20 (open circles); L. plantarum TMW1.460Δlah was grown in mMRS-Tween80 (closed triangle) or mMRS-Tween 20 (open triangle) during treatment. Values obtained at the same treatment time that do not share common superscripts are significantly different (P < 0.05). Data represent mean ± standard deviation of three independent experiments with duplicate determinations of cell counts.
The Effect of lah on Ethanol-Dependent Membrane Fluidity in L. plantarum
The ethanol-dependent membrane phase behavior of strains grown in mMRS-Tween80 or -Tween 20 was analyzed (Figure 5). The GP values decreased with increasing ethanol concentration, indicating that ethanol increased the fluidity of the membrane. The response of the membrane fluidity of L. plantarum TMW1.460 and TMW1.460Δlah to ethanol was similar. Tween 80 supplemented medium induced a more fluid membrane in both strains.
FIGURE 5.
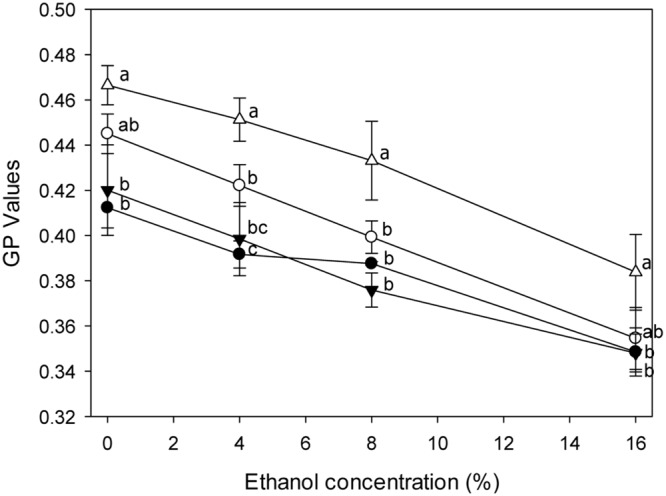
General polarization (GP) values of L. plantarum and its Δlah derivative stained with Laurdan under ethanol stress. L. plantarum TMW1.460 was cultivated in mMRS-Tween80 (closed circles) or mMRS-Tween 20 (open circles) prior to staining; L. plantarum TMW1.460Δlah was cultivated in mMRS-Tween80 (closed triangle) or mMRS-Tween 20 (open triangle) prior to staining. Values obtained at the same treatment time that do not share common superscripts are significantly different (P < 0.05). Data represent mean ± standard deviation of three independent experiments with duplicate determinations of cell counts.
Influence of lah on Cell Surface Properties in L. plantarum
The MATS of L. plantarum TMW1.460 and TMW1.460Δlah cultivated in mMRS-Tween20, or mMRS-Tween20 supple mented with oleic or linoleic acid are shown in Figure 6. The deletion of lah modified the properties of the cell surface. The adhesion of L. plantarum TMW1.460Δlah was more solvent-dependent when compared to the wild-type strain and the affinity of cells to the four solvents was generally higher for Δlah mutant than for wild-type strain. Similar trends were noted with strains grown in mMRS with different supplements, suggesting that the differences between wild-type and mutant strains are attributable to 10-LAH, not the product of this enzyme. Similar behavior was observed when strains cultivated in Tween 80 (Data not shown).
FIGURE 6.
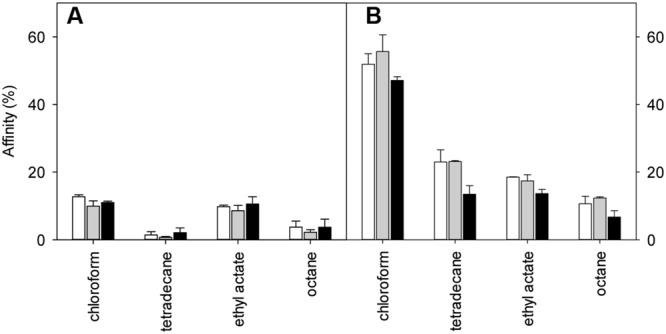
Effect of 10-lah on cell surface properties of L. plantarum grown in different media. Cell surface hydrophobicity was measured using the MATS method. (A) L. plantarum TMW1.460; (B) L. plantarum TMW1.460Δlah. White bar indicates % affinity to solvents when cells were grown in mMRS -Tween 20; gray bar indicated in mMRS (Tween 20) supplemented with 1 g/L oleic acid; black bar indicated in mMRS (Tween 20) supplemented with 1 g/L linoleic acid. Data represent mean ± standard deviation of three independent experiments.
In this study, both wild-type and mutant strains exhibited low affinity for tetradecane and octane (non-polar solvents), indicating that the cell surface of both strains was hydrophilic rather than hydrophobic. To determine the effect of deletion of lah on the Lewis electron donor/electron acceptor property, the bacterial affinity to chloroform and ethyl acetate between different strains were also compared (Figure 6). L. plantarum wild-type strain showed similar adhesion ability to chloroform and ethyl acetate. However, regardless of the different supplement in medium, the adhesion of the mutant strain was always higher to chloroform (electron acceptor and acidic solvent) than to ethyl acetate (electron donor and basic solvent).
Antifungal Properties of Purified Hydroxy Fatty Acids
13-HOE and 10-HOE were purified from organic extracts of cultures of L. plantarum TMW1.460Δlah and L. hammesii, respectively, when 4 g/L linoleic acid was used as substrate. The major impurity detected in the organic phase was linoleic acid; other more oxidized forms of hydroxy C18 fatty acids were also present. The compounds were purified by silica solid phase microextraction and analyzed by LC-APPI-MS/MS. 10-HOE or 13-HOE were eluted in the 1% isopropanol fraction and a single peak was observed in the LC-APPI-MS/MS chromatogram after purification. The antifungal activity of purified 13-HOE and 10-HOE against A. niger and P. roqueforti is shown in Table 3, and compared to reference lipids differing in the number and position of hydroxyl groups or double bonds. Both coriolic acid and ricinoleic acid were active against all fungi indicators with MICs between 0.26 and 0.29. Linoleic acid showed the lowest antifungal activity.
Table 3.
Minimum inhibitory concentrations (MICs) of hydroxy fatty acids extracted from cultures of L. hammesii and L. plantarum TMA1.460Δlah and reference fatty acids (n = 3).
 |
Discussion
Linoleate hydratases are highly conserved in both Gram-positive and Gram-negative bacteria (Volkov et al., 2010). Our study revealed that 10-LAH and 13-LAH, or enzymes that produce both 10-HOE and 13-HOE are not distinguished by phylogenetic and sequence analysis. All hydratases from lactobacilli examined in this study were determined as 10-LAH, however, they are distributed in two different clusters which also contain 13-LAH. Moreover, the topology of the protein tree disagreed with the evolutionary relationship of the organisms (Zheng et al., 2015), indicating that LAHs are accessory proteins (Wyllie et al., 2000).
Linoleate 10-hydratase is a FAD-containing enzyme and exhibits flavin-like UV absorbance (Rosberg-Cody et al., 2011). FAD cofactor is bound to the conserved FAD binding motif of 10-LAH and stabilizes the active conformation of the enzyme but it is not directly involved in catalysis (Volkov et al., 2010; Yang et al., 2013). The non-covalently bound FAD is easily lost in the purification process (Volkov et al., 2013). This study confirmed that 10-LAH is a FAD-dependent enzyme (Joo et al., 2012) and demonstrated that purification of active 10-LAH from inclusion bodies required addition of FAD. Different from the LAH of S. pyogenes, which catalyzes formation of 10-HOE and 13-HOE (Volkov et al., 2010), the 10-LAH characterized in this study formed exclusively 10-HOE from linoleate. The comparison of products formed by L. plantarum TMS1.460, the 10-LAH deficient mutant of this strain, and the 10-LAH of this strain strongly suggest that 13-HOE and 10,13 dihydroxy octadecanoic acid formation by this strain is attributable to a linoleate 13-hydratase that was recently characterized in L. acidophilus (Park et al., 2015).
Unsaturated fatty acids are essential for growth of many LAB (Johnsson et al., 1995; Gänzle et al., 2000) but high concentrations may inhibit growth of LAB (Guerrini et al., 2002). In contrast, in the present work oleic and linoleic acids stimulated growth of L. reuteri, L. hammesii, and L. sanfranciscensis. The bacteriostatic and bactericidal activities exerted by UFAs against lactobacilli is strain dependent (Jenkins and Courtney, 2003); the observation that the LAH-negative L. sanfranciscensis was the only strain that was inhibited by oleic and linoleic acids suggests that LAH contributes to these strain-specific differences.
Oleic acid modulated membrane fluidity, and influenced the ethanol tolerance of L. plantarum TMW1.460 and TMW1.460Δlah. Consistent with our results, addition of Tween 80 to the growth medium increased viability of Oenococcus oeni in wine (Guerrini et al., 2002) and supplementation of UFAs to Saccharomyces cerevisiae protected against stress (Mishra and Prasad, 1989). However, we observed no difference in ethanol resistance between L. plantarum TMW1.460 and TMW1.460Δlah. The protective effect of 10-LAH that was previously observed in exponential phase bifidobacteria may relate to the presence of the enzyme rather than its products (Rosberg-Cody et al., 2011; O’Connell et al., 2013).
The physiochemical properties of cell surface play a critical role in adhesion of pathogens and probiotics to intestinal surfaces. An evaluation of surface properties is achieved by determination of MATS (Bellon-Fontaine et al., 1996; Rosenberg, 2006). Both L. plantarum wild-type and lah mutant strains displayed a hydrophilic surface character with weak adhesion to non-polar solvents. Similar results were obtained in other Lactobacillus spp. and Lactococcus spp. (Pelletier et al., 1997; Boonaert and Rouxhet, 2000; Ly et al., 2006). The hydrophobicity of the cell surface changed when the bacteria were cultivated in medium with addition of (UFAs; Kankaanpää et al., 2004; Muller et al., 2011), which was not observed in our study. Bacteria grown in mMRS medium with or without different supplement exhibited similar affinity to solvents. However, lah deficiency resulted in a fundamental change in the profile of solvent affinity. Compared with wild-type strain, L. plantarum TMW1.460Δlah presented more basic and electron-donating properties. The bacteria with basic character are considered to possess COO- and HSO3- chemical groups on their cell surface (Pelletier et al., 1997). Lactobacillus casei BL83, BL208 and BL229 also displayed low cell adhesion that was associated with their basic surface property (Muñoz-Provencio et al., 2009). Indeed, the deletion of 10-LAH was involved in the reduced adherence to human keratinocytes by S. pyogenes and to human intestinal epithelial cells by L. acidophilus (O’Flaherty and Klaenhammer, 2010; Volkov et al., 2010). In our study, 10-LAH mediated differences in cell surface properties may explain the changed cell adhesion to human cells in the previous reports.
Linoleic acid possesses antifungal activity against the plant pathogenic fungi, especially for Crinipellis perniciosa at the concentration of 100 μM (Walters et al., 2004). In our study, linoleic acid was less active against A. niger and P. roqueforti, fungi that are commonly found in cereals and cereal products (Legan, 1993; Sjögren et al., 2003). Remarkably, the MIC of 13-HOE was approximately 15 times lower than that of linoleic acid. Therefore, 13-HOE may be as suitable as 10-HOE as an antifungal agent in foods (Black et al., 2013b). The antifungal activity of hydroxy fatty acids is likely related to their partitioning into lipid bilayers, thus increasing membrane permeability (Sjögren et al., 2003). The MICs of 13-HOE, 10-HOE along with coriolic acid and ricinoleic acid were all comparable, suggesting that all unsaturated monohydroxy fatty acids of C18 varying in their hydroxyl group position or degree of unsaturation exert antifungal property. Similar inhibition activities of Aspergillus and Penicillium spp. were detected among 9- and 13-hydroxy of C18 UFA analogs of plant oxylipins (Prost et al., 2005). In contrast, dihydroxy saturated C18 fatty acids do not display antifungal activity (Black et al., 2013b).
Conclusion
Our study revealed that the differentiation of accessory proteins between 10-LAH and 13-LAH cannot be achieved by phylogenetic analysis. Thus the LAHs from lactobacilli were characterized by heterologous expression and identified as FAD-dependent 10-LAH. Generation of a 10-LAH deficient mutant of L. plantarum demonstrated that 13-HOE generated by a different and dedicated hydratase, and is a novel antifungal hydroxy fatty acid. The most prominent physiological difference of the 10-LAH deficient mutant and the wild-type strain was the altered surface hydrophobicity of the bacterial cells. L. plantarum is part of the phyllosphere of many plants (Minervini et al., 2015) and oxylipids are an important component of the plant defense against pathogens (Prost et al., 2005). It is possible that the lipid-converting properties of LAHs and their influence on cell surface properties are components of host–microbe interactions.
Author Contributions
YC and NL conducted experiments, YC and MG wrote the manuscript, JC contributed to experimental design and writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Alberta Agriculture Funding Consortium (project number 2014F126R) is acknowledged for funding. YC acknowledges support from the China Scholarship Council; NL acknowledges support from Mitacs. Dr. Euna Oh is acknowledged for support with cloning and expression of enzymes.
Footnotes
References
- Bellon-Fontaine M. N., Rault J., Van Oss C. J. (1996). Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf. B Biointerfaces 7 47–53. 10.1016/0927-7765(96)01272-6 [DOI] [Google Scholar]
- Black B. A., Sun C., Zhao Y. Y., Gänzle M. G., Curtis J. M. (2013a). Antifungal lipids produced by lactobacilli and their structural identification by normal phase LC/atmospheric pressure photoionization-MS/MS. J. Agric. Food Chem. 61 5338–5346. 10.1021/jf400932g [DOI] [PubMed] [Google Scholar]
- Black B. A., Zannini E., Curtis J. M., Gänzle M. G. (2013b). Antifungal hydroxy fatty acids produced during sourdough fermentation: microbial and enzymatic pathways, and antifungal activity in bread. Appl. Environ. Microbiol. 79 1866–1873. 10.1128/AEM.03784-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonaert C. J., Rouxhet P. G. (2000). Surface of lactic acid bacteria: relationships between chemical composition and physicochemical properties. Appl. Environ. Microbiol. 66 2548–2554. 10.1128/AEM.66.6.2548-2554.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois A. P., Smith V. J. (2010). Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 85 1629–1642. 10.1007/s00253-009-2355-3 [DOI] [PubMed] [Google Scholar]
- Gänzle M. G., Höltzel A., Walter J., Jung G., Hammes W. P. (2000). Characterization of reutericyclin produced by Lactobacillus reuteri LTH2584. Appl. Environ. Microbiol. 66 4325–4333. 10.1128/AEM.66.10.4325-4333.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini S., Bastianini A., Granchi L., Vincenzini M. (2002). Effect of oleic acid on Oenococcus oeni strains and galolactic fermentation in wine. Curr. Microbiol. 44 5–9. 10.1007/s00284-001-0066-9 [DOI] [PubMed] [Google Scholar]
- Hou C. T. (2008). New bioactive fatty acids. Asia Pac. J. Clin. Nutr. 17(Suppl. 1) 192–195. [PubMed] [Google Scholar]
- Hou C. T., Forman Iii R. J. (2000). Growth inhibition of plant pathogenic fungi by hydroxy fatty acids. J. Ind. Microbiol. Biotechnol. 24 275–276. 10.1038/sj.jim.2900816 [DOI] [Google Scholar]
- Jenkins J. K., Courtney P. D. (2003). Lactobacillus growth and membrane composition in the presence of linoleic or conjugated linoleic acid. Can. J. Microbiol. 49 51–57. 10.1139/w03-003 [DOI] [PubMed] [Google Scholar]
- Johnsson T., Nikkilä P., Toivonen L., Rosenqvist H., Laakso S. (1995). Cellular fatty acid profiles of Lactobacillus and Lactococcus strains in relation to the oleic acid content of the cultivation medium. Appl. Environ. Microbiol. 61 4497–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo Y. C., Jeong K. W., Yeom S. J., Kim Y. S., Kim Y., Oh D. K. (2012). Biochemical characterization and FAD-binding analysis of oleate hydratase from Macrococcus caseolyticus. Biochimie 94 907–915. 10.1016/j.biochi.2011.12.011 [DOI] [PubMed] [Google Scholar]
- Kankaanpää P., Yang B., Kallio H., Isolauri E., Salminen S. (2004). Effects of polyunsaturated fatty acids in growth medium on lipid composition and on physicochemical surface properties of lactobacilli. Appl. Environ. Microbiol. 70 129–136. 10.1128/AEM.70.1.129-136.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil K. S., Cunningham M. W., Barnett L. A. (1994). Cloning and sequence analysis of a gene encoding a 67-kilodalton myosin-cross-reactive antigen of Streptococcus pyogenes reveals its similarity with class II major histocompatibility antigens. Infect. Immun. 62 2440–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. R., Oh D. K. (2013). Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 31 1473–1485. 10.1016/j.biotechadv.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Kim K. R., Oh H. J., Park C. S., Hong S. H., Park J. Y., Oh D. K. (2015). Unveiling of novel regio-selective fatty acid double bond hydratases from Lactobacillus acidophilus involved in the selective oxyfunctionalization of mono- and di-hydroxy fatty acids. Biotechnol. Bioeng. 112 2206–2213. 10.1002/bit.25643 [DOI] [PubMed] [Google Scholar]
- Kishino S., Park S. B., Takeuchi M., Yokozeki K., Shimizu S., Ogawa J. (2011). Novel multi-component enzyme machinery in lactic acid bacteria catalyzing C = C double bond migration useful for conjugated fatty acid synthesis. Biochem. Biophys. Res. Commun. 416 188–193. 10.1016/j.bbrc.2011.11.022 [DOI] [PubMed] [Google Scholar]
- Koritala S., Bagby M. O. (1992). Microbial conversion of linoleic and linolenic acids to unsaturated hydroxy fatty acids. J. Am. Oil Chem. Soc. 69 575–578. 10.1007/BF02636111 [DOI] [Google Scholar]
- Legan J. D. (1993). Mould spoilage of bread: the problem and some solutions. Int. Biodeterior. Biodegr. 32 33–53. 10.1016/0964-8305(93)90038-4 [DOI] [Google Scholar]
- Ly M. H., Vo N. H., Le T. M., Belin J. M., Waché Y. (2006). Diversity of the surface properties of lactococci and consequences on adhesion to food components. Colloids Surf. B Biointerfaces 52 149–153. 10.1016/j.colsurfb.2006.04.015 [DOI] [PubMed] [Google Scholar]
- Magnusson J., Ström K., Roos S., Sjögren J., Schnürer J. (2003). Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 219 129–135. 10.1016/S0378-1097(02)01207-7 [DOI] [PubMed] [Google Scholar]
- Minervini F., Celano G., Lattanzi A., Tedone L., De Mastro G., Gobbetti M., et al. (2015). Lactic acid bacteria in durum wheat flour are endophytic components of the plant during its entire life cycle. Appl. Environ. Microbiol. 81 6736–6748. 10.1128/AEM.01852-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Prasad R. (1989). Relationship between ethanol tolerance and fatty acyl composition of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 30 294–298. 10.1007/BF00256221 [DOI] [Google Scholar]
- Molina-Höppner A., Doster W., Vogel R. F., Gänzle M. G. (2004). Protective effect of sucrose and sodium chloride for Lactococcus lactis during sublethal and lethal high-pressure treatments. Appl. Environ. Microbiol. 70 2013–2020. 10.1128/AEM.70.4.2013-2020.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J. A., Ross R. P., Sybesma W. F., Fitzgerald G. F., Stanton C. (2011). Modification of the technical properties of Lactobacillus johnsonii NCC 533 by supplementing the growth medium with unsaturated fatty acids. Appl. Environ. Microbiol. 77 6889–6898. 10.1128/AEM.05213-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Provencio D., Llopis M., Antolín M., de Torres I., Guarner F., Pérez-Martínez G., et al. (2009). Adhesion properties of Lactobacillus casei strains to resected intestinal fragments and components of the extracellular matrix. Arch. Microbiol. 191 153–161. 10.1007/s00203-008-0436-9 [DOI] [PubMed] [Google Scholar]
- O’Connell K. J., Motherway M. O., Hennessey A. A., Brodhun F., Ross R. P., Feussner I., et al. (2013). Identification and characterization of an oleate hydratase-encoding gene from Bifidobacterium breve. Bioengineered 4 313–321. 10.4161/bioe.24159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Flaherty S. J., Klaenhammer T. R. (2010). Functional and phenotypic characterization of a protein from Lactobacillus acidophilus involved in cell morphology, stress tolerance and adherence to intestinal cells. Microbiology 156 3360–3367. 10.1099/mic.0.043158-0 [DOI] [PubMed] [Google Scholar]
- Oliveira P. M., Zannini E., Arendt E. K. (2014). Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: from crop farming to cereal products. Food Microbiol. 37 78–95. 10.1016/j.fm.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Park J. Y., Lee S. H., Kim K. R., Park J. B., Oh D. K. (2015). Production of 13S-hydroxy-9(Z)-octadecenoic acid from linoleic acid by whole recombinant cells expressing linoleate 13-hydratase from Lactobacillus acidophilus. J. Biotechnol. 208 1–10. 10.1016/j.jbiotec.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Pelletier C., Bouley C., Cayuela C., Bouttier S., Bourlioux P., Bellon-Fontaine M. N. (1997). Cell surface characteristics of Lactobacillus casei subsp. casei, Lactobacillus paracasei subsp. paracasei, and Lactobacillus rhamnosus strains. Appl. Environ. Microbiol. 63 1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost I., Dhondt S., Rothe G., Vicente J., Rodriguez M. J., Kift N., et al. (2005). Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 139 1902–1913. 10.1104/pp.105.066274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rosberg-Cody E., Liavonchanka A., Göbel C., Ross R. P., O’Sullivan O., Fitzgerald G. F., et al. (2011). Myosin-cross-reactive antigen (MCRA) protein from Bifidobacterium breve is a FAD-dependent fatty acid hydratase which has a function in stress protection. BMC Biochem. 12:9 10.1186/1471-2091-12-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. (2006). Microbial adhesion to hydrocarbons: twenty-five years of doing MATH. FEMS Microbiol. Lett. 262 129–134. 10.1111/j.1574-6968.2006.00291.x [DOI] [PubMed] [Google Scholar]
- Sjögren J., Magnusson J., Broberg A., Schnürer J., Kenne L. (2003). Antifungal 3-hydroxy fatty acids from Lactobacillus plantarum MiLAB 14. Appl. Environ. Microbiol. 69 7554–7557. 10.1128/AEM.69.12.7554-7557.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M. S., Schlicht S., Gänzle M. G. (2011). Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Fact. 10(Suppl. 1):S8 10.1186/1475-2859-10-S1-S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov A., Khoshnevis S., Neumann P., Herrfurth C., Wohlwend D., Ficner R., et al. (2013). Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus. Acta Crystallogr. D Biol. Crystallogr. 69 648–657. 10.1107/S0907444913000991 [DOI] [PubMed] [Google Scholar]
- Volkov A., Liavonchanka A., Kamneva O., Fiedler T., Goebel C., Kreikemeyer B., et al. (2010). Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J. Biol. Chem. 285 10353–10361. 10.1074/jbc.M109.081851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D., Raynor L., Mitchell A., Walker R., Walker K. (2004). Antifungal activities of four fatty acids against plant pathogenic fungi. Mycopathologia 157 87–90. 10.1023/B:MYCO.0000012222.68156.2c [DOI] [PubMed] [Google Scholar]
- Wyllie D. H., Kiss-Toth E., Visintin A., Smith S. C., Boussouf S., Segal D. M., et al. (2000). Evidence for an accessory protein function for toll-like receptor 1 in anti-bacterial responses. J. Immunol. 165 7125–7132. 10.4049/jimmunol.165.12.7125 [DOI] [PubMed] [Google Scholar]
- Yang B., Chen H., Song Y., Chen Y. Q., Zhang H., Chen W. (2013). Myosin-cross-reactive antigens from four different lactic acid bacteria are fatty acid hydratases. Biotechnol. Lett. 35 75–81. 10.1007/s10529-012-1044-y [DOI] [PubMed] [Google Scholar]
- Zhang C., Brandt M. J., Schwab C., Gänzle M. G. (2010). Propionic acid production by cofermentation of Lactobacillus buchneri and Lactobacillus diolivorans in sourdough. Food Microbiol. 27 390–395. 10.1016/j.fm.2009.11.019 [DOI] [PubMed] [Google Scholar]
- Zheng J., Ruan L., Sun M., Gänzle M. (2015). A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 81 7233–7243. 10.1128/AEM.02116-15 [DOI] [PMC free article] [PubMed] [Google Scholar]


