Abstract
Fluoroquinolone (FQ) resistance is highly prevalent among clinical strains of Pseudomonas aeruginosa, limiting treatment options. We have reported previously that highly virulent strains containing the exoU gene of the type III secretion system are more likely to be FQ-resistant than strains containing the exoS gene, as well as more likely to acquire resistance-conferring mutations in gyrA/B and parC/E. We hypothesize that FQ-resistance imposes a lower fitness cost on exoU compared to exoS strains, thus allowing for better adaptation to the FQ-rich clinical environment. We created isogenic mutants containing a common FQ-resistance conferring point mutation in parC from three exoU to three exoS clinical isolates and tested fitness in vitro using head-to-head competition assays. The mutation differentially affected fitness in the exoU and exoS strains tested. While the addition of the parC mutation dramatically increased fitness in one of the exoU strains leaving the other two unaffected, all three exoS strains displayed a general decrease in fitness. In addition, we found that exoU strains may be able to compensate for the fitness costs associated with the mutation through better regulation of supercoiling compared to the exoS strains. These results may provide a biological explanation for the observed predominance of the virulent exoU genotype in FQ-resistant clinical subpopulations and represent the first investigation into potential differences in fitness costs of FQ-resistance that are linked to the virulence genotype of P. aeruginosa. Understanding the fitness costs of antibiotic resistance and possibilities of compensation for these costs is essential for the rational development of strategies to combat the problem of antibiotic resistance.
Keywords: Pseudomonas aeruginosa, fluoroquinolone resistance, type III secretion, fitness, virulence
Introduction
Pseudomonas aeruginosa is a Gram-negative pathogen that is the leading cause of nosocomial pneumonia (Restrepo and Anzueto, 2009; Quartin et al., 2013). Resistance to the fluoroquinolone (FQ) antibiotics has risen dramatically due to the widespread prescribing of this class of drug, limiting treatment options for P. aeruginosa infections (Linder et al., 2005). P. aeruginosa acquires resistance to the FQs through mutations in genes regulating the expression of efflux pumps and through point mutations in target site genes. The target enzymes of the FQs are the type II topoisomerases, GyrA/B and ParC/E (Hooper, 2001). Resistance-conferring mutations in these genes, known as target site mutations, have been well described in P. aeruginosa (Nakano et al., 1997; Mouneimné et al., 1999; Higgins et al., 2003).
Pseudomonas aeruginosa has the ability to cause a variety of severe infections due to its many virulence factors. Specifically, P. aeruginosa utilizes the type III secretion system (TTSS) during acute infections to evade phagocytosis, invade host cells, and cause cell death (Veesenmeyer et al., 2009). The TTSS consists of a molecular syringe-like apparatus that extends through the inner and outer membranes and directly contacts the host cell. This allows effector toxins (ExoU, ExoS, ExoY, and/or ExoT) to be directly injected into the cytoplasm of host cells. Although residing at entirely separate loci, the genes encoding the toxins ExoU and ExoS are mutually exclusive in most strains, with the exoS genotype accounting for about 70% of clinical and environmental strains (Feltman et al., 2001; Garey et al., 2008). While less prevalent in general, exoU strains are more virulent than exoS strains, as has been shown in animal models of acute pneumonia (Shaver and Hauser, 2004). Importantly, infection with these strains leads to poor outcomes in patients with ventilator-associated pneumonia (Roy-Burman et al., 2001; El Solh et al., 2008) as well as increased persistence and severity of disease (Schulert et al., 2003).
Alarmingly, clinical studies have shown a correlation between FQ-resistance and virulence. We have previously reported that patients infected with FQ-resistant strains of P. aeruginosa had threefold higher mortality or prolonged illness by an additional 5 days compared to those infected with FQ-susceptible strains (Hsu et al., 2005). In addition, clinical isolates with the more virulent exoU genotype were shown to more likely be FQ-resistant than exoS strains (Wong-Beringer et al., 2008; Agnello and Wong-Beringer, 2012). Others have reported similar results in isolates from various infection sites (Zhang et al., 2014; Peña et al., 2015). Furthermore, in a separate study, we found that the combined traits of FQ-resistance and exoU genotype among respiratory isolates of P. aeruginosa were significantly associated with the development of pneumonia rather than bronchitis or colonization (Sullivan et al., 2014), suggesting that resistance and virulence traits may be linked, negatively impacting disease severity.
In a large study of 270 clinical isolates, we found that significantly more exoU strains were FQ-resistant, compared to exoS strains (63% vs. 49%, p = 0.03). Sequencing of the FQ target site genes gyrA/B and parC/E revealed that exoU strains were more likely than exoS strains to acquire two or more resistance-conferring target site mutations (Agnello and Wong-Beringer, 2012). Specifically, we found that while FQ-resistant exoU and exoS strains were similarly likely to possess a mutation in gyrA, exoU isolates were more likely to also have acquired an additional mutation in parC, resulting in greatly increased levels of resistance. While there is mounting evidence for the co-selection of resistance and virulence traits among pathogenic bacteria (Beceiro et al., 2013), the biological mechanisms underlying these observations are not well understood. Since the identification of the ExoU and ExoS toxins of the P. aeruginosa type III secretion system, many studies have investigated the roles of each toxin during infection (Allewelt et al., 2000; Schulert et al., 2003; Shaver and Hauser, 2004, 2006) while others have described the association of exoU strains with increased FQ resistance (Lakkis and Fleiszig, 2001; Maatallah et al., 2011) supporting our own observations. The aim of the current study was to gain insights into the biological basis underlying the differential development of resistance in exoU vs. exoS strains, which previous studies have not explored. We hypothesize that the greater propensity of exoU strains to acquire multiple target site mutations compared to exoS strains reflects a difference in the fitness costs associated with FQ-resistance mutations, favoring the strains with the exoU genetic background.
A recent meta-analysis of studies conducted on fitness costs of resistance to a variety of antibiotic classes in different organisms showed that most point mutations are generally costly (Melnyk et al., 2015). However, there is great variability depending on organism, drug, and mechanism of resistance. Furthermore, fitness costs can be mitigated through the accumulation of compensatory mutations that restore fitness to wild-type levels without loss of resistance (Andersson and Hughes, 2010). FQ-conferring mutations have been shown to impose variable fitness costs, depending on strain background and specific combinations of mutations (Komp Lindgren et al., 2005; Luo et al., 2005; Marcusson et al., 2009; Machuca et al., 2014; Wasels et al., 2015).
In the current study, we compared the fitness effects of a target site mutation in parC using isogenic mutants created from clinical exoU and exoS isolates. We investigated the fitness effects of this mutation using in vitro head-to-head competition experiments. Changes in supercoiling level and mutation frequency between mutants and parent strains were also assessed to explore potential mechanisms for the difference in fitness observed.
Our results suggest that the FQ resistance-conferring point mutation in parC studied here may confer less of a fitness cost to exoU strains than to exoS strains, with exoU strains showing evidence of compensation for the fitness costs. These results provide a potential biological explanation for the observed predominance of exoU strains in the clinical FQ-resistant population. The co-selection of FQ-resistance in highly virulent exoU strains will likely have a negative impact on patient outcomes, which underscores the importance to gain a better understanding of the biological basis for this observation.
Materials and Methods
Ethics Statement
The institutional review boards at both Huntington Hospital and the University of Southern California have approved this study. Informed consent was waived from all participants since bacteria cultures were saved as part of a longitudinal epidemiological surveillance study. All data from respiratory cultures were analyzed anonymously to protect patient privacy.
Bacterial Strains and Culture Conditions
Strains of P. aeruginosa used in this study (Table 1) were selected from a collection of isolates obtained from the respiratory tract of hospitalized patients at Huntington Hospital, Pasadena, CA, USA from 2005 to 2009 and were stored in cryovials at -80°C in 30% glycerol. All strains had been previously characterized for: the presence of the exoU or exoS gene, resistance to levofloxacin, presence of mutations in the quinolone-resistance determining regions of gyrA/B and parC/E, and clonal relatedness by RAPD PCR, as described in Agnello and Wong-Beringer (2012). Strains were routinely grown as 5 ml cultures in Luria-Bertani (LB) broth in 12 ml polypropylene culture tubes at 37°C and shaking at 250 rpm for liquid culture, or grown on Pseudomonas Isolation Agar (PIA) plates at 37°C. Strains were routinely sub-cultured twice after inoculation from the frozen stock by 1:100 dilution (vol:vol) before use in any experiment.
Table 1.
Characteristics of Strains.
| Name | LVXa MIC (μg/ml) | LVX + EPIb MIC (μg/ml) | Amino Acid Change | Doubling Time, h (-/+ SEM)c | |
|---|---|---|---|---|---|
| exoU | U-37 | 16 | 0.5 | GyrA: T83I | 2.0 (1.7–2.4) |
| U-37PC∗ | 16 | 0.5 | GyrA: T83I ParC: S87L |
1.7 (1.5–1.9) | |
| U-91 | 2 | 2 | GyrA: T83I | 3.9 (3.78–3.93) | |
| U-91PC∗ | 16 | 2 | GyrA: T83I ParC: S87L |
3.9 (3.87–3.96) | |
| U-92 | 16 | 0.5 | GyrA: T83I | 2.5 (2.3–2.7) | |
| U-92PC∗ | 16 | 0.5 | GyrA: T83I ParC: S87L |
2.8 (2.6–3.0) | |
| exoS | S-139 | 4 | 0.5 | GyrA: T83I | 3.4 (3.3–3.6) |
| S-139PC∗ | 32 | 0.25 | GyrA: T83I ParC: S87L |
3.1 (2.9–3.3) | |
| S-247 | 16 | 1 | GyrA: T83I | 2.5 (2.4–2.6) | |
| S-247PC∗ | 16 | 8 | GyrA: T83I ParC: S87L |
2.5 (2.4–2.7) | |
| S-215 | 16 | 1 | GyrA: T83I | 2.4 (2.2–2.7) | |
| S-215PC∗ | 16 | 1 | GyrA: T83I ParC: S87L |
2.3 (2.2–2.5) |
Mutants with Ser87Leu substitution in ParC denoted PC∗. aLVX, Levofloxacin; bEPI, efflux pump inhibitor; cDoubling time during exponential phase.
Susceptibility Testing and Sequencing
Susceptibility testing to levofloxacin and rifampicin was performed by broth microdilution in twofold dilutions at concentrations ranging from 0.25 to 128 μg/ml according to guidelines recommended by CLSI (Clinical and Laboratory Standards Institute, 2007). In order to assess the involvement of the multidrug Mex efflux pumps to resistance, MIC of levofloxacin was also measured with the addition of an efflux pump inhibitor (EPI, MC-0228; Sigma) at 20 μg/ml (Kriengkauykiat et al., 2005).
For sequencing, genomic DNA was extracted and purified from isolates using the DNeasy Mini Kit (Qiagen). The quinolone-resistance determining regions of target genes gyrA, gyrB, parC, and parE were amplified by PCR using previously published primers and conditions and sequenced to identify mutations compared to wild-type strain PAO1 (Table 2) (Jalal and Wretlind, 1998; Lee et al., 2005; Winsor et al., 2011).
Table 2.
Genetic elements used.
| Sequence | Reference | |
|---|---|---|
| Primer name PCR | ||
| glmS-up | CTGTGCGACTGCTGGAGCTGA | Choi and Schweizer, 2006 |
| glmS-down | GCACATCGGCGACGTGCTCTC | Choi and Schweizer, 2006 |
| Tn7-R | CACAGCATAACTGGACTGATTTC | Choi and Schweizer, 2006 |
| Tn7-L | ATTAGCTTACGACGCTACACC | Choi and Schweizer, 2006 |
| Oligo for recombination | ||
| PC∗ | TGCTCGGCAAGTTCCACCCGCACGGCGACTTGGCCTGCTACGAGGCCATGGTGCTGATGG | Agnello and Wong-Beringer, 2012 |
| Plasmid | Function | References |
| pTNS3 | Helper plasmid, encodes Tn-7 transposition pathway | Choi et al., 2005 |
| pUC18T-mini-Tn7T-Gm-eyfp | Delivery plasmid for GmR marker and YFP | Choi and Schweizer, 2006 |
| pUC18T-mini-Tn7T-Gm-ecfp | Delivery plasmid for GmR marker and CFP | Choi and Schweizer, 2006 |
| TOP10/mini-Tn7-PA0614 promoter-Gm-luxCDABE | Delivery plasmid for GmR marker and lux operon | Choi and Schweizer, 2006 |
Creation of Target Site Mutants
Since most FQ-resistant isolates have gyrA mutations, three exoU and three exoS isolates with a pre-existing gyrA FQ-resistance mutation (Thr83→Ile substitution) were selected for mutagenesis. A site-directed point mutation in the parC gene leading to the amino acid substitution Ser87→Leu was introduced via electroporation of an oligonucleotide in order to create isogenic mutants, using the technique of oligonucleotide recombination as described previously (Swingle et al., 2010; Agnello and Wong-Beringer, 2014). Briefly, a single-stranded oligonucleotide 60 bases in length was designed using the parC gene sequence of reference strain PAO1 (Winsor et al., 2011) (Table 2). The oligo is identical to the PAO1 sequence from nucleotides 230 to 289, save for the point mutation TCG→TTG at location 31 of the oligo, corresponding to nucleotide 260 in the parC gene. This point mutation, which is the most common parC mutation observed among fluoroquinolone-resistant clinical strains, gives rise to the Ser87→Leu amino acid change in the ParC protein.
Strains were made electro-competent by several washes with 300 mM sucrose solution as described in Choi et al. (2006) and transformed by electroporation at 2.5 kV in a 0.2 cm cuvette using a Micropulser (Bio-Rad, Hercules, CA, USA). SOC medium (1 ml) was immediately added and the cells were outgrown overnight at 37°C on PIA plus levofloxacin at concentrations two and fourfold above the original minimum inhibitory concentration (MIC) of the isolates for selection. Single colonies were collected and the parC gene was sequenced as described above to confirm that recombination had occurred.
Growth Rate Measurements
Independent overnight cultures were diluted to OD600 ~0.1 and grown at 37°C with shaking in 50 ml LB broth in a 100 ml flask. An aliquot of the culture (150 μl) was sampled every 30 min for 8 h and turbidity was measured at OD600 using a Tecan Sunrise microplate reader (Tecan Group Ltd., Switzerland). Results were reported as an average of at least three independent experiments in terms of doubling time per hour.
Insertion of Fluorescent Tag
Mini-Tn7 vectors developed and generously shared with us by Dr. Herbert Schweizer (Choi et al., 2005) were utilized to insert cassettes containing the genes encoding YFP or CFP as well as a cassette containing the lux operon into strains used for competition experiments and supercoiling experiments, respectively. The cassette inserts into the P. aeruginosa genome at a single location (attTn7 site downstream of the glmS gene) and contains the gene for the fluorescent protein under the control of a constitutive promoter or the lux operon under the control of a supercoiling-sensitive promoter (Moir et al., 2007), as well as a gentamicin-resistance marker for selection. The delivery plasmid was co-electroporated with a helper plasmid (pTNS3) encoding the necessary transposase function for insertion. The electroporation protocol used for transformation is described in detail in Choi and Schweizer (2006); briefly, strains were made electro-competent through a series of washes with 300 mM sucrose, subjected to electroporation as described above, and plated on LB plates containing 30 μg/ml gentamicin for selection. To confirm insertion had occurred, PCR was performed to amplify the insertion region using primers listed in Table 2 and protocol as previously described (Choi and Schweizer, 2006) (data not shown).
In vitro Competition Assays
To investigate the effects of the parC mutation (PC∗) on fitness, mutants were directly competed against isogenic parent strains in co-culture. Strains were tagged with either CFP or YFP for differentiation as described above. PC∗ mutant strains were initially tagged with CFP, and competed against the respective isogenic parent strain tagged with YFP. To confirm neutrality of the tags, experiments were also performed with YFP-mutants and CFP-parents.
Strains were grown overnight in LB, and 105 CFU of each strain was co-inoculated into 10 ml LB in a 50 ml flask, and grown with shaking at 37°C. Every 24 h, 10 μl of the culture was transferred to a new flask containing 10 ml fresh LB, and a sample of the culture was serially diluted then plated on PIA for CFU enumeration. Colonies of each color were counted using a fluorescent wide-field microscope (Zeiss Axio Zoom.V16). Experiment duration ranged from 4 to 7 days. The ratio of the number of CFUs of the mutant-to-parent strain on each day of the experiment was calculated and used to determine the relative fitness of the mutant. Experiments were repeated a minimum of five times. Assays were also performed with the addition of levofloxacin equal to 1/8 the MIC (2 or 0.5 μg/ml). At day 7 of each competition experiment, colonies were selected from plates, inoculated into LB, grown overnight, and stored at -80°C in 30% glycerol until ready for use in subsequent experiments. These were described as ‘aged’ strains. Aged strains were subjected to a secondary competition experiment vs. the original strains. Secondary competition with aged strains was performed as described above, and lasted for 4 days.
Supercoiling Assay
We adapted a reporter assay to estimate the ability to maintain supercoiling levels in mutants compared to parent strains by inserting a Tn7 cassette containing the lux operon under the control of a supercoiling-sensitive P. aeruginosa promoter from the gene PA0614 (Moir et al., 2007). The cassette was inserted as described above. Luminescent strains were grown to mid-exponential phase, then diluted 1:4 in LB+ levofloxacin at 1/4, 1/2, and 1x the measured MIC, and grown in triplicate in a deep well 96-well plate for 7 h. Luminescence was measured using the Envision Multi-Label plate reader (Perkin-Elmer) as well as OD600 as described above. Relative Luminescence Units (RLU) was normalized to OD600 for all comparisons.
Mutation Frequency Assay
Mutation frequency was estimated during competition experiments using the spontaneous rifampicin resistance method (Oliver et al., 2000). At each 24-h interval, competition cultures were plated on PIA+ rifampicin at 5x the MIC (ranging from 40 to 500 μg/ml depending on the strain) in addition to plating on PIA. The mutation frequency reported is the number of colonies that grew on PIA + rifampicin after 48 h divided by the number of colonies on PIA alone. Colonies of each strain were differentiated and counted based on fluorescence as described above. For strain 139, 5x the MIC of rifampicin corresponds to 500 μg/ml, which turned the plates a dark shade of red and prevented the differentiation of the fluorescence of the colonies. Thus, for this strain, each rifampicin-containing plate was replica-plated using sterile velvets onto a fresh PIA plate and incubated for an additional 24 h before colonies were counted.
Statistical Analysis
GraphPad Prism v.6 (GraphPad.com, San Diego, CA, USA) was used to perform statistical analysis. ANOVA was used to compare continuous variables where appropriate, and student’s t-test was used to compare mean ratios to the null hypothesis of 1 (indicating no difference). A p value ≤ 0.05 denotes significance.
Results
We previously created isogenic mutants from 6 clinical isolates of P. aeruginosa (three exoU and three exoS) by inserting a point mutation commonly observed in fluoroquinolone-resistant isolates into the parC gene (Agnello and Wong-Beringer, 2012). The goal of the current study was to investigate the biological effects of this target site mutation in a controlled genetic background. Mutants are denoted PC∗. Interestingly, the addition of the parC mutation increased the levofloxacin MIC in only two of the six isolates. Characteristics of mutant and parent strains used in this study are summarized in Table 1.
Growth Rate in Monoculture
As an initial comparison of the relative fitness of mutant vs. parent strains, we tested the doubling time in independent cultures by measuring optical density every 30 min over an 8-h period; results are an average of at least three independent experiments (Table 1). Doubling time during exponential phase was calculated by dividing the time interval over the number of generations. Growth rates of all six mutants were comparable to parent strains.
parC Mutation Differentially Affects Competitive Fitness of exoU vs. exoS Strains
To investigate the fitness costs of the parC mutation, we performed head-to-head competition assays. Each PC∗ mutant was directly competed against its isogenic parent strain by growth in co-culture for 4 days. Because the strains competed are identical save for the point mutation, any fitness cost is attributable to the mutation. In each individual competition experiment, one strain is tagged with CFP (cyan fluorescent protein), and the other with YFP (yellow fluorescent protein). This allows for the direct enumeration of the number of colonies of each strain after serial dilution onto agar plates. Colonies were counted every 24 h, and the relative fitness of the mutant is determined from the ratio of mutant-to-parent CFU/ml. Each experiment was independently repeated at least five times. We confirmed the neutrality of the YFP and CFP tags by repeating each experiment with each strain carrying the opposite tag. No difference was found, and therefore results from all experiments were combined.
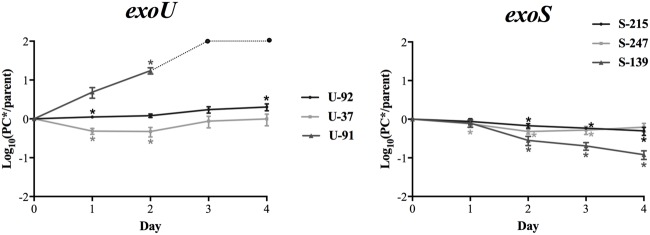
Figure 1 shows the average mutant-to-parent ratios over time for each strain. Each strain has a unique pattern of fitness costs associated with this mutation; however, in general, there appears to be a higher fitness cost for the exoS strains. The exoS strains S-139PC∗, S-247PC∗, and S-215PC∗ show a consistent fitness defect over all time points, with strain S-139PC∗ the most affected. On the other hand, strain U-92 has the smallest fitness effect, with the mutant/parent ratios ranging from 1.2 to 2.0 over 2–4 days of competition. Strain U-37 shows a similar pattern except the mutant starts with lower fitness (ratio 0.5 at day 2), but catches up to the parent strain by day 4 with a ratio of 0.99. Strain U-91 is unique and interestingly shows a striking increase in fitness due to the mutation; the mutant rapidly outcompetes the parent strain and completely takes over the culture by day 3.
FIGURE 1.
In vitro fitness of exoU and exoS-PC∗ mutants. The average log10 CFU/ml ratio of PC∗ vs. parent strains per day of in vitro competition are shown. Ratios above zero indicate the mutant is more fit, while ratios below zero indicate a fitness cost. The ratio for exoS strains generally decreases over the course of the experiment, while the ratios for exoU strains either increases or remains stable. Results are an average of at least five independent experiments. Error bars = SEM. For strain U-91, the dotted line and circles indicate ratios after day 2 are beyond the limit of detection. ∗Indicates the PC∗/parent CFU ratio is significantly different than 1 at p < 0.05.
exoU-PC∗ Mutants can Better Maintain Wild-Type Supercoiling Levels than exoS-PC∗ Mutants
We assessed supercoiling of parent vs. mutants in exoU strains U-92 and U-37 and exoS strains S-215 and S-139 by inserting a Tn7 genetic element in which a supercoiling sensitive promoter controls the lux operon (Moir et al., 2007). Due to challenges inserting the reporter, only two strains of each background were chosen as representative strains. Strains with the insertion were grown under levels of levofloxacin at and below the MIC in order to maximally induce expression of the reporter, and luminescence values were normalized to OD600. We measured the level of luminescence in the parent strain as an indirect reporter of the baseline level of supercoiling; any difference in luminescence expression for the PC∗ strain reflects a change in supercoiling due to the parC mutation. Results show that the PC∗ mutants in general have decreased luminescence compared to parent strains; however, this decrease was significantly more pronounced in exoS-PC⋆∗ strains (Figure 2), suggesting that exoS strains with the PC∗ mutation are somehow less able to maintain the supercoiling level of the parent strains, which may have negative effects on fitness. This is surprising given that the primary role of topoisomerase IV (ParC) is decatenation of daughter replicons and not regulation of supercoiling (Hooper, 2001); however, in this context it may suggest an overall difference in supercoiling regulation in exoU vs. exoS strains.
FIGURE 2.
Supercoiling changes in PC∗ mutants compared to parent strains. Each strain contains a chromosomal reporter construct in which the lux operon is under the control of a supercoiling-sensitive promoter. Strains were grown in concentrations of levofloxacin equal to 1/4, 1/2, and 1x the MIC. Luminescence was normalized to cell count using OD600. Results are an average of three independent experiments, and error bars represent SEM. ∗p < 0.05; ∗∗p < 0.01.
Analysis of Aged Strains
Because, we observed that the fitness of exoU-PC∗ mutants tended to increase after 24 h of competition, we investigated whether stable changes were occurring during competition that allowed the strains to compensate for fitness costs associated with the parC mutation. We collected colonies of parent and PC∗ strains from a primary competition experiment (parent vs. mutant) after 7 days and labeled these ‘aged’ strains. In addition, we investigated the effect of sub-inhibitory levels of fluoroquinolone exposure on competitive fitness and ability to compensate for fitness costs by collecting aged strains from primary competition experiments in which 1/8 MIC of levofloxacin was added to the media.
We were not able to collect aged colonies from the primary competition experiments of strain 91 due to the vast difference in fitness between the PC∗ mutant and parent strains; the PC∗ strain completely overtook the culture by day 3. Because this left us with only two exoU strains (U-92 and U-37), we chose two exoS strains in order to have an equal number in each group for comparison. We chose strains 215 and 139 because these represent both ends of the spectrum in terms of fitness of the PC∗ mutants in vitro; strain S-215PC∗ shows the least fitness cost of all the exoS strains, while strain S-139PC∗ exhibits the greatest fitness cost.
Sequencing the FQ-target site genes of the aged strains revealed that no changes occurred in the genes associated with FQ-resistance except for exoU strain U-92, in which the parent acquired the Ser87Leu substitution in ParC during competition under levofloxacin exposure (+LVX) at 1/8 MIC. All PC∗ strains maintained the parC mutation. The MIC to levofloxacin did not change in any of the aged strains (data not shown).
Aged PC∗ Mutant exoU vs. exoS Strains Show Dramatic Difference in Fitness
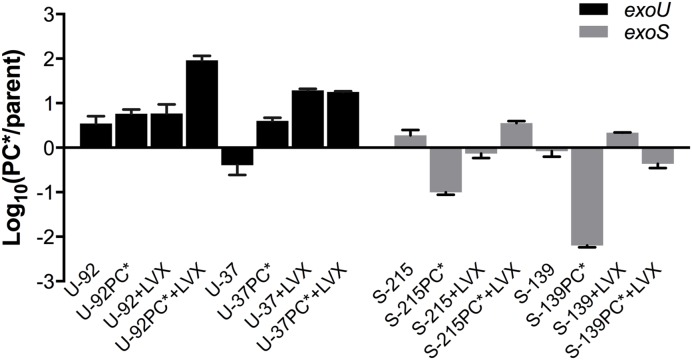
To investigate whether the aged strains had acquired adaptations during competition that rendered them more fit, we compared the fitness of each aged strain to its un-aged counterpart. Aged strains were subjected to a secondary competition experiment in which they were competed against the original, un-aged strain that contained the opposite fluorescent tag, enabling the discrimination and enumeration of each strain during competition. Results reflect the aged-to-un-aged ratio at Day 2 (Figure 3). Because each aged strain was competed directly against an un-aged version of itself, we were able to directly observe if fitness had changed in the aged strains. All parent strains had a negligible change in fitness. Both exoU-PC∗ aged strains increased in fitness greater than fourfold, while the exoS-PC∗ aged strains showed a 50% decrease in fitness; S-139PC∗ was greater than 100 times less fit than before aging.
FIGURE 3.
Fitness of aged strains. Strains were collected at the end of a 7-day primary competition experiment (PC∗ vs. parent) with and without the addition of sub-inhibitory concentration of levofloxacin (+LVX) and subsequently re-competed in a secondary competition experiment (aged vs. un-aged) with or without levofloxacin at the same concentration. Results shown reflect the average fitness of aged strains after 48 h of competition. Experiments were performed in duplicate. Error bars = SEM.
We also investigated whether growth under sub-inhibitory levels of levofloxacin affected fitness, and if mechanisms to compensate for fitness costs evolved under these conditions. The addition of levofloxacin did not affect the results of primary competition experiments (data not shown), but notable results were seen in the secondary competition experiments of the aged vs. un-aged strains, which also included 1/8 MIC of levofloxacin. Upon subsequent exposure to the drug, exoU-PC∗ strains were much more fit, outcompeting the un-aged strain rapidly. Specifically, the aged PC∗ mutants of exoU strains U-37 and U-92 outcompeted the un-aged strains significantly and rapidly, much more so than in the competition experiments without the drug. Interestingly, the measured MICs of the aged and un-aged strains remain unchanged. exoS-PC∗ strains showed conflicting results; S-215PC∗ had gained fitness, while S-139PC∗ had slightly decreased fitness (Figure 3).
Mutation Frequency
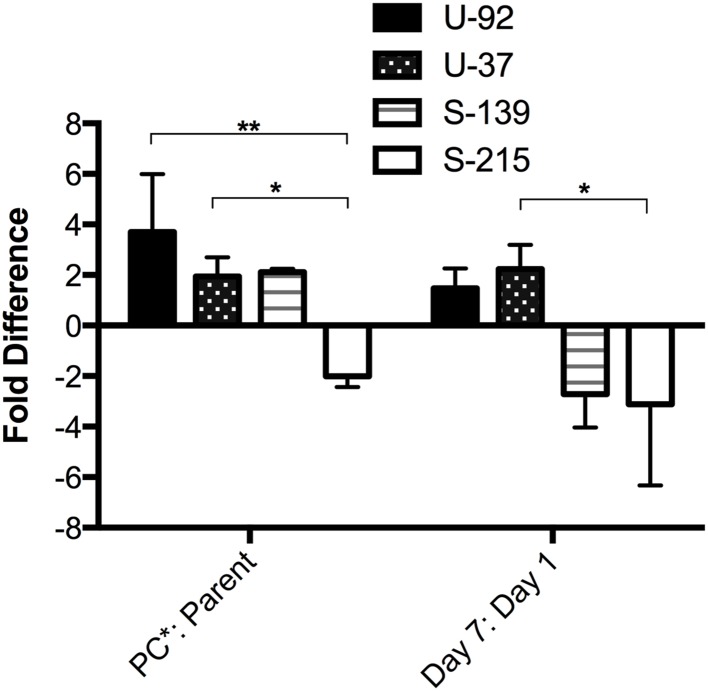
Compensation for potential fitness costs of a resistance-conferring mutation can evolve through the accumulation of beneficial mutations. Therefore, we investigated whether the rate of spontaneous mutation frequency could explain the increase in fitness during competition that was seen in exoU-PC∗ strains. Mutation frequency was estimated by calculating the frequency of spontaneous resistance to rifampicin (RifR) (Oliver et al., 2000). We estimated mutation frequency during the primary competition experiments for exoU strains U-92 and U-37, and exoS strains S-215 and S-139 by plating the mixed competition culture on agar containing 5x the MIC of rifampicin at each time point (Days 1–7), and counting the number of colonies based on fluorescence of each strain that were able to grow on the rifampicin-containing plates compared to the overall CFU counts. At Day 7, the exoU-PC∗ strains have both a higher mutation frequency than the parent strains as well as a higher mutation frequency compared to Day 1. exoS strain S-215PC∗ had a lower frequency at Day 7 compared to its parent strain as well as a lower frequency compared to Day 1. Surprisingly, exoS strain S-139PC∗, which is significantly less fit than its parent strain, has a twofold higher mutation frequency than the parent strain at Day 7. However, its mutation frequency at Day 7 decreased almost threefold from Day 1 (Figure 4). Therefore, mutation frequencies seem to generally correlate with fitness results.
FIGURE 4.
Mutation frequency of PC∗ mutants vs. parent strains measured during competition experiments. The frequency of spontaneous rifampicin resistance was used to estimate mutation frequency at each time point during a 7-day competition experiment. Bars represent fold difference of the PC∗ vs. parent at Day 7 and the fold difference at Day 7 vs. Day 1 for each PC∗ strain. Error bars = SEM. ∗p < 0.05; ∗∗p < 0.01.
Discussion
Pseudomonas aeruginosa is considered one of six bacteria that pose an immediate threat to public health, according to the Infectious Diseases Society of America, due to its increasing prevalence, broad arsenal of virulence factors, and emergence of resistance to all available antibiotics (Boucher et al., 2009). In its recent report entitled “Antibiotic Resistance Threats in the United States” the Centers for Disease Control named P. aeruginosa a serious threat that requires prompt action and continual monitoring to prevent a worsening of the problem of resistance (Centers for Disease Control [CDC], 2013). Notably, resistance rates of P. aeruginosa to the once-effective fluoroquinolone (FQ) antibiotics are now greater than 30% in the United States, with many of those strains also multi-drug resistant (Rosenthal et al., 2012).
During acute disease, P. aeruginosa utilizes the toxins of the type III secretion system to circumvent the host immune system and establish infection. Of the four exotoxins, exoU is the most virulent, encoding a potent phospholipase that disrupts the plasma membrane and leads to rapid cell death (Sato et al., 2003). ExoU activity induces excessive inflammation and tissue damage in the host as well as increased bacterial dissemination that can lead to septic shock (Kurahashi et al., 1999) and higher rates of mortality compared to infections with ExoS-secreting strains (Shaver and Hauser, 2004; Zhang et al., 2014; Peña et al., 2015).
As the problem of antibiotic resistance continues to worsen, it is becoming apparent that the evolution of virulence factors and antibiotic resistance cannot be considered strictly independent processes (Beceiro et al., 2013; Hauser, 2014). Previously, we found that infection by FQ-resistant P. aeruginosa was an independent predictor for threefold higher mortality or prolonged illness by an additional 5 days compared to those infected with FQ-susceptible strains (Hsu et al., 2005). In a separate study, we reported that among patients from whom P. aeruginosa was isolated from the respiratory tract, strains with the combined traits of exoU and FQ-resistance were much more likely to cause pneumonia than exoS, FQ-resistant strains or either trait alone (Sullivan et al., 2014).
Analysis of 270 of our P. aeruginosa clinical isolates led to the observation that exoU strains are not only enriched in the FQ resistant sub-population, but are also more likely to have acquired two or more target site mutations than exoS strains, especially in the parC gene (Agnello and Wong-Beringer, 2012). In order to study the biological effects of a resistance-conferring mutation in a controlled, isogenic background, we adapted a technique for inserting point mutations into bacterial genomes for use in our clinical isolates (Swingle et al., 2010). Using the technique of oligonucleotide recombination, we created 6 FQ-resistant mutants (three exoU and three exoS) (Agnello and Wong-Beringer, 2014) from clinical strains by inserting a mutation into the parC gene.
Because the target site mutation, we inserted in parC rarely occurs in clinical isolates without a pre-existing mutation in gyrA (Jalal and Wretlind, 1998), we chose clinical strains that had a pre-existing gyrA mutation (Thr83Ile) in order to mimic FQ-resistant strains encountered in the clinical setting. These specific point mutations in gyrA and parC are commonly found in FQ-resistant clinical strains of P. aeruginosa (Nakano et al., 1997; Mouneimné et al., 1999; Higgins et al., 2003; Agnello and Wong-Beringer, 2012). The goal of the current study was to assess the biological effects on fitness of the parC mutation and determine if the magnitude of the effects differ in exoU vs. exoS strains.
The acquisition of resistance is generally thought to be accompanied by a fitness cost to the bacterium (Andersson and Levin, 1999); however, there are many reports of neutral fitness effects or enhanced fitness due to resistance-conferring mutations. Evidence from FQ-resistant E. coli, S. pneumoniae, and N. gonorrhoeae suggest that a secondary resistance mutation does not further decrease fitness and may restore the low fitness of primary mutants back to wild-type levels (Komp Lindgren et al., 2005; Rozen et al., 2007; Marcusson et al., 2009; Kunz et al., 2012; Machuca et al., 2014). Furthermore, many fitness effects seem to be strain dependent, as was shown in a study of ciprofloxacin-resistant Campylobacter jejuni, in which a gyrA mutation conferred a high cost to one strain but actually increased fitness for a different strain (Luo et al., 2005). Similar results were seen in S. pneumoniae, in which some FQ-resistant mutant strains had increased fitness while others had a significant cost (Balsalobre and de la Campa, 2008). This suggests that fitness effects are highly dependent on strain genetic background, which may explain the differences, we have seen between exoU and exoS strains.
Head-to-head competition assays are a standard method for investigating the relative fitness of a mutant strain compared to its isogenic parent strain, and it is possible to detect differences in fitness as small as 1% (Andersson and Levin, 1999; Corzett et al., 2013). Strains are mixed together in co-culture, and must compete for the limited available resources (Finkel, 2006; Kraigsley and Finkel, 2009). In most studies, strains are differentiated based on selective growth on antibiotic plates. However, since the parC mutation, we inserted did not increase the levofloxacin MIC in all strains, we had to develop a different approach. We took advantage of a Tn7-based system developed by Schweizer (Choi et al., 2005) in order to insert a cassette containing either YFP or CFP under the control of a strong promoter.
The growth of the PC∗ strains did not differ dramatically from those of the parents when grown in monoculture; however, differences were seen when strains had to compete for resources in the same culture flask during competition experiments. Overall, the parC mutation imposed a moderate to considerable fitness cost on exoS strains, while exoU strains were able to better tolerate the mutation. Notably, fitness of exoU-PC∗ strains ranged from maintaining the wild-type level of fitness to outcompeting the parent strain by more than 10-fold, whereas exoS-PC∗ strains were consistently less fit than parent strains.
The FQ target site mutation investigated in the current study occurs in a topoisomerase enzyme and therefore may have an effect on the regulation of supercoiling. Common methods for investigating supercoiling levels rely on reporter plasmids that need to be selected for and maintained. Because, we are using clinical isolates, there is a high level of multi-drug resistance that precludes the use of standard selection antibiotics. To circumvent this, we inserted a Tn7 element (Moir et al., 2007) onto the chromosome in which a supercoiling sensitive promoter controls lux expression. exoU-PC∗ mutants were better able to maintain the supercoiling levels of the parent strains, while exoS-PC∗ mutants showed a more drastic change in supercoiling, as observed by decreased lux expression. Other studies have shown that changes in supercoiling are correlated with changes in fitness (Kugelberg et al., 2005; Marcusson et al., 2009), and therefore the regulation of supercoiling may reflect the level of overall fitness and may explain the fitness differences seen in our strains. The ability of exoU-PC∗ strains to better regulate supercoiling levels under the stress of FQ exposure reflects increased fitness of these strains overall. The supercoiling level of the cell affects global gene expression and replication efficiency, and changes in supercoiling may alter response to environmental stressors, or even modulate pathogenesis in the host (Redgrave et al., 2014).
In the primary competition experiments, exoU-PC∗ mutants tended to increase in fitness over the course of the 4-day experiment. This suggests that exoU strains may be acquiring beneficial adaptations that allow for the compensation of the fitness costs associated with the PC∗ mutation. Bacteria have the ability to rapidly evolve compensatory mechanisms to mitigate the fitness costs associated with antibiotic resistance, and compensatory mechanisms can reverse fitness costs without any loss of resistance (Andersson and Hughes, 2010). The exoU-PC∗ strains that had been ‘aged’ through a primary competition experiment for 7 days have increased fitness compared to the un-aged strains, while aged exoS-PC∗ strains have decreased fitness. The results suggest that the compensatory mechanisms in exoU strains most likely are acting as a repressor of the negative fitness effects of the parC mutation, as opposed to just conferring a general gain in fitness, based on the observation that the aged parent strains did not show as much of an increase in fitness as the PC∗ mutants. The observed ability of exoU strains to compensate for the costs of FQ-resistance has many clinically negative consequences. Compensation in clinical populations leads to the stabilization of resistant populations without the presence of drug (Andersson and Hughes, 2010). Increased fitness after acquisition of FQ-resistance adds to the already complicated problem of treating infections caused by highly virulent exoU strains.
The inability of exoS-PC∗ strains to regulate supercoiling suggests that the compensatory mechanisms in exoU strains are acting to maintain wild-type supercoiling levels in the PC∗ mutants, as has been reported in other studies of compensation of the costs of FQ-resistance (Kugelberg et al., 2005; Marcusson et al., 2009). Although, we did not identify the exact mechanisms responsible for the compensatory effects, we have shown that they are stable since strains were frozen and grown before testing for compensation in secondary competition experiments, and the results were reproducible in repeated experiments. Sequencing of the quinolone-resistance determining regions of gyrA/B and parC/E revealed no additional mutations had occurred during competition; however, beneficial mutations may have occurred elsewhere. For both exoU strains tested, the mutation frequencies correlated with the increase in fitness observed during competition experiments, suggesting that higher mutation frequency may facilitate the acquisition of beneficial mutations that mitigate the fitness costs of the parC mutation.
A dramatic increase in fitness occurred in the aged exoU-PC∗ strains taken from a primary competition experiment in which sub-inhibitory levels (1/8 the MIC) of levofloxacin were added to culture. Although this low level of drug did not affect the growth of strains in the primary competition experiment, when the aged exoU-PC∗ strains were re-competed under the same concentration of levofloxacin, they were 20–100 times more fit than before aging. In contrast, exoS-PC∗ strains were dramatically less fit under these circumstances. However, none of the aged PC∗ strains had an increase in MIC, nor any additional FQ-resistance mutations in target site genes. These results suggest that although the level of levofloxacin was much below the MIC, highly fit strains were selected for rapidly, more so than in conditions without drug. Also, the highly fit strains were not more resistant, suggesting that perhaps the presence of levofloxacin accelerated the process of compensation for the already present resistance mutations. As sub-inhibitory concentrations of antibiotics are routinely present during treatment due to insufficient dosing or inadequate penetration to certain areas of the body (Baquero and Negri, 1997; Andersson and Hughes, 2014), the implications of our results are alarming and suggest that low levels of antibiotic can rapidly select for highly fit strains, preferentially the highly virulent exoU-containing strains. Furthermore, when these highly fit, highly virulent strains are re-introduced to the antibiotic, they will rapidly outcompete all other strains. The sub-inhibitory concentration was also able to select for a parC mutation in an exoU parent strain, emphasizing the known phenomenon that selection for resistance mutations can occur at sub-inhibitory levels of antibiotic (Baquero et al., 1998; Andersson and Levin, 1999; Gullberg et al., 2011; Andersson and Hughes, 2014).
Overall, these results suggest a potentially lower fitness burden associated with FQ-resistance for exoU strains than for exoS strains, which in part provides an initial biological explanation for exoU strains’ greater tendency to acquire FQ-resistance in the clinical setting. Although, we have yet to identify the specific genetic components underlying the fitness differences, we suspect that genes unique to strains with the exoU genetic background may contribute to the fitness benefits seen in this study. The P. aeruginosa genome consists of a highly conserved core genome, but variability is introduced in the form of genomic islands, which make up the accessory genome. Genes within the islands usually encode accessory activities such as specific pathogenicity or symbiosis factors (Harrison et al., 2010). The exoU gene, along with its chaperone spcU, is located on a pathogenicity island, a specialized genomic island. exoU has been identified as part of a few different pathogenicity islands. The most highly studied is from the reference strain PA14, in which the exoU gene is on an island named PAPI-2 (Kulasekara et al., 2006). Studies with PA14 have shown that virulence is dependent on the presence of the entire island and not just the exoU gene alone; therefore, other as yet unknown genes contained on the pathogenicity islands contribute combinatorially to the increased virulence of exoU strains (Harrison et al., 2010). Furthermore, pathogenicity in strain PA14 requires the coordinated action of multiple virulence factors, associated with both the core and accessory genomes (Lee et al., 2006). Therefore, it is possible that other genes in the accessory genome in combination with exoU may provide fitness benefits to exoU strains that allow for increased ability to adapt to the fitness costs of FQ-resistance. Genomic islands have been shown to confer fitness benefits, and the accessory genome of P. aeruginosa is an important driver of the ability of strains to persist in a particular environment (Hacker and Carniel, 2001; Ozer et al., 2014).
This study has several limitations. We acknowledge that a limited number of clinical strains and their isogenic mutants were evaluated in the study. Despite having access to a large collection of clinical strains, the selection of strains to create isogenic target site mutants to study the effect of FQ-resistance on fitness proved challenging, as most of the isolates in our collection were found to already contain the resistance-conferring mutation that, we are investigating, due to the high prevalence of FQ resistance in clinical isolates of P. aeruginosa. Therefore, we were not able to use those to create isogenic pairs. Furthermore, genetic manipulation of these clinical isolates proved to be difficult, as these strains were not universally amenable to the creation of isogenic mutants via oligonucleotide recombination. Nonetheless, it is worth nothing that the strains evaluated in this study were carefully selected to represent a broad range of characteristics such as varying degrees of FQ resistance, virulence, and clinical outcomes. While the limited number of strains evaluated in this study may not necessarily represent the genetic variability present among all exoU and exoS clinical strains, strain-specific differences in fitness effects were observed even among strains with the same exoU or exoS genetic background, though the trend observed appears to follow similar pattern for strains with the same exoU or exoS genetic background.
Understanding the fitness costs of antibiotic resistance and possibilities of compensation for these costs is essential for rationally combating the problem of antibiotic resistance. Importantly, this study outlines a useful model with the creation of isogenic clinical strains for investigating the biological costs of resistance in a medically important pathogen and can be applied to other drug-organism pairs.
Author Contributions
MA, AW-B, and SF designed the experiments. MA and AW-B wrote the manuscript. SF edited and provided critical review of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Herbert Schweizer and Dr. Donald Moir for generously sharing plasmids and strains, and Juliana Brondani and Nicole Schrad for assistance with growth and competition experiments.
Footnotes
Funding. This work was supported by award number TL1TR000132 from NIH/NCRR/NCATS to MA and award number R21AI073467 from the National Institute of Allergy and Infectious Diseases to AWB. SF was supported in part by U.S. Army Research Office grants W911NF1010444 and W911NF1210321.
References
- Agnello M., Wong-Beringer A. (2012). Differentiation in quinolone resistance by virulence genotype in Pseudomonas aeruginosa. PLoS ONE 7:e42973 10.1371/journal.pone.0042973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnello M., Wong-Beringer A. (2014). The use of oligonucleotide recombination to generate isogenic mutants of clinical isolates of Pseudomonas aeruginosa. J. Microbiol. Methods 98 23–25. 10.1016/j.mimet.2013.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allewelt M., Coleman F. T., Grout M., Priebe G. P., Pier G. B. (2000). Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect. Immun. 68 3998–4004. 10.1128/IAI.68.7.3998-4004.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8 260–271. 10.1038/nrmicro2319 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Hughes D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12 465–478. 10.1038/nrmicro3270 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Levin B. R. (1999). The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2 489–493. 10.1016/S1369-5274(99)00005-3 [DOI] [PubMed] [Google Scholar]
- Balsalobre L., de la Campa A. G. (2008). Fitness of Streptococcus pneumoniae fluoroquinolone-resistant strains with topoisomerase IV recombinant genes. Antimicrob. Agents Chemother. 52 822–830. 10.1128/AAC.00731-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F., Negri M. C. (1997). Selective compartments for resistant microorganisms in antibiotic gradients. Bioessays 19 731–736. 10.1002/bies.950190814 [DOI] [PubMed] [Google Scholar]
- Baquero F., Negri M. C., Morosini M. I., Blázquez J. (1998). Antibiotic-selective environments. Clin. Infect. Dis. 27(Suppl. 1), S5–S11. 10.1086/514916 [DOI] [PubMed] [Google Scholar]
- Beceiro A., Tomás M., Bou G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26 185–230. 10.1128/CMR.00059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control [CDC] (2013). Antibiotic Resistance Threats in the United States. Atlanta, GA: Centers for Disease Control. [Google Scholar]
- Choi K. H., Gaynor J. B., White K. G., Lopez C., Bosio C. M., Karkhoff-Schweizer R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2 443–448. 10.1038/nmeth765 [DOI] [PubMed] [Google Scholar]
- Choi K. H., Kumar A., Schweizer H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64 391–397. 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Choi K. H., Schweizer H. P. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1 153–161. 10.1038/nprot.2006.26 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2007). Performance Standards for Antimicrobial Susceptibility Testing: 17th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute, M100–M117. [Google Scholar]
- Corzett C. H., Goodman M. F., Finkel S. E. (2013). Competitive fitness during feast and famine: how SOS DNA polymerases influence physiology and evolution in Escherichia coli. Genetics 194 409–420. 10.1534/genetics.113.151837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Solh A. A., Akinnusi M. E., Wiener-Kronish J. P., Lynch S. V., Pineda L. A., Szarpa K. (2008). Persistent infection with Pseudomonas aeruginosa in ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 178 513–519. 10.1164/rccm.200802-239OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltman H., Schulert G., Khan S., Jain M., Peterson L., Hauser A. R. (2001). Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147 2659–2669. 10.1099/00221287-147-10-2659 [DOI] [PubMed] [Google Scholar]
- Finkel S. E. (2006). Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4 113–120. 10.1038/nrmicro1340 [DOI] [PubMed] [Google Scholar]
- Garey K. W., Vo Q. P., Larocco M. T., Gentry L. O., Tam V. H. (2008). Prevalence of type III secretion protein exoenzymes and antimicrobial susceptibility patterns from bloodstream isolates of patients with Pseudomonas aeruginosa bacteremia. J. Chemother. 20 714–720. 10.1179/joc.2008.20.6.714 [DOI] [PubMed] [Google Scholar]
- Gullberg E., Cao S., Berg O. G., Ilbäck C., Sandegren L., Hughes D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158 10.1371/journal.ppat.1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J., Carniel E. (2001). Ecological fitness, genomic islands and bacterial pathogenicity. a Darwinian view of the evolution of microbes. EMBO Rep. 2 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E. M., Carter M. E., Luck S., Ou H. Y., He X., Deng Z., et al. (2010). Pathogenicity islands PAPI-1 and PAPI-2 contribute individually and synergistically to the virulence of Pseudomonas aeruginosa strain PA14. Infect. Immun. 78 1437–1446. 10.1128/IAI.00621-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A. R. (2014). Pseudomonas aeruginosa virulence and antimicrobial resistance: two sides of the same coin? Crit. Care Med. 42 201–202. 10.1097/CCM.0b013e3182a120cd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins P. G., Fluit A. C., Milatovic D., Verhoef J., Schmitz F. J. (2003). Mutations in GyrA, ParC, MexR and NfxB in clinical isolates of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 21 409–413. 10.1016/S0924-8579(03)00009-8 [DOI] [PubMed] [Google Scholar]
- Hooper D. C. (2001). Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7 337–341. 10.3201/eid0702.010239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu D. I., Okamoto M. P., Murthy R., Wong-Beringer A. (2005). Fluoroquinolone-resistant Pseudomonas aeruginosa: risk factors for acquisition and impact on outcomes. J. Antimicrob. Chemother. 55 535–541. 10.1093/jac/dki026 [DOI] [PubMed] [Google Scholar]
- Jalal S., Wretlind B. (1998). Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb. Drug Resist. 4 257–261. 10.1089/mdr.1998.4.257 [DOI] [PubMed] [Google Scholar]
- Komp Lindgren P., Marcusson L. L., Sandvang D., Frimodt-Møller N., Hughes D. (2005). Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob. Agents Chemother. 49 2343–2351. 10.1128/AAC.49.6.2343-2351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraigsley A. M., Finkel S. E. (2009). Adaptive evolution in single species bacterial biofilms. FEMS Microbiol. Lett. 293 135–140. 10.1111/j.1574-6968.2009.01526.x [DOI] [PubMed] [Google Scholar]
- Kriengkauykiat J., Porter E., Lomovskaya O., Wong-Beringer A. (2005). Use of an efflux pump inhibitor to determine the prevalence of efflux pump-mediated fluoroquinolone resistance and multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49 565–570. 10.1128/AAC.49.2.565-570.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E., Löfmark S., Wretlind B., Andersson D. I. (2005). Reduction of the fitness burden of quinolone resistance in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 55 22–30. 10.1093/jac/dkh505 [DOI] [PubMed] [Google Scholar]
- Kulasekara B. R., Kulasekara H. D., Wolfgang M. C., Stevens L., Frank D. W., Lory S. (2006). Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J. Bacteriol. 188 4037–4050. 10.1128/JB.02000-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz A. N., Begum A. A., Wu H., D’Ambrozio J. A., Robinson J. M., Shafer W. M., et al. (2012). Impact of fluoroquinolone resistance mutations on gonococcal fitness and in vivo selection for compensatory mutations. J. Infect. Dis. 205 1821–1829. 10.1093/infdis/jis277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi K., Kajikawa O., Sawa T., Ohara M., Gropper M. A., Frank D. W., et al. (1999). Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 104 743–750. 10.1172/JCI7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkis C., Fleiszig S. M. (2001). Resistance of Pseudomonas aeruginosa isolates to hydrogel contact lens disinfection correlates with cytotoxic activity. J. Clin. Microbiol. 39 1477–1486. 10.1128/JCM.39.4.1477-1486.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. G., Urbach J. M., Wu G., Liberati N. T., Feinbaum R. L., Miyata S., et al. (2006). Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7:R90 10.1186/gb-2006-7-10-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K., Lee Y. S., Park Y. K., Kim B. S. (2005). Alterations in the GyrA and GyrB subunits of topoisomerase II and the ParC and ParE subunits of topoisomerase IV in ciprofloxacin-resistant clinical isolates of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 25 290–295. 10.1016/j.ijantimicag.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Linder J. A., Huang E. S., Steinman M. A., Gonzales R., Stafford R. S. (2005). Fluoroquinolone prescribing in the United States: 1995 to 2002. Am. J. Med. 118 259–268. 10.1016/j.amjmed.2004.09.015 [DOI] [PubMed] [Google Scholar]
- Luo N., Pereira S., Sahin O., Lin J., Huang S., Michel L., et al. (2005). Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U.S.A. 102 541–546. 10.1073/pnas.0408966102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatallah M., Cheriaa J., Backhrouf A., Iversen A., Grundmann H., Do T., et al. (2011). Population structure of Pseudomonas aeruginosa from five mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS ONE 6:e25617 10.1371/journal.pone.0025617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuca J., Briales A., Labrador G., Díaz-de-Alba P., López-Rojas R., Docobo-Pérez F., et al. (2014). Interplay between plasmid-mediated and chromosomal-mediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J. Antimicrob. Chemother. 69 3203–3215. 10.1093/jac/dku308 [DOI] [PubMed] [Google Scholar]
- Marcusson L. L., Frimodt-Møller N., Hughes D. (2009). Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 5:e1000541 10.1371/journal.ppat.1000541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk A. H., Wong A., Kassen R. (2015). The fitness costs of antibiotic resistance mutations. Evol. Appl. 8 273–283. 10.1111/eva.12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D. T., Ming Di, Opperman T., Schweizer H. P., Bowlin T. L. (2007). A high-throughput, homogeneous, bioluminescent assay for Pseudomonas aeruginosa gyrase inhibitors and other DNA-damaging agents. J. Biomol. Screen. 12 855–864. 10.1177/1087057107304729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimné H., Robert J., Jarlier V., Cambau E. (1999). Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43 62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Deguchi T., Kawamura T., Yasuda M., Kimura M., Okano Y., et al. (1997). Mutations in the gyrA and parC genes in fluoroquinolone-resistant clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41 2289–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Cantón R., Campo P., Baquero F., Blázquez J. (2000). High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288 1251–1254. 10.1126/science.288.5469.1251 [DOI] [PubMed] [Google Scholar]
- Ozer E. A., Allen J. P., Hauser A. R. (2014). Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics 15:737 10.1186/1471-2164-15-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C., Cabot G., Gómez-Zorrilla S., Zamorano L., Ocampo-Sosa A., Murillas J., et al. (2015). Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin. Infect. Dis. 60 539–548. 10.1093/cid/ciu866 [DOI] [PubMed] [Google Scholar]
- Quartin A. A., Scerpella E. G., Puttagunta S., Kett D. H. (2013). A comparison of microbiology and demographics among patients with healthcare-associated, hospital-acquired, and ventilator-associated pneumonia: a retrospective analysis of 1184 patients from a large, international study. BMC Infect. Dis. 13:561 10.1186/1471-2334-13-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redgrave L. S., Sutton S. B., Webber M. A., Piddock L. J. (2014). Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 22 438–445. 10.1016/j.tim.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Restrepo M. I., Anzueto A. (2009). The role of gram-negative bacteria in healthcare-associated pneumonia. Semin. Respir. Crit. Care Med. 30 61–66. 10.1055/s-0028-1119810 [DOI] [PubMed] [Google Scholar]
- Rosenthal V. D., Bijie H., Maki D. G., Mehta Y., Apisarnthanarak A., Medeiros E. A., et al. (2012). International Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for 2004-2009. Am. J. Infect. Control 40 396–407. 10.1016/j.ajic.2011.05.020 [DOI] [PubMed] [Google Scholar]
- Roy-Burman A., Savel R. H., Racine S., Swanson B. L., Revadigar N. S., Fujimoto J., et al. (2001). Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183 1767–1774. 10.1086/320737 [DOI] [PubMed] [Google Scholar]
- Rozen D. E., McGee L., Levin B. R., Klugman K. P. (2007). Fitness costs of fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51 412–416. 10.1128/AAC.01161-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Frank D. W., Hillard C. J., Feix J. B., Pankhaniya R. R., Moriyama K., et al. (2003). The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin ExoU. EMBO J. 22 2959–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulert G. S., Feltman H., Rabin S. D., Martin C. G., Battle S. E., Rello J., et al. (2003). Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188 1695–1706. 10.1086/379372 [DOI] [PubMed] [Google Scholar]
- Shaver C. M., Hauser A. R. (2004). Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 72 6969–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver C. M., Hauser A. R. (2006). Interactions between effector proteins of the Pseudomonas aeruginosa type III secretion system do not significantly affect several measures of disease severity in mammals. Microbiology 152 143–152. 10.1099/mic.0.28368-0 [DOI] [PubMed] [Google Scholar]
- Sullivan E., Bensman J., Lou M., Agnello M., Shriner K., Wong-Beringer A. (2014). Risk of developing pneumonia is enhanced by the combined traits of fluoroquinolone resistance and type III secretion virulence in respiratory isolates of Pseudomonas aeruginosa. Crit. Care Med. 42 48–56. 10.1097/CCM.0b013e318298a86f [DOI] [PubMed] [Google Scholar]
- Swingle B., Markel E., Costantino N., Bubunenko M. G., Cartinhour S., Court D. L. (2010). Oligonucleotide recombination in Gram-negative bacteria. Mol. Microbiol. 75 138–148. 10.1111/j.1365-2958.2009.06976.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veesenmeyer J. L., Hauser A. R., Lisboa T., Rello J. (2009). Pseudomonas aeruginosa virulence and therapy: evolving translational strategies. Crit. Care Med. 37 1777–1786. 10.1097/CCM.0b013e31819ff137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasels F., Kuehne S. A., Cartman S. T., Spigaglia P., Barbanti F., Minton N. P., et al. (2015). Fluoroquinolone resistance does not impose a cost on the fitness of Clostridium difficile in vitro. Antimicrob. Agents Chemother. 59 1794–1796. 10.1128/AAC.04503-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L., Lam D. K., Fleming L., Lo R., Whiteside M. D., Yu N. Y., et al. (2011). Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39 D596–D600. 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Beringer A., Wiener-Kronish J., Lynch S., Flanagan J. (2008). Comparison of type III secretion system virulence among fluoroquinolone-susceptible and -resistant clinical isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 14 330–336. 10.1111/j.1469-0691.2007.01939.x [DOI] [PubMed] [Google Scholar]
- Zhang J., Chu Y., Wang P., Ji X., Li X., Wang C., et al. (2014). Clinical outcomes of multidrug resistant Pseudomonas aeruginosa infection and the relationship with type III secretion system in patients with diabetic foot. Int. J. Low. Extrem. Wounds 13 205–210. 10.1177/1534734614545878 [DOI] [PubMed] [Google Scholar]