Abstract
Motivation: The identification of ligand-binding sites from a protein structure facilitates computational drug design and optimization, and protein function assignment. We introduce AutoSite: an efficient software tool for identifying ligand-binding sites and predicting pseudo ligand corresponding to each binding site identified. Binding sites are reported as clusters of 3D points called fills in which every point is labelled as hydrophobic or as hydrogen bond donor or acceptor. From these fills AutoSite derives feature points: a set of putative positions of hydrophobic-, and hydrogen-bond forming ligand atoms.
Results: We show that AutoSite identifies ligand-binding sites with higher accuracy than other leading methods, and produces fills that better matches the ligand shape and properties, than the fills obtained with a software program with similar capabilities, AutoLigand. In addition, we demonstrate that for the Astex Diverse Set, the feature points identify 79% of hydrophobic ligand atoms, and 81% and 62% of the hydrogen acceptor and donor hydrogen ligand atoms interacting with the receptor, and predict 81.2% of water molecules mediating interactions between ligand and receptor. Finally, we illustrate potential uses of the predicted feature points in the context of lead optimization in drug discovery projects.
Availability and Implementation: http://adfr.scripps.edu/AutoDockFR/autosite.html
Contact: sanner@scripps.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Proteins are involved in a wide variety of biological processes such as signalling pathways and enzymatic reactions. While structural genomics projects have revealed structures of a large number of proteins, most of them lack reliable information about their biochemical functions (Mills et al., 2015). The identification and characterization of protein-binding sites can help decipher the function of proteins of unknown function. The interactions made by protein with ligands are often exploited in the design of small molecules to inhibit pathogenic, or overexpressed proteins involved in a biological process. Binding site characterization has various potential applications including: filtering databases of ligands, lead compound optimization, template definition for ligand search and automated docking.
Methods for identifying the locations where ligand molecules are likely to bind a receptor molecule of known 3D structure can be classified into three broad categories: (i) methods analyzing evolutionary information (Brylinski and Skolnick, 2008; Stark et al., 2004), which exploit the idea that important binding site residues are conserved for functional reasons; (ii) geometry-based methods (Dundas et al., 2006; Hendlich et al., 1997) which identify the clefts/cavities on the protein surface according to the size and depth of the pockets; and (iii) energy-based methods (Halgren, 2009; Harris et al., 2008; Hernandez et al., 2009; Laurie and Jackson, 2005), which rely on the potential generated by probe atoms or chemical moieties to determine ligand-binding sites. Surfnet (Laskowski, 1995) identifies pocket regions in proteins by fitting spheres in the spaces between protein atoms resulting in groups of interpenetrating spheres that correspond to the protein-binding sites. Ligsite (Hendlich et al., 1997) places the protein in a Cartesian grid and scans along the x, y, and z axes and the cube’s diagonals for solvent accessible areas that are enclosed by protein atoms on both sides. PocketFinder (An et al., 2004) uses van der Waals affinity map computed with a carbon probe of radius 1.7Å. The regions with larger cumulative values are identified and contoured with a threshold to identify binding sites. Concavity (Capra et al., 2009) provides its own implementation of the LigSite, Surfnet, and Pocketfinder algorithms and combines them with evolutionary sequence conservation data to improve the accuracy of binding site prediction. Concavity’s implementations of these algorithms when evolutionary information is not included are named LigSite+, Surfnet+ and Pocketfinder+. These programs report the binding sites as a collection of 3D points selected from a regular 3D grid. Binding site characterization methods analyze an identified binding site to provide more detailed information about the physicochemical properties of potential ligands. Several review papers (Ghersi and Sanchez, 2011; Henrich et al., 2010; Perot et al., 2010) provide a detailed analysis of computational approaches for identifying and characterizing protein-binding sites. These methods often characterize binding sites based on the amino acid composition of the receptor around the cavity, and properties computed at the binding site such as solvation, hydrophobicity, and electrostatics. A common approach to characterize the binding site is by generating affinity grids(Goodford, 1985; Huey et al., 2007) using different probe atoms at the binding site—a concept exploited by energy-based approaches in identifying binding sites, and analysing the binding site by 1) docking fragments (Jain, 2003) 2) condensing the interactions into discrete pharmacophore points (Baroni et al., 2007; Halgren, 2009) or 3) applying geometric rules (Lower et al., 2011; Lower and Proschak, 2011). The characterization heavily relies on the binding site information and hence these techniques are often combined with custom or existing binding site identification algorithms with or without emphasis on the shape of the ligand-binding region. The obtained pharmacophore points are used to compare and annotate the function of proteins, or used in virtual screening experiments for drug design. AutoLigand (Harris et al., 2008) is a software program that places emphasis on chemically detailed ligand shape prediction when identifying the ligand-binding sites. It works on the hypothesis that protein-binding sites have evolved to create a region of maximal affinity for the given size and shape of a binding ligand. It identifies a contiguous region of maximal-binding affinity by growing a cluster of contiguous, high-energy points from on a grid of potential values obtained by combining affinity maps from multiple atom-types. The resulting cluster of grid points yields a prediction of ligand shape and each fill point of the predicted binding sites is associated with an AutoDock atom type. Although the above described approaches attempts to characterize the binding site to provide information about governing interactions between protein and ligand at the binding site, there is no available study to the best of our knowledge that assess their quality of prediction with existing ligand(s) that binds to the respective binding site.
Here, we present AutoSite, a new energy-based method for identifying ligand-binding sites and predicting potential pseudo-ligand in each of the predicted binding site of a protein structure. Contrary to AutoLigand, AutoSite filters out low affinity points from affinity maps computed with hydrophobic (carbon) and hydrophilic (oxygen, hydrogen) atomtypes to select high affinity points, merges the selected points, and predicts feature points corresponding to potential ligand atoms by applying knowledge-based and geometric rules. We show that AutoSite performs equally to or slightly better than the state-of-the-art energy- and geometry-based binding site identification methods. We further demonstrate that AutoSite outperforms AutoLigand in the accuracy of labeling fill points as hydrophobic, hydrogen bond donor/acceptor atoms and in the coverage of ligand atoms. Finally, we introduce putative ligand atomic centres called feature points derived from the AutoSite fill points. We show that for the Astex Diverse Set, these feature points correctly identify 79.3% of hydrophobic ligand atoms as well as 81.4% and 62.9% of the ligand hydrogen acceptor and donor hydrogen atoms that interact with the receptor, and predict 81.2% of water molecules that interact both with ligand and receptor. In addition, we illustrate potential uses of the predicted feature points in the context of drug discovery projects.
2 Methods
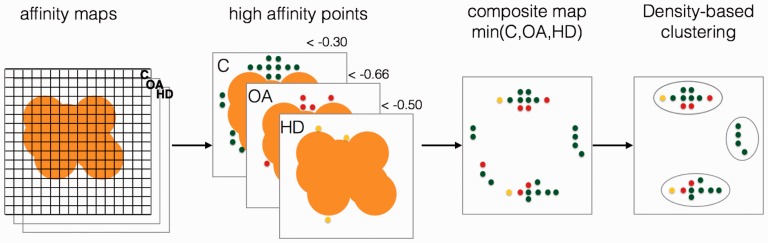
AutoSite is an energy-based method for identifying and characterizing ligand-binding pockets on receptors and deriving feature points corresponding to putative ligand atoms. It relies on potentials generated by receptor atoms on grid points to identify clusters corresponding to potential binding sites, and geometric measures for ranking these binding sites. AutoSite uses AutoDock (Morris et al., 2009) affinity maps computed using AutoGrid4 (Huey et al., 2007) for carbon (AutoDock atom type C, hydrophobic), oxygen (AutoDock atom type OA, hydrogen bond acceptor) and hydrogen (AutoDock atom type HD, hydrogen bond donor) atom types to identify binding sites. These maps are regularly spaced grids where each grid point yields the sum of the pairwise interaction energies between a probe-atom of a given type with all receptor atoms. The maps include atom-specific affinities and do not include electrostatics and charge-based desolvation. AutoSite computes maps covering the entire receptor, and selects high affinity points from each map based on probe-specific affinity cutoffs. It then merges the three sets of high affinity points into a composite map by selecting the minimum value at each grid position (Fig. 1) . The selected grid points are then clustered to define potential binding sites.
Fig. 1.
High affinity points in the hydrophobic affinity map (C), and hydrogen bond forming affinity maps (acceptor – OA; donor – HD) are selected, combined and clustered to yield putative ligand binding sites (Color version of this figure is available at Bioinformatics online.)
Affinity cutoffs: The AutoSite algorithm relies on affinity cut-off values used to identify high affinity grid points. The cutoff value of −0.3 kcal/mol for the carbon affinity map has been used by us (Ravindranath et al., 2015) and others (Ghersi and Sanchez, 2009) as it selects grid points covering ligands atoms. The donor (HD) and acceptor cutoffs (OA), −0.66 kcal/mol and −0.5 kcal/mol respectively, were obtained by analyzing affinity maps computed for the 189 receptor-ligand complexes from the AutoDock calibration dataset (Huey et al., 2007) as follows. First, hydrogen (HD) and oxygen (OA) affinity maps were computed using AutoGrid4. The maps were sized to cover the entire receptor and computed using a grid spacing of 1Å. Next, hydrogen donor atoms (HD) and hydrogen acceptor atoms (OA-oxygen, SA-sulphur, and NA-nitrogen) contributing to the solvent excluded surface computed using MSMS (Sanner et al., 1996) were identified. For each of solvent-accessible hydrogen-bond capable receptor atom, the best affinity grid point within 2.5Å of the atom was recorded in the partner affinity map (i.e. the OA map for hydrogen donor atoms and the HD map for hydrogen acceptor atoms) yielding a distribution of affinity values in the vicinity of surface receptor donor and acceptor atoms. The averages of these two distributions are −0.66 kcal/mol for the OA grid points in the vicinity of a receptor hydrogen donor atom, and −0.50 kcal/mol for the HD grids points in the vicinity of a receptor hydrogen acceptor OA atom. The vdW term of the AutoDock energy function used by AutoGrid to compute affinity maps for non-hydrogen ligand atoms provides an average of about −0.3 to −0.5 kcal/mol. For hydrogen bonding atoms the value is approximately −0.6 kcal/mol for a single hydrogen bond and approximately twice of −0.6 kcal/mol for oxygen atoms that accept two hydrogen bonds (Huey et al., 2007). The values can be anywhere less than or equal to the reported value on a grid. Thus the considered cutoff for hydrophobic (−0.3 kcal/mol), hydrogen acceptor (−0.66 kcal/mol) and donor (−0.50 kcal/mol) respects the magnitude of energy terms that corresponds to the hydrophobic and hydrogen-bonding interactions.
The composite map is obtained by merging the highest affinity grid points from the three maps into a single set of grid points. If a grid point has an identical high affinity in more than one map, the atom type assignment follows the order: C, OA, HD. The rationale behind this order is that hydrophobic groups govern the shape of the ligand and hence it is given the highest priority; followed by hydrogen acceptor that are specific and directional and hence expected to be highly accurate, and finally the donor whose contribution is essential for hydrogen bonding but becomes uncertain on designed ligands due to their physicochemical properties.
Density-based clustering: High affinity grid points are clustered to partition the points into contiguous sets, each set corresponding to a potential binding site. This clustering is performed using a modified implementation of the DBSCAN algorithm (Ester et al., 1996). DBSCAN uses a local density cutoff value to grow clusters, and is known for its ability to handle noise (i.e. isolated points). AutoSite exploits the fact that the points to be clustered are located on a grid to replace the DBSCAN local density calculation by a simple count of neighbouring grid points. A given grid point will be added to a growing cluster if it has at least N neighbouring grid points in the set of high affinity grid points. Intuitively, N controls the minimum width of channels connecting a collection of compact points. Lower values of N allow cluster to have more protrusions and grow larger, while larger values of N generate more compact clusters. The clustering of a set P of high affinity grid points, for a minimum number of neighbours N is performed as follows. A point S is selected randomly from P as a seed point. If S has at least N neighbours in the set, S and all its neighbours are added to the cluster seeded by S. S is marked as treated and removed from the set P, and the seed S and its neighbours that do not yet belong to a cluster are added to the cluster. The test for number of neighbours is performed for every un-treated point in the cluster, potentially adding more points to the cluster, until all points in the cluster have been treated. After a cluster is completed, a new point S is selected from P as a seed for a new cluster, until P is empty. On a cubic grid, a point has a maximum of 26 neighbours. We identified 14 as the best value for the minimum number of neighbours N by maximizing the Jaccard/Tanimoto coefficient (Jaccard, 1901) between the 189 ligands of the AutoDock calibration set and fills obtained for values of N ranging from 12 to 17. The Jaccard coefficient is used to compare the similarity of two shapes as the ratio of the intersection over the union of the two shapes. As the fills are clusters of high affinity points located on a 3D grid, we discretized the ligand atomic-spheres onto the same grid using atomic radii from the AutoDock4 parameters set to obtain ligand shape as a set of grid points. The Jaccard coefficient is calculated as the ratio of the total of intersecting grid points over the total of union of intersecting and non-intersecting points from discretized ligand and reported fill.
Pocket ranking: The fills identified by clustering are ranked using a geometry-based score. Like most site-finding software programs, AutoSite computes a variety of numerical descriptors for each fill, including: affinity, number of points, efficiency (i.e. affinity/volume), and radius of gyration. In addition, we compute fill buriedness by dividing the fill’s buried surface area by the fill’s total surface area. This value ranges from 1.0 for a fill entirely buried by the receptor (i.e. an enclosed cavity) to 0.0 for a fill entirely exposed to solvent. The buriedness is calculated numerically in AutoSite as follows. First, we identify the set of grid points covered by ligand atomic spheres augmented by 1.0Å. Next the grid points covered by the ligand atomic spheres with their original radii are removed from this set, yielding a shell of grid points around the ligand. The number of grid points in this shell, Nt is used as a numerical approximation of the fill’s total surface area. Next, the receptor atomic spheres, with a radius augmented by 1.0, are used to tag as “buried” grid points in the surface shell. The buriedness is calculated as the ratio of the number of buried surface shell grid points over Nt. We rank the fills using an empirical composite score in which the fill size (i.e. number of points) is multiplied by the square of its buriedness and divided by the radius of gyration. This metric was designed to favour fills that have large volumes with compact buried cavities.
AutoSite’s ability to identify the binding site was evaluated with the Astex Diverse Set (Hartshorn et al., 2007). This dataset contains 85 well-curated proteins-ligand complexes with ligands that have drug-like properties. This set has no overlap with the AutoDock’s calibration dataset and thus the predictions have no training bias. The proteins from the dataset were converted to the AutoDock’s PDBQT format and gasteiger charges were assigned. AutoSite’s performance was compared with LigSite+, Surfnet+ and Pocketfinder+. These programs were run with their default parameters as documented by Concavity and the resulting predictions were ranked by volume which is the default ranking metric for these three programs. The success rates of all programs were assessed with Jaccard coefficient at cutoffs 0.0, 0.1, 0.25 and 0.5.
Binding site characterization: While LigSite+, Surfnet+ and Pocketfinder+ report pockets as a collection of un-typed 3D points, AutoSite fill points are labelled as hydrophobic, hydrogen bond donor and acceptor points. AutoLigand and FLAP (Baroni et al., 2007) are other software programs that predict typed fills. We were unable to obtain a license to test FLAP, hence we evaluated the merit of our method by comparing the typed fills predicted by AutoSite with the ones predicted by AutoLigand on the Astex Diverse Set. The fills were generated for both programs using the same carbon, oxygen and hydrogen affinity maps. These maps were calculated using AutoGrid4 as cubic grids of 27 Å on a side (including a padding of 2Å) and centred on the ligand, with a grid spacing of 1Å. By default, AutoLigand initiates fill starting from 3D points randomly picked on the grid. As we are interested in comparing fills that overlap with the ligand, we ran AutoLigand using the ligand geometric centre as the starting point for the fill. We ran the program for numbers of fill points ranging from 10 to 1500 in increments of 10. The fill with the best energy-per-volume was picked as the AutoLigand prediction. The AutoSite program was executed with the default parameters (i.e. affinity cutoffs of -0.3 kcal/mol for C, −0.66 kcal/mol for O and −0.5 kcal/mol for H, and 14 minimum number of neighbours for clustering) and the predicted fill closest to the ligand geometric centre was considered for comparison.
Feature Points
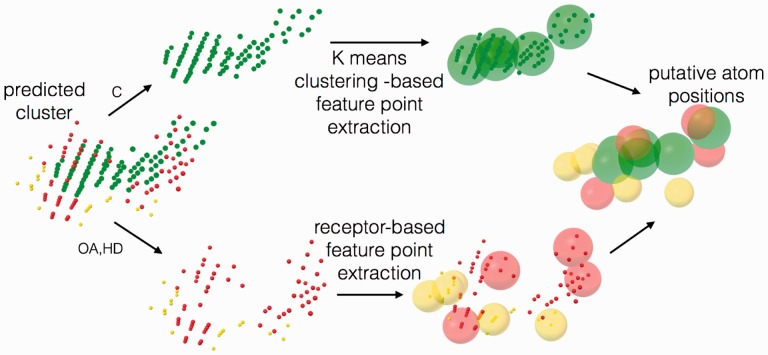
AutoSite converts the fills points into putative hydrophobic, or hydrogen-bond forming ligand atoms. For hydrophobic fill points we use k-means clustering for dividing the fill points into compact clusters of fill points representing a pre-defined number of carbon atoms. A survey of the ligands of the Astex Diverse Set revealed that on average carbon atoms cover 14.3 grid points for grids of spacing 1Å. This average was found to be highly stable when the grid was translated by small amounts in random directions (data not shown). We cluster the hydrophobic fill points to form Nhp/14 clusters, where Nhp is the number of hydrophobic fill points. The centres of the resulting clusters are used as atomic centres for putative hydrophobic atoms. Putative hydrogen bond donors (HD) and hydrogen-bond acceptor (OA) ligand atoms are identified as follows. Every fill point of type HD is associated with all receptor OA located within 2.5 Å and every fill point of type OA is associated with all receptor HD atoms located within 2.5 Å. Next, for each of these receptor atoms the associated fill point with the best affinity (i.e. putative atom centre) is selected as a feature point. The result of this process is a set of putative ligand atoms. Figure 2 illustrates this process using an actual fill generated for the streptavidin receptor.
Fig. 2.
Fill points are segregated into hydrophobic points (C-green), and hydrogen bond acceptor (OA-red) and donor hydrogen points (HD-yellow). K-means clustering is performed on the C points and the resulting cluster centroids are kept as the hydrophobic feature points. Feature point extraction for O and H is performed using the receptor surface hydrogen-bonding atoms (Color version of this figure is available at Bioinformatics online.)
3 Results
The binding site identification results summarized in Table 1 show that AutoSite performs comparably or better than the other four programs. With fill overlap ratio (Jaccard coefficient) > 0.1 as the cutoff, AutoSite identified 58 binding sites in the top prediction correctly out of 85 compared to 54 for LigSite+, 43 for Surfnet+, 44 for PocketFinder+ and 56 for SiteHound. Considering the top three binding site predictions for each system, AutoSite identified 76 binding sites compared to 74 for LigSite+, 61 for Surfnet+, 62 for Pocketfinder+ and 72 for SiteHound.
Table 1.
Binding site identification performance comparison between LigSite+, Surfnet+, Pocketfinder+, SiteHound and AutoSite
|
Top1
|
Top3
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Jaccard coefficient | >0.0 | >0.1 | >0.25 | >0.5 | >0.0 | >0.1 | >0.25 | >0.5 |
| LigSite+ | 57 | 54 | 38 | 6 | 78 | 74 | 56 | 10 |
| Pocketfinder+ | 52 | 44 | 29 | 1 | 71 | 62 | 41 | 2 |
| Surfnet+ | 49 | 43 | 36 | 8 | 69 | 61 | 49 | 11 |
| SiteHound | 56 | 56 | 41 | 2 | 72 | 72 | 52 | 2 |
| AutoSite | 60 | 58 | 45 | 3 | 78 | 76 | 59 | 3 |
The first column shows number of systems (out of 85) for which the programs correctly identify the ligand-binding site as the top prediction. The second column shows the number when top 3 predictions are considered.
The programs were tested using different Jaccard coefficient thresholds, and AutoSite showed to generate more fills with Jaccard Coefficients greater than 0.0, 0.1 and 0.25 than other programs, indicating better coverage of the ligand both in top prediction as well as when considering Top 3 predictions. SiteHound was found to do better than LigSite+ in Top 1 at both threshold >0.1 and >0.25. LigSite+ and Surfnet+ found 6 and 8 systems in Top1, and 10 and 11 systems in Top3 out of 85 respectively, with Jaccard coefficient >0.5 compared to 3 by AutoSite both in Top1 and Top3 predictions. Visual analysis of AutoSite predicted fills corresponding to the 12 systems that did not get >0.5 Jaccard coefficient, showed that they overlap well with the ligand except in 2 cases (1l7f – 0.17; 1vcj – 0.21), where AutoSite predicts only part of the ligand. The consistency demonstrated by AutoSite in success rate shows that the fills produced by AutoSite have better overlap with the ligand in addition to identifying the correct binding sites.
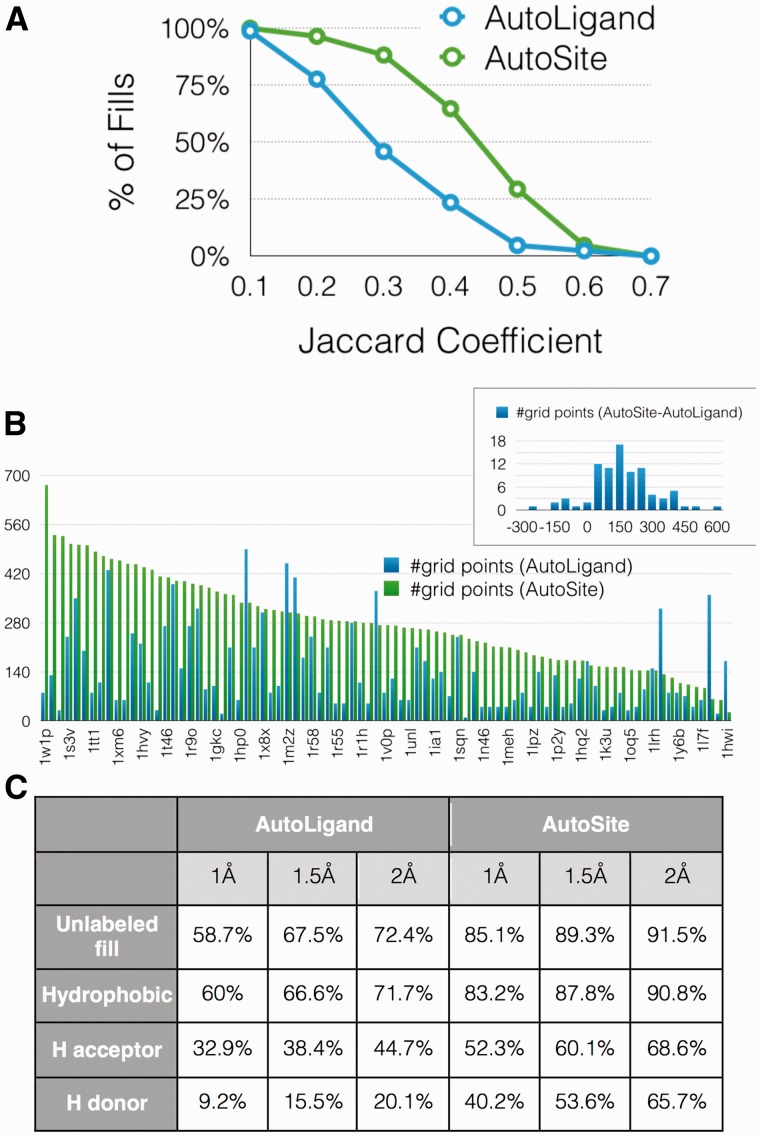
The comparison of the binding site characterization performed by AutoLigand and AutoSite is shown in Figure 3. Figure 3A plots the percentage of systems as a function of the Jaccard coefficient of the fills produced by both programs. We observe a substantial increase in the number of fills with better Jaccard coefficients for AutoSite, with 64.7% of the fills having a Jaccard coefficient > 0.3 compared to 23.5% for AutoLigand fills. The average Jaccard coefficient for AutoSite and AutoLigand were found to be 0.34 and 0.21, respectively. The relatively low values of the Jaccard coefficients can be explained by the fact that ligands do not always exploit the entire binding pocket, hence, the predicted fills tend to extend beyond the ligand as discussed below in the discussion where the predicted fill cover regions corresponding to different known ligands. Small Jaccard coefficients can arise when the overlap between the fill and the ligand is small or when the fill extends far beyond the ligand, or both. To investigate the results further, we analyzed the fill sizes and the overlap with the ligands separately. It should be noted that while the Jaccard coefficient is computed using a ligand discretized on a grid identical to the affinity maps, the percentage of ligand overlap is computed with the ligand atom coordinates. Figure 3B shows the volume of the fills sorted by decreasing size of the AutoSite fills. For 77 out of 85 ligands (90%), AutoSite fills are larger than the AutoLigand fills with an average of 150 more points per fill as indicated by the distribution of fill size differences in the inset. To analyze the overlap of the fill with the ligand, we computed the distance between every ligand atom and the closest fill point. Figure 3C shows that 91.5% of ligand atoms are within 2.0 Å of a fill point, compared to 72.4% for AutoLigand. We further segregated ligand atoms into hydrophobic (C, A, N, S, P), hydrogen acceptors (OA, NA, SA) and hydrogen donor (HD) atoms and computed the distance of each ligand atom to the closest fill point of the appropriate type. The ligands have 90.8% hydrophobic, 68.6% hydrogen acceptors, and 65.7% for hydrogen donor atoms within 2 Å of an AutoSite fill point of the proper type, compared to the 71.7%, 44.7% and 20.1% for AutoLigand fills, respectively. These results indicate that AutoSite fills, while larger, have a better overlap with the ligand and as such offer a more accurate depiction of the ligand as predicted from the receptor structure.
Fig. 3.
(A) Percentage of fills as a function of their fill-ligand Jaccard coefficients. (B) AutoSite and AutoLigand fill sizes comparison for the 85 systems. The inset plot shows the histogram of the differences in fill sizes. (C) Percentage of ligand atoms that find a fill point within 1Å, 1.5Å and 2Å. The ‘Unlabeled fill’ row provides the percentage of ligand atoms with a fill point with any label within the distance cut-offs. The rows below provide the percentages when fill labels are considered (Color version of this figure is available at Bioinformatics online.)
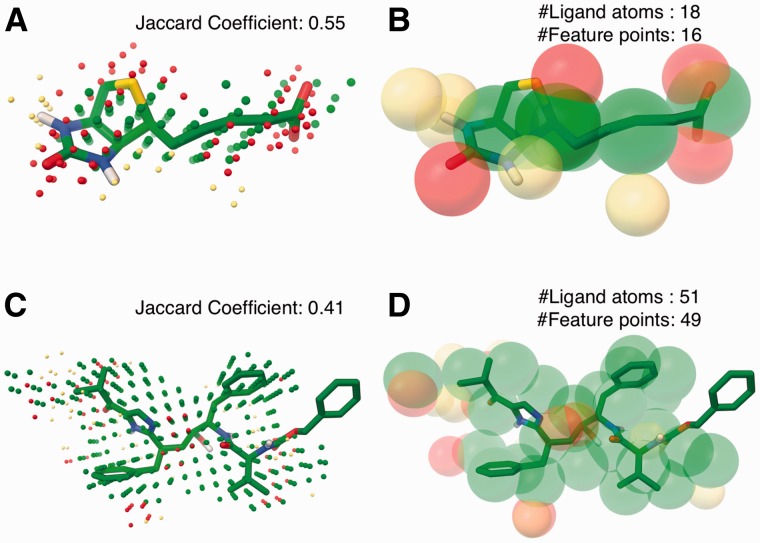
Beyond the labeling of fill points in the predicted binding sites, AutoSite predicts the number and positions of potential ligand atoms interacting with the receptor in this binding site. Examples of such predicted feature points from fill points are shown in Figure 4 for PDB ids 1stp and 1hps. Fig. 4A and 4C shows the starting fill points for streptavidin and HIV-1 protease. Figure 4B shows that the predicted hydrogen bond acceptor feature points (red spheres) overlap with the oxygen and sulphur atoms, hydrogen bond donor feature points (yellow spheres) are in close proximity to nitrogen atoms that are known to be protonated in the bound form, and the hydrophobic feature points overlay well with the carbon atoms. Our method performs particularly well on this ligand, as it is highly potent with a binding affinity of 40 fM. Figure 4D shows another example of feature points for the site predicted for the binding site of an HIV-1 protease inhibitor (PDB id 1hps). The cross-shaped peptide geometry found in many HIV protease inhibitors and most hydrogen bonding ligand atoms are identified by the feature points. In particular the strong hydrogen bond acceptor preference identified at the centre of the fill is a common interaction observed in HIV-1 proteases. This position is known to accommodate a water molecule, which is displaced in cyclic urea inhibitors by an oxygen atom (Harris et al., 2008). The phenyl group in the ligand that is not covered by the feature points doesn’t show any strong interaction with the receptor in the crystal structure. The feature points predicted for 20 other protease structures yielded the cross-shaped geometry as described above and tends to identify the key interactions (Supplementary information SFig.1). Figure 4 shows ligands that have excellent overlap with the predicted fills. Hence we see remarkably good agreement between the number of atoms in ligands and the predicted number of feature points. In Table 2, we report the correlation between number of ligand atoms and the number of predicted feature points for the Astex diverse dataset at increasing cutoff values of the Jaccard Coefficient. The correlation increases with the increase in the overlap ratio, and with the cutoff of the average Jaccard coefficient obtained by AutoSite (0.34) we get a correlation coefficient of 0.74. The analysis includes ligand hydrogen atoms, and it is noteworthy that the Astex diverse set contains manually curated ligands for corresponding proteins.
Fig. 4.
Predicted cluster and extracted feature points for streptavidin (PDB: 1stp; A, B) and HIV-1 protease (PDB: 1hps; C, D) overlaid with their respective experimentally determined bound ligand (balls and sticks; carbon - green). The potential hydrogen acceptor positions are shown as red spheres, hydrogen positions are shown in yellow spheres, and the hydrophobic positions are shown as green spheres (Color version of this figure is available at Bioinformatics online.)
Table 2.
Correlation between the number of atoms in crystallographic ligands and predicted pseudo ligands
| Jaccard Coefficient | >0.1 | >0.2 | >0.3 | >0.33(avg.) | >0.4 | >0.5 |
|---|---|---|---|---|---|---|
| Correlation Coefficient | 0.11 | 0.21 | 0.5 | 0.74 | 0.88 | 0.96 |
In order to quantify the ability of AutoSite’s predicted feature points to match with actual ligand atoms we computed the distance from each ligand atom to the closest matching feature point. Table 3 shows that over 79% of all hydrophobic ligand atoms have a corresponding hydrophobic feature point within 2Å. The percentages for all ligand hydrogen-bond forming atoms are 41% for acceptor atoms and 46% for donor hydrogen atoms respectively. Given that the feature points are derived from fill points, which are high affinity points for the receptor, the method is expected to only predict the subset of ligand hydrogen bond forming atoms that actually interact with the receptor. Performing the analysis considering only the ligand atoms involved in a hydrogen bond with a receptor atom (i.e. distance < 2.5Å) the percentages of ligand atoms with a fill point within 2Å of a feature point increase to 81.4% for acceptor atoms and to 62.9% for donor hydrogen atoms. Although a decrease in the percentage ligand overlap is expected when extracting representative feature points from fill points, the result provides scope for further optimization to the extraction procedure. In this analysis, we considered the AutoDock4 atom types (C, A, N, S, P) as hydrophobic, (OA, NA, SA) as acceptor atoms, and HD as donor atoms. In the Astex Diverse Set, 72% of ligands atoms are hydrophobic, 15.1% acceptor atoms, and 10.9% hydrogen bond donor atoms (HD). Another 2% of ligand atoms are halogen atoms (Cl, Br, F) and were ignored for this analysis.
Table 3.
Ligand coverage by feature points
| All ligand atoms |
Ligand atoms H-bonding with receptor |
|||
|---|---|---|---|---|
| 1.5Å | 2.0Å | 1.5Å | 2.0Å | |
| Hydrophobic | 58.9% | 79.3% | – | – |
| H acceptor | 33.2% | 41.1% | 73.6% | 81.4% |
| H donor | 30.1% | 46.4% | 30.8% | 62.9% |
The first column reports the percentage of ligand atoms within 1.5Å and 2Å of a predicted feature point of the appropriate type. The second column shows the percentages when only ligand atoms interacting with the receptor are considered.
Predicted hydrogen bonding feature points should also identify crystallographic water positions. To verify this hypothesis, we protonated the receptors of the Astex Diverse Set that included water molecules using What If web server (Vriend, 1990), and selected water molecules within 2.5Å of the ligand. We found that 38.3% of the oxygen atoms, 30.8% of the hydrogen atoms of these selected water molecules found corresponding feature point within 2Å (Table 4A). When including only water molecules within 2.5Å of both the receptors and ligand, AutoSite predicts 58% of the oxygen and 41.3% of the hydrogen atoms of these water molecules (Table 4A). Since the prediction gets only hydrogen bond interactions between the receptor and atoms that are at a distance of 2.5Å, we further analyzed (Table 4B) the percentage of oxygen atoms in water molecules that finds either a hydrogen or an oxygen feature point within 3.5Å (maximum interaction distance between hydrogen bonding heavy atoms). The result showed that 81.2% percentage of the oxygen atoms from the water molecules mediating interaction between a receptor and ligand, have a feature point within 3.5Å. Examples in Supplementary information (SFig. 2)
Table 4.
Water molecules within 2.5Å of the ligand identified as feature points
| A | Water molecules within 2.5Å of ligand |
Water molecules within 2.5Å of ligand and receptor |
|||
|---|---|---|---|---|---|
| 1.5Å | 2.0Å | 1.5Å | 2.0Å | ||
| H acceptor | 29.9% | 38.3% | 45.0% | 58.0% | |
| H donor | 19.2% | 30.8% | 26.1% | 41.3% | |
| B | Water molecules within 2.5Å of ligand and receptor (oxygen) | ||||
| 1.5Å | 2.0Å | 2.5Å | 3.0Å | 3.5Å | |
| H acceptor or H donor | 50.7% | 65.2% | 68.1% | 72.5% | 81.2% |
A. The first column reports the percentage of oxygen and hydrogen atoms from water molecules within 1.5Å and 2Å of a predicted feature point of the appropriate type. The second column shows the percentages when water molecules mediate interaction between the receptor and the ligand. B. Percentage of water molecules’ oxygen finding a hydrogen bond acceptor or donor within cutoffs ranging from 1.5Å to 3.5Å.
Timing: On a Intel Xeon E5-1620 3.5GHz processor, starting from 1.0 Å spacing carbon, oxygen and hydrogen maps covering the entire receptor, AutoSite takes on average 5.7 seconds to compute all fills with 50 or more points, extract the feature points for all fills, rank the fills, and write them out to file along with feature points in the PDB file format. Using AutoGrid4 for computing the carbon, oxygen and hydrogen maps takes 34.5 seconds on average for the Astex diverse set receptors. A custom version of AutoGrid that allows the exclusion of electrostatics and desolvation maps computes these maps in 4.1 seconds on an average. Thus, AutoSite takes on average less than 10 seconds to compute the maps, identify the binding sites with 50 or more fill points and obtain the pseudo ligand for each of the predicted sites.
4 Discussion
AutoSite identifies binding sites with higher accuracy than other leading methods. While it only performs marginally better than LigSite+ for binding site identification, it produces labelled points which can further be used to characterize the binding site. In addition, the Astex Diverse Set is a collection of diverse receptors with geometrically well-defined binding sites, often buried cavities, which overall favours geometry based methods like LigSite+. Similar to AutoLigand, AutoSite fill points are annotated as hydrophobic, hydrogen-bond acceptor or donor hydrogen atoms. The comparison of labelled fills produced by these two programs was performed using AutoLigand fills seeded at the ligand geometric centre to ensure an overlap between the fill and the ligand and the AutoSite fill with the point closest to the ligand geometric centre. It is noteworthy that 59 out of the 85 AutoSite fills selected for this analysis also are the top ranking fills obtained for a grid spanning the entire receptor. Moreover, 81 of the AutoSite fills selected for this analysis are the top ranking fills obtained for the smaller 27x27x27Å3 boxes. AutoSite was shown to outperform AutoLigand by covering more ligand atoms. The fills identified by AutoSite tend to be larger than the ligand bound to the receptor. This is because ligands exploit a subset of the possible interactions with the receptor as exemplified by Chitinase, which binds the cyclic GLY-PRO fragment (PDB id 1w1p) as well as a natural-product cyclopentapeptide (argadin) (PDB id 1h0g) and the CI4 inhibitor (PDB id 1o6i). The cyclic GLY-PRO fragment bound structure was part of the study to understand the mechanism of inhibition by cyclic dipeptide inhibitors of chitinase, which is involved in chitin degradation(Houston et al., 2004) and only occupies a small portion of the fill. Argidin binds to the (+) sub-sites of the enzymes (Houston et al., 2002) and the CI4 inhibitor prefers the (–) sub-sites of the enzyme (Houston et al., 2004) thus each inhibitor occupying one half of the fill (Supplementary information SFig. 3). This illustrates the reason for the fills identified by AutoSite to be larger than certain ligands. On the other hand, the fill predicted by AutoLigand only identifies the (–) sub-sites of chitinase. For another class of receptors, we observe that AutoSite generates multiple smaller adjacent clusters. For example, in PDB id 1hwi - HMG-CoA reductase with Fluvastatin, the five-membered ring in the ligand acts as a scaffold, which positions three hydrophobic functional groups that are increasing this ligand’s potency (Istvan and Deisenhofer, 2001). While fills covering the HMG-like moiety and the hydrophobic groups are identified, they are disconnected because the five-membered ring does not interact strongly with the receptor creating a gap in high affinity points preventing the clustering algorithm from connecting these fills (Supplementary information SFig. 4).
The feature points predicted by AutoSite for a given binding site have several potential applications relevant to medicinal chemistry, some of which we discuss and illustrate below. Feature points predict crystallographic water molecules, suggesting that they can potentially be used for designing and optimizing drugs that make use of the water interactions or that replace the water positions for binding the receptor.
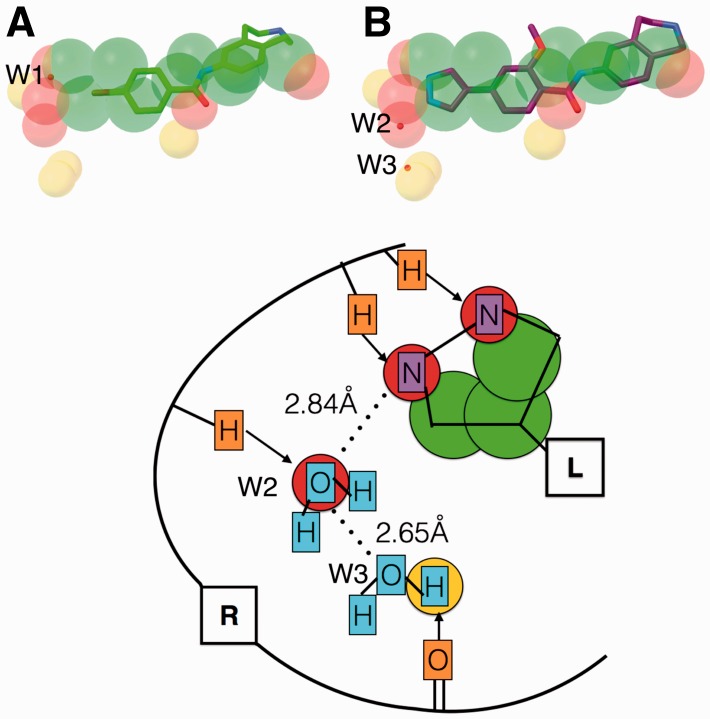
A recent study by Johnson et al. (Johnson et al., 2015) on lead optimization of inhibitors for MELK (maternal embryonic leucine zipper kinase) using fragment-based discovery illustrates the potential use of feature points for lead optimization. The authors optimized a fragment with 160 micromolar affinity using structure based drug design into a 37 nanomolar inhibitor. We obtained feature points using AutoSite on the fragment-bound crystal structure of the receptor (PDB id 4d2w) (Fig. 5A). The predicted putative ligand atoms identified the crystallographic water (W1) location, as well as additional hydrogen bond forming positions. The optimized nanomolar inhibitor, when superimposed over the AutoSite feature points (Fig. 5B) occupies two of the predicted hydrogen bond acceptor spots near nitrogen atoms, leaving one more hydrogen bond acceptor spot and a few donor spots available. It is interesting to notice that the crystal structure of the nanomolar inhibitor bound protein (PDB id 4d2v) has a water molecule (W2) interacting with the ligand nitrogen atom (2.84 Å) as well as a second crystallographic water molecule (W3) (2.65 Å). Our method predicts a hydrogen acceptor atom at the first water molecule (W2) location and a donor hydrogen atom close by the second water molecule (W3) (Fig. 5C), indicating that predicted feature points could be used to further optimize the ligand.
Fig. 5.
(A) Predicted feature points represented as spheres (carbon – green; oxygen – red; and hydrogen – yellow) overlaid with 4d2w bound fragment (stick and balls; carbon – green). B) Fragment optimized inhibitor (stick and balls; carbon – purple) overlaid with the fragment and the predicted feature points for 4d2w. The crystallographic water molecules are shown as small red spheres. C) Schematic illustration of (B): Receptor (R: atoms - orange) interactions (arrows) are used to extract the feature points represented as circles (green - carbon; red - oxygen; yellow- hydrogen). Predicted feature points capture the ligand atoms (L: atoms - purple) and water molecules (cyan) (Color version of this figure is available at Bioinformatics online.)
Feature points could also potentially be used for protein function annotation. This could be achieved by keeping track of the receptor amino acids interacting with feature points and comparing them with the binding site databases.
Automated docking is another area that could benefit from the feature points produced by AutoSite. Some docking programs such as SLIDE (Schnecke et al., 1998), DOCK (Allen et al., 2015), and Surflex (Jain, 2003) place the ligand into the receptor using templates of interactions. These programs could benefit from using feature points generated by AutoSite. Programs such as AutoDockFR (Ravindranath et al., 2015) allow for the explicit representation of receptor side chains as flexible during docking but require the a-priori identification of these side chains. Feature points could be used to select receptor side chains (the ones interacting with the feature points) to be made flexible. Finally, feature points could be used to filter large databases of ligands for compounds matching the feature points.
AutoSite like any energy-based methods will be sensitive to the translation and orientation of coarse grids. This limitation is attenuated when using smaller grid spacing. Pseudo-ligands derived by AutoSite for 85 proteins in Astex diverse set, using a grid spacing of 0.375Å on cubic boxes of 26.625Å on a side and centred on the respective ligands shows improvement in the ligand atoms coverage by feature points (SFig. 5). AutoSite takes on an average 19 seconds on Astex diverse set when maps are provided. Hence we recommend smaller grid spacing only when studying pockets of interest.
5 Conclusion
We have introduced AutoSite, a software program to reliably and efficiently identify ligand-binding sites for the receptors of known 3D structure, and characterize them by labeling the fill points as hydrophobic, or hydrogen bond donors or acceptors, as well as by deriving a set of putative ligand atomic positions called feature points. We have demonstrated that this method identifies binding sites comparably or better than other popular binding site identification software programs. We also demonstrated that the labelled fill points produced by AutoSite are larger than the ones produced by AutoLigand and also have a better overlap with the ligand. Finally, we have shown that the predicted putative ligand atoms capture around 80% of ligand hydrophobic and hydrogen acceptor atoms that interact with the receptor and over 60% of ligand hydrogen donor atoms, and illustrated and discussed applications of feature points for rational drug design and optimization. The software is available under the LGPL Open Source license at: http://adfr.scripps.edu/AutoDockFR/autosite.html.
Supplementary Material
Acknowledgements
We thank Drs. Daniel Santiago, Ludovic Autin and Stefano Forli for the valuable discussions. Drs. David S. Goodsell, and Arthur J. Olson for their input and careful reading of the manuscript. We thank all our colleagues in The Molecular Graphics Laboratory for their continuous motivation and support. This work was performed with support from grant R01 GM096888-05 from the NIH to Dr. Michel F. Sanner and a sub-contract to Dr. Michel Sanner from grant 2P41GM103426-21 from the National Institute of Health to Dr. Rommie E. Amaro. This is manuscript 29275 from The Scripps Research Institute.
Funding
This work was supported by grant R01 GM096888-05 from the National Institute of Health to Michel F. Sanner and Dr. Michel F. Sanner and a sub-contract to Dr. Michel Sanner from grant 2P41GM103426-21 from the National Institute of Health to Rommie E. Amaro.
Conflict of Interest: none declared.
References
- Allen W.J. et al. (2015) DOCK 6: Impact of new features and current docking performance. J Comput Chem, 36, 1132–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. et al. (2004) Comprehensive identification of “druggable” protein ligand binding sites. Genome Inform, 15, 31–41. [PubMed] [Google Scholar]
- Baroni M. et al. (2007) A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): theory and application. J Chem Inf Model, 47, 279–294. [DOI] [PubMed] [Google Scholar]
- Brylinski M., Skolnick J. (2008) A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. P Natl Acad Sci USA, 105, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra J.A. et al. (2009) Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput Biol, 5, e1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J. et al. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res, 34, W116–W118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester M. et al. (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. KDD-96 Proceedings, AAAI,. In, [Google Scholar]
- Ghersi D., Sanchez R. (2009) Improving accuracy and efficiency of blind protein-ligand docking by focusing on predicted binding sites. Proteins, 74, 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi D., Sanchez R. (2011) Beyond structural genomics: computational approaches for the identification of ligand binding sites in protein structures. J Struct Funct Genomics, 12, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodford P.J. (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem, 28, 849–857. [DOI] [PubMed] [Google Scholar]
- Halgren T.A. (2009) Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model, 49, 377–389. [DOI] [PubMed] [Google Scholar]
- Harris R. et al. (2008) Automated prediction of ligand-binding sites in proteins. Proteins, 70, 1506–1517. [DOI] [PubMed] [Google Scholar]
- Hartshorn M.J. et al. (2007) Diverse, high-quality test set for the validation of protein-ligand docking performance. J Med Chem, 50, 726–741. [DOI] [PubMed] [Google Scholar]
- Hendlich M. et al. (1997) LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model, 15, 359363–359389. [DOI] [PubMed] [Google Scholar]
- Henrich S. et al. (2010) Computational approaches to identifying and characterizing protein binding sites for ligand design. J Mol Recognit, 23, 209–219. [DOI] [PubMed] [Google Scholar]
- Hernandez M. et al. (2009) SITEHOUND-web: a server for ligand binding site identification in protein structures. Nucleic Acids Res, 37, W413–W416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston D.R. et al. (2002) High-resolution structures of a chitinase complexed with natural product cyclopentapeptide inhibitors: mimicry of carbohydrate substrate. Proc Natl Acad Sci U S A, 99, 9127–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston D.R. et al. (2004) Structure-based exploration of cyclic dipeptide chitinase inhibitors. J Med Chem, 47, 5713–5720. [DOI] [PubMed] [Google Scholar]
- Huey R. et al. (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem, 28, 1145–1152. [DOI] [PubMed] [Google Scholar]
- Istvan E.S., Deisenhofer J. (2001) Structural mechanism for statin inhibition of HMG-CoA reductase. Science, 292, 1160–1164. [DOI] [PubMed] [Google Scholar]
- Jaccard P. (1901) Distribution de la flore alpine dans le bassin des Dranses et dans quelques régions voisines. Bulletin De La Société Vaudoise Des Sciences Naturelles, 37, 241–272. [Google Scholar]
- Jain A.N. (2003) Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem, 46, 499–511. [DOI] [PubMed] [Google Scholar]
- Johnson C.N. et al. (2015) Fragment-based discovery of type I inhibitors of maternal embryonic leucine zipper kinase. ACS Med Chem Lett, 6, 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A. (1995) SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J Mol Graph, 13, 323–330. 307–328. [DOI] [PubMed] [Google Scholar]
- Laurie A.T.R., Jackson R.M. (2005) Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics, 21, 1908–1916. [DOI] [PubMed] [Google Scholar]
- Lower M. et al. (2011) Inhibitors of Helicobacter pylori protease HtrA found by ‘virtual ligand' screening combat bacterial invasion of epithelia. PLoS One, 6, e17986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower M., Proschak E. (2011) Structure-Based Pharmacophores for Virtual Screening. Mol Inform, 30, 398–404. [DOI] [PubMed] [Google Scholar]
- Mills C.L. et al. (2015) Biochemical functional predictions for protein structures of unknown or uncertain function. Comput Struct Biotechnol J, 13, 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M. et al. (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem, 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perot S. et al. (2010) Druggable pockets and binding site centric chemical space: a paradigm shift in drug discovery. Drug. Discov Today, 15, 656–667. [DOI] [PubMed] [Google Scholar]
- Ravindranath P.A. et al. (2015) AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Comput Biol, 11, e1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner M.F. et al. (1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers, 38, 305–320. [DOI] [PubMed] [Google Scholar]
- Schnecke V. et al. (1998) Screening a peptidyl database for potential ligands to proteins with side-chain flexibility. Proteins, 33, 74–87. [PubMed] [Google Scholar]
- Stark A. et al. (2004) Finding functional sites in structural genomics proteins. Structure, 12, 1405–1412. [DOI] [PubMed] [Google Scholar]
- Vriend G. (1990) What If - a Molecular Modeling and Drug Design Program. J Mol Graphics, 8, 52. &. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.