Abstract
Motivation: Next generation sequencing technologies have revolutionized our ability to rapidly and affordably generate vast quantities of sequence data. Once generated, raw sequences are assembled into contigs or scaffolds. However, these assemblies are mostly fragmented and inaccurate at the whole genome scale, largely due to the inability to integrate additional informative datasets (e.g. physical, optical and genetic maps). To address this problem, we developed a semi-automated software tool—Genome Puzzle Master (GPM)—that enables the integration of additional genomic signposts to edit and build ‘new-gen-assemblies’ that result in high-quality ‘annotation-ready’ pseudomolecules.
Results: With GPM, loaded datasets can be connected to each other via their logical relationships which accomplishes tasks to ‘group,’ ‘merge,’ ‘order and orient’ sequences in a draft assembly. Manual editing can also be performed with a user-friendly graphical interface. Final pseudomolecules reflect a user’s total data package and are available for long-term project management. GPM is a web-based pipeline and an important part of a Laboratory Information Management System (LIMS) which can be easily deployed on local servers for any genome research laboratory.
Availability and Implementation: The GPM (with LIMS) package is available at https://github.com/Jianwei-Zhang/LIMS
Contacts: jzhang@mail.hzau.edu.cn or rwing@mail.arizona.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Illumina and PacBio SMRT sequencing technologies are the two most widely accepted sequencing platforms currently used for large scale genomics-driven data generation. Illumina, representing the most widely used second-generation sequencing technology, produces short reads (35–150 base read lengths), that are highly accurate, with base call outputs that can yield hundreds of millions of bases from a single lane over several days (depending upon complexity) (Schatz et al., 2010). PacBio produces hundreds of thousands of long-read error-corrected sequences (up to 20 kb average read lengths) that can be produced in 3–6 h per SMRT cell. Data generated by either platform can be used independently, or in combination, to successfully assemble genomes de novo (Alkan et al., 2011; Chin et al., 2013; Kajitani et al., 2014). Many assemblers have been developed to assemble raw sequence reads into sequence contigs (i.e. minimum sequence units for an assembly), such as SOAPdenovo (Luo et al., 2012), Allpaths (Butler et al., 2008; MacCallum et al., 2009), HGAP (Chin et al., 2013), or Falcon (https://github.com/PacificBiosciences/FALCON-integrate). A few programs or packages (e.g. Bambus, ABACAS, Mauve Aligner, ALLMAPS, etc.) are also available for scaffolding contigs (Assefa et al., 2009; Hunt et al., 2014; Pop et al., 2004; Rissman et al., 2009; Tang et al., 2015). However, software tools that can be used to inspect/edit NGS sequence assemblies, as well as integrate other evidence types (i.e. physical and genetics maps) to produce an assembly that more accurately and completely reflects the native structure of a given genome, are currently lacking.
To analyze, manage and incorporate genome datasets for diverse sequencing projects, such as pseudomolecule construction for the maize and several wild Oryza genome sequencing projects (Schnable et al., 2009; J. Stein et al., submitted for publication; Wei et al., 2009), we developed a software tool called ‘Genome Puzzle Master’ (GPM). GPM does not require sophisticated bioinformatics skills or support, and the final products are ready to use in the form of annotation-ready pseudomolecules. GPM can also facilitate the incorporation of additional datasets as new refinements are generated. Here we make GPM available to researchers who have NGS genome assemblies and other unlinked genomic datasets, and are struggling to generate ‘annotation-ready’ or ‘submission-ready’ pseudomolecules.
2 Methods
GPM is a key part of a web-based Laboratory Information Management System (LIMS) that we developed to manage and analyze genomic data at different levels from both wet and dry lab experiments. The LIMS is set up in a LAMP environment (Linux operating system, Apache HTTP Server, MySQL database software and Perl programing language) and requires additional libraries and software listed in Supplementary Table S1. We used jQuery (plus UI), a fast and concise JavaScript library, to build GPM’s highly interactive web applications.
A database schema was designed which contains one main table called ‘matrix’ to record most types of data from ‘wet’ or ‘dry’ lab experiments (Supplementary Table S2), and several extra tables (e.g. ‘link’, ‘alignment’) to store relationship data. Combined with these tables, we can extensively handle all types of datasets and connect them logically. In our database, the expansibility is flexible, as we can either add a new type of ‘container’ in the ‘matrix’ table or create another table for new data if needed, depending on the data type. For example, huge amounts of individual clone information can potentially cause process servers to slow down while performing complicated search operations on the ‘matrix’ table; hence, we specifically created a ‘clone’ table to store this type of data and separately can run queries on it to avoid concurrent queries on the ‘matrix’ table. To include additional information for certain existing data types, we extended the ability to store more informative data by adopting the JavaScript Object Notation (JSON), a lightweight data-interchange format, in the ‘note’ field of the ‘matrix’ table.
GPM can currently utilize data categorized into different types, such as sequence contigs/scaffolds, Bacterial Artificial Chromosome (BAC) clone end-sequences (BESs), and reference genome sequences (RefSeq) that are the basic input elements for GPM assemblies. Different data types can be connected to one another via their sequence relationships. For example, AGP (a golden path, https://www.ncbi.nlm.nih.gov/assembly/agp/AGP_Specification/) information can describe the assembly of a larger sequence object (e.g. a contig, a scaffold, or a chromosome) from smaller objects and list their relationships. Most importantly, these kinds of relationships are used to link multiple datasets and to logically integrate genomic data with accuracy.
To demonstrate a GPM assembly result, here we specifically defined two terms, ‘assemblyCtg’ for a GPM assembly contig and ‘assemblySeq’ for a sequence component that belongs to an assemblyCtg. Both data types are stored in the ‘matrix’ table as individual containers. In the database, a record of assemblyCtg describes its constituent members (assemblySeqs) and attributes (e.g. length, chromosome number and position); and an assemblySeq records the original sequence and the actual part and status of the component sequence (e.g. coordinates and orientation) that contributes to its corresponding assemblyCtg. A complete GPM assembly contains a set of assemblyCtgs, which are formed by various numbers of assemblySeqs.
3 Results
3.1 Information-guided assembly
To build pseudomolecules from sequence contigs with reference information, a GPM ‘assemblyRun’ execution (Fig. 1) can be divided into 11 optional operations:
Fig. 1.
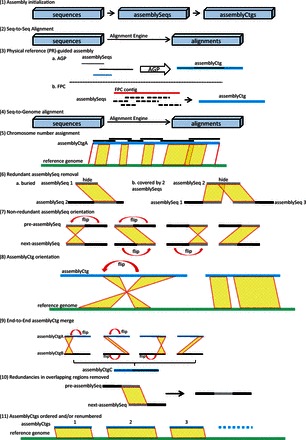
GPM assemblyRun operations
Assembly initialization. First, to initialize a new assembly, GPM converts all de novo assembled sequences into assemblySeqs and each assemblySeq is assigned to a single assemblyCtg. (Note: This operation can be skipped for an existing assembly to avoid losing any manual edits.)
Seq-to-Seq alignment. Depending on the potential relationship of all sequence elements, GPM can prepare overlapping alignments among all sequences in an assembly by running a ‘Seq-to-Seq’ pre-calculation with ‘blastn’ (NCBI BLAST 2.2.29+, E-value =1e-200) (Camacho et al., 2008) as the default alignment engine. Pre-calculation parameters can be customized as needed. Overlap information between sequence contigs can be used in later processing steps, e.g. to determine the orientation of two neighboring sequences.
Physical reference (PR)-guided assembly. Based on an existing physical reference, such as a physical map (PM) or an AGP file, sequences will be assigned to assemblyCtgs. If an AGP file is available, object-component information is used to build links between sequences. If a FingerPrinted Contig (FPC) file (Nelson et al., 2005) is available, we use PM contig-clone information to connect potentially neighboring sequences and merge two assemblySeqs from neighboring BACs on PMs into an assemblyCtg. AssemblyCtgs will only be merged when overlapping evidence is detected. For a non-PR-based whole genome shotgun (WGS) project, the original sequences would be loaded into GPM, and this operation would be skipped. To build assemblyCtgs, the default parameters for merging two sequences are ‘minOverlapSeqToSeq = 1000 bp’ and ‘identitySeqToSeq = 99%,’ plus the overlap should be at both ends of each sequence.
Seq-to-Genome alignment. This operation allows the user to run the alignment engine (‘blastn’ as default, E-value =1e-200) to search against reference genome sequences by using the original sequences of all assemblySeqs. The alignment information can be used to ‘group,’ ‘order,’ and ‘orient’ assemblyCtgs at a chromosomal scale. Depending on the similarity between the RefSeq and the to-be-assembled sequence, alignment parameters can be varied and tested by the user. GPM currently supports blastn, megablast (Camacho et al., 2008) and BLAT (Kent et al., 2002) as alignment engines for sequence comparison. Users can choose a proper alignment engine to accelerate GPM’s performance. For example, megablast or BLAT can be chosen for aligning sequences with high similarities since they run faster than blastn (in most situations). Importantly, to minimize the misleading impacts of repetitive sequences in both to-be-assembled and reference datasets during this step, users can (i) activate the ‘Mark Repeat Region’ option to filter out any non-unique alignments, which will not be considered as evidence to guide a GPM assembly; or (ii) select the alternative ‘Soft Masking’ option for the blastn/megablast alignment processing if repetitive regions in reference sequences have already been soft-masked for an assembly.
Chromosome number assignment. If a high-quality reference genome is available, we prefer to use it as a guide to automatically assign chromosome numbers to assemblyCtgs. Alternatively, chromosome assignments can also be performed manually in a batch mode.
Redundant assemblySeq removal. GPM will mask and remove a buried or redundant sequence according to the alignment information processed in operation 2. Here a redundant sequence is defined as a sequence that can be fully aligned by two neighboring sequences and the two neighboring sequences have end-overlapped alignment (default parameters ‘minOverlapSeqToSeq = 1000 bp’ and ‘identitySeqToSeq = 99%’). Such assemblySeqs would be marked as redundant and are hidden in the final assembly. Masking redundancies can reduce the computational complexity of orienting assemblySeqs and assemblyCtgs.
Non-redundant assemblySeq orientation. In a multi-assemblySeq contig, GPM can orient sequences based on overlap data. This operation is performed on the premise that an overlapping region of two neighboring sequences should be at the end of each sequence, specifically at the right end of a pre-assemblySeq, and at the left end of the next-assemblySeq. GPM adjusts the orientation of these assemblySeqs to fit the premise accordingly with the proper extension of both non-overlapping parts that will elongate an expanding contig. GPM determines the orientation of each initial seed assemblySeq and then the subsequent assemblySeq one-by-one.
AssemblyCtg orientation. Using a reference genome, GPM can orient assemblyCtgs according to the linear information suggested by the ‘Seq-to-Genome’ alignment data.
End-to-End assemblyCtg merge. In some cases, physically overlapping sequences can be separated into two different assemblyCtgs due to the lack of strong evidence derived from the guide data, for example, when there is weak evidence to merge two contigs during a PM construction step. These kinds of situations usually arise from BACs located at the ends of PM contigs, and as a consequence, assemblySeqs of these BACs are also found at assemblyCtg ends. Detection of end-to-end overlaps between neighboring assemblyCtgs facilitates merging into larger assemblyCtgs.
Redundancies in overlapping regions removed. Overlapping sequences between two neighboring assemblySeqs are considered redundant, and one redundant portion of either assemblySeq is removed from the final pseudomolecule. Here GPM makes no preferences on which overlapping sequence is retained. However, if evidence is provided that one overlapping sequence is of a higher quality than the other, then the highest quality sequence will be retained.
AssemblyCtgs ordered and/or renumbered. GPM can sort assemblyCtgs based on their chromosomal number and position and provide an option to renumber assemblyCtgs in their proper order.
After running an automated GPM assembly, the user is also able to manually check and edit the assembly (Fig. 2). With the visualization function, GPM can provide a convenient way to manually check and edit an assembly, thereby eliminating the need to rely solely on automated assembly results. Relationships among datasets (including BAC sequences, BESs, physical maps, reference sequences, etc.) can logically guide assembly operations, including but not limited to grouping, merging, ordering and orienting. All manual editing steps are saved automatically, so the entire editing process can be reproduced and quality checked. Upon completion of an editing step, GPM can be used to export contig sequences, chromosome-based pseudomolecules and AGP files in real time.
Fig. 2.
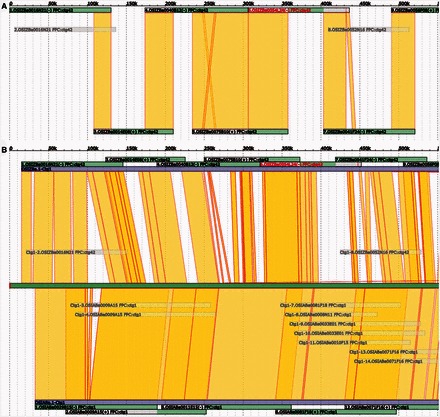
Visualization of typical available data in GPM. (A) GPM assemblyCtg view of a 500-KB region. AssemblySeqs, top and bottom, are shown as overlapping (yellow) and fully redundant assemblySeqs are gray. The retained (green) and removed (gray) portions of assemblySeqs are indicated. (B) Chromosome-scale view of a 500-KB region that compares two genome assemblies to a Reference sequence. The Reference Sequence is shown in the middle (bright green) with alignments (yellow) to each assemblyCtg (violet) at the top and bottom. The assemblyCtg order can be changed by drag-and-drop (Color version of this figure is available at Bioinformatics online.)
3.2 Application of the GPM pipeline to assemble two high-quality reference genome sequences for indica rice: a case study
Recently, our consortia published two high-quality reference genome sequences for the two main varietal groups of indica rice—Zhenshan 97 (ZS97) and Minghui 63 (MH63) (Zhang et al., 2016a,b). These genomes were primarily sequenced using PacBio long-read sequencing of minimum tiling path BAC pools, combined with Illumina WGS assembled contigs to fill gaps. Once individual BAC sequences were assembled they were loaded into GPM for assembly editing and pseudomolecule construction.
Following is a summary of how these data were used to assemble two of the highest quality indica rice genome assemblies produced to date:
To assemble the ZS97 and MH63 genomes, the following datasets were loaded in to GPM: (i) Whole Genome Profiling (WGP, van Oeveren et al., 2011) sequence-based PMs for each genome; and (ii) 5363 assembled BAC sequences (including duplicates for the same BAC clones sequenced in multiple jobs or pools) from 188 HGAP jobs for ZS97, and 6,801 from 313 jobs for MH63. Here we take ZS97 as an example to demonstrate the assembly procedure once these data were loaded (Supplementary Fig. S1). To start a new assembly, we set the ‘FPC: ZS97 v.1’ PM as the physical reference and the ‘IRGSP-MSU’ (i.e. O.sativa subsp. japonica cv. Nipponbare genome sequence, Kawahara et al., 2013) as the reference genome. We checked ‘Assign chromosome number for contigs’ and ‘Orient contigs based-on reference genome’ since both ZS97 and Nipponbare belong to the same genus and species (i.e. O.sativa), and minor differences between the to-be-assembled and reference genomes would not mislead the results because they are so closely related. ‘Seq-to-Seq Alignment’ was used to pre-build all possible overlapping relationships among all BAC sequences since it was expected that those should be connected. ‘Seq-to-Genome Alignment’ was also used to map all BAC sequences to the reference genome. We also utilized the ‘End-to-End Merge,’ ‘Auto-Orient Sequences’ and ‘Filter Redundant Sequences and Overlaps’ options. (Note: A similar process was used to assemble the MH63 genome, except we used the ‘FPC: MH63 v.1’ PM as the physical reference.)
After manual checking, editing and removing redundancies, the final assembly products yielded 318 (ZS97, composed of 3862 assemblySeqs) and 216 (MH63, composed of 3256 assemblySeqs) assemblyCtgs that were ordered, oriented, and assigned to their appropriate chromosomes. The ‘assemblyCtg’ sequences were used as the primary frameworks to build pseudomolecules after gap-filling sequences were integrated during the second round of GPM assembly (Zhang et al., 2016b).
4 Discussion
GPM is an integrated pipeline for generating and editing pseudomolecules from existing next gen sequence assembles using evidence-based guides such as reference sequences, physical maps, genetic maps and paired BESs. We demonstrated how GPM can be used to generate high-quality submission-ready pseudomolecules for two indica rice accessions, ZS97 and MH63, by the integration of BAC-based physical maps and reference sequence guides. To perform an assembly with GPM, not all the resources like those used for ZS97 and MH63 are required, but GPM does require at least one guide type (e.g. genetic, physical or genome reference). Since GPM does not reassemble sequence data from the original short/long reads, the kind and quality of the evidence-based guides will greatly affect the amount of time required for editing and will impact the final quality of an assembly. Using a reference genome in a GPM assembly is quite valuable, however, the utilization of more evidence guides can greatly enhance the ability to build high-quality pseudomolecules and to avoid the overuse of the reference sequences in conforming a GPM assembly to mimic a RefSeq. If a reference genome is the only evidence to guide an assembly, then reference sequence overuse mistakes can’t be avoided. However, using other information that is available could detect overuse errors which would be corrected during a manual check step (which is not labor-intensive and strongly recommended) with GPM. Hence, evidence data are key factors used to build high-quality GPM assemblies.
In the current version of our LIMS, we can track each processing step from BAC library construction to the final genome assembly seamlessly during the entire phase of a sequencing project. BAC library resources and wet lab sequencing runs are recorded in our system. As a part of the LIMS, GPM supports FASTA-formatted sequences. However, the ability to deal with raw reads has been limited due to their huge data size. Some additional features, such as paired end/mate and MTP information, are partially adapted for assembly manipulation in the current version of GPM. Pseudomolecules in FASTA format with an AGP file can be generated as the final output for further downstream analysis and public repository sequence submissions (e.g. GenBank).
The LIMS can be expanded to integrate new functions for a specific project as needed. For example, we built a pipeline called ‘postHGAP’ to perform circularization and identification of BAC sequences for the two rice genome sequencing projects described above (Fig. 3). As reported in our data descriptor (Zhang et al., 2016b), we used a map-based BAC-pool sequencing strategy to produce sequence data that were assembled into a set of sequences (termed unitigs by HGAP) for each pool. We were able to process the output of each corresponding HGAP job to circularize and identify BAC sequences properly using related data information in the LIMS as follows: (i) postHGAP first filtered unitigs with lengths shorter than 10 kb, then (ii) trimmed vectors and circularized plasmid or BAC sequences according to pairs of sequences at specific breakpoints and (iii) assigned BAC IDs to each sequence according to WGP tags or BES information. During the postHGAP processing step, two parameters (default: minOverlap = 500 bp, overlapIdentity = 95%) could be set for sequence circularization and four optional parameters (default: minCloneTagNumber = 5, tagMatchIdentity = 100%, tagMatchPercent = 80%, besMatchIdentity = 98% if no WGP tags available) for BAC ID assignment. The program ‘blastn’ (NCBI BLAST 2.2.29+) was used to perform sequence comparison and alignment analysis in postHGAP. As a result, a total of 501 HGAP jobs (multiple runs for the same pool count multiple times) for 375 pools were run through postHGAP in this study and produced 12 164 BAC-ID-ready sequences (including duplicates for the same BAC processed in multiple jobs or pools) for both ZS97 and MH63. All BAC-ID-ready sequences were seamlessly converted as input datasets for both GPM assemblies.
Fig. 3.
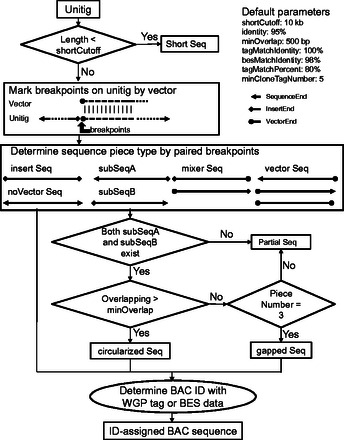
Flowchart for processing unitigs with postHGAP
Technically, the use of a one-main-table database schema design may simplify query processing. However, one possible disadvantage for this schema is that the response time to query might be longer with increasing amounts of data. Based upon our LIMS structure, we used the new ‘container’ option for less complicated data and created new tables for large datasets (e.g. clones). To date, we are able to operate GPM smoothly on a 4× 12-core cluster with more than 1.7 million data records. These kinds of strategies are recommended for other users. We can provide free host services to academic projects of medium-size genomes (up to 1 Gb) with full technical support.
Several WGS scaffolding packages (e.g. Bambus, Mauve Aligner, ABACAS, ALLMAPS, etc.) treat assembled contig sequences as non-overlapping and single-copy sequences. Unfortunately, these packages are not focused toward the handling of inherent sequence redundancies and are not programmed to produce incremental assemblies. Further, these scaffolding tools are all file-based and do not provide a flexible process for manual checking and editing, especially if the input data has been modified or updated. In contrast, GPM is a relationship-based pipeline, which has the flexibility to edit and visualize assembled data, not only by showing the order and orientation of contig sequences and sequence redundancies but also by displaying the necessary guide information for easy user confirmation. One thing GPM cannot do is to assemble a genome de novo from raw reads. This deficit can be compensated for by its ability to import results (sequences and AGPs) from other assembly programs to guide new and improved assemblies. The final quality of a GPM assembly relies heavily on the quality and richness of the guide information used. For example, if a draft genome assembly is used as the guide rather than a map-based BAC-by-BAC assembly, the quality of the GPM assembly will suffer.
DNA sequencing technologies and assembly programs change rapidly, and the GPM pipeline presented here is no exception. We will continue to improve the interactive functionalities of GPM, as well as integrate additional tools to support more data types. With GPM, data types are seamlessly linked and logically integrated into an encompassing LIMS for all genomic data. GPM is an open source software dynamically developed for the genomics research community and can be extensively adapted/improved by different research groups for their own applications.
Supplementary Material
Funding
This study was supported by the Start-up Fund of the National Key Laboratory of Crop Genetic Improvement to J.Z. and the National Science Foundation Grant (#1026200) to R.A.W.
Conflict of Interest: none declared.
References
- Alkan C. et al. (2011) Limitations of next-generation genome sequence assembly. Nat. Methods, 8, 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa S. et al. (2009) ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics, 25, 1968–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. et al. (2008) ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res., 18, 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. et al. (2008) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C.S. et al. (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods, 10, 563–569. [DOI] [PubMed] [Google Scholar]
- Hunt M. et al. (2014) A comprehensive evaluation of assembly scaffolding tools. Genome Biol., 15, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R. et al. (2014) Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res., 24, 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y. et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice, 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. et al. (2002) BLAT – the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R. et al. (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience, 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum I. et al. (2009) ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol., 10, R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W. et al. (2005) Whole-genome validation of high-information-content fingerprinting. Plant Physiol., 139, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M. et al. (2004) Hierarchical scaffolding with Bambus. Genome Res., 14, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman A. et al. (2009) Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics, 25, 2071–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz M. et al. (2010) Assembly of large genomes using second-generation sequencing. Genome Res., 20, 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science, 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Tang H. et al. (2015) ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol., 16, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oeveren J. et al. (2011) Sequence-based physical mapping of complex genomes by whole genome profiling. Genome Res., 21, 618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F. et al. (2009) The physical and genetic framework of the maize B73 genome. PLoS Genet., 5, e1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. (2016a) Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan97 and Minghui 63. Proc. Natl. Acad. Sci. USA, doi:10.1073/pnas.1611012113. [DOI] [PMC free article] [PubMed]
- Zhang J. et al. (2016b) Building two indica rice reference genomes with PacBio long-read and Illumina paired-end sequencing data. Sci. Data, 10.1038/sdata.2016.76. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


