Abstract
PRDM9 is currently the sole speciation gene found in vertebrates causing hybrid sterility probably due to incompatible alleles. Its role in defining the double strand break loci during the meiotic prophase I is crucial for proper chromosome segregation. Therefore, the rapid turnover of the loci determining zinc finger array seems to be causative for incompatibilities. We here investigated the zinc finger domain-containing exon of PRDM9 in 23 tarsiers. Tarsiers, the most basal extant haplorhine primates, exhibit two frameshifting indels at the 5′-end of the array. The first mutation event interrupts the reading frame and function while the second compensates both. The fixation of this allele variant in tarsiers led to hypothesize that de- and reactivation of the zinc finger domain drove the speciation in early haplorhine or tarsiiform primates. Moreover, the high allelic diversity within Tarsius points to multiple effects of genetic drift reflecting their phylogeographic history since the Miocene.
Meiosis is a fundamental process to generate hereditary variability, thus being a driving force in evolutionary biology. Meiotic abruption is a cause for hybrid sterility which is a postzygotic barrier leading to speciation1,2. The question now increasingly being asked is therefore what type of genetic modification underlies species formation. In vertebrates PRDM9 is, at present, the top candidate gene associated with hybrid sterility1,3. The gene is exclusively expressed by germ cells in the meiotic prophase I and encodes a zinc finger protein that specifies hotspots of recombination4,5,6. The zinc finger domain evolves rapidly, both as to the number and sequence of repetitive motifs resulting in altered DNA-binding specificity, which in turn may lead to genetic incompatibilities promoting species divergence3. A recently published study on the evolutionary dynamics of the primate PRDM9 zinc finger array revealed taxon-specific alleles across 18 haplorhine species, further strengthening its role in speciation processes7. However, tarsiers, the sole extant representatives of non-anthropoid haplorhine primates8, were not included. As the deepest offshoot within haplorhini they have a long independent history, possibly covering about 80 million years of primate evolution9. Fossils of Tarsius are scarcely reported and restricted to Asia and North Africa spanning a period from the middle Eocene to the middle Miocene10. Assuming the fossil families of Omomyidae, Archicebidae and Afrotarsiidae as primitive tarsiiforms, extinct ancestors of tarsiers were widespread holarctic Eocene-Oligocene species12,13,14 and occurred as far back as 55 Mya15. Extant tarsiers, however, are endemic to insular Southeast Asia, where they fall into the three geographically and evolutionary distinct lineages of Western, Philippine, and Sulawesi tarsiers, with the latter constituting the most species-rich clade16,17. Even considering only the radiation of this ancient primate lineage in the Malay-Archipelago since the Miocene, allopatric speciation both between and within extant tarsier clades has left traceable molecular signatures of biogeographic events18,19,20,21.
Due to their independent evolution within haplorhine primates and their marked diversification, especially evident over the last 2.5 million years on the Indonesian island of Sulawesi21, tarsiers represent the unique opportunity for exploring the evolutionary dynamics of the PRDM9 gene and its role in speciation processes within primates and tarsiers in particular. We therefore sequenced the PRDM9 exon encoding the zinc finger array of 23 tarsier individuals, including 21 specimen from Sulawesi and two non-Sulawesi specimen, one each representing the Western and the Philippine tarsier clade. Based on these data we investigated the functionality and allelic diversity of the zinc finger domain in tarsiers. In addition we inferred evolutionary relationships between zinc fingers of anthropoid primates7 and tarsiers, and discussed the possible function of PRDM9 as driving force behind the anthropoid-tarsiiform split and the divergence of the tarsiiform lineage.
Results
Tarsier PRDM9 sequence characterization
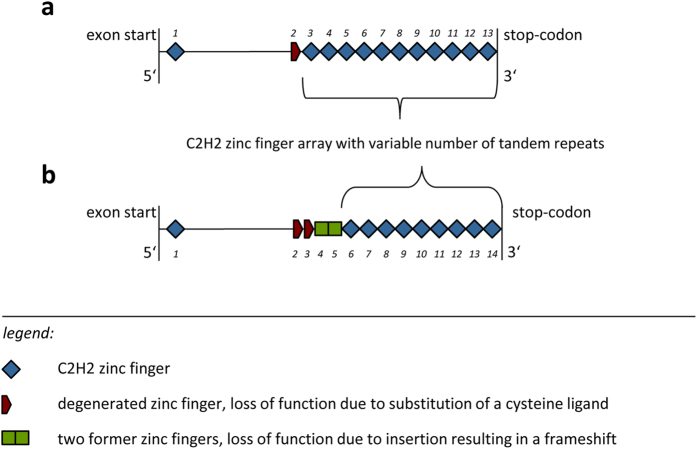
The examined exon of Western, Philippine and Sulawesi tarsiers was homologous to exon 11 of the human PRDM9 gene and was about 1037–1793 bp long depending on the number of zinc finger repeats. It could be divided into two parts, the 5′ sequence and the 3′ C2H2 zinc finger array. Structural and functional properties of the zinc finger protein encoding exon are shown in Fig. 1.
Figure 1. Schematic example of the PRDM9 exon with the C-terminal zinc finger array.
of [a] human allele A as representative for anthropoid primates and of [b] allele TD1 for tarsiers. The comparison between the two exons shows the impact of the insertion and deletion on the zinc finger array, i.e. the loss of three functional C2H2 zinc fingers. Zinc finger numbers are written in italics.
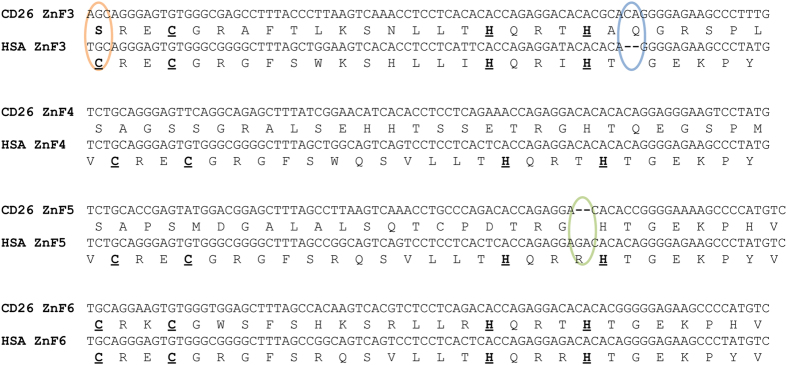
In contrast to the variable C2H2 zinc finger domain which was used for the analyses, the 5′ sequence of the exon was more conserved. This part of the sequence contained a C2H2 zinc finger (ZnF1) at the beginning of the exon and four presumably former functional zinc fingers. The second and third zinc fingers were both degenerated by the loss of a zinc ion binding cysteine ligand, see Fig. 2, which is in principal not uncommon in zinc finger arrays22. The sequence of the third zinc finger was also interrupted by a 2 bp-insertion that caused a framing error affecting the next two zinc fingers which we refer to as former zinc fingers. A deletion in the second altered zinc finger, which would be the fifth overall, restored the reading frame. The array of classical 84 bp C2H2 zinc fingers started with the end of this fifth zinc finger (Figs 1 and 2).
Figure 2. Frameshift mutations in tarsier PRDM9.
Shown are aligned nucleic acid and peptide sequences comprising zinc fingers 3–6 (ZnF3–ZnF6) of Homo sapiens (HSA) and Tarsius (represented by a Sulawesian individual, CD26) and illustrating the frameshifting nature of insertion (blue) and deletion (green) mutations. Note the nonsynonymous substitution in the very first triplet which turns cysteine to serine (orange) and degrades ZnF3 of the tarsier. Amino acid sequences of human ZnF3–ZnF6 and ZnF6 of the tarsier show the classical C2H2 zinc finger motif (indicated by bold and underlined amino acids).
Indel events are quite rare in coding regions due to purifying selection23,24, especially if their length is not a multiple of three25. While the insertion results in a different amino acid sequence and may alter protein function, the deletion alone generates premature stop codons which lead to a faster pseudogenization respectively gene loss26. We therefore consider it likely that the insertion predates the deletion mutation which then in turn acted as a compensatory mutation. Compensatory mutations are twice as common as reversions to the original state and are often found in close proximity of the initial mutation27,28 being consistent with our findings. As only both indels jointly restore protein integrity we further assume that the temporal offset between indel mutations resulted in a temporary loss or limited protein function.
Allelic variation of the zinc finger array in tarsiers
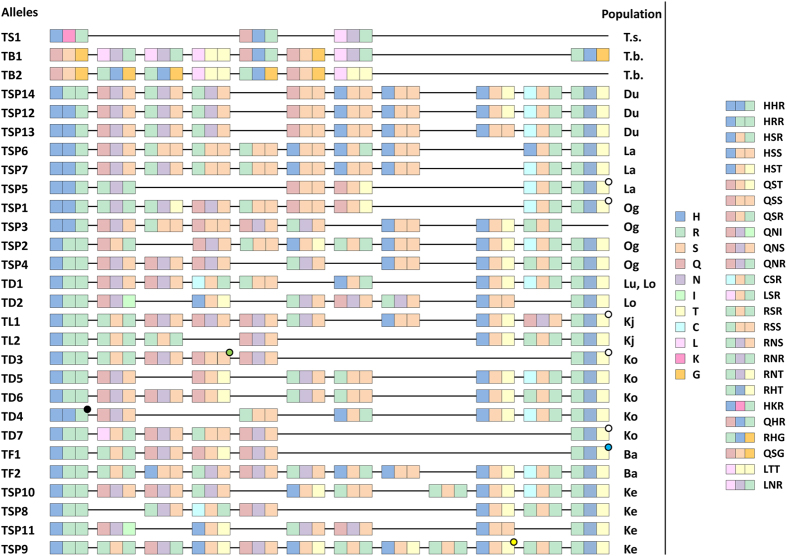
The alleles are defined on amino acid level, i.e. synonymous nucleotide changes are excluded, and are restricted to the C2H2 array. We found 28 alleles in 23 individuals with 15 individuals being heterozygous, (Table 1, Fig. 3).
Table 1. Allelic diversity of the zinc finger array in tarsiers.
| Individuals | Origin | Alleles (Number of C2H2-ZnFs) | |||
|---|---|---|---|---|---|
| Species | Sample ID | Biogeographic Region | Sample Site | 1 | 2 |
| Tarsius syrichta | TSY | Philippines | N/A | TS1 (3) | |
| Tarsius bancanus | TBA | Sundaland | N/A | TB1 (8) | TB2 (7) |
| Tarsius dentatus | T111 | Wallacea | Laone, Central Sulawesi | TD1 (9) | TD2 (8) |
| Tarsius lariang | T47 | Wallacea | Koja, Central Sulawesi | TL1 (10) | TL2 (7) |
| Tarsius sp. | CD02 | Wallacea | Ogatemuku, North-Sulawesi | TSP1 (9) | TSP2 (10) |
| Tarsius sp. | CD05 | TSP3 (9) | TSP4 (9) | ||
| Tarsius sp. | CD10 | TSP1 (9) | TSP4 (9) | ||
| Tarsius dentatus | CD14 | Wallacea | Korosule, Central Sulawesi | TD3 (6) | TD4 (7) |
| Tarsius dentatus | CD16 | TD5 (8) | TD6 (9) | ||
| Tarsius dentatus | CD17 | TD3 (6) | TD7 (6) | ||
| Tarsius dentatus | CD19 | Wallacea | Luwuk, Central Sulawesi | TD1 (9) | |
| Tarsius dentatus | CD24 | TD1 (9) | |||
| Tarsius dentatus | CD26 | TD1 (9) | |||
| Tarsius dentatus | CD27 | TD1 (9) | |||
| Tarsius sp. | CD34 | Wallacea | Labanu, North Sulawesi | TSP5 (6) | |
| Tarsius sp. | CD36 | TSP6 (10) | TSP7 (10) | ||
| Tarsius sp. | CD41 | Wallacea | Kendari, South Sulawesi | TSP8 (7) | TSP9 (12) |
| Tarsius sp. | CD43 | TSP10 (10) | TSP11 (7) | ||
| Tarsius sp. | CD46 | Wallacea | Duasaudara, North Sulawesi | TSP12 (10) | |
| Tarsius sp. | CD48 | TSP13 (10) | |||
| Tarsius sp. | CD51 | TSP14 (10) | TSP12 (10) | ||
| Tarsius fuscus | CD64 | Wallacea | Bantimurung, South Sulawesi | TF1 (6) | TF2 (11) |
| Tarsius fuscus | CD65 | TF1 (6) | TF2 (11) | ||
Each individual is listed with its species, sample ID, origin, allele identification and the number of C2H2 zinc fingers per allele. TSY: Tarsius syrichta; TBA: Tarsius bancanus; Tarsius sp.: Taxon yet unclassified and currently affiliated with the Tarsius tarsier-population17.
Figure 3. Alignment of C2H2 zinc finger alleles.
The numbers identifying the alleles are identical to those in Table 1. Each zinc finger is depicted with a triplet composed of its binding amino acids (−1, 3, 6). Each amino acid is marked with a distinct colour. Dots hint towards one or more nonsynonymous substitutions apart from the shown amino acids. Abbreviations: T.s., Tarsius syrichta; T.b., Tarsius bancanus; Du, Duasaudara; La, Labanu; Og, Ogatemuku; Lu, Luwuk; Lo, Laone; Kj, Koja; Ko, Korosule; Ba, Bantimurung; Ke, Kendari.
The 25 alleles exclusively found in Sulawesi tarsiers show similarities, most often within populations (see Materials and Methods for sample sites). They mainly differ in the number of zinc fingers but hardly in their sequence like the alleles 9 and 10. Alleles 4, 5 and 6 have each ten zinc fingers with minor changes in the amino acid sequence (Fig. 3). Only allele 14 is shared between two Sulawesian populations (Luwuk and Laone), both belonging to Tarsius dentatus.
There are two types of 5′-most C2H2 zinc fingers differing in one binding amino acid (HHR, HRR). One variant can be found on the northern peninsula (Duasaudara, Labanu, Ogatemuku) and in Korosule. The other is specific to populations inhabiting the central, eastern and southern parts of Sulawesi, with the exception of one copy also occurring in north-eastern Sulawesi. The 5′-most C2H2 zinc fingers accord to some extend with the two lineages found in Driller et al.21. Lineage 1 comprises the populations on the northern peninsula and Kendari, lineage 2 all other populations.
The 3′-most C2H2 zinc fingers are, with one exception, identical regarding the three binding amino acids RHT (Fig. 3). Five of them show a nonsynonymous substitution which, however, does not affect the DNA-binding sites. They were observed in the populations of Labanu, Korosule and Koja. Three out of 19 zinc fingers with key codon positions QSR, LSR, and RNT are unique. Some are only found in few populations like the motifs specifying the binding triplet QNI or RNR and being restricted to Laone and Kendari, or Labanu and Ogatemuku, respectively.
Especially the hints on the relationship of the populations on the northern peninsula among each other and between Laone and Luwuk are comparable with results of Driller et al.21. The relatively conserved 5′-most and 3′-most C2H2 zinc finger, the more variable ZnFs in between and the appearance of similar ZnFs in different species are also seen in mice29.
Alleles of T. bancanus and T. syrichta are species-specific encoding zinc finger motifs not present in Sulawesi tarsiers. Despite their long geographic isolation they still share two zinc fingers (Fig. 3). Both alleles of the Western tarsier end with a zinc finger degenerated by the exchange of the second zinc binding histidine ligand with asparagine. With only three repeats the zinc finger domain of the homozygous Philippine specimen is unusually short, but according to Stubbs et al.30 capable to bind DNA. However, more individuals or rather more alleles of both species could reveal further and better insights regarding the relationship of PRDM9 zinc fingers between the three main tarsier groups.
Phylogenic analysis
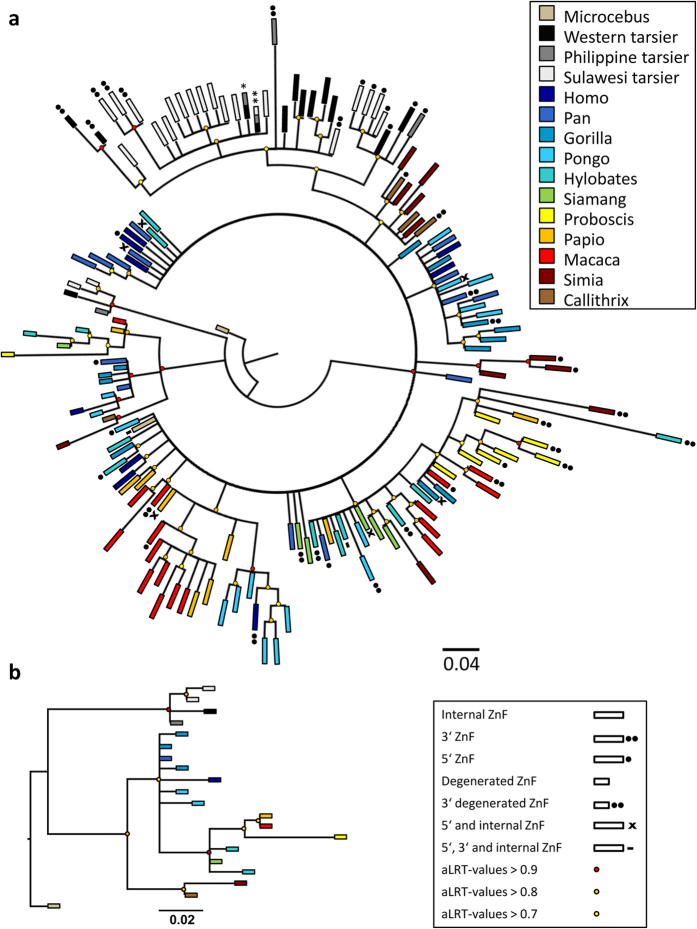
To estimate the phylogenetic affiliations of the sequenced tarsier zinc fingers within the haplorhine divergence we reconstructed a phylogeny with Microcebus murinus as strepsirhine outgroup. Including all first degenerated and C2H2 zinc fingers (Fig. 4a) it completes the results of Schwartz et al.7 from a haplorhine perspective. The basal trifurcation separates degenerated zinc fingers of tarsiers, degenerated zinc fingers of anthropoid primates and all C2H2 zinc fingers with node support values above 0.9 (Fig. 4a). Within the C2H2 cluster tarsier zinc fingers form a well-supported monophyletic group. Within this group 5′ most and 3′ most C2H2 zinc fingers are clustered reflecting the biogeography of the individuals tested. Apart from these findings our tree topologies (Fig. 4) are congruent with those obtained for anthropoids as presented by Schwartz et al.7.
Figure 4. Phylogeny reconstructions based on Schwartz et al.7, including tarsier zinc fingers.
[a] Shows a tree topology including the first degenerated zinc finger and all C2H2 zinc fingers of the array. [b] Depicts only the first degenerated zinc finger. The genera can be identified by colour, different kinds of or particular zinc fingers can be distinguished by their form, see key. Branches with aLRT-values above 0.9, 0.8 and 0.7 are indicated with a red, orange and yellow dot, respectively. The three main branches towards tarsier degenerated zinc fingers, anthropoid degenerated zinc fingers and towards C2H2 zinc finger show the highest aLRT-values of 0.992, 0.966 and 0.98, respectively. One asterisk indicates identical zinc fingers of Western and Philippine tarsier. Two asterisks denote ZnFs shared by Western, Philippine and Sulawesi tarsier including the 3′-most C2H2 ZnFs of the Western tarsier.
The second phylogeny (Fig. 4b) is based exclusively on first degenerated zinc fingers. Here again tarsiers constitute a monophyletic group basal to the anthropoid clade. Within Tarsius Sulawesi tarsiers are monophyletic with respect to Western and Philippine tarsiers.
Positive and negative selected sites
Zinc finger genes are known to alter quickly in their zinc finger array and in particular at the DNA-binding codons (positions −1, 3 and 6 relative to the α-helix) which show signatures of positive selection31,32,33. The PRDM9 zinc finger domain is no exception as shown by positively selected DNA-binding amino acids in rodents and primates3,34,35. The high mutation rates at codon positions directing the interaction of PRDM9 with DNA is deemed responsible for the formation of new hotspots and thus antagonizing hotspot erosion5,36,37.
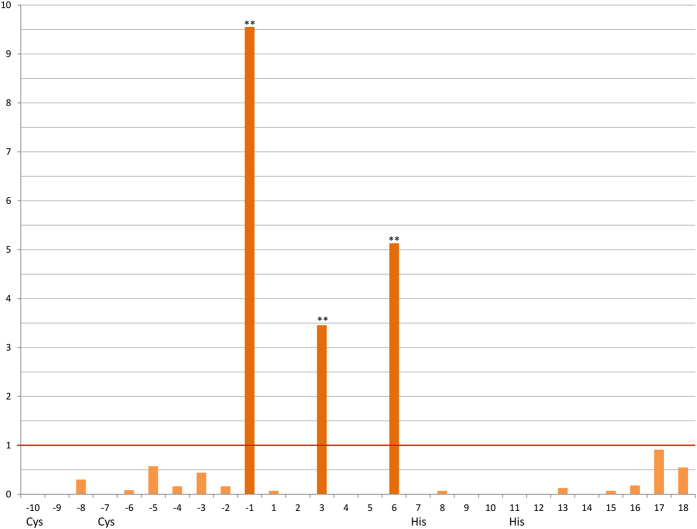
We found positive selection at the three DNA-binding codons, i.e. positions −1, 3, 6 relative to the α-helix (see Fig. 5). Structural important sites like the zinc ion ligands are conserved by negative selection. We consider the ongoing selective pressure – positive as well as negative – acting on tarsier PRDM9 as relevant proof for gene integrity being important in the context of a probable temporary loss of function which was subsequently abolished by a compensatory mutation.
Figure 5. ω-values of C2H2 zinc finger sites in tarsiers.
Sites on the x-axis are indicated with their position relative to the beginning of the α-helix. Ligands - cysteine and histidine - are also labeled. The y-axis shows the ω-values with one being the value for neutral selection highlighted by a red line. The three DNA-binding sites (−1, 3, 6) are positively selected witch a p-value < 0.01 after Benjamini-Hochberg correction (**). The other sites show ω-values revealing negative selection with p-values < 0.05 after Benjamini-Hochberg correction (except for the sites −5 to −2, 2, 17 and 18).
Discussion
With four non-functional zinc fingers at the 5′ end of the tarsier zinc finger array (Fig. 1), degenerated by missing cysteine ligands and frameshifting indels, we detected a completely new PRDM9 allelic variant in primates and mammals in general. This sequence singularity is also reflected in our inferred phylogenies including first degenerated and all functional C2H2 zinc fingers of the array and clearly designating tarsiers as monophyletic group within haplorhine primates (Fig. 4a,b).
All these sequence autapomorphies, but especially those associated with a temporary loss of allelic integrity (see results) might possibly indicate an active role of PRDM9 in the divergence process between anthropoid and tarsiiform primates or alternatively, along the tarsiid lineage. A possible speciation scenario could be as follows:
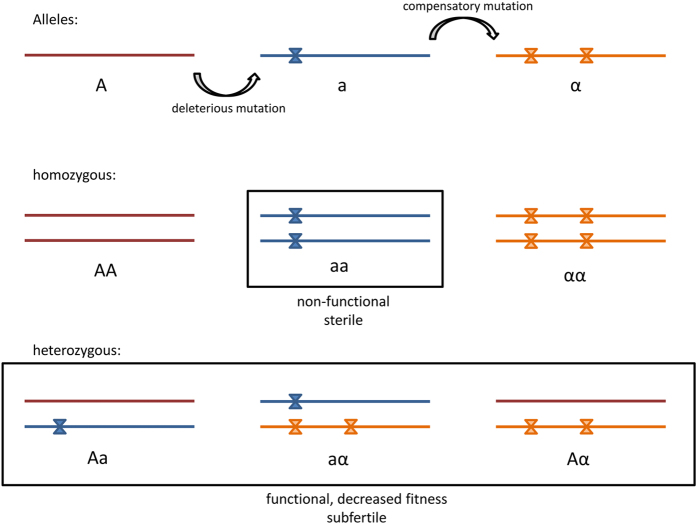
In a population of haplorhine or tarsiid ancestors a damaging, frameshifting mutation occurs in one allele (Figs 2 and 6). Heterozygous individuals may be subfertile because gene function is reduced due to one non-functional allele. However, Brick et al.6 showed that one allele can determine 75% of the hotspots while the other is responsible for 22% [1% of all hotspots are shared and 2% are new in comparison to parental homozygote constellations]. Furthermore, male mice which are heterozygote null for Prdm9 are not sterile but reproduce at a later stage for the first time and father fewer offspring, both in comparison to the wild-type38. Individuals homozygous for the damaged allele are probably infertile due to a considerably reduced or loss of gene function and comparable to Prdm9 knockout mice where a meiotic arrest at pachytene stage leads to sterility4. The second frameshifting mutation restores the functionality of the former erroneous allele (Figs 2 and 6) and is therefore positively selected and fixated39,40.
Figure 6. Schematic demonstration of the mutational events in tarsier PRDM9 zinc finger array.
The insertion and deletion create two more alleles, a and α, besides the wild-type allele A. All possible genotypes are shown where all heterozygous are possibly functional but subfertile and two out of three homozygous are functional without constraints and the third is non-functional and sterile, comparable to Prdm9 knockout mice6.
Two of the now three possible allelic variants in homozygotes are functional (Fig. 6), the one with two original or wild-type alleles and the other with two alleles carrying both indel mutations. By contrast, all heterozygotes shown in Fig. 6 are possibly less fit or rather subfertile resulting from a decreased recombination activity affected by either one damaged allele or different hotspot usage among the two types of functional PRDM9 alleles41. The novel allele finally get fixed in a precursor of tarsiiform primates but in any case no later than 20 MYA, the time to the most recent common ancestor (MRCA) of the three extant tarsier clades17,19,21, all exclusively carrying the new allelic variant. Explanations for the fixation of the mutant allele in an ancestral population of modern tarsiers may include directed and/or random evolutionary processes like biased gene conversion or genetic drift. With regard to the former it has been shown that a newly introduced PRDM9 allele increased hotspot activity in mice through altered and haplotype-biased DNA-binding, thus counteracting fitness loss during the process of hotspot erosion by allelic gene conversion36. The mutant allele could have had a similar hotspot enhancing and therewith beneficial fitness effect in a population of ancient tarsiids or their progenitors. Given that its selective advantage was sufficiently large the novel allele could also have overcome the effects of genetic drift where typically rare beneficial mutations are more likely to go extinct42,43. On the other hand, we could suppose that it was this process of random change in genetic composition that maintained the newly arisen allele resulting in the structural uniformity of the PRDM9 zinc finger domain and facilitating divergence of an ancestral lineage finally leading to modern tarsiers.
Extinct relatives of living tarsiids branched away from anthropoid primates very shortly after the haplorhine-strepsirhine split9,19,44. Depending on the data, either paleontological or molecular, first primates evolved sometime between 55–87 MYA9,15,19,45. Within this period two drastic climate changes occurred and both induced mass extinctions44,46,47 creating conditions promoting adaptive radiation44. Proto-tarsiiforms could have survived or emerged from these evolutionary bottlenecks possibly also because PRDM9 produced new recombination landscapes allowing allele combinations more favourable or adaptive to the changing environmental conditions and/or newly vacant ecological niches. Another extinction event associated with Eocene-Oligocene cooling48 largely decreased primate diversity with the disappearance of omomyids, a controversial fossil tarsiiform primate49,50,51. This event presumably caused a geographic shift in tarsier distribution from mainland to insular Southeast Asia52,53 exposing tarsiids of modern aspects to another population bottleneck and thus providing a further option for the fixation of the novel tarsier-specific PRDM9 allele. A heightened relevance of genetic drift in tarsier molecular evolution was already discussed54, as extant tarsiers, and especially those endemic to the Indonesian island of Sulawesi, have been subject to significant climatic and tectonic change55,56 that triggered allopatric speciation in the Malay Archipelago since the Miocene17,18,20,21,56,57,58. Hence, the much longer history of independent evolution in environmental instability may gives more credence to the theory that the salient molecular changes of the PRDM9 zinc finger array described here have developed along the tarsiid lineage rather than initialized the anthropoid-tarsiid split.
PRDM9 variation in alleles, zinc finger motifs and especially at the key codon positions reflect the different levels of evolutionary independence of the three major tarsier clades (Western, Philippine, and Sulawesi tarsiers), provided the single non-Sulawesi specimens are representative for the respective biogeographic region. Like it has been observed in mice, chimpanzee and humans, 5′-most and 3′-most C2H2 zinc fingers are more conserved or even identical within or at least between closely related species29,41,59,60 even if positive selected sites are factored in. In tarsiers this appears to apply mainly to species endemic to the same biogeographic region, but is also fairly reflected in lineage-specific constraints on 5′-most C2H2 zinc fingers of Sulawesi tarsiers21. Considering the three DNA binding amino acids at internal zinc fingers of Sulawesi tarsiers we found several duplicated zinc finger motifs in each species/subspecies indicating their recent origin61 for most populations being estimated at less than 500,000 years ago21. Particularly striking are the blocks of zinc finger motifs in northern Sulawesi tarsier populations (Fig. 3). The order of their occurrence very plastically depicts how these populations very recently evolved from a single common ancestor21.
In conclusion, the high mutation rate of the PRDM9 zinc finger domain3,7,29 and multiple events of genetic drift produced an enormous diversity of PRDM9 alleles in tarsiers. The geographic distribution of zinc finger alleles and especially the occurrence and enrichment of specific zinc finger motifs reflect phylogeographic patterns of extant tarsiers further strengthening an involvement of PRDM9 in population differentiation. More intriguing, however, is our discovery of a hitherto unknown and, in addition, tarsier-specific PRDM9 allele variant that arose from two indel mutations and played a perhaps decisive role in tarsiid evolution.
Material and Methods
Sample set
The sampling of 23 individuals comprises 21 specimens of nine Sulawesi tarsier populations (1–4 individuals/population, see Fig. 7) and two non-Sulawesi tarsiers (Tarsius bancanus and T. syrichta). Samples from Sulawesi were obtained from previous studies18,21, while the Western and the Philippine specimen were provided by Y. Rumpler (Les Hôpitaux Universitaires de Strasbourg, France) and J. Brosius (University of Muenster, Germany), respectively. Whole genome amplifications (WGA) of each sample (40–80 ng/μl) were used as a template in PCR to amplify the exonic region of the PRDM9 gene containing the zinc finger domain.
Figure 7. Maps of Southeast Asia and Sulawesi.
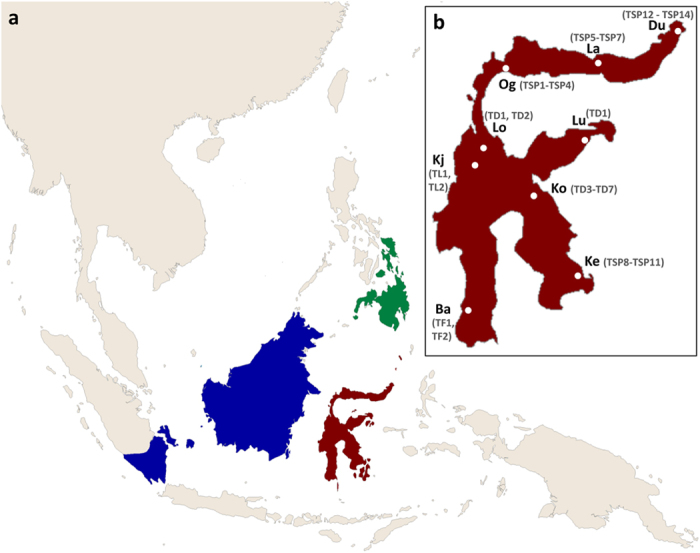
[a] Shows the region of Southeast Asia with ranges of Tarsius bancanus in blue, of Tarsius syrichta in green and of Sulawesi tarsiers in red16. [b] Is a close-up of Sulawesi with sample sites highlighted by white dots. Alleles per population are written in parenthesis. Abbreviations: Du, Duasaudara; La, Labanu; Og, Ogatemuku; Lu, Luwuk; Lo, Laone; Kj, Koja; Ko, Korosule; Ba, Bantimurung; Ke, Kendari. Base map was created in ArcGIS (ArcMap 10.0; http://www.esri.com/software/arcgis).
PCR and Sequencing
We PCR-amplified and sequenced the exon which encodes the zinc finger domain of the PRDM9 gene in three parts: the 5′ and 3′ flanking regions and the repetitive elements in between (see Table 2).
Table 2. PCR primer specifications.
| Primer | Sequence 5′-3′ [bp] | Combinations | T (°C) | Product length (bp) |
|---|---|---|---|---|
| PRDM9-F1 | GGGGTAGGAATCTCAAGAATTG[22] | F3-R1 | 54 | 594 (5′ end) |
| PRDM9-F2 | AAGAATTGCAAGGGTCAAGTATG[23] | F1/F2-R3 | 53 | 977–1481/963–1467 |
| PRDM9-F3 | TGGTGGGAGACAGCTTGC[18] | F3-R3 | 53 | 1501–2005 |
| PRDM9-F4 | GCCAAGAGGACGAGTAAGTCAATAG[25] | F5-R4 | 56 | 1152–1656 |
| PRDM9-F5 | CAAAGATCCACCCATGTTC[19] | F4-R2 | 57 | 198 (3′ end) |
| PRDM9-R1 | TGACTCATCACTGAGGCCTTG[21] | |||
| PRDM9-R2 | CCTCATCTCTAGTCATGAAAGTGG[24] | |||
| PRDM9-R3 | GAAGCACCCTCCAAGCTG[18] | |||
| PRDM9-R4 | GGTCTCTTTACACTCTTGSAGTC[23] |
Given are applied primer combinations, annealing temperatures and product lengths.
To reduce amplification of unspecific products we conducted wax-mediated hotstart PCR. Each PCR-reaction contained 30 μl with final concentrations of 200 μM dNTPs, 2.5 units Taq DNA polymerase, 1x PCR buffer including 1.5 mM MgCl2 (QIAGEN Taq PCR Core Kit), and 0.33 pM per primer. PCRs were run under the following conditions: 3 minutes of initial template denaturation at 94 °C was followed by 35 cycles of denaturation (40 sec at 94 °C), primer annealing (1 min at primer-specific temperatures) and DNA elongation (1–1.5 min at 72 °C). A final elongation step of 5 minutes at 72 °C finished the PCR. Product sizes were estimated on ethidium bromide stained 1.5% agarose gels together with O’RangeRuler 100 bp DNA Ladder and GeneRuler 100 bp Plus DNA Ladder (both Fermentas/Thermo Scientific).
In general PCR products were enzyme purified before sequencing. In some individuals PCR reactions yielded multiple different sized products, either because of size variation between the two alleles or unspecific amplicons. The separation of the two alleles or the isolation of the product from non-specific sequences was done by gel extraction with the QIAquick Gel Extraction Kit (QIAGEN). Purified PCR products were sequenced on both strands using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and the corresponding PCR primer pair. After an SDS/heat treatment for elimination of unincorporated dye terminators sequences were processed on an ABI 3130xl genetic analyzer (Applied Biosystems).
For individuals with ambiguous sites present in their DNA sequences PCR products were purified by ethanol precipitation, ligated into a plasmid vector (pGem-T Vector System I, Promega) and transformed into One Shot TOP10 Chemically Competent E. coli cells (Invitrogen). Where gel extraction failed to separate alleles of different lengths sequences were also isolated by cloning.
At least six positive clones, three of each allele, were PCR amplified and sequenced. Colony PCR was carried out in 20 μl reaction volumes according to standard protocols using the Taq PCR Core Kit (QIAGEN) and vector-specific primers. For long amplicons where sequencing read length was not sufficient we used the internal primer PRDM9-F2 (see Table 2) to get the middle portion of the target sequence.
Sequence Analyses
Raw sequences were edited and consensus sequences (KU948327 - KU948367) were generated in BioEdit 7.1.3.062. To reveal the structure and integrity of the tarsier PRDM9 gene we created alignments in BioEdit with the following approach. We compared 5′-terminal sequences including the degenerated zinc fingers in front of the C2H2 array 1) within Tarsius, and 2) between tarsiers, anthropoids (Homo sapiens, ENST00000296682; Pan troglodytes, GU166820.1) and rodents (Rattus norvegicus, ENSRNOT00000066370; Mus musculus, ENSMUST00000167994). We further examined the zinc finger domain by comparison of isolated zinc fingers within and between taxa of Tarsius, anthropoids and rodents. Structural and functional properties of tarsier PRDM9, in particular with regard to the type of zinc finger motifs, was also validated by peptide sequences obtained with EMBOSS Transeq v. 6.3.163,64.
Zinc finger Phylogeny
We generated two tree topologies, one including all 5′-most degenerated zinc fingers and one including both, functional C2H2-zinc fingers and degenerated first zinc fingers of the array. Haplorhine taxa represented in each data set comprised Tarsius (this study), Homo sapiens (ENST00000296682) and several other anthropoid primates, whose sequence data were adopted from Schwartz et al.7. Microcebus murinus served as outgroup. Corresponding sequence information was extracted from the Ensembl data base (assembly micMur1, database version 77) and by using BLAST+2.2.2865. The zinc fingers were isolated with the first codon for cysteine as starting point. All except 5′-most degenerated and several 3′-most zinc fingers had a length of 84 bp. Lacking the first cysteine-triplet 5′-most degenerated zinc fingers start with the second codon for cysteine, and therefore being nine bp shorter. 3′-most zinc fingers can lack base pairs when interrupted by a stop codon, but they contain at least both cysteine and histidine ligands. Two 3′ degenerated zinc fingers of Tarsius bancanus were included. Like Schwartz et al.7 we excluded the four binding triplets. To reduce the calculation time we removed identical zinc fingers within genera with the Fabox DNAcollapser66. Models of nucleotide substitution were determined with jModelTest 2.1.667,68. Phylogenetic trees were generated with the Maximum Likelihood method (PhyML 3.067) including an approximate likelihood ratio test (aLRT) and SH-like supports. The degenerated zinc finger of Microcebus murinus served as outgroup in both tree topologies. Nodes with support values below 0.5 were collapsed in TreeGraph 2.2.0–407 beta69. The final phylogenetic trees were visualized in FigTree 1.3.170.
Detecting selective pressure
All functional C2H2 zinc fingers (84 bp) were tested for selective pressure on codon sites. We inferred a phylogenetic tree based on all non-identical sequences determined by Fabox DNAcollapser66 using the Maximum Likelihood method [PhyML 3.0, settings other than default: K80, bootstrap: 100]67. The best fitting substitution model was selected using jModelTest 2.1.667,68. Tests of selection were performed with the sitewise-likelihood ratio method (SLR71) using default settings.
Additional Information
Accession codes: The DNA sequences have been deposited in GenBank under the accession codes: KU948327-KU948367.
How to cite this article: Heerschop, S. et al. The pioneering role of PRDM9 indel mutations in tarsier evolution. Sci. Rep. 6, 34618; doi: 10.1038/srep34618 (2016).
Acknowledgments
This work was supported by grants (ZI568/6-1 and ZI568/6-2) from the Deutsche Forschungsgemeinschaft (DFG, http://www.dfg.de) allocated to H.Z.
Footnotes
Author Contributions H.Z. and C.D. conceived the study; S.H. performed experiments; S.H. analysed data; H.Z. and D.P.-F. contributed reagents, materials and analysis tools; C.D. and S.M. provided samples; S.H. and C.D. wrote the manuscript; H.Z., S.M. and D.P.-F. revised the manuscript.
References
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C. & Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323, 373–375 (2009). [DOI] [PubMed] [Google Scholar]
- Forejt J. Hybrid sterility in the mouse. Trends Genet. 12, 412–417 (1996). [DOI] [PubMed] [Google Scholar]
- Oliver P. L. et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5, e1000753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Yoshida K. & Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374–378 (2005). [DOI] [PubMed] [Google Scholar]
- Grey C. et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 9, e1001176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K., Smagulova F., Khil P., Camerini-Otero R. D. & Petukhova G. V. Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J. J., Roach D. J., Thomas J. H. & Shendure J. Primate evolution of the recombination regulator PRDM9. Nat. Commun. 5, 4370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Ohme M. & Zischler H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics 157, 777–784 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P. et al. A molecular phylogeny of living primates. PLoS Genet. 7, e1001342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard K. C., Qi T., Dawson M. R., Wang B. & Li C. A diverse new primate fauna from middle Eocene fissure-fillings in southeastern China. Nature 368, 604–609 (1994). [DOI] [PubMed] [Google Scholar]
- Chaimanee Y., Lebrun R., Yamee C. & Jaeger J.-J. A new Middle Miocene tarsier from Thailand and the reconstruction of its orbital morphology using a geometric-morphometric method. Proc. R. Soc. B 278, 1956–1963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen D. T., Conroy G. C. & Simons E. L. Tarsier-like locomotor specializations in the Oligocene primate Afrotarsius. Proc. Natl. Acad. Sci. USA 95, 14848–14850 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J.-J. et al. Late middle Eocene epoch of Libya yields earliest known radiation of African anthropoids. Nature 467, 1095–1098 (2010). [DOI] [PubMed] [Google Scholar]
- Szalay F. S. Systematics of the Omomyidae (Tarsiiformes, Primates): Taxonomy, phylogeny, and adaptations. Bull. Am. Mus. Nat. Hist. 156, 157–450 (1976). [Google Scholar]
- Ni X. et al. The oldest known primate skeleton and early haplorhine evolution. Nature 498, 60–64 (2013). [DOI] [PubMed] [Google Scholar]
- Groves C. & Shekelle M. The genera and species of tarsiidae. Int. J. Primatol. 31, 1071–1082 (2010). [Google Scholar]
- Shekelle M., Meier R., Wahyu I., Wirdateti & Ting N. Molecular phylogenetics and chronometrics of Tarsiidae based on 12S mtDNA haplotypes. Evidence for Miocene origins of crown tarsiers and numerous species within the Sulawesian clade. Int. J. Primatol. 31, 1083–1106 (2010). [Google Scholar]
- Merker S., Driller C., Perwitasari-Farajallah D., Pamungkas J. & Zischler H. Elucidating geological and biological processes underlying the diversification of Sulawesi tarsiers. Proc. Natl. Acad. Sci. USA 106, 8459–8464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S. et al. Macroevolutionary dynamics and historical biogeography of primate diversification inferred from a species supermatrix. PLoS ONE 7, e49521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelbrink B., Albrecht C., Hall R. & Rintelen T. von. The biogeography of Sulawesi revisited. Is there evidence for a vicariant origin of taxa on wallace’s “anomalous island”? Evolution 66, 2252–2271 (2012). [DOI] [PubMed] [Google Scholar]
- Driller C. et al. Stop and Go – Waves of tarsier dispersal mirror the genesis of Sulawesi island. PLoS ONE 10, e0141212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looman C., Abrink M., Mark C. & Hellman L. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 19, 2118–2130 (2002). [DOI] [PubMed] [Google Scholar]
- Ng P. C. et al. Genetic variation in an individual human exome. PLoS Genet. 4, e1000160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth-Petroczy A. & Tawfik D. S. Protein Insertions and Deletions Enabled by Neutral Roaming in Sequence Space. Mol. Biol. Evol. 30, 761–771 (2013). [DOI] [PubMed] [Google Scholar]
- Williams L. E. & Wernegreen J. J. Sequence context of indel mutations and their effect on protein evolution in a bacterial endosymbiont. Genome Biol. Evol. 5, 599–605 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. D., Frankish A., Hunt T., Harrow J. & Gerstein M. Identification and analysis of unitary pseudogenes. Historic and contemporary gene losses in humans and other primates. Genome Biol. 11, R26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. R., Perrot V. & Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154, 985–997 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon A. The rate of compensatory mutation in the DNA bacteriophage X174. Genetics 170, 989–999 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J. et al. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS ONE 9, e85021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs L., Sun Y. & Caetano-Anolles D. Function and evolution of C2H2 zinc finger arrays. Subcell. Biochem. 52, 75–94 (2011). [DOI] [PubMed] [Google Scholar]
- Shannon M., Hamilton A. T., Gordon L., Branscomb E. & Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 13, 1097–1110 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A. T. et al. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 16, 584–594 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson R. O., Thomas J. H. & Myers S. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 5, e1000325 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H., Emerson R. O. & Shendure J. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS ONE 4, e8505 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K. et al. Gain, loss and divergence in primate zinc-finger genes. A rich resource for evolution of gene regulatory differences between species. PLoS ONE 6, e21553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L. et al. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 11, e1004916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton A., Myers R. S. & Redfield R. J. The hotspot conversion paradox and the evolution of meiotic recombination. Proc. Natl. Acad. Sci. USA 94, 8058–8063 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L. et al. Multimer formation explains allelic suppression of PRDM9 recombination hotspots. PLoS Genet. 11, e1005512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A. S., Sunyaev S. & Kondrashov F. A. Dobzhansky-Muller incompatibilities in protein evolution. Proc. Natl. Acad. Sci. USA 99, 14878–14883 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Weinreich D. M. & Hartl D. L. Missense meanderings in sequence space. A biophysical view of protein evolution. Nat. Rev. Genet. 6, 678–687 (2005). [DOI] [PubMed] [Google Scholar]
- Berg I. L. et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 42, 859–863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwa Z. & Wahl L. M. The fixation probability of beneficial mutations. J. R. Soc. Interface 5, 1279–1289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uecker H. & Hermisson J. On the fixation process of a beneficial mutation in a variable environment. Genetics 188, 915–930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith R. W. et al. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524 (2011). [DOI] [PubMed] [Google Scholar]
- Tavaré S., Marshall C. R., Will O., Soligo C. & Martin R. D. Using the fossil record to estimate the age of the last common ancestor of extant primates. Nature 416, 726–729 (2002). [DOI] [PubMed] [Google Scholar]
- Keller G. et al. Chicxulub impact predates the K-T boundary mass extinction. Proc. Natl. Acad. Sci. USA 101, 3753–3758 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich P. D. Environment and evolution through the Paleocene-Eocene thermal maximum. Trends Ecol. Evol. 21, 246–253 (2006). [DOI] [PubMed] [Google Scholar]
- Prothero D. R. The late Eocene-Oligocene extinctions. Annu. Rev. Earth Planet Sci. 22, 145–165 (1994). [Google Scholar]
- Beard K. C., Krishtalka L. & Stucky R. K. First skulls of the early Eocene primate Shoshonius cooperi and the anthropoid-tarsier dichotomy. Nature 349, 64–67 (1991). [DOI] [PubMed] [Google Scholar]
- Kay R. F., Ross C. & Williams B. A. Anthropoid origins. Science 275, 797–804 (1997). [DOI] [PubMed] [Google Scholar]
- Williams B. A., Kay R. F. & Kirk E. C. New perspectives on anthropoid origins. Proc. Natl. Acad. Sci. USA 107, 4797–4804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski N. G. The evolution of the tarsiid niche in Tarsiers (eds Wright P. C., Simons E. L. & Gursky S.) 35–49 (Rutgers University Press, 2003).
- Jablonski N. G. Primate homeland: Forests and the evolution of primates during the Tertiary and Quaternary in Asia. Anthropol. Sci. 113, 117–122 (2005). [Google Scholar]
- Merker S. et al. Control region length dynamics potentially drives amino acid evolution in tarsier mitochondrial genomes. J. Mol. Evol. 79, 40–51 (2014). [DOI] [PubMed] [Google Scholar]
- Haq B. U., Hardenbol J. & Vail P. R. Chronology of fluctuating sea levels since the Triassic. Science 235, 1156–1167 (1987). [DOI] [PubMed] [Google Scholar]
- Hall R. Southeast Asia’s changing palaeogeography. Blumea 54, 148–161 (2009). [Google Scholar]
- Evans B. J. et al. Monkeys and toads define areas of endemism on Sulawesi. Evolution 57, 1436–1443 (2003). [DOI] [PubMed] [Google Scholar]
- Merker S. et al. Tarsius wallacei. a new tarsier species from central sulawesi occupies a discontinuous range. Int. J. Primatol. 31, 1107–1122 (2010). [Google Scholar]
- Baudat F. et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld L. F., Atencia R., Garriga R. M., Vigilant L. & Mailund T. High diversity at PRDM9 in chimpanzees and bonobos. PLoS ONE 7, e39064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P. What are the genomic drivers of the rapid evolution of PRDM9? Trends Genet. 27, 165–171 (2011). [DOI] [PubMed] [Google Scholar]
- Hall T. A. BioEdit:. a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- Rice P., Longden I. & Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- Goujon M. et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. et al. BLAST+. Architecture and applications. BMC bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P. FaBox: an online toolbox for fasta sequences. Mol. Ecol. Notes 7, 965–968 (2007). [Google Scholar]
- Guindon S. & Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R. & Posada D. jModelTest 2. More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöver B. C. & Müller K. F. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC bioinformatics 11, 7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. FigTree Version 1.3.1. Available at http://tree.bio.ed.ac.uk/software/figtree. (Accessed: 1st December 2014) (2009).
- Massingham T. & Goldman N. Detecting amino acid sites under positive selection and purifying selection. Genetics 169, 1753–1762 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]