Abstract
The tRNA specific for methionine (tRNAM et) of human mitochondria contains a formyl‐cytosine at the wobble position of the anticodon to facilitate its binding to AUG, AUA and (in one instance) to AUU. In this issue of The EMBO Journal, Haag et al identify a two‐step enzyme pathway facilitating the modification of the tRNA. Sequential reactions of the methyltransferase NSUN3 and the dioxygenase ALKBH1/ABH1 are important to render the tRNA as able to recognize the non‐canonical methionine codons AUA and AUUs, a property critical for efficient protein synthesis in human mitochondria.
Subject Categories: Protein Biosynthesis & Quality Control, RNA Biology
Eukaryotic cells contain two distinct translation machineries: one in the cytosol and one in mitochondria. While cytosolic ribosomes synthesize thousands of different proteins, the mitochondrial translation system is specialized on the production of only a handful of proteins. The size of the mitochondrial genome, particularly in animals (including humans), was reduced to almost the theoretical minimum. The 16,569 base pairs of human mitochondrial genome code for 13 proteins (all being core constituents of the respiratory chain complexes), two ribosomal RNAs (one forming the core of the large subunit and one the core of the small subunit), and 22 tRNAs (Gustafsson et al, 2016). Except for some small regulatory elements controlling transcription and replication, all other sequences were removed during evolution. Even the equivalent of the 5S rRNA, a structural element found in the large subunit of all other ribosomes, was lost in mammalian mitochondria and replaced by the valine tRNA that was incorporated into the body of the ribosome (Brown et al, 2014). This drastic reduction in the mitochondrial genome size was presumably driven by Muller's ratchet, that is, the drift to irretrievably loose genetic information in genomes lacking sexual recombination.
As a consequence to their reduced genome size, human mitochondria employ a minimalistic set of tRNAs, which as compensation often decodes an increased number of codons. This change is apparent in the tRNAMet, which in the canonical codon usage only recognizes AUG codons. However, in mammalian mitochondria the tRNAMet also binds to AUA codons and, in the unconventional context of the start codon of the NADH dehydrogenase subunit 2 (ND2), also to AUU, both normally coding for isoleucine. Thus, a single tRNAIle is sufficient to incorporate all isoleucines of mitochondrially encoded proteins. The larger number of codons recognized by the tRNAMet resulted in an increased methionine content in mitochondrial proteins. High levels of this easily oxidized amino acid residue might have helped to counteract the oxidative conditions at the inner membrane (Bender et al, 2008).
The mammalian mitochondrial genome has a strong bias toward a very high AT content. Presumably as a consequence of this trend, 167 of the 207 mitochondrially encoded methionine residues use AUA and only 40 use the canonical AUG codon. This increase in the codon spectrum of the mitochondrial tRNAMet is made possible due to posttranscriptional modifications of the cytosine residue in the wobble position of the anticodon (Bilbille et al, 2011) (Fig 1). Three recent studies identified two sequentially acting enzymes, which are responsible for the modification of this cytosine 34 (Haag et al, 2016; Nakano et al, 2016; Van Haute et al, 2016). Initially a methyl group is added to this nucleoside and then further converted to a formyl group.
Figure 1. The anticodon of the mitochondrial tRNAMet is modified by two sequential enzymatic reactions.
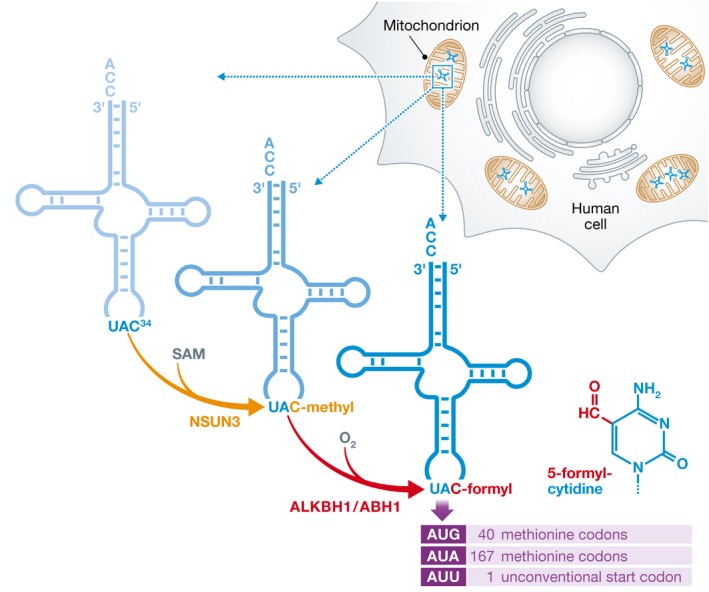
In a first step the cytosine at position 34 in the wobble position of the anticodon is methylated by NSUN3 in a reaction using S‐adenosylmethionine (SAM). Subsequently, the oxygenase ALKBH1/ABH1 converts the methyl‐cytosine to formyl‐cytosine, thereby increasing the spectrum of recognized codons to AUG, AUA and AUU.
In the first step, cytosine 34 is methylated on carbon 5 (Fig 1) by NSUN3, a mitochondrial representative of the Nol1/Nop2/Sun domain (NSUN) family of putative m5C RNA methyltransferases. Mammalian genomes encode at least seven highly conserved members of this protein family to modify different RNAs, predominantly tRNAs and rRNAs, in a highly site‐specific manner. Using in vivo crosslinking with UV light or the chemical 5‐azacytidine, Haag et al (2016) identified NSUN3 as a direct binder of the tRNAMet of mitochondria. They could even reconstitute the methylation reaction in vitro by incubation of recombinant NSUN3 in the presence of the methyl‐group donor S‐adenosylmethionine (SAM) with freshly transcribed methyl‐tRNA, which was efficiently methylated at its cytosine 34 position (Haag et al, 2016). In the parallel study by Van Haute et al, the authors designed an NSUN3 trapping mutant which contains the catalytic cysteine but lacks the resolving cysteine required for cleavage of the covalent adduct and thus for release of the methylated RNA from the enzyme. Using this elegant tool, NSUN3 was caught red‐handed in the process of binding to the anticodon loop of the mitochondrial tRNAMet (Van Haute et al, 2016). Van Haute et al initially had become interested in NSUN3 when they identified a patient with a loss‐of‐function variant of NSUN3. This patient developed mitochondrial disease symptoms such as a combined OXPHOS deficiency in muscle cells at the age of 3 months. The non‐modified anticodon of the mitochondrial tRNAMet (Haag et al, 2016; Nakano et al, 2016; Van Haute et al, 2016) results in strongly reduced translation rates in mitochondria, which in turn leads to a decrease in their respiratory activity (Nakano et al, 2016).
The Haag et al (2016) study also identified the enzyme responsible for the second reaction in which the methyl‐cytosine at position 34 of the tRNAMet is further converted to formyl‐cytosine. This enzyme, ALKBH1/ABH1, is a member of the AlkB‐like Fe2+/α‐ketoglutarate‐dependent dioxygenases (ALKBH), which they reported to be largely located in the mitochondria of HEK293 cells. In the reconstitution experiments, the addition of ALKBH1/ABH1 together with Fe2+ and α‐ketoglutarate converted the methylated tRNAMet into the formylated species that is prevalent under physiological conditions. It is currently not clear whether in vivo, the entire pool of mitochondrial tRNAMet is formylated or whether differentially modified states are used to modulate mitochondrial translation activity.
Over the past few years, more than 100 different species of modified nucleosides have been identified in RNAs from all domains of life (Machnicka et al, 2013). Many of these modifications alter the first (wobble) position of the anticodon of tRNA molecules to modulate codon recognition. The identification of the enzymes that exhibit these modifications now offers an exciting opportunity to unravel how changes in the tRNA modification patterns translate into different codon preferences. Differential modification patterns on tRNAs might represent an additional important level of regulation by which cellular translation activity can be adapted to the needs of different tissues or developmental stages (Roundtree & He, 2016). It will be exciting to explore this “RNA epigenetics” in greater detail in the future.
See also: S Haag et al (October 2016)
References
- Bender A, Hajieva P, Moosmann B (2008) Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc Natl Acad Sci USA 105: 16496–16501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbille Y, Gustilo EM, Harris KA, Jones CN, Lusic H, Kaiser RJ, Delaney MO, Spremulli LL, Deiters A, Agris PF (2011) The human mitochondrial tRNAMet: structure/function relationship of a unique modification in the decoding of unconventional codons. J Mol Biol 406: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, Scheres SH, Ramakrishnan V (2014) Structure of the large ribosomal subunit from human mitochondria. Science 346: 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson CM, Falkenberg M, Larsson NG (2016) Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem 85: 133–160 [DOI] [PubMed] [Google Scholar]
- Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S, Rehling P, Rodnina MV, Höbartner C, Bohnsack MT (2016) NSUN3 and ABH1 modify the wobble position of mt‐tRNAMet to expand codon recognition in mitochondrial translation. EMBO J 35: 2104–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin‐Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H (2013) MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 41: D262–D267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T (2016) NSUN3 methylase initiates 5‐formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol 12: 546–551 [DOI] [PubMed] [Google Scholar]
- Roundtree IA, He C (2016) RNA epigenetics–chemical messages for posttranscriptional gene regulation. Curr Opin Chem Biol 30: 46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, Kotzaeridou U, Hoffmann GF, Memari Y, Kolb‐Kokocinski A, Durbin R, Mayr JA, Frye M, Prokisch H, Minczuk M (2016) Deficient methylation and formylation of mt‐tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun 7: 12039 [DOI] [PMC free article] [PubMed] [Google Scholar]


