Abstract
Upon DNA damage, cyclin‐dependent kinases (CDKs) are typically inhibited to block cell division. In many organisms, however, it has been found that CDK activity is required for DNA repair, especially for homology‐dependent repair (HR), resulting in the conundrum how mitotic arrest and repair can be reconciled. Here, we show that Arabidopsis thaliana solves this dilemma by a division of labor strategy. We identify the plant‐specific B1‐type CDKs (CDKB1s) and the class of B1‐type cyclins (CYCB1s) as major regulators of HR in plants. We find that RADIATION SENSITIVE 51 (RAD51), a core mediator of HR, is a substrate of CDKB1‐CYCB1 complexes. Conversely, mutants in CDKB1 and CYCB1 fail to recruit RAD51 to damaged DNA. CYCB1;1 is specifically activated after DNA damage and we show that this activation is directly controlled by SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1), a transcription factor that acts similarly to p53 in animals. Thus, while the major mitotic cell‐cycle activity is blocked after DNA damage, CDKB1‐CYCB1 complexes are specifically activated to mediate HR.
Keywords: CDK, cell cycle, cyclin, DNA damage, homologous recombination
Subject Categories: Cell Cycle; DNA Replication, Repair & Recombination; Plant Biology
Introduction
DNA damage is a crucial problem for every organism and many repair pathways exist to recover from the different types of DNA damage. Of key importance after DNA damage is an arrest of cell division to allow sufficient time for repair and to prevent that mutated daughter cells are generated that will propagate incorrect genetic information. One severe type of DNA damage often caused by irradiation or chemical mutagens is double‐strand breaks (DSBs), and the signaling cascades from DSBs to cell division arrest are well understood in yeast and animals. In essence, DSBs induce the activity of the kinase ataxia‐telangiectasia mutated (ATM) that phosphorylates and activates checkpoint kinase 2 (Chk2). Chk2 in turn inhibits the Cdc25 phosphatase, a central activator of the main cell‐cycle regulators Cdk1 and Cdk2 in animals. In addition, the ATM pathway activates Wee1, a negative regulator of Cdk1 and Cdk2 providing a parallel block of the cell cycle (Kastan & Bartek, 2004; Harper & Elledge, 2007; Yata & Esashi, 2009).
Remarkably, plants can cope with very high concentrations of harmful agents in comparison with animals. For instance, a comparative study of tobacco BY‐2 and Chinese hamster ovary cells showed that plant cells yielded one‐third less double‐strand breaks after the same dose of ionizing radiation (IR). Furthermore, the plant cells also tolerated a much higher number of DSBs before they died (Yokota et al, 2005). Despite the apparent power and their relevance for agriculture under changing environmental conditions, the plant DNA repair pathways are not very well understood. Moreover, the canonical response pathways of yeast and animals appear to be only partially conserved. While homologs of ATM and its sister kinase ATR (ATM‐ and Rad3‐related) predominantly involved in replication stress response by sensing single‐stranded DNA have also been identified in Arabidopsis (Garcia et al, 2000; Culligan et al, 2004; Culligan & Britt, 2008), no homologs of Chk2 or its sister kinase Chk1 could be found in plants to date. Furthermore, even though a homolog of the yeast Wee1 kinase exists in Arabidopsis and other plants, its function appears to be different as Arabidopsis WEE1 was found to act during S phase after hydroxyurea (HU)‐induced replication stress and not in repressing CDK activity during mitosis or blocking cell division after DSB formation (De Schutter et al, 2007; Cools et al, 2011). Moreover, transgenic plants expressing a mutant version of CDKA;1, the Arabidopsis homolog of mammalian Cdk1 and Cdk2, in which the putative WEE1 target sites were replaced with non‐phosphorylatable amino acids, were not hypersensitive to HU indicated that cell‐cycle arrest after DNA damage is differently regulated in plants (Dissmeyer et al, 2009, 2010).
Besides CDKA;1, plants contain B‐type CDKs that have been implicated in cell‐cycle control. While there appears to be only a single B‐type CDK in the unicellular algae Chlamydomonas rheinhardii that is essential for mitosis (Bisova et al, 2005; Tulin & Cross, 2014), B‐type CDKs are divided into a B1 and B2 class in Arabidopsis and other multicellular plants. B2‐type CDKs appear to be major regulators of mitosis in Arabidopsis and their loss as well as their overexpression interferes with cell proliferation hinting at a strong dose‐dependent action (Andersen et al, 2008). However, due to the lack of mutants, a detailed analysis of B2‐type kinases is still pending. In contrast, B1‐type CDKs have been functionally analyzed, but these studies revealed so far that they apparently act as axillary kinases to A1‐type kinases contributing to the refinement of developmental decisions (Xie et al, 2010; Cruz‐Ramirez et al, 2012; Nowack et al, 2012; Weimer et al, 2012).
Another obvious difference between plants and other well‐studied eukaryotes is the presence of a large groups of cyclins, for example, more than 30 cyclins in Arabidopsis, most of which are still uncharacterized (Harashima et al, 2013). Very little is known about the regulation of these cyclins but remarkably, previous studies have revealed that CYCB1;1 is upregulated during various treatments of DNA damage‐inducing agents or in mutants affected in chromatin organization, DNA metabolism, and/or repair such as fasciata 1 (fas1), jing he sheng 1 (jhs1), and dna replication factor c1 (rfc1) (Chen et al, 2003; Culligan et al, 2004; Endo et al, 2006; Liu et al, 2010; Adachi et al, 2011; Jia et al, 2016). This upregulation is remarkable due to the predicted role of these cyclins in promoting cell division. Up to now, it was not clear what the role of B1‐type cyclins in DNA damage response is, especially in which DNA damage pathway they could act.
In plants as well as in other organisms, two major DNA repair pathways are responsible for genomic integrity after DNA double‐strand breaks: non‐homologous end‐joining (NHEJ) and homology‐dependent repair, also called homologous recombination repair (HR). With NHEJ, the damaged DNA is repaired by direct ligation of the broken ends. The double‐strand break is recognized by a KU70/KU80 heterodimer and then processed by the MRN complex that is composed of MRE11 (MEIOTIC RECOMBINATION 11), RAD50 (RADIATION SENSITIVE 50), and NBS1 (NIJMEGEN‐BREAKAGE SYNDROME 1) (Amiard et al, 2013). DNA ends are ligated by LIG4 (DNA LIGASE 4) and XRCC4 (X‐RAY REPAIR CROSS‐COMPLEMENTATION PROTEIN 4) (Bray & West, 2005). Consistently, ku70 and ku80 mutants are hypersensitive to the DSB‐inducing agents bleomycin (BLM) and methyl methane sulfonate (MMS) (Riha et al, 2002). However, NHEJ can be imprecise, leading to the loss of nucleotides when overlaps are not compatible (Takata et al, 1998).
In contrast to NHEJ, HR is highly accurate since it exactly replaces the defective DNA (Shrivastav et al, 2008). HR requires a homologous template to repair the damaged DNA and can therefore only occur after DNA replication in S phase and the subsequent G2 phase of the cell cycle when sister chromatids are available. This pathway is initiated by the resection of DNA, also executed by the MRN complex, and formation of long 3′ tails, which are coated by RPA (REPLICATION PROTEIN A) in order to prevent winding of the DNA. Homology search and strand invasion are performed by RAD51 family members (Serra et al, 2013), the eukaryotic homolog of the E. coli recA protein (Mengiste & Paszkowski, 1999). RAD51 has five paralogs in Arabidopsis (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3), all of which function in HR in somatic or meiotic cells and show fewer homologous recombination events after DNA damage (Abe et al, 2005; Da Ines et al, 2013a,b; Serra et al, 2013), which is in line with studies in other eukaryotes (Hosoya & Miyagawa, 2014).
A key question is how cells can decide whether to follow NHEJ or enter an HR pathway. While this decision appears to be complex and likely also involves developmental factors, many studies have revealed that CDKs play an important role in the choice of the repair pathway based on the observation that mitotic CDK activity is rising after S phase and hence allowing a cell to discriminate between a G1 and a G2 phase (Wohlbold & Fisher, 2009; Yata & Esashi, 2009; Trovesi et al, 2013). Moreover, CDKs were found to be directly involved in promoting HR. However, the requirement of active CDKs for HR causes an apparent dilemma for a cell since mitotic CDK activity needs to be shut down to arrest the cell division program as a first measure to DNA damage.
Here, we show that plants solve this problem by specifically activating B1‐type CDKs at a transcriptional and posttranslational level after DNA damage. With this, we reveal a previously not recognized key function of B1‐type CDKs as central regulators of DNA damage response in plants. We show that CYCB1s are the specific partner of CDKB1 during DNA damage and both form active complexes that can phosphorylate RAD51. Moreover, we show that HR and NHEJ pathways act at least partially redundantly on DSB, possibly contributing to the powerful DNA damage repair system of plants.
Results
Mutants for B1‐type cyclins are specifically hypersensitive to DNA cross‐links
Based on the observation that CYCB1;1 is upregulated during treatments with DNA damage‐inducing agents (Chen et al, 2003; Culligan et al, 2004, 2006; Ricaud et al, 2007; Adachi et al, 2011), we isolated mutants in all four B1‐type cyclins to address a possible role of these cyclins in DNA stress (Fig EV1A–C). To this end, we monitored root growth of Arabidopsis plants on agar plates containing different DNA‐damaging drugs (see below). On control plates without DNA damage agents, none of the cycb1 mutants showed altered root growth in comparison with the wild type (Figs 1A and B, and EV2A–D and I).
Figure EV1. T‐DNA insertion mutants of the Arabidopsis thaliana CYCB1 genes.
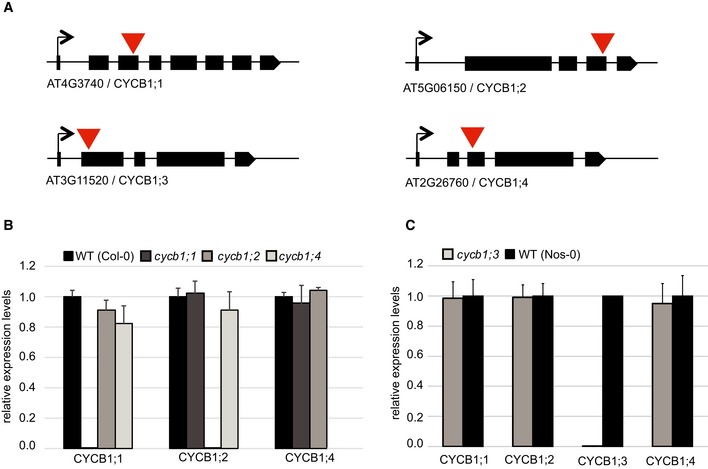
- Schematic overview of T‐DNA insertions for cycb1;1, cycb1;2, cycb1;3, and cycb1;4. Black boxes display exons, and red arrowheads indicate the positions of the T‐DNA insertion.
- Quantitative PCR for relative expression levels in the wild type (Col‐0), cycb1;1, cycb1;2 and cycb1;4. EXP, SAND, and ACT7 were used as reference genes for normalization. Each value represents the mean ± standard deviation of three independent experiments.
- Quantitative PCR for relative expression levels in the wild type (Nos‐0) and cycb1;3. EXP, SAND, and ACT7 were used as reference genes for normalization. Each value represents the mean ± standard deviation of three independent experiments.
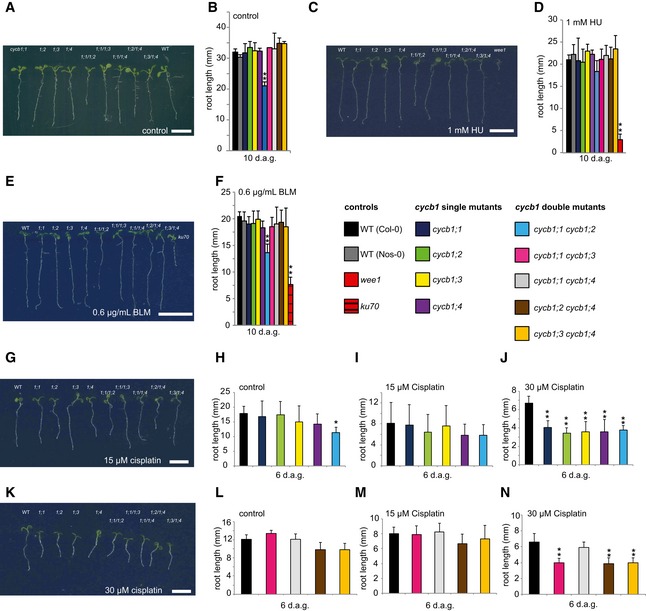
Figure 1. Mutants of B1‐type cyclins are hypersensitive to cisplatin.
-
Acycb1;1, cycb1;2, cycb1;3, cycb1;4, cycb1;1/1;2, cycb1;1/1;3, cycb1;1/1;4, cycb1;2/1;4, cycb1;3/1;4, and the wild type (from left to right) on control plates without genotoxic agent 10 days after germination.
-
BThe wild type, single, and double mutants of cycb1 were grown on control plates without genotoxic agent. Root lengths were measured 10 days after germination.
-
CThe wild type, cycb1;1, cycb1;2, cycb1;3, cycb1;4, cycb1;1/1;2, cycb1;1/1;3, cycb1;1/1;4, cycb1;2/1;4, cycb1;3/1;4 (from left to right) on plates containing 1 mM hydroxyurea (HU) 10 days after germination. The rightmost plant is the wee1 mutant that shows high sensitivity to HU.
-
DThe wild type, single, and double mutants of cycb1 were grown on plates supplemented with 1 mM HU. Root lengths were measured 10 days after germination.
-
EThe wild type, cycb1;1, cycb1;2, cycb1;3, cycb1;4, cycb1;1/1;2, cycb1;1/1;3, cycb1;1/1;4, cycb1;2/1;4, cycb1;3/1;4 (from left to right) on plates containing 0.6 μg/ml bleomycin (BLM) 10 days after germination. The rightmost plant is the ku70 mutant that shows high sensitivity to BLM.
-
FThe wild type, single, and double mutants of cycb1 were grown on plates supplemented with 0.6 μg/ml BLM. Root lengths were measured 10 days after germination.
-
GThe wild type, cycb1;1, cycb1;2, cycb1;3, cycb1;4, cycb1;1/1;2, cycb1;1/1;3, cycb1;1/1;4, cycb1;2/1;4, cycb1;3/1;4 (from left to right) on plates containing 15 μM cisplatin 6 days after germination, that is, 3 days after transfer from control plates.
-
H–Jcycb1 mutants were germinated on control plates and were transferred to new control plates (H) or plates supplemented with 15 μM (I) or 30 μM (J) cisplatin 3 days after germination. Root lengths were measured 3 days after transfer and the net root growth of 3 days is shown in the graphs.
-
KThe wild type, cycb1;1, cycb1;2, cycb1;3, cycb1;4, cycb1;1/1;2, cycb1;1/1;3, cycb1;1/1;4, cycb1;2/1;4, cycb1;3/1;4 (from left to right) on plates containing 30 μM cisplatin 6 days after germination, that is, 3 days after transfer from control plates.
-
L–Ncycb1 double mutants germinated on control plates and were transferred to new control plates (L) or plates supplemented with 15 μM (M) or 30 μM (N) cisplatin 3 days after germination. Root lengths were measured 3 days after transfer and the net root growth of 3 days is shown in the graphs.
Figure EV2. Phenotype of cycb1 mutants under greenhouse conditions.
-
A–DSingle mutants cycb1;1 (A), cycb1;2 (B), cycb1;3 (C), and cycb1;4 (D).
-
E–GDouble mutants cycb1;1 cycb1;2 (E), cycb1;1 ku70 (F), and cycb1;2 ku70 (G). Seedlings of cycb1;1 cycb1;2 are smaller and more diverse than wild‐type plants.
-
HThe triple mutant cycb1;1 cycb1;2 ku70 displays a similar phenotype as the double mutant cycb1;1 cycb1;2.
-
IWild type.
-
JSingle mutant ku70.
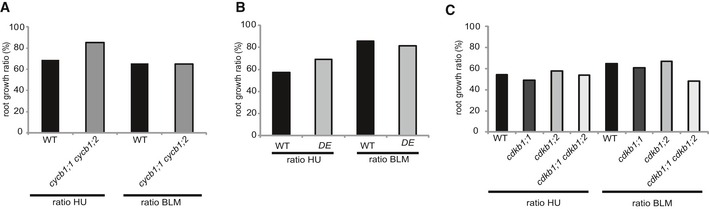
First, we tested root growth on media containing HU, which causes intra‐S‐phase stress due to the inhibition of the enzyme ribonucleotide reductase and thus a decrease in production of deoxyribonucleotides (Yarbro, 1992). For this analysis, wee1 was used as a positive control and, consistent with previous data, was found to be highly sensitive to HU, whereas it shows no growth abnormalities on control medium (De Schutter et al, 2007; Cools et al, 2011). In contrast, root growth on HU of all tested cycb1 mutants was comparable to the growth of wild‐type plants (Fig 1C and D). To address a possible redundant function among the CYCB1 group, we generated the double mutants cycb1;1 cycb1;2, cycb1;1 cycb1;3, cycb1;1 cycb1;4, cycb1;2 cycb1;4, and cycb1;3 cycb1;4. With the exception of cycb1;1 cycb1;2, all double mutants grew indistinguishably from the wild type on media with and without HU (Figs 1A–D and EV3A). The double mutant cycb1;1 cycb1;2 had shorter roots than the wild type on both media with and without DNA stress‐inducing drugs (Fig 1A–D). Comparing the root growth ratios of plants grown on media without and with HU, it became obvious that this double mutant was not more sensitive than the wild type to HU (Fig EV3A).
Figure EV3. Root growth ratios of cycb1 and cdkb1 mutants.
-
A, BGraphs represent the ratio of the mean growth rate on 1 mM hydroxyurea (HU) and 0.6 μg/ml bleomycin (BLM) compared to control experiments on plates lacking genotoxins for the wild type and the double mutant cycb1;1 cycb1;2 (A) or the weak CDKA;1 allele DE (CDKA;1 T15D;Y15E) (B).
-
CGraphs represent the ratio of the mean growth rate on 1 mM HU and 0.6 μg/ml BLM compared to control experiments on plates lacking genotoxins for the wild type, the single mutants cdkb1;1 and cdkb1;2, and the double mutant cdkb1;1 cdkb1;2.
Next, root growth of single and double mutant combinations of cycb1s was tested on media containing the DSB‐inducing drug BLM. As a positive control, we used mutants in ku70 that were shown to grow as the wild type on medium without drugs (Cools et al, 2011). Whereas ku70 mutants were sensitive to BLM and grew only very little consistent with previous reports (Tamura et al, 2002; West et al, 2002; Cools et al, 2011), no significant difference was found between cycb1 single and double mutants versus the wild type again with the exception of cycb1;1 cycb1;2 (Fig 1E and F). Comparing root growth ratios on plates with and without BLM indicated that cycb1;1 cycb1;2 is also not hypersensitive to this drug (Fig EV3A).
As a third drug, the hypersensitivity of cycb1 mutants to cisplatin was tested. Cisplatin causes in addition to DNA breaks also intra‐ and interstrand DNA links that require repair by HR in contrast to damage caused by BLM and HU that can also be repaired by NHEJ (Kartalou & Essigmann, 2001; Belenkov et al, 2002; De Silva et al, 2002; Fuertes et al, 2002; Crul et al, 2003; Siddik, 2003; Pinato et al, 2014). Since cisplatin is unstable in solution, seedlings were germinated on media without the drug and then transferred to plates containing two concentrations of cisplatin (15 and 30 μM) 3 days after germination. On plates with 15 μM cisplatin, the net root growth of the cycb1 mutants at 3 days after the transfer appeared to be reduced but was not statistically significantly different from the growth of wild‐type plants (Fig 1G–I, L and M). However, at 30 μM cisplatin, the roots of all B1‐type cyclin mutants were significantly shorter than the roots of wild‐type plants (Student's t‐test P < 0.01) (Fig 1J and K). The observation that root growth of the cycb1 double mutants was not further reduced in comparison with the growth of the single mutants suggested that all four cyclins contribute in a non‐additive manner to growth on media with cisplatin (Fig 1G, J, K and N). One exception was the double mutant cycb1;1 cycb1;4 that, while being shorter than the wild type, grew better than the other double mutants. However, since all single mutants including cycb1;1 and cycb1;4 as well as all other double mutant combinations are significantly shorter, we conclude that an indirect effect, for example, a compensatory action, in the cycb1;1 cycb1;4 double mutant triggers this response and that the general theme of mutants in B1‐type cyclins is a hypersensitivity against cisplatin.
Next, we asked whether the hypersensitivity of the cycb1 mutants and their reduced growth on cisplatin was due to increased cell death. To this end, we stained the wild type and cycb1 mutants with propidium iodide to visualize dying cells. Under control conditions, no cell death occurred in all tested genotypes. After cisplatin treatment, dead cells were observed in close proximity to the quiescent center. However, we did not see obvious difference between the wild type and cycb1 mutants indicating a higher level of cisplatin‐induced DNA damage in the cycb1 mutants (Fig EV4).
Figure EV4. Cell death in cycb1 mutants after cisplatin treatment.
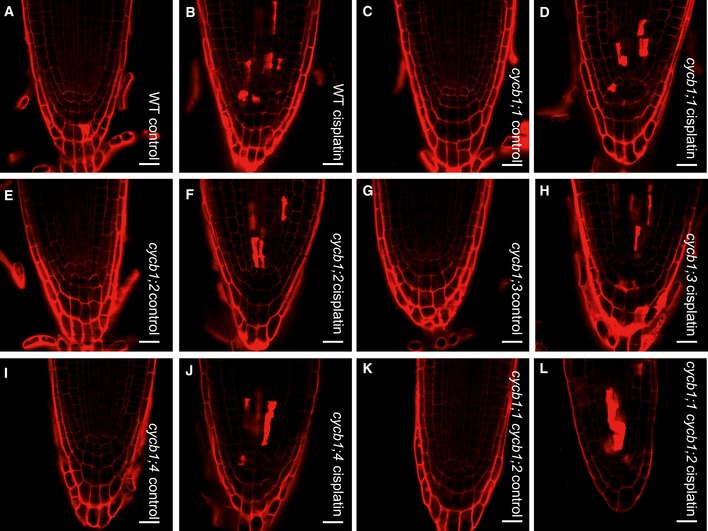
-
A–LPropidium iodide staining of the apical root meristem in wild type grown on control plates (A) or plates supplemented with 50 μM cisplatin for 24 h (B); cycb1;1 grown on control plates (C) or plates supplemented with 50 μM cisplatin for 24 h (D); cycb1;2 grown on control plates (E) or plates supplemented with 50 μM cisplatin for 24 h (F); cycb1;3 grown on control plates (G) or plates supplemented with 50 μM cisplatin for 24 h (H); cycb1;4 grown on control plates (I) or plates supplemented with 50 μM cisplatin for 24 h (J); cycb1;1 cycb1;2 grown on control plates (K) or plates supplemented with 50 μM cisplatin for 24 h (L). Red spots show cell death. Scale bars: 20 μm.
Previously, it was reported that Arabidopsis root cells entered an endoreplication cycle in which the nuclear DNA is amplified without subsequent cell division as a response to zeocin‐induced DNA damage (Adachi et al, 2011; De Veylder et al, 2011; Edgar et al, 2014). Although we cannot exclude long‐term effects of cisplatin to promote endoreplication, we did not see a major increase in endoreplication levels in comparison with control plants when we analyzed cells of the root tips of 5‐day‐old wild‐type plants grown for 24 h on media with 50 μM cisplatin. Likewise, a strong increase in endoreplication was not observed in cycb1;1 cycb1;3 double mutants when treated with cisplatin for 24 h (Fig EV5).
Figure EV5. Ploidy levels in the wild type, cdkb1, and cycb1 mutants.
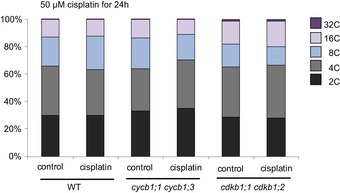
Flow cytometry analysis of root tips of the wild type and the double mutants cdkb1;1 cdkb1;2 and cycb1;1 cycb1;3 under control conditions without genotoxins and transferred 5 days after germination to plates supplemented with 50 μM cisplatin for 24 h.
Next, we tested the response to genotoxic stresses of other mutants in cyclins with a mitotic function. To this end, we investigated the group of A2‐type cyclins that build a small gene family in Arabidopsis with four members (Vanneste et al, 2011). The loss of all four members leads to very slow and impaired postembryonic growth, but the triple mutant cyca2;2 cyca2;3 cyca2;4 (named in the following cyca2;234) is viable and was analyzed on media containing HU, BLM, and cisplatin. In contrast to mutants in B1‐type cyclins, cyca2;234 triple mutants were neither sensitive to HU nor BLM and, notably, also not to cisplatin (Fig EV6A–C). Thus, the hypersensitivity to cisplatin is a specific feature of cycb1 mutants.
Figure EV6. The triple mutant cyca2;2 cyca2;3 cyca2;4 is not hypersensitive to hydroxyurea, bleomycin, and cisplatin.
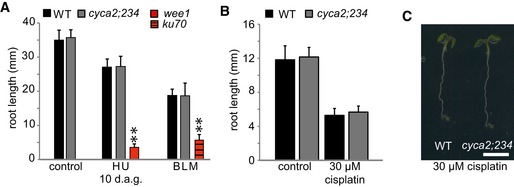
- The wild type and the triple mutant cyca2;234 were grown on control plates or plates containing 1 mM hydroxyurea (HU) or 0.6 μg/ml bleomycin (BLM) for 10 days. Root lengths were measured 10 days after germination. The mutants wee1 and ku70 were used as positive controls for hydroxyurea and bleomycin sensitivity, respectively.
- The wild type and cyca2;234 triple mutant germinated on control plates without cisplatin and were transferred to plates containing 30 μM cisplatin 3 days after germination. Roots were measured 3 days after transfer and the net growth of 3 days is shown in the graph.
- Images show the wild type and cyca2;234 triple mutant on plates supplemented with 30 μM cisplatin. Images were taken 6 days after germination, that is, 3 days after transfer to cisplatin. Scale bar: 0.5 cm.
To assess whether cycb1 mutants indeed accumulate more DNA damage after cisplatin treatment, we performed comet assays, which allow the visualization of DNA DSBs through the formation of a DNA “tail” after electrophoresis of isolated nuclei. For this analysis, the double mutant cycb1;1 cycb1;3 was selected as a representative genotype and assayed after cisplatin treatment and after a recovery phase of 30 min. Quantification of the DNA in the tail showed that cycb1;1 cycb1;3 double mutants contained significantly more DNA breaks than the wild type during and after withdrawal from cisplatin media (Student's t‐test P < 0.01) (Fig 2A and B).
Figure 2. Mutants of cdkb1 and cycb1 show increased number of DSBs and delayed DNA repair upon cisplatin treatment.
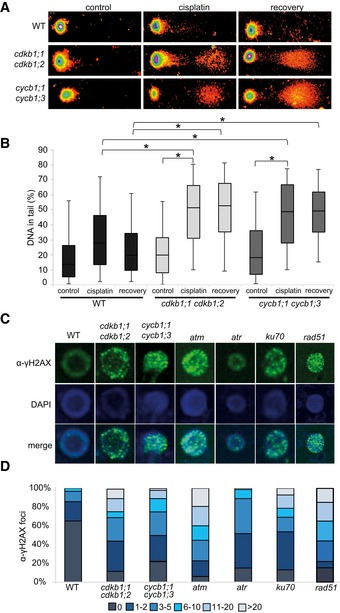
- Representative examples of comets of 21‐day‐old wild‐type plants, cdkb1;1 cdkb1;2 and cycb1;1 cycb1;3 double mutant seedlings in full spectrum view of the TriTek Comet Score software. Shown are comets of plants incubated with 50 μM cisplatin for 1 h and then transferred to medium without cisplatin for 30 min (recovery) and plants incubated without cisplatin for 1 h (control), respectively.
- Box plot of percentage of tail DNA of wild‐type cells, cdkb1;1 cdkb1;2 and cycb1;1 cycb1;3 double mutants under cisplatin treatment. Plots are based on analyses of 200 cells per sample from random microscopic fields of three independent biological replicates. The percentage of DNA fragments in the comet tail was calculated by the TriTek Comet Score software. The box represents the interquartile range, the line across the box indicates the median values, and whiskers represent 5–95 percentile values. Brackets connect plots of sample groups that are significantly different with a confidence level higher than 99.99% calculated with Student's t‐test.
- Immunostaining of γ‐H2AX foci in wild‐type plants and mutant cells after 2 h of treatment with 50 μM cisplatin.
- Counted numbers of γ‐H2AX foci per cell detected after 2 h of treatment with 50 μM cisplatin in wild‐type and mutant plants. For each sample, the γ‐H2AX foci of 100 cells were counted and grouped into six categories: cells with no, 1–2, 3–5, 6–10, 11–20, and more than 20 foci per cell.
As a response to DSBs, the histone variant H2AX becomes phosphorylated at serine 139 (designated gamma‐H2AX) at the break site (Kuo & Yang, 2008). In mutants impaired in DNA damage repair, such as atm, atr, ku70, and rad51, many more gamma‐H2AX can be observed. Consistent with a higher rate of DNA lesions, we found that cycb1;1 cycb1;3 double mutants also showed more gamma‐H2AX foci in immunohistological stainings than the wild type after growth on cisplatin‐containing media (Student's t‐test P < 0.0001) (Fig 2C and D).
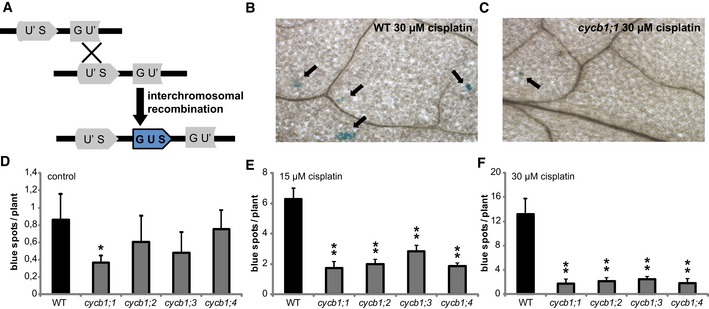
To investigate whether cycb1 mutants are indeed compromised in HR, we deployed a previously described assay to detect homologous recombination events (Swoboda et al, 1994). This assay system makes use of a disrupted gene encoding beta‐glucuronidase (uidA/GUS) gene, which serves as a substrate for homologous recombination (Fig 3A). In cells, where homologous recombination events occur, the uidA gene is restored and subsequently, GUS activity can be detected as blue spots after histochemical staining (Fig 3B and C). The line with the disrupted uidA was crossed to all four cycb1 single mutants and plants homozygous for the reporter and the respective mutant were identified. Already under control conditions, cycb1 mutants appeared to have significantly fewer blue sectors, average of 0.5 per seedling plant, in comparison with the wild type with on average 1 spot (Student's t‐test P < 0.001) (Fig 3D). After incubation on 15 and 30 μM cisplatin for 3 days, recombination rates increased in the wild type reaching 6 and 13 spots per plant, respectively (Fig 3B, E and F). However, all cycb1 mutants showed fewer blue spots indicative for reduced number of homologous recombination events (Student's t‐test P < 0.001) (Fig 3C, E and F). We therefore concluded that B1‐type cyclins are required for homologous recombination repair in Arabidopsis.
Figure 3. Homologous recombination frequencies are strongly reduced in cycb1 mutants.
-
ASchematic drawing of homologous recombination assay. Restoring the functional GUS gene from two disrupted parts (GU' and US') is restricted to an intermolecular homologous recombination event. Homologous events occur only when a sister chromatid or homolog is available as a template, that is, in G2 phase of the cell cycle.
-
BWild‐type plants show blue spots on the leaves after 3 days of incubation on 30 μM cisplatin. Arrows indicate representative blue sectors.
-
Ccycb1;1 plants show blue spots on the leaves after 3 days of incubation on 30 μM cisplatin. Arrows indicate representative blue sectors.
-
D–FGraphs show numbers of blue sectors per plant grown without drug treatment (D) or after incubation on 15 μM (E) or 30 μM (F) cisplatin for 3 days. One or two asterisks indicate significant differences within a 5 and 1% confidence interval, respectively (Student's t‐test). Three biological replicates, each containing at least 15 plants, were analyzed. The mean of the root length of each individual experiment was determined and again averaged for the three biological replicates. Graphs represent mean ± SD.
Mutants for B1‐type CDKs are specifically hypersensitive to DNA cross‐links
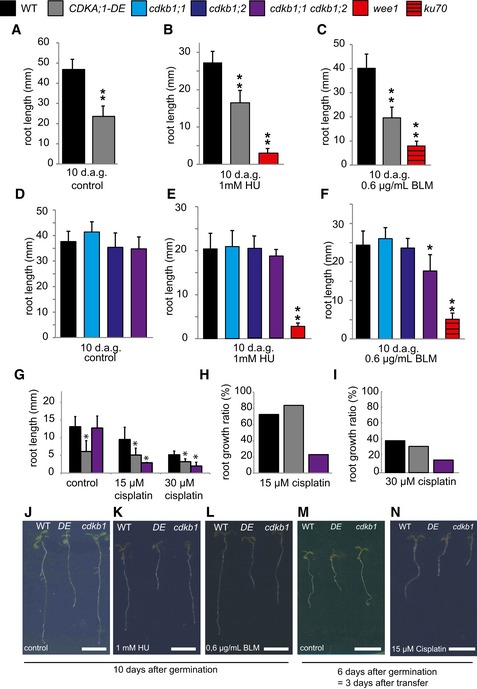
Cyclins usually act together with a CDK partner. Among the five CDKs in Arabidopsis that have been implicated in direct regulation of cell‐cycle progression (i.e. CDKA;1, CDKB1;1, CDKB1;2; CDKB2;1, and CDKB2;2), CDKA;1 has been found to be of central importance controlling both the G1‐S and G2‐M transition (Nowack et al, 2012). Although homozygous cdka;1 mutants exist, they are so severely compromised that an analysis, especially under stress conditions, faces the danger of giving ambiguous results due to pleiotropic and indirect effects (Nowack et al, 2012). We therefore made use of a previously described weak loss‐of‐function allele, designated CDKA;1 T14D;Y15E or short DE (Dissmeyer et al, 2009), to address whether CDKA;1 plays a role in the response to cisplatin. DE displayed shorter roots than the wild type on plates without DNA damage‐inducing agents (Fig 4A, G, J and M) as well as on media supplemented with BLM (Fig 4C and L). Previously, we found that DE is not sensitive to HU (Dissmeyer et al, 2009) (Fig 4B and K). Taking the root growth ratio into account, we determined that DE was neither more sensitive to HU nor to BLM than the wild type (Fig EV3B). Mutants in ku70 and wee1 were used as a control for BLM and HU treatments, respectively, and, consistent with previous reports, showed hypersensitivity on the respective drugs (Fig 4B, C, E and F). In addition, analyzing the growth ratio of DE roots on 15 and 30 μM cisplatin‐containing medium compared to roots grown on control plates without DNA damage‐inducing drugs, it became obvious that DE was also not hypersensitive to cisplatin (Fig 4G–I, M and N). This result suggested that other CDKs might be involved in the response to cisplatin damages and operate together with B1‐type cyclins.
Figure 4. Mutants of cdkb1 but not cdka;1 are hypersensitive to cisplatin.
-
A–CThe wild type and CDKA;1‐DE mutants were grown on control plates (A) or containing 1 mM hydroxyurea (B) or 0.6 μg/ml bleomycin (C) for 10 days. The mutants wee1 and ku70 were used as positive controls for hydroxyurea or bleomycin sensitivity, respectively. Root lengths were measured 10 days after germination.
-
D–FThe wild type, cdkb1;1, cdkb1;2, and the double mutant cdkb1;1 cdkb1;2 were grown on control plates (D) or plates containing 1 mM hydroxyurea (E) or 0.6 μg/ml bleomycin (F) for 10 days. The mutants wee1 and ku70 were used as positive controls for hydroxyurea and bleomycin sensitivity, respectively. Root lengths were measured 10 days after germination.
-
GThe wild type, CDKA;1‐DE, and the double mutant cdkb1;1 cdkb1;2 were grown on control plates and were transferred to plates containing 15 or 30 μM cisplatin 3 days after germination. Root lengths were measured 3 days after transfer and the net root growth of 3 days is shown in the graphs.
-
H, IGraphs represent the ratio of the mean growth rate on 15 μM (H) or 30 μM (I) cisplatin compared to control experiments on plates lacking cisplatin for the wild type, CDKA;1‐DE, and cdkb1;1 cdkb1;2.
-
J–NImages show a wild‐type plant, CDKA;1‐DE, and the double mutant cdkb1;1 cdkb1;2 (from left to right) on the indicated day after germination and the indicated drug treatment. Scale bars: 1 cm.
Putative candidates are the plant‐specific CDKB1s since the expression pattern of B1‐type cyclins in G2 phase overlaps with the expression pattern of CDKB1s during the cell cycle (Menges et al, 2005). In addition, it was shown that CYCB1;2 has kinase activity in complex with CDKB1s in vitro (Harashima & Schnittger, 2012). Since CDKB1;1 and CDKB1;2 were found to have overlapping functions (Xie et al, 2010; Cruz‐Ramirez et al, 2012; Nowack et al, 2012; Weimer et al, 2012), we used the previously generated double mutant cdkb1;1 cdkb1;2 in the following studies, referred to as cdkb1. We first tested cdkb1 mutants on HU but the single as well as the double mutants grew indistinguishably from the wild type (Fig 4D, E and K). As a control, we monitored again wee1 mutants that were found to be highly susceptible to HU in the media. Next, we analyzed root growth on BLM‐containing media with ku70 mutants as a control (Fig 4F and L). While both cdkb1 single mutants were not hypersensitive, the double mutant cdkb1;1 cdkb1;2 showed a reduction by approximately 25% in root growth when compared to wild‐type plants grown on BLM (Student's t‐test P < 0.05). This indicated a minor but distinguishable role of CDKB1s in DSB repair induced by BLM (see below).
Next, we assayed root growth of cdkb1 mutants on two different concentrations of cisplatin. Under these conditions, the roots of cdkb1;1 cdkb1;2 were dramatically compromised and grew only up to a third of the size of the wild type (Student's t‐test P < 0.001) (Fig 4G–I, M and N). Consistent with this growth reduction, we could detect very high levels of DNA damage in cdkb1;1 cdkb1;2 during cisplatin treatment and 30 min after recovery (Fig 2A and B). Quantification of gamma‐H2AX foci even showed higher levels of DNA damage in cdkb1;1 cdkb1;2 double mutants than in cycb1;1 cycb1;3 double mutants reaching a level comparable to the one seen in atm mutants (Fig 2C and D). Similar to cycb1 mutants, we also did not observe strongly increased endoreplication levels in cdkb1 mutants grown on media with cisplatin in comparison with mutants grown on media without cisplatin (Fig EV5).
To test whether CDKB1s operate in the same or in a parallel genetic pathway as CYCB1s, we generated the homozygous triple mutant cycb1;1 cdkb1;1 cdkb1,2 and analyzed its response to the DNA‐damaging drugs HU, BLM, and cisplatin. Our hypothesis was that, if CDKB1s and CYCB1s act in different pathways, cycb1;1 cdkb1;1 cdkb1;2 triple mutants should be more compromised on media containing DNA‐damaging drugs than cdkb1 mutants. However, the triple mutant was not significantly different from cdkb1;1 cdkb1;2 double mutants grown on control plates and plates supplemented with 1 mM HU, 0.6 μg/ml BLM, and 30 μM cisplatin (Fig EV7).
Figure EV7. The triple mutant cycb1;1 cdkb1;1 cdkb1;2 shows no additive effect in DNA damage sensitivity.
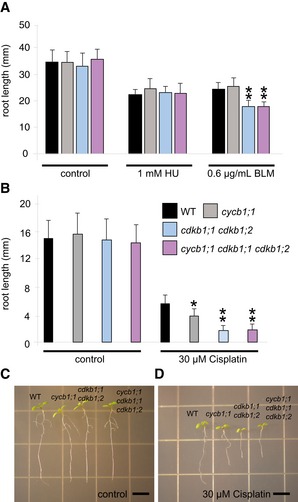
- The wild type, the cycb1;1 single mutant, the cdkb1;1 cdkb1;2 double mutant, and the cycb1;1 cdkb1;1 cdkb1;2 triple mutant were grown on control plates without genotoxins and plates supplemented with 1 mM hydroxyurea (HU) or 0.6 μg/ml bleomycin (BLM). Root lengths were measured 10 days after germination.
- The wild type, the cycb1;1 single mutant, the cdkb1;1 cdkb1;2 double mutant, and the cycb1;1 cdkb1;1 cdkb1;2 triple mutant germinated on control plates and were transferred to plates supplemented with 30 μM cisplatin 3 days after germination. Root lengths were measured 3 days after transfer and the net root growth of 3 days is shown in the graphs.
- Images show the wild type, the cycb1;1 single mutant, the cdkb1;1 cdkb1;2 double mutant, and the cycb1;1 cdkb1;1 cdkb1;2 triple mutant (from left to right) 6 days after germination (= 3 days after transfer to 30 μM cisplatin plates).
Taken together, we conclude that B1‐type CDKs are involved in the response after DNA damage, especially damage induced by cisplatin. Our data show that CDKB1s do not carry out this function in combination with A2‐type cyclins (Fig EV6) that were previously identified as specific partners of B1‐type CDKs (Boudolf et al, 2009) but largely in conjunction with B1‐type cyclins.
RAD51 localization depends on CDKB1s and CYCB1s
RAD51 is a homolog of the bacterial RecA recombinase and a key factor in HR in eukaryotes. RAD51 and its homologs build a small gene family in Arabidopsis similar to other eukaryotes (Lin et al, 2006). Previous studies revealed hypersensitivity of mutants in Arabidopsis RAD51 family members to DNA‐cross‐linking agents such as cisplatin and mitomycin C, as seen, for example, by a reduced number of true leaves formed in rad51b and rad51c mutants in comparison with the wild type (Osakabe et al, 2002, 2005; Bleuyard & White, 2004; Abe et al, 2005; Bleuyard et al, 2005; Li et al, 2005; Charbonnel et al, 2010). Consistent with these reports, we found that the root growth of rad51 mutants is impaired in the presence of cisplatin (Fig 5A–C). Correspondingly, we found that rad51 mutants accumulated a large number of gamma‐H2AX foci after treatment with cisplatin (Fig 2C and D).
Figure 5. The CDKB1;1‐CYCB1;1 complex phosphorylates RAD51.
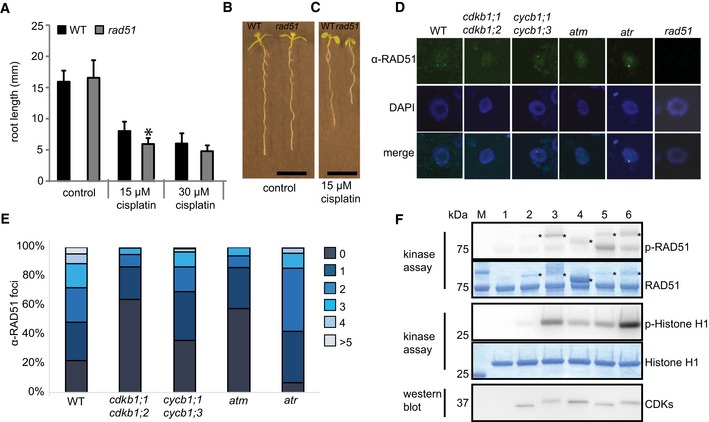
- WT and rad51 mutants were grown on control plates or transferred to plates supplemented with 15 or 30 μM cisplatin, respectively, 3 days after germination. Root lengths were measured 3 days after transfer and the net root growth of 3 days is shown in the graph. Asterisk indicates significant differences within a 5% confidence interval (Student's t‐test). Three biological replicates, each containing at least 15 plants, were analyzed. The mean of the root length of each individual experiment was determined and again averaged for the three biological replicates. Graphs represent mean ± SD.
- Image shows a wild‐type plant (left) and rad51 mutant (right) grown on control plates. Images were taken 6 days after germination. Scale bar: 1 cm.
- Image shows a wild‐type plant (left) and rad51 mutant (right) germinated on control plates and transferred to plates supplemented with 15 μM cisplatin 3 days after germination. Images were taken 6 days after germination, that is, 3 days after transfer to cisplatin. Scale bar: 1 cm.
- Immunostaining of RAD51 foci in the wild‐type and indicated mutant cells after 2 h of treatment with 50 μM cisplatin.
- Counted numbers of RAD51 foci per cell detected after 2 h of treatment with 50 μM cisplatin in wild‐type and mutant plants. For each sample, the RAD51 foci of 100 cells were counted and grouped into six categories: cells with 0, 1, 2, 3, 4, or > 5 foci per cell.
- In vitro kinase assay of purified CDK complexes phosphorylating RAD51. RAD51 and histone H1 kinase assays were performed with [γ‐32P]ATP as a phosphate donor. Proteins were subjected to SDS–PAGE after the kinase reaction and stained with Coomassie Brilliant Blue demonstrating the equal loading of the substrates (lower panels). Phosphorylated proteins were detected by autoradiography (upper panels). The reactions were normalized by using equal amounts of CDKs assuring equal levels of active CDK‐cyclin complexes; the protein blot indicates the relative amounts of the CDKs in the reaction (bottom panel). Abbreviation: p‐RAD51 and p‐histone H1 for [32P]‐phosphorylated MBP‐RAD51‐His6 and recombinant human histone H1, respectively, resulting from kinase assays with radiolabeled ATP. Asterisks indicate varying amounts of cyclins that can be in the reaction due to purification procedure. 1: without kinase, 2: CDKA;1‐CYCA2;3, 3: CDKB1;1‐CYCA2;3, 4: CDKB1;1‐CYCD2;1, 5: CDKA;1‐CYCB1;1, 6: CDKB1;1‐CYCB1;1.
Matching the previously reported localization patterns after DNA damage (Da Ines et al, 2013b), we found that RAD51 builds foci in the nuclei of root cells after treatment with cisplatin as seen with an antibody raised against RAD51 (Fig 5D and E). These foci depend on the presence of the checkpoint kinase ATM and to a lesser degree on ATR activity while they are absent, as expected, in the rad51 mutant (Fig 5D). In accordance with their key role in HR, we found that RAD51 foci are strongly reduced in cycb1;1 cycb1;3 double mutants. A reduction of RAD51 foci was even more pronounced in the cdkb1;1 cdkb1;2 double mutant consistent with its severe mutant phenotype on cisplatin‐containing media (Fig 5D and E).
Next, we asked whether RAD51 could be a direct target of CDKB1‐CYCB1 complexes. To this end, we expressed and purified from bacterial extracts next to CDKB1;1‐CYCB1;1 several other CDK‐cyclin complexes as controls, that is, CDKA;1‐CYCA2;3, CDKA;1‐CYCB1;1, CDKB1;1‐CYCA2;3, CDKB1;1‐CYCD2;1 (Fig 5F). Protein blots confirmed that all complexes contained similar amount of CDKs (Fig 5F). All kinase reactions were incubated with comparable amounts of RAD51 and histone H1 as an alternative substrate.
In general, all CDK complexes tested here were active although some complexes had a much higher activity against histone H1 than others with CDKB1;1‐CYCA2;3 and CDKB1;1‐CYCB1;1 performing the best and CDKA;1‐CYCA2;3 the poorest (Fig 5F). Importantly, we found that CDKB1;1‐CYCB1;1 could phosphorylate RAD51 in vitro. Moreover, the substrate preference toward RAD51 was different with CDKA;1‐CYCB1;1 and CDKB1;1‐CYCB1;1 now having the highest activity (Fig 5F). Thus, it appears that especially CYCB1;1‐containing complexes are active against RAD51.
Currently, it remains difficult to further test the functional relevance of RAD51 phosphorylation by CDKB1‐CYCB1 complexes since a genomic RAD51 reporter was not functional during DNA damage response (Da Ines et al, 2013b). Nevertheless, the in vitro phosphorylation identified here suggests a direct regulation of RAD51 in vivo by mitotic kinases and corroborates the conclusion that CDKB1 and CYCB1 act together in the control of HR in Arabidopsis.
CYCB1;1 upregulation upon DNA damage is directly controlled by SOG1
Seeing that CDKB1 and CYCB1 are involved in the control of HR, we explored next how this complex is regulated under DNA damage conditions. To obtain a cellular resolution, we created reporter lines for all four CYCB1 genes. For this purpose, the N‐terminal parts of the respective cyclin (CYCB1;1 to CYCB1;4) were cloned, including the destruction box that was fused to GFP. These reporter genes were placed under the control of ~1.2 kb 5′ region of the respective cyclin. We found more cells expressing GFP in the root tips of the CYCB1;1 reporter line compared to plants carrying the reporter grown on medium without cisplatin (Fig 6A and D). Noteworthy, only cells in the root tip, that is, cells with mitotic potential, showed the accumulation of the CYCB1;1 reporter. This effect was specific to CYCB1;1 as the other B1‐type cyclins, CYCB1;2, CYCB1;3, and CYCB1;4 were not upregulated based on our reporter lines (Figs 6B and E, and EV8).
Figure 6. CYCB1;1 is upregulated after cisplatin treatment.
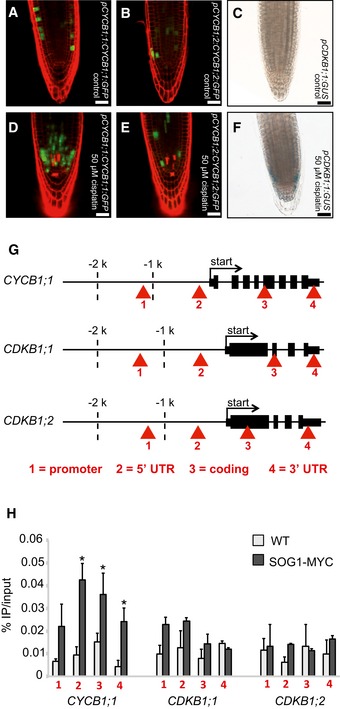
- Transgenic Arabidopsis plants harboring the CYCB1;1 promoter and a GFP fused to the N‐terminal part of CYCB1;1 including the destruction box grown on control plates and imaged 5 days after germination.
- Transgenic Arabidopsis plants harboring the CYCB1;2 promoter and a GFP fused to the N‐terminal part of CYCB1;2 including the destruction box grown on control plates and imaged 5 days after germination.
- Transgenic Arabidopsis plants harboring CDKB1;1 promoter fused to GUS grown on control plates and were stained and imaged 5 days after germination.
- Transgenic Arabidopsis plants harboring the CYCB1;1 promoter and a GFP fused to the N‐terminal part of CYCB1;1 including the destruction box germinated on control plates and were transferred 4 days after germination to plates supplemented with 50 μM cisplatin and were imaged 24 h after drug application.
- Transgenic Arabidopsis plants harboring the CYCB1;2 promoter and a GFP fused to the N‐terminal part of CYCB1;2 including the destruction box germinated on control plates and were transferred 4 days after germination to plates supplemented with 50 μM cisplatin and were imaged 24 h after drug application.
- Transgenic Arabidopsis plants harboring CDKB1;1 promoter fused to GUS germinated on control plates and were transferred 4 days after germination on plates supplemented with 50 μM cisplatin and were stained and imaged 24 h after drug application.
- Structure of genes tested by ChIP with an anti‐MYC antibody in PRO SOG1 : SOG1‐Myc and wild‐type plants. A total of four regions were tested as indicated by arrowheads. Asterisks indicate significant differences within a 5% confidence interval (Student's t‐test).
- Chromatin immunoprecipitation (ChIP) of wild‐type plants and PRO SOG1 : SOG1‐Myc lines and anti‐MYC antibody. The promoter region of CYCB1;1 is enriched in SOG1‐Myc after cisplatin treatment. Red arrowheads indicate the primer binding sites for PCR.
Figure EV8. CYCB1;3:GFP and CYCB1;4:GFP are not upregulated after cisplatin treatment.
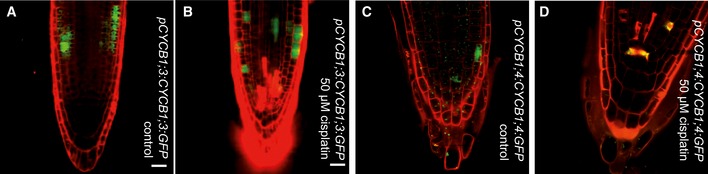
- Transgenic Arabidopsis plants harboring the CYCB1;3 promoter fused to GFP grown on control plates and imaged 4 days after germination.
- Transgenic Arabidopsis plants harboring the CYCB1;3 promoter fused to GFP germinated on control plates and were transferred 3 days after germination to plates supplemented with 50 μM cisplatin and were imaged 24 h after drug application.
- Transgenic Arabidopsis plants harboring the CYCB1;4 promoter fused to GFP grown on control plates and imaged 4 days after germination.
- Transgenic Arabidopsis plants harboring the CYCB1;4 promoter fused to GFP germinated on control plates and were transferred 3 days after germination to plates supplemented with 50 μM cisplatin and were imaged 24 h after drug application.
The fact that the CYCB1;2, CYCB1;3, and CYCB1;4, although showing the typical patchy pattern of mitotically expressed genes, were not altered in their expression pattern already indicated that the upregulation of CYCB1;1 cannot be largely due to a cell‐cycle phase effect, that is, a potential enrichment of G2 cells due to an active G2‐M checkpoint. This conclusion was consistent with our flow cytometry analyses of wild‐type plants on media with and without cisplatin and in comparison with cycb1;1 cycb1;3 double mutants (Fig EV5). Next, we analyzed the expression of CDKB1;1 using a previously generated promoter reporter line (Boudolf et al, 2004). Similar to CYCB1;1, we found that the promoter of CDKB1;1 is activated upon cisplatin treatment (Fig 6C and F).
SOG1 is a central regulator of DNA damage response in plants that mediates checkpoint signaling (Yoshiyama et al, 2009, 2013). Previously, it was found that CYCB1;1 upregulation depends on ATM and SOG1 (Culligan et al, 2006; Yoshiyama et al, 2009). To test therefore whether CYCB1;1 and CDKB1s are directly regulated by SOG1, we performed ChIP experiments with plants expressing a tagged SOG1 version grown on cisplatin. While we could not detect SOG1 on the CDKB1;1 nor CDKB1;2 genomic region, SOG1 was found to bind to the CYCB1;1 genomic region, especially the 5′ and 3′ UTR (Fig 6G and H). Thus, we conclude that SOG1 directly controls CYCB1;1 expression where CDKB1 appears to be controlled by another, yet unknown pathway.
Genetic interaction between NHEJ and HR pathways
The transcriptional upregulation of CYCB1;1 not only on cisplatin but also on medium containing other DNA‐damaging drugs raised the hypothesis that CYCB1;1 also promotes HR in response to DSB formation. A second hint for a role of HR in DSB repair comes from the observation that the cdkb1;1 cdkb1;2 double mutant was also sensitive to BLM (see above, Fig 4F and L). In addition, mutants in the plant RAD51 family are well known to be sensitive to DSB‐inducing drugs such as BLM and MMC (Bleuyard et al, 2005; Da Ines et al, 2013b; Wang et al, 2014). Previous studies have shown that NHEJ is the most prominent repair mechanism in plants and hence a role of CDKB1‐CYCB1 complexes in DSB repair might be largely masked by a very efficient execution of NHEJ (Knoll et al, 2014). We therefore asked which role CYCB1‐mediated HR plays in a genetic background in which the NHEJ pathway is severely compromised, that is, in mutants of the DNA binding protein KU70 that is required during NHEJ. Therefore, ku70 was introgressed into cycb1 mutants (Fig EV2) resulting in lines compromised in both NHEJ and presumably also in HR. Root growth of cycb1;1 cycb1;2 ku70 on medium without BLM was similar to cycb1;1 cycb1;2 (Fig 7A and B). The cycb1;1 ku70 and cycb1;2 ku70 double mutants were as sensitive to BLM as ku70. However, the generated triple mutant cycb1;1 cycb1;2 ku70 was much more sensitive to BLM than ku70 by itself with roots growing approximately only one‐tenth as long as the wild‐type control plants (Fig 7B, C and E). These results suggest that some BLM‐induced damage in ku70 is repaired by homologous recombination in a CYCB1‐dependent manner. Thus, the presence of KU70 might drive repair toward NHEJ but a HR pathway is activated at the same time and functions in parallel to back up NHEJ. Likely, this setup also contributes to the power of DNA repair in plants.
Figure 7. Mutants of cycb1 ku70 are hypersensitive to BLM: genetic interaction between NHEJ und HR.
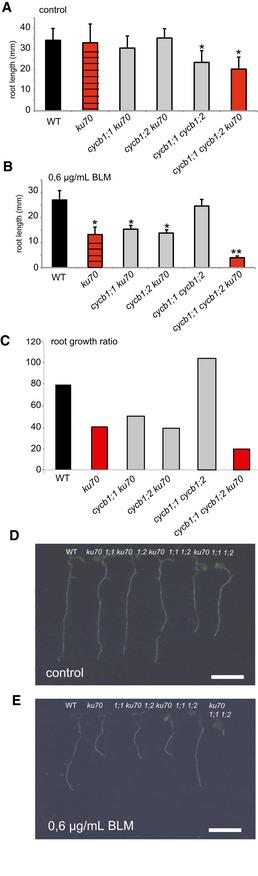
-
A, BThe wild type, ku70, the double mutants cycb1;1 ku70, cycb1;2 ku70, and cycb1;1 cycb1;2, and the triple mutant cycb1;1 cycb1;2 ku70 were grown on control plates (A) or plates containing 0.6 μg/ml BLM (B) and root lengths were measured 10 days after germination.
-
CGraph represents the ratio of the mean growth rate on 0.6 μg/ml BLM compared to control experiments on plates lacking BLM for the wild type, ku70, cycb1;1 ku70, cycb1;2 ku70, and cycb1;1 cycb1;2, and the triple mutant cycb1;1 cycb1;2 ku70.
-
D, EImage shows the wild type, ku70, the double mutants cycb1;1 ku70, cycb1;2 ku70, and cycb1;1 cycb1;2, and the triple mutant cycb1;1 cycb1;2 ku70 (from left to right) grown on control plates (D) or plates containing 0.6 μg/ml BLM (E) 10 days after germination.
Discussion
In this study, we have shown that B1‐type cyclins together with B1‐type CDKs play a major role in HR‐dependent DSB repair in plants. Our work assigns a new and important function to B1‐type CDKs, which have been previously thought to just act as auxiliary kinases with a minor and largely redundant role with the major cell division kinase CDKA;1. Seeing the impact of CDKB1s during DNA damage, as visualized by the growth retardation, the accumulation of gamma‐H2AX foci, and the pronounced formation of a tail of fragmented DNA in comet assays of the mutant, puts these kinases in the group of major DNA damage response regulators in Arabidopsis such as ATM, ATR, and SOG1 (Culligan et al, 2004, 2006; Yoshiyama et al, 2009).
Inhibition versus activation of CDKs under DNA damage
The requirement of CDK activity for DNA damage pathways stands in apparent contradiction to the general theme in DNA damage repair to arrest cell division. Indeed, CycB1 expression was reported to be suppressed in HeLa cells after DNA damage resulting in a G2 delay (Muschel et al, 1991; Maity et al, 1995). Conversely, DNA damage response has been found to be repressed by high levels of Cdk1 activity in dividing mammalian cells (Zhang et al, 2011). However, a key role for active CDK complexes in DNA damage response matches recent reports in yeast and metazoans (Wohlbold & Fisher, 2009; Yata & Esashi, 2009; Trovesi et al, 2013).
In S. cerevisiae, cell‐cycle arrest is executed with high CDC28p activity (Sorger & Murray, 1992; Enserink et al, 2009). Furthermore, high CDK activity is needed in G2 to concurrently promote HR and to repress NHEJ (Zhang et al, 2009). In particular, it was shown that CDC28p is required for processing of DNA ends to produce single‐stranded DNA essential for HR (Aylon et al, 2004; Ira et al, 2004). Conversely, S. cerevisiae cells with reduced CDK activity are highly sensitive to DNA‐damaging agents (Enserink et al, 2009). Thus, it appears that DNA damage repair and the subsequent choice of repair pathways in S. cerevisiae is dependent on the activity level of CDC28p as HR can operate exclusively after DNA replication in S and G2 phase of the cell cycle (Zhang et al, 2009). However, S. cerevisiae has only one cell‐cycle CDK and it is still an open question how inhibition of mitosis can be reconciled with an activation of CDKs to promote DNA damage repair.
In metazoans, CDK activity has also been found to be important for DNA repair and the application of chemical CDK inhibitors such as roscovitine was reported to increase sensitivity of cells to DNA‐damaging compounds such as ionizing radiation and cisplatin (Ongkeko et al, 1995; Maggiorella et al, 2003). The closest homologs of CDC28p in metazoans are Cdk1 and Cdk2. The general dogma is that Cdk2 complexes promote entry and progression through S phase while Cdk1 controls mitosis although Cdk1 can almost completely compensate for the loss of Cdk2 (Santamaría et al, 2007; Malumbres et al, 2009). In particular, Cdk2 was found to play an active role in DNA repair (Deans et al, 2006; Wohlbold et al, 2012). Likewise, CycA1 and CycA2, two cyclin partners of Cdk2, have been reported to promote DNA DSB repair by HR in mice (Müller‐Tidow et al, 2004). However, Cdk2 is inhibited by the CDK inhibitor p21 after occurrence of DNA damage (Bartek & Lukas, 2001). Thus, it is not clear how Cdk2 complexes could execute DNA repair when they are in an inhibited state as a response to DNA damage. It is also not clear how HR can be promoted in late G2 and early M phase when presumably only Cdk1 is active.
Arabidopsis has only one Cdk1/Cdk2 homolog, namely CDKA;1, which also contains functional aspects of both CDKs in metazoans, that is, CDKA;1 function in S and M phase control (Nowack et al, 2012). Here, we have shown that plants with a reduced activity level of CDKA;1 are neither hypersensitive to BLM nor cisplatin. In addition, we have previously found that a weak loss‐of‐function allele of CDKA;1 also does not sensitize plants to HU (Dissmeyer et al, 2009). Although we cannot fully exclude a role of CDKA;1 in DNA repair, our combined data suggest that this kinase is largely not required for a proper DNA damage response in Arabidopsis. Instead, we found that especially CDKB1 complexes control HR.
Remarkably, A2‐type cyclins, which are well‐known partners of CDKB1 activity, for instance, during stomata development (Vanneste et al, 2011), were not found to play a major role during DNA repair on cisplatin media in Arabidopsis. Instead, we found that B1‐type cyclins, which can also form an active complex with CDKB1s, are hypersensitive to cisplatin. However, Arabidopsis has eight D‐type cyclins next to six further A‐ and six B‐type cyclins that all belong to the core cell division machinery. The slightly weaker mutant phenotype of cycb1 mutants in comparison with the double mutant cdkb1;1 cdkb1;2 suggests that one or a combination of different cyclins acts partially redundantly with the class of B1‐type cyclins. Further studies are required to work out the minor role of other cyclins in HR.
B1‐type cyclins also form active complexes with CDKB2 (Harashima & Schnittger, 2012). Based on their expression pattern and the phenotype of their downregulation, it is conceivable that CDKB2s are the major mitotic regulators in Arabidopsis (Menges et al, 2003; Andersen et al, 2008). It has been observed that CDKB2s are degraded and transcriptionally downregulated upon DNA damage in Arabidopsis (Adachi et al, 2011). However, whether the function and regulation of CDKB2 is conserved in plants is not clear yet since the only member of the CDKB2 group in rice, CDKB2;1, accumulates upon DNA damage (Endo et al, 2012). Moreover, CDKB2;1 knockdown lines in rice were more sensitive to radiation possibly hinting at a conserved role for mitotic CDKs in plants for HR. However, it remains to be seen whether CDKB2;1 from rice is functionally more related to CDKB2s than CDKB1s from Arabidopsis, especially since expression of CDKB2;1 in rice is not restricted to mitosis resembling more the CDKB1 expression pattern in Arabidopsis (Menges et al, 2003; Endo et al, 2012).
In Arabidopsis, the transcriptional repression of CDKB2s depends on the putative transcription factor SOG1 that plays a key role in DNA damage response in Arabidopsis (Yoshiyama et al, 2009). At the same time, it was previously found that upregulation of CYCB1;1 after DNA damage requires SOG1 (Yoshiyama et al, 2009). Here, we have shown that SOG1 binds to the promoter of CYCB1;1 and hence appears to directly activate its expression presumably promoting CDKB1 activity and the execution of HR.
Taken together, we propose a division of labor among the CDKs in Arabidopsis (Fig 8). The major mitotic force represented by CDKB2 and possible other kinases might be shut down in a SOG1‐dependent mechanism, while SOG1 promotes the expression of CYCB1 and hence stimulates the activity of CDKB1 complexes required for HR.
Figure 8. Division of labor model of the regulation of cell proliferation and DNA damage response in Arabidopsis .
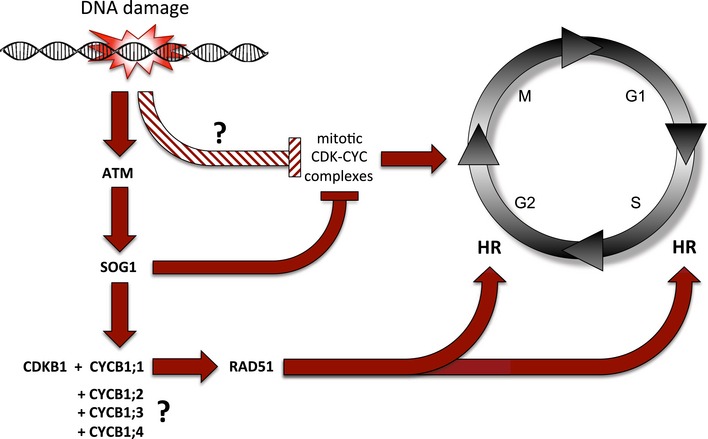
DNA damage, for example induced by chemical mutagens, is followed by the activation of the checkpoint kinase ATM (ataxia‐telangiectasia mutated) that activates the transcription factor SOG1 (SUPPRESSOR OF GAMMA RESPONSE 1). On the one hand, SOG1 represses, directly or indirectly, the expression CDKB2 and possible other CDKs as the major driving force of mitosis to allow a cell time for DNA repair. On the other hand, SOG1 directly binds to the promoter of CYCB1;1 and activates its expression. CYCB1;1 builds an active complex together with CDKB1. This complex phosphorylates the DNA binding protein RAD51 (RADIATION SENSITIVE 1) that gets recruited to the DNA damage site. This cascade is required for HR in S and G2 phases of the cell cycle in meristematic cells. To trigger this response, the action of all four B1‐type cyclins is necessary, possibly by providing a threshold of mitotic CDK activity that then gets amplified through SOG1‐dependent stimulation of CYCB1;1 expression.
Targets of CDK‐CYC complexes in HR versus NHEJ
How do active CDK complexes, such as CDKB1‐CYCB1, control HR? A first step to answer this question is the identification of substrates of CDK complexes during DNA damage conditions. An early and decisive step in HR is the resection of the 3′ ends at the site of the DSB to allow subsequent strand invasion of the damaged into the undamaged chromatid. In animals and yeast, it has been found that CDKs phosphorylate proteins in the MRN complex that process 3′ ends, for example, Nbs1, and proteins that work in concert with this complex, such as the nuclease Sae2/CtIP/Com1 (Huertas et al, 2008; Huertas & Jackson, 2009; Wohlbold et al, 2012; Simoneau et al, 2014). Conversely, mutants of Nbs1 in which the phosphorylation site of Cdk2 was eliminated were hypersensitive to radiation in a similar manner as cells in which Cdk2 was chemically inhibited (Wohlbold et al, 2012).
Here, we identified RAD51 as a possible target of DNA damage‐induced CDK activity. To our knowledge, RAD51 has not been identified outside of plants as a possible target of CDKs under DNA damage conditions. However, animal RAD51 also contains a consensus CDK phosphorylation site S/T‐X‐R/K and it has been previously observed in animals and yeast that the formation of Rad51 foci after DNA damage indirectly depends on CDK activity via BRCA1 and BRCA2 (Scully et al, 1997; Ira et al, 2004; Johnson et al, 2009; Quennet et al, 2011). At least in plants, recruitment of RAD51 to damaged DNA might not only be an indication for HR but could directly be dependent on CDK activity.
Arabidopsis also contains COM1 and NBS1 homologs and it remains to be seen whether these proteins are also phosphorylated by CDKB1 complexes upon DNA damage, at least both proteins contain several CDK consensus phosphorylation sites. Hence, one alternative possibility to explain the failure to recruit RAD51 in cdkb1 mutants is an insufficient resection of DSBs due to compromised MRN and/or COM1 activity. In addition, there could also be other plant‐specific phospho‐targets of CDKB1 during HR. The few targets of CDK action identified in Arabidopsis so far suggest a rather large number of species‐specific CDK targets (Pusch et al, 2012). Hence, an unbiased forward approach to identify additional CDK targets under DNA damage conditions might be powerful to further understand how HR is controlled by CDKs.
Several components of the NHEJ pathway have also been identified as targets of CDKs. However, CDK phosphorylation inhibits the recruitment of NHEJ factors, for instance, YKu70 (the yeast KU70 homolog) and Lif1 to DNA breaks in S. cerevisiae (Zhang et al, 2009) and XRCC4/XLF1 in S. pombe (Hentges et al, 2014). Thus, there appears to a reciprocal relationship between these two repair pathways where NHEJ is high in G1 and low in G2 phase, whereas HR is not possible in G1 and high in G2 phase (Ferreira & Cooper, 2004). Whether such an antagonistic relationship between these two different classes of DNA repair mechanisms exists in plants as well needs to be shown.
Previous experiments suggested that NHEJ and not HR is the predominant type of DNA repair in plants, a situation reminiscent to mammals but different to yeast in which HR is preferred over NHEJ (Puchta et al, 1996; Kempin et al, 1997; Gisler et al, 2002). However, our work clearly indicates that HR is executed under DNA damage conditions in plants and depends on the presence of CDKB1s and CYCB1s. This is consistent with previous studies that have reported an upregulation of CYCB1;1 after DNA damage (Chen et al, 2003; Culligan et al, 2006; Ricaud et al, 2007). Our finding that CYCB1;1 is upregulated outside of mitosis but still limited to the meristematic region of the root where cells are in a proliferative mode suggests a developmental stage dependent choice of DNA repair pathways. We postulate that cells with mitotic potential prefer the error‐free HR over NHEJ after progression through S phase.
The execution of HR might be very sensitive to the dosage of mitotic activity as seen by the reduced growth and decreased blue spots in our recombination assays already in all single cycb1 mutants. We speculate that a threshold level of mitotic activity is needed for HR after DNA damage and all CYCB1 proteins contribute to this threshold in a non‐redundant manner. A second, yet not mutually exclusive explanation for the sensitivity of all cyclins could by that different CYCB1 complexes have to some degree non‐overlapping substrates that are still required for HR, for example, RAD51 by CDKB1s‐CYCB1;1 and a different substrate, possibly COM1, by another CDKB1s‐CYCB1 complex. Alternatively, HR substrates might require the concomitant phosphorylation of different CDKB1‐CYCB1 complexes.
Nonetheless, loss of mitotic activity in cycb1 or cdkb1 mutants does not immediately result in reduced repair activity and reduced growth upon the formation of DSBs presumably due to remaining NHEJ activity as seen by the unperturbed growth of cycb1 mutants and only a slight reduction of cdkb1 mutants on BLM‐containing media. Such a parallel role of NHEJ and HR matches the observation that the concomitant inactivation of both NHEJ and HR as presented in ku70 cycb1;1 cycb1;2 triple mutant results in severe growth reduction on BLM.
An apparent dominance of NHEJ over HR could further be explained if terminal differentiated cells or endoreplicating cells that comprise the vast majority of all cells of a plant might preferentially execute NHEJ. One reason for this could simply be the fact that mitotic genes are generally inactive in differentiated cells and hence mitotic CDK activity required for HR is missing. The importance of the developmental state in DNA damage response has already been observed by the preferential cell death of stem cells that is not found in other cells of the root after mild induction of DSBs (Fulcher & Sablowski, 2009).
However, DNA repair in plants appears to be even more complex. The fact that cycb1;1 cycb1;2 ku70 showed severe reduction in root growth on BLM but not a complete growth stagnancy, suggests that a third pathway might back up the DNA repair to a certain extent. This could be the KU70‐independent NHEJ repair pathway (Charbonnel et al, 2010), which depends on the action of XRCC1 (XRAY REPAIR CROSS‐COMPLEMENTATION PROTEIN 1). KU80 has a similar function as KU70 and xrcc1 ku80 double mutants display more defects after irradiation than the single mutants, which leads to the conclusion that both proteins act in independent repair pathways (Charbonnel et al, 2010). Most likely, this pathway remains functional in cycb1;1 cycb1;2 ku70 after BLM treatment but has only a minor contribution due to the severe root growth defects of the triple mutant. Further work is now required to understand how an XRCC1‐dependent pathway is integrated with HR and NHEJ. Likely, the interplay of all these pathways is the reason for the enormous power of plant DNA repair systems.
Materials and Methods
Plant material and growth conditions
The Arabidopsis thaliana plants used in this study were either grown on soil (16 h light) or in vitro on half‐strength (½) Murashige and Skoog (MS) medium (Sigma‐Aldrich) containing 0.5% sucrose (16 h light) in a growth chamber. The accessions Columbia (Col‐0) and Nossen (No‐0) were used as the wild‐type control. cycb1;1 and cycb1;2 T‐DNA insertion lines were isolated from the Koncz collection (Rios et al, 2002). T‐DNA insertion line cycb1;3 (pst15850) was obtained from the RIKEN collection (Yokohama, Japan) and cycb1;4 from the GABI‐KAT collection (Bielefeld, Germany) (Kleinboelting et al, 2012). The ku70 (Riha et al, 2002), wee1 (De Schutter et al, 2007), and rad51 (Li et al, 2004) mutants were described previously. The reporter line uidA/IC9C was used as described in Molinier et al (2004) and the pCDKB1;1:GUS line as in Boudolf et al (2004). The mutant CDKA;1 T14D;Y15E is published in Dissmeyer et al (2009), the cdkb1;1 cdkb1;2 double mutant in Nowack et al (2012). All genotypes were determined by PCR and primers are indicated in Table EV1.
Root growth analysis
Elongation of the roots was marked daily for 10 days after germination on vertical plates in 16 h/8 h growth chamber and measured with ImageJ. Root length was measured from the root tip to the root–hypocotyl border. Three biological replicates, each containing at least 15 plants, were analyzed. The mean of the root length of each individual experiment was determined and again averaged for the three biological replicates. In order to measure root growth on DNA damage‐inducing media, plants germinated on 1 mM hydroxyurea (Sigma‐Aldrich) or 0.6 μg/ml bleomycin (Duchefa) and were measured daily for 10 days. For analyzing the sensitivity on cisplatin, plants were sown on ½ Murashige and Skoog medium with 0.5% sugar and transferred to either control plates or plates containing 15 or 30 μM cisplatin (Sigma‐Aldrich) 3 days after germination. Roots were measured 3 days after transfer (figures show elongation of the roots within 3 days).
Cloning of CYCB1 marker lines
The fragments were first cloned in pGEM‐T easy and sequence validated, then excised and cloned into pTV50‐GUS or pTV50‐GFP: These two vectors are derived from pBIB (Becker, 1990): First, the NOS terminator in pBIB was replaced by the OCS terminator, and then all sequences between the unique HindIII and SacI sites in pBIB were excised and replaced by the modified uidA gene from pRAJ260 or GFP6, respectively, with the following polylinker preceding the respective start codon: aagcttgaggtcgactctagA, giving rise to pTV50‐GUS or pTV50‐GFP, respectively. Cyclin promoter‐gene fragments were cloned in frame using either SalI or HindIII on the 5′ end and an XbaI‐compatible restriction site at the 3′ end. Primer sequences are indicated in Table EV1. The CYCB1;1 construct was described in Ubeda‐Tomás et al (2009).
Root cell wall staining
Entire seedlings were stained with propidium iodide (Invitrogen, 1 mg/ml, 100× dilution) in H2O for 3–4 min and rinsed afterward two times in H2O.
Confocal laser scanning microscopy
Confocal laser scanning microscopy was performed on an inverted Zeiss LSM 510 confocal microscope. Excitation and detection windows were set as follows: GFP excitation at 488 nm, emission at 500–600 nm; propidium iodide excitation at 488 nm, emission at 500–550 nm.
Homologous recombination assay
For the homologous recombination assay, all cycb1 single mutants were crossed to the IC9C reporter line (kindly provided by Holger Puchta, KIT, Karlsruhe, Germany) and double homozygous lines were obtained in the F3 generation. Plants were germinated on ½ Murashige and Skoog medium with 0.5% sugar. Five days after germination seedlings were transferred to plates containing either 15 or 30 μM cisplatin (Sigma‐Aldrich) and control plates without cisplatin. Three days after transferring, seedlings were incubated in staining solution for 48 h and afterward destained in 70% ethanol at 60–70°C. Staining solution for 5 ml: 100 μl of 10% Triton X‐100, 250 μl 1M NaPO4 (pH 7.2, make 1 M NaH2PO4 and titrate with NaOH), 100 μl 100 mM potassium ferrocyanide, 100 μl potassium ferricyanide, 400 μl 25 mM X‐Gluc, 4,050 μl dH2O. Blue sectors were counted using a binocular Leica S4E. Images of leaves with blue spots were taken with an Olympus BX51 light microscope.
Immunostaining
Ten‐day‐old seedlings were treated with 50 μM cisplatin for 2 h. γ‐H2AX immunostaining of root tip spreads was performed as described earlier (Friesner et al, 2005). A rabbit anti‐plant γ‐H2AX antibody (kindly provided by Charles White, CNRS, Clermont‐Ferrand, France) was used in a 1:600 dilution (Charbonnel et al, 2010). As secondary antibody, a goat Alexa Fluor 488 anti‐rabbit (Life Technologies, Carlsbad, CA, USA) was used in a 1:300 dilution. Imaging of the nuclei was done with a LSM700 confocal microscope (Carl Zeiss, Jena, Germany). Immunostaining of RAD51 foci was performed with the Rad51 (H‐92) sc‐8349 antibody (Santa Cruz Biotechnology, Texas, USA) with a dilution of 1:400. A goat Alexa Fluor 488 anti‐rabbit (Life Technologies, Carlsbad, CA, USA) was used in a 1:300 dilution as a secondary antibody.
Comet assay
The evaluation of DNA damage in cisplatin treated plants was done by an N‐methyl‐N‐nitrosourea (N/N) comet assay. Therefore, seedlings were grown for 21 days under sterile conditions on ½ MS medium, 0.5% sugar. The plantlets were transferred to ½ MS liquid medium (control) or ½ MS liquid medium containing 50 μM cisplatin. After 1 h of incubation, a fraction of the cisplatin treated plants were separated. The remaining plants were shortly dried on paper towels and immediately frozen in liquid nitrogen. The separated plants were washed three times with ½ MS and transferred to ½ MS liquid medium without cisplatin for recovery. After 30 min of incubation, these plants were also briefly dried and frozen. The preparation of the comet slides was performed according to Menke et al (2001) and stained with 3× GelRed Nucleic Acid stain (Biotium, Hayward, CA, USA) diluted in 0.1 M NaCl. The comets were observed and images were taken on an AXIO Imager Z1 fluorescence microscope with an AXIOCam MRm (Carl Zeiss, Jena, Germany). The analysis of the images was done utilizing the TriTek Comet Score software and 200 comets per sample were measured.
Ploidy analysis
Root tips of 5‐day‐old plants treated and untreated with 50 μM cisplatin for 24 h were chopped with a razor blade in 200 ml of Cystain UV Precise P nuclei extraction buffer, supplemented with 800 ml staining buffer (Partec). The mix was filtered through a 50‐mm column and read by the Cyflow MB flow cytometer (Partec). Nuclei were analyzed using the Cyflogic software.
Kinase assay
To clone RAD51, total RNA was extracted from inflorescences by using the NucleoSpin RNA plant kit (Macherey‐Nagel). First‐stranded cDNA was synthesized by SuperScript III reverse transcriptase (Life Technologies) with oligo dT‐AP_M13 according to the manufacturer's instruction. RAD51 cDNA was amplified first with primers RAD51_s1 and M13 forward, followed by primers RAD51_s2 and RAD51_as with Phusion DNA polymerase (Thermo Scientific). The PCR product was cloned into pJET1.2 (Thermo Scientific), followed by sequence confirmation. After RAD51 was subcloned into pDONR223 (Invitrogen), a recombination reaction was performed between the resulting entry clone and a destination vector pMGWA (Busso et al, 2005). CDKB1;1‐CYCB1;1 complexes were expressed and purified as described previously (Harashima & Schnittger, 2012). To express recombinant proteins, E. coli BL21‐AI cells (Invitrogen) were transformed with the resulting vector. Escherichia coli cells were grown in LB medium containing 100 mg/l ampicillin at 37°C until OD600 = 0.6 and the production of the fusion protein was induced by adding 0.3 mM IPTG (Nacalai Tesque) and 0.2%(w/v) L‐arabinose (Wako) overnight at 18°C. Cells were harvested by centrifugation and re‐suspended in Ni‐NTA binding buffer (50 mM NaH2PO4, 100 mM NaCl, 10% (v/v) glycerol, 25 mM imidazole, pH 8.0) and lysed by sonication (Digital Sonifier 450D, BRANSON). After addition of Triton X‐100 to 0.2% (w/v), the cell slurry was incubated at 4°C and clarified by centrifugation. The supernatant was passed through a column with Ni‐NTA resin (Qiagen), which was washed sequentially with Ni‐NTA binding buffer, and eluted with Ni‐NTA elution buffer (Ni‐NTA binding buffer containing 200 mM imidazole). The eluate was sequentially purified with a column packed with amylose resin (NEB), which had been equilibrated with Ni‐NTA binding buffer. The column was washed with Ni‐NTA binding buffer followed by kinase buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 1 mM EGTA). MBP fusion proteins were eluted with kinase buffer containing 10 mM maltose (Wako).
Kinase reactions were normalized so that equal amount of the respective CDKs (and hence functional CDK‐cyclin complexes) were present in the reaction mixture. The assays were carried out with equal amounts of MBP‐RAD51‐His6 as a substrate and 92.5 kBq of [γ‐32P]ATP (PerkinElmer) as previously described (Harashima & Sekine, 2011; Harashima & Schnittger, 2012). After the kinase reaction, proteins were subjected to SDS–PAGE on a 10% TGX gel (Bio‐Rad) and stained with Bio‐Safe Coomassie G‐250 Stain (Bio‐Rad). The gel was dried with HydroTech Gel Drying System (Bio‐Rad). The radioisotopic signals were detected with Typhoon FLA 7000 (GE Healthcare).
ChIP
ChIP experiments were carried out as previously described with minor modifications (Saleh et al, 2008). Seeds of PRO SOG1 :SOG1‐Myc, described in Yoshiyama et al (2013), were germinated and cultured in ½ MS medium containing 0.5% sucrose (pH 5.7) under continuous light at 22°C with gentle shaking (50 rpm). The 2‐week‐old seedlings were treated with 50 μM cisplatin for 2 h. Sonicated chromatin solution (corresponding to 0.7 g tissue) was used for immunoprecipitation with anti‐Myc antibodies (Santa Cruz Biotechnology). The ChIP products were used for real‐time quantitative PCR analysis. qPCR was performed using primers (Table EV1) on a LightCycler system (Roche) with Thunderbird SYBR qPCR Mix (Toyobo) according to the following reaction conditions: 95°C for 1 min; 70 cycles at 95°C for 10 s, at 60°C for 10 s, and at 72°C for 20 s.
qPCR
The entire inflorescences of Col‐0, Nos, cycb1;1, cycb1;2, cycb1;3, and cycb1;4 were immediately frozen in liquid nitrogen after collection and stored temporarily at −80°C. RNA was extracted using NucleoSpin RNA Kit (MACHEREY‐NAGEL). RNA concentration and purity was tested using nanodrop‐photometric quantification (Thermo Scientific). RNA integrity was verified by running 1 μl of total RNA on 1.5% agarose TBE gels to detect the 28S and 16S rRNA bands. 1 μg of total RNA was processed to obtain cDNA using polyT primer and SuperScript III RNase H reverse transcriptase. As negative control, all steps were followed in the same manner, except for adding the reverse transcriptase. The resulting cDNA was used for quantitative real‐time PCR (qRT–PCR) using the Roche LightCycler 480 system. Three to four biological with three technical replicates each were processed. Cq calling was done using the Second Derivative Maximum method. Reaction‐specific efficiencies were deduced using LinRegPCR 7.4 (http://LinRegPCR.nl). Data were quality‐controlled, normalized against 3 reference genes, and statistically evaluated (unpaired t‐test) using qbasePLUS 2.3 (http://biogazelle-qbaseplus.software.informer.com/2.3/).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CYCB1;1 (AT4G37490); CYCB1;2 (AT5G06150); CYCB1;3 (AT3G11520); CYCB1;4 (AT2G26760); RAD51 (AT5G20850); KU70 (AT1G16970); WEE1 (AT1G02970); CYCA2;2 (AT5G11300); CYCA2;3 (AT1G15570); CYCA2;4 (AT1G80370); CDKB1;1 (AT3G54180); CDKB1;2 (AT2G38620); CDKA;1 (AT3G48750); SOG1 (AT1G25580).
Author contributions
AKW, SB, HH, FR, NT, CK, PD, MU, and AS conceived and designed the experiments. AKW, SB, HH FR, NT, JF, YG, and GP performed the experiments. DVD, KS, CK, PD, MU, and AS contributed material and reagents. AKW, SB, HH, NT, MH, PD, MU, and AS analyzed the data. AKW and AS wrote the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We thank Holger Puchta (KIT, Karlsruhe, Germany) for the IC9C reporter line seeds, Steffen Vanneste (VIB/University of Gent, Belgium) for cyca2;2 cyca2;3 cyca2;4 triple mutant and pCDKB1;1:GUS seeds, and Bernd Reiss (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for the rad51 line. We are thanking Charles White (CNRS, Clermont‐Ferrand, France) for providing us with an anti‐plant γ‐H2AX antibody. The GABI‐Kat collection (Bielefeld, Germany) and the Koncz collection (Cologne, Germany) are acknowledged for obtaining cycb mutants. We also thank Nico Dissmeyer for critical reading and helpful comments on the article. This work was supported by a postdoctoral fellowship of the German Research Foundation (DFG) to S.B., a fellowship of the German Academic Exchange Program (DAAD) to F.R., and a grant from the German Research Foundation (DFG) (SCHN 736/2‐2), and an European Research Council starting grant to A.S.
The EMBO Journal (2016) 35: 2068–2086
See also: B Desvoyes & C Gutierrez (October 2016)
References
- Abe K, Osakabe K, Nakayama S, Endo M, Tagiri A, Todoriki S, Ichikawa H, Toki S (2005) Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol 139: 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, Kurihara D, Matsunaga S, Umeda M (2011) Programmed induction of endoreduplication by DNA double‐strand breaks in Arabidopsis . Proc Natl Acad Sci USA 108: 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S, Gallego ME, White CI (2013) Signaling of double strand breaks and deprotected telomeres in Arabidopsis . Front Plant Sci 4: 405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SU, Buechel S, Zhao Z, Ljung K, Novak O, Busch W, Schuster C, Lohmann JU (2008) Requirement of B2‐type cyclin‐dependent kinases for meristem integrity in Arabidopsis thaliana . Plant Cell 20: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double‐strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas J (2001) Pathways governing G1/S transition and their response to DNA damage. FEBS Lett 490: 117–122 [DOI] [PubMed] [Google Scholar]
- Becker D (1990) Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res 18: 203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkov AI, Paiement JP, Panasci LC, Monia BP, Chow TY (2002) An antisense oligonucleotide targeted to human Ku86 messenger RNA sensitizes M059K malignant glioma cells to ionizing radiation, bleomycin, and etoposide but not DNA cross‐linking agents. Cancer Res 62: 5888–5896 [PubMed] [Google Scholar]
- Bisova K, Krylov DM, Umen JG (2005) Genome‐wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii . Plant Physiol 137: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard JY, White CI (2004) The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J 23: 439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI (2005) Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J 41: 533–545 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Barroco R, Engler Jde A, Verkest A, Beeckman T, Naudts M, Inze D, De Veylder L (2004) B1‐type cyclin‐dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana . Plant Cell 16: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, Van Leene J, Van Den Daele H, Maes S, Van Isterdael G, Russinova E, Kondorosi E, Witters E, De Jaeger G, Inze D, De Veylder L (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray CM, West CE (2005) DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol 168: 511–528 [DOI] [PubMed] [Google Scholar]
- Busso D, Delagoutte‐Busso B, Moras D (2005) Construction of a set Gateway‐based destination vectors for high‐throughput cloning and expression screening in Escherichia coli . Anal Biochem 343: 313–321 [DOI] [PubMed] [Google Scholar]
- Charbonnel C, Gallego ME, White CI (2010) Xrcc1‐dependent and Ku‐dependent DNA double‐strand break repair kinetics in Arabidopsis plants. Plant J 64: 280–290 [DOI] [PubMed] [Google Scholar]
- Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H (2003) The transcriptional response of Arabidopsis to genotoxic stress – a high‐density colony array study (HDCA). Plant J 35: 771–786 [DOI] [PubMed] [Google Scholar]
- Cools T, Iantcheva A, Weimer AK, Boens S, Takahashi N, Maes S, Van den Daele H, Van Isterdael G, Schnittger A, De Veylder L (2011) The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crul M, van Waardenburg RC, Bocxe S, van Eijndhoven MA, Pluim D, Beijnen JH, Schellens JH (2003) DNA repair mechanisms involved in gemcitabine cytotoxicity and in the interaction between gemcitabine and cisplatin. Biochem Pharmacol 65: 275–282 [DOI] [PubMed] [Google Scholar]
- Cruz‐Ramirez A, Diaz‐Trivino S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, Caballero‐Perez J, Horvath B, Long Y, Mahonen AP, Zhang H, Xu J, Murray JA, Benfey PN, Bako L, Maree AF et al (2012) A bistable circuit involving SCARECROW‐RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K, Tissier A, Britt A (2004) ATR regulates a G2‐phase cell‐cycle checkpoint in Arabidopsis thaliana . Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB (2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Culligan KM, Britt AB (2008) Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double‐strand breaks. Plant J 55: 629–638 [DOI] [PubMed] [Google Scholar]
- Da Ines O, Degroote F, Amiard S, Goubely C, Gallego ME, White CI (2013a) Effects of XRCC2 and RAD51B mutations on somatic and meiotic recombination in Arabidopsis thaliana . Plant J 74: 959–970 [DOI] [PubMed] [Google Scholar]
- Da Ines O, Degroote F, Goubely C, Amiard S, Gallego ME, White CI (2013b) Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet 9: e1003787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Joubes J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir S, Inze D, De Veylder L (2007) Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA (2002) Defects in interstrand cross‐link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res 30: 3848–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Deans AJ, Khanna KK, McNees CJ, Mercurio C, Heierhorst J, McArthur GA (2006) Cyclin‐dependent kinase 2 functions in normal DNA repair and is a therapeutic target in BRCA1‐deficient cancers. Cancer Res 66: 8219–8226 [DOI] [PubMed] [Google Scholar]
- Dissmeyer N, Weimer AK, Pusch S, De Schutter K, Kamei CL, Nowack M, Novak B, Duan GL, Zhu YG, De Veylder L, Schnittger A (2009) Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21: 3641–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Weimer AK, De Veylder L, Novak B, Schnittger A (2010) The regulatory network of cell‐cycle progression is fundamentally different in plants versus yeast or metazoans. Plant Signal Behav 5: 1613–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C (2014) Endocycles: a recurrent evolutionary innovation for post‐mitotic cell growth. Nat Rev Mol Cell Biol 15: 197–210 [DOI] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, Shibahara K, Abe K, Ichikawa H, Valentine L, Hohn B, Toki S (2006) Increased frequency of homologous recombination and T‐DNA integration in Arabidopsis CAF‐1 mutants. EMBO J 25: 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Nakayama S, Umeda‐Hara C, Ohtsuki N, Saika H, Umeda M, Toki S (2012) CDKB2 is involved in mitosis and DNA damage response in rice. Plant J 69: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink JM, Hombauer H, Huang ME, Kolodner RD (2009) Cdc28/Cdk1 positively and negatively affects genome stability in S. cerevisiae . J Cell Biol 185: 423–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP (2004) Two modes of DNA double‐strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev 18: 2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB (2005) Ionizing radiation‐dependent gamma‐H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3‐related. Mol Biol Cell 16: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MA, Castilla J, Alonso C, Pérez JM (2002) Novel concepts in the development of platinum antitumor drugs. Curr Med Chem Anticancer Agents 2: 539–551 [DOI] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R (2009) Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Salanoubat M, Choisne N, Tissier A (2000) An ATM homologue from Arabidopsis thaliana: complete genomic organisation and expression analysis. Nucleic Acids Res 28: 1692–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisler B, Salomon S, Puchta H (2002) The role of double‐strand break‐induced allelic homologous recombination in somatic plant cells. Plant J 32: 277–284 [DOI] [PubMed] [Google Scholar]
- Harashima H, Sekine M (2011) Measurement of plant cyclin‐dependent kinase activity using immunoprecipitation‐coupled and affinity purification‐based kinase assays and the baculovirus expression system. Methods Mol Biol 779: 65–78 [DOI] [PubMed] [Google Scholar]
- Harashima H, Schnittger A (2012) Robust reconstitution of active cell‐cycle control complexes from co‐expressed proteins in bacteria. Plant Methods 8: 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A (2013) Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 23: 345–356 [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Hentges P, Waller H, Reis CC, Ferreira MG, Doherty AJ (2014) Cdk1 restrains NHEJ through phosphorylation of XRCC4‐like factor Xlf1. Cell Rep 9: 2011–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya N, Miyagawa K (2014) Targeting DNA damage response in cancer therapy. Cancer Sci 105: 370–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortés‐Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA‐end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP (2009) Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Liu X, Gao H (2016) A DNA2 homolog is required for DNA damage repair, cell cycle regulation, and meristem maintenance in plants. Plant Physiol 171: 318–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N, Cai D, Kennedy RD, Pathania S, Arora M, Li YC, D'Andrea AD, Parvin JD, Shapiro GI (2009) Cdk1 participates in BRCA1‐dependent S phase checkpoint control in response to DNA damage. Mol Cell 35: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartalou M, Essigmann JM (2001) Mechanisms of resistance to cisplatin. Mutat Res 478: 23–43 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell‐cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Kempin SA, Liljegren SJ, Block LM, Rounsley SD, Yanofsky MF, Lam E (1997) Targeted disruption in Arabidopsis . Nature 389: 802–803 [DOI] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI‐Kat simplesearch: new features of the Arabidopsis thaliana T‐DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A, Fauser F, Puchta H (2014) DNA recombination in somatic plant cells: mechanisms and evolutionary consequences. Chromosome Res 22: 191–201 [DOI] [PubMed] [Google Scholar]
- Kuo LJ, Yang LX (2008) Gamma‐H2AX – a novel biomarker for DNA double‐strand breaks. In Vivo 22: 305–309 [PubMed] [Google Scholar]
- Li W, Chen C, Markmann‐Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang X, Lin Z, Timofejeva L, Xiao R, Makaroff CA, Ma H (2005) The AtRAD51C gene is required for normal meiotic chromosome synapsis and double‐stranded break repair in Arabidopsis . Plant Physiol 138: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H (2006) Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA 103: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang J, Miki D, Xia R, Yu W, He J, Zheng Z, Zhu JK, Gong Z (2010) DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis . Plant Cell 22: 2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggiorella L, Deutsch E, Frascogna V, Chavaudra N, Jeanson L, Milliat F, Eschwege F, Bourhis J (2003) Enhancement of radiation response by roscovitine in human breast carcinoma in vitro and in vivo . Cancer Res 63: 2513–2517 [PubMed] [Google Scholar]
- Maity A, McKenna WG, Muschel RJ (1995) Evidence for post‐transcriptional regulation of cyclin B1 mRNA in the cell cycle and following irradiation in HeLa cells. EMBO J 14: 603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, Morgan DO, Tsai LH, Wolgemuth DJ (2009) Cyclin‐dependent kinases: a family portrait. Nat Cell Biol 11: 1275–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA (2003) Genome‐wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JA (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Mengiste T, Paszkowski J (1999) Prospects for the precise engineering of plant genomes by homologous recombination. Biol Chem 380: 749–758 [DOI] [PubMed] [Google Scholar]
- Menke M, Chen I, Angelis KJ, Schubert I (2001) DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat Res 493: 87–93 [DOI] [PubMed] [Google Scholar]
- Molinier J, Ries G, Bonhoeffer S, Hohn B (2004) Interchromatid and interhomolog recombination in Arabidopsis thaliana . Plant Cell 16: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller‐Tidow C, Ji P, Diederichs S, Potratz J, Bäumer N, Köhler G, Cauvet T, Choudary C, van der Meer T, Chan WY, Nieduszynski C, Colledge WH, Carrington M, Koeffler HP, Restle A, Wiesmüller L, Sobczak‐Thépot J, Berdel WE, Serve H (2004) The cyclin A1‐CDK2 complex regulates DNA double‐strand break repair. Mol Cell Biol 24: 8917–8928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschel RJ, Zhang HB, Iliakis G, McKenna WG (1991) Cyclin B expression in HeLa cells during the G2 block induced by ionizing radiation. Cancer Res 51: 5113–5117 [PubMed] [Google Scholar]
- Nowack MK, Harashima H, Dissmeyer N, Zhao X, Bouyer D, Weimer AK, De Winter F, Yang F, Schnittger A (2012) Genetic framework of cyclin‐dependent kinase function in Arabidopsis . Dev Cell 22: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Ongkeko W, Ferguson DJ, Harris AL, Norbury C (1995) Inactivation of Cdc2 increases the level of apoptosis induced by DNA damage. J Cell Sci 108: 2897–2904 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Yoshioka T, Ichikawa H, Toki S (2002) Molecular cloning and characterization of RAD51‐like genes from Arabidopsis thaliana . Plant Mol Biol 50: 71–81 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yamanouchi H, Takyuu T, Yoshioka T, Ito Y, Kato T, Tabata S, Kurei S, Yoshioka Y, Machida Y, Seki M, Kobayashi M, Shinozaki K, Ichikawa H, Toki S (2005) Arabidopsis Rad51B is important for double‐strand DNA breaks repair in somatic cells. Plant Mol Biol 57: 819–833 [DOI] [PubMed] [Google Scholar]
- Pinato O, Musetti C, Sissi C (2014) Pt‐based drugs: the spotlight will be on proteins. Metallomics 6: 380–395 [DOI] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B (1996) Two different but related mechanisms are used in plants for the repair of genomic double‐strand breaks by homologous recombination. Proc Natl Acad Sci USA 93: 5055–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch S, Harashima H, Schnittger A (2012) Identification of kinase substrates by bimolecular complementation assays. Plant J 70: 348–356 [DOI] [PubMed] [Google Scholar]
- Quennet V, Beucher A, Barton O, Takeda S, Löbrich M (2011) CtIP and MRN promote non‐homologous end‐joining of etoposide‐induced DNA double‐strand breaks in G1. Nucleic Acids Res 39: 2144–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montane MH (2007) ATM‐mediated transcriptional and developmental responses to gamma‐rays in Arabidopsis . PLoS ONE 2: e430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Watson JM, Parkey J, Shippen DE (2002) Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J 21: 2819–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios G, Lossow A, Hertel B, Breuer F, Schaefer S, Broich M, Kleinow T, Jasik J, Winter J, Ferrando A, Farras R, Panicot M, Henriques R, Mariaux JB, Oberschall A, Molnar G, Berendzen K, Shukla V, Lafos M, Koncz Z et al (2002) Rapid identification of Arabidopsis insertion mutants by non‐radioactive detection of T‐DNA tagged genes. Plant J 32: 243–253 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez‐Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M (2007) Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448: 811–815 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM (1997) Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90: 425–435 [DOI] [PubMed] [Google Scholar]
- Serra H, Da Ines O, Degroote F, Gallego ME, White CI (2013) Roles of XRCC2, RAD51B and RAD51D in RAD51‐independent SSA recombination. PLoS Genet 9: e1003971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double‐strand break repair pathway choice. Cell Res 18: 134–147 [DOI] [PubMed] [Google Scholar]
- Siddik ZH (2003) Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22: 7265–7279 [DOI] [PubMed] [Google Scholar]
- Simoneau A, Robellet X, Ladouceur AM, D'Amours D (2014) Cdk1‐dependent regulation of the Mre11 complex couples DNA repair pathways to cell cycle progression. Cell Cycle 13: 1078–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Murray AW (1992) S‐phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature 355: 365–368 [DOI] [PubMed] [Google Scholar]
- Swoboda P, Gal S, Hohn B, Puchta H (1994) Intrachromosomal homologous recombination in whole plants. EMBO J 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi‐Iwai Y, Shinohara A, Takeda S (1998) Homologous recombination and non‐homologous end‐joining pathways of DNA double‐strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J 17: 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H (2002) Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double‐strand breaks. Plant J 29: 771–781 [DOI] [PubMed] [Google Scholar]
- Trovesi C, Manfrini N, Falcettoni M, Longhese MP (2013) Regulation of the DNA damage response by cyclin‐dependent kinases. J Mol Biol 425: 4756–4766 [DOI] [PubMed] [Google Scholar]
- Tulin F, Cross FR (2014) A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26: 4019–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda‐Tomás S, Federici F, Casimiro I, Beemster GT, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ (2009) Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol 19: 1194–1199 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Coppens F, Lee E, Donner TJ, Xie Z, Van Isterdael G, Dhondt S, De Winter F, De Rybel B, Vuylsteke M, De Veylder L, Friml J, Inze D, Grotewold E, Scarpella E, Sack F, Beemster GT, Beeckman T (2011) Developmental regulation of CYCA2s contributes to tissue‐specific proliferation in Arabidopsis . EMBO J 30: 3430–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao R, Wang H, Cheng Z, Li W, Zhu G, Wang Y, Ma H (2014) The Arabidopsis RAD51 paralogs RAD51B, RAD51D and XRCC2 play partially redundant roles in somatic DNA repair and gene regulation. New Phytol 201: 292–304 [DOI] [PubMed] [Google Scholar]
- Weimer AK, Nowack MK, Bouyer D, Zhao X, Harashima H, Naseer S, De Winter F, Dissmeyer N, Geldner N, Schnittger A (2012) Retinoblastoma related1 regulates asymmetric cell divisions in Arabidopsis . Plant Cell 24: 4083–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Story GW, Sunderland PA, Jiang Q, Bray CM (2002) Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double‐strand breaks in vivo . Plant J 31: 517–528 [DOI] [PubMed] [Google Scholar]
- Wohlbold L, Fisher RP (2009) Behind the wheel and under the hood: functions of cyclin‐dependent kinases in response to DNA damage. DNA Repair 8: 1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold L, Merrick KA, De S, Amat R, Kim JH, Larochelle S, Allen JJ, Zhang C, Shokat KM, Petrini JH, Fisher RP (2012) Chemical genetics reveals a specific requirement for Cdk2 activity in the DNA damage response and identifies Nbs1 as a Cdk2 substrate in human cells. PLoS Genet 8: e1002935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Lee E, Lucas JR, Morohashi K, Li D, Murray JA, Sack FD, Grotewold E (2010) Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22: 2306–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbro JW (1992) Mechanism of action of hydroxyurea. Semin Oncol 19: 1–10 [PubMed] [Google Scholar]
- Yata K, Esashi F (2009) Dual role of CDKs in DNA repair: to be, or not to be. DNA Repair 8: 6–18 [DOI] [PubMed] [Google Scholar]
- Yokota Y, Shikazono N, Tanaka A, Hase Y, Funayama T, Wada S, Inoue M (2005) Comparative radiation tolerance based on the induction of DNA double‐strand breaks in tobacco BY‐2 cells and CHO‐K1 cells irradiated with gamma rays. Radiat Res 163: 520–525 [DOI] [PubMed] [Google Scholar]
- Yoshiyama K, Conklin PA, Huefner ND, Britt AB (2009) Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M (2013) ATM‐mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis . EMBO Rep 14: 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shim EY, Davis M, Lee SE (2009) Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks. DNA Repair 8: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Peng G, Lin SY, Zhang P (2011) DNA damage response is suppressed by the high cyclin‐dependent kinase 1 activity in mitotic mammalian cells. J Biol Chem 286: 35899–35905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Review Process File


