Abstract
In budding yeast, the monopolin complex mediates sister kinetochore cross‐linking and co‐orientation in meiosis I. The CK1δ kinase Hrr25 is critical for sister kinetochore co‐orientation, but its roles are not well understood. Here, we present the structures of Hrr25 and its complex with the monopolin subunit Mam1. Hrr25 possesses a “central domain” that packs tightly against the kinase C‐lobe, adjacent to the binding site for Mam1. Together, the Hrr25 central domain and Mam1 form a novel, contiguous embellishment to the Hrr25 kinase domain that affects Hrr25 conformational dynamics and enzyme kinetics. Mam1 binds a hydrophobic surface on the Hrr25 N‐lobe that is conserved in CK1δ‐family kinases, suggesting a role for this surface in recruitment and/or regulation of these enzymes throughout eukaryotes. Finally, using purified proteins, we find that Hrr25 phosphorylates the kinetochore receptor for monopolin, Dsn1. Together with our new structural insights into the fully assembled monopolin complex, this finding suggests that tightly localized Hrr25 activity modulates monopolin complex–kinetochore interactions through phosphorylation of both kinetochore and monopolin complex components.
Keywords: monopolin complex, meiosis, kinetochore, kinase
Subject Categories: Cell Cycle, Structural Biology
Introduction
Sexual reproduction in eukaryotes depends on meiosis, a specialized two‐stage cell division program that reduces a cell's ploidy by half to generate haploid gametes. In the first meiotic division, termed meiosis I, homologous chromosomes become linked by reciprocal DNA exchanges called crossovers. These links allow the kinetochores of each homolog pair to bi‐orient, or attach to microtubules extending from opposite spindle poles, enabling their subsequent segregation from one another in the meiosis I division. As each homolog is made up of two sister chromosomes, proper homolog bi‐orientation also requires that sister kinetochores co‐orient, or attach to the same spindle pole. In budding yeast, sister kinetochore co‐orientation is mediated by the monopolin complex, which cross‐links each pair of sister kinetochores to fuse them into a single complex that binds one microtubule in meiosis I (Toth et al, 2000; Rabitsch et al, 2003; Winey et al, 2005; Petronczki et al, 2006; Corbett et al, 2010; Sarangapani et al, 2014).
The structural core of the monopolin complex comprises the Csm1 and Lrs4 proteins, which are found throughout fungi and assemble into a “V”‐shaped complex with two “heads” that each contains at least one binding site for the kinetochore protein Dsn1 (Corbett et al, 2010; Corbett & Harrison, 2012; Sarkar et al, 2013). In S. pombe, this complex localizes to kinetochores in mitosis and suppresses the attachment of single kinetochores to microtubules extending from both spindle poles, called merotelic attachments (Gregan et al, 2007; Rumpf et al, 2010b; Tada et al, 2011; Burrack et al, 2013). In budding yeast, whose kinetochores cannot form merotelic attachments as they bind a single microtubule, Csm1 and Lrs4 play only a minor role in supporting mitotic chromosome segregation accuracy (Brito et al, 2010). Budding yeast Csm1 and Lrs4 are critically important in meiosis I, however, where they mediate sister kinetochore co‐orientation along with two regulatory subunits, Mam1 and Hrr25 (Toth et al, 2000; Rabitsch et al, 2003; Petronczki et al, 2006).
The budding yeast‐specific Mam1 protein is expressed only in meiosis and contains binding sites for both Csm1 and Hrr25 (Toth et al, 2000; Rabitsch et al, 2003; Petronczki et al, 2006; Corbett & Harrison, 2012). Hrr25 is a CK1δ‐family kinase, and while its kinase activity is dispensable for monopolin's kinetochore localization, it is required for specific sister kinetochore co‐orientation in meiosis I (Petronczki et al, 2006). Hrr25 kinase activity is not required for cross‐linking of purified kinetochore particles in vitro (Sarangapani et al, 2014), perhaps due to high protein concentration in these experiments, and/or a lack of geometric constraints imposed by chromosome structure. Overall, these data imply that Hrr25 likely contributes to meiosis I sister kinetochore co‐orientation by phosphorylating its targets to either reinforce correct monopolin–kinetochore interactions or suppress incorrect interactions. Hrr25's specific roles remain unclear, however, due largely to a lack of mechanistic information on its recruitment to kinetochores, its differential regulation when part of the monopolin complex, and its specific substrates in this context.
CK1 kinases comprise a large, functionally diverse kinase family that regulates numerous processes including circadian rhythms (Gallego & Virshup, 2007), Wnt signaling (Price, 2006), and many other pathways (Knippschild et al, 2005). Major CK1 isoforms in mammals include CK1α, CK1δ, CK1ε, and CK1γ (Knippschild et al, 2005). Saccharomyces cerevisiae possesses four CK1 family members (Yck1, Yck2, Yck3, and Hrr25), of which three (Yck1, Yck2, and Yck3) are C‐terminally prenylated and membrane‐localized (Vancura et al, 1994; Wang et al, 1996). Hrr25 is the sole soluble CK1 ortholog in S. cerevisiae and is most closely related to mammalian CK1δ. In addition to its role in the monopolin complex, S. cerevisiae Hrr25 has been implicated in DNA repair (Hoekstra et al, 1991), ribosome biogenesis (Schäfer et al, 2006; Ghalei et al, 2015), vesicle trafficking and autophagy (Lord et al, 2011; Bhandari et al, 2013; Mochida et al, 2014; Pfaffenwimmer et al, 2014; Tanaka et al, 2014; Wang et al, 2015), clathrin‐mediated endocytosis (Peng et al, 2015a), and most recently microtubule assembly (Peng et al, 2015b). Hrr25 and its fission‐yeast homologs Hhp1 and Hhp2 have also been implicated in several monopolin‐independent meiotic functions, including controlling cleavage of the meiotic cohesin subunit Rec8 (Ishiguro et al, 2010; Katis et al, 2010; Rumpf et al, 2010a) and assembly of the meiotic chromosome axis (Phadnis et al, 2015; Sakuno & Watanabe, 2015). In most of these diverse contexts, the mechanistic basis for Hrr25 recruitment and regulation, if any, remains largely unknown.
Here, we present the structures of Hrr25 from two species of budding yeast, S. cerevisiae and Candida glabrata, and the structure of the S. cerevisiae Hrr25:Mam1 complex. The structures reveal that Hrr25 bears close structural similarity to other CK1δ kinases, but unique features of both Hrr25 and its complex with Mam1 reveal a novel mode for kinase recruitment and regulation. Mam1 binds a conserved hydrophobic surface on the Hrr25 N‐lobe, suggesting that CK1δ‐family kinases throughout eukaryotes may be regulated via this surface. Along with our prior structures of the Csm1:Lrs4 and Csm1:Mam1 complexes, we can now assemble a nearly complete atomic model for the budding yeast monopolin complex. Based on this model, we propose a scheme in which highly localized Hrr25 activity modulates the binding affinity of the monopolin complex for kinetochores, thereby mediating the specific cross‐linking of sister kinetochores in meiosis I.
Results
CK1δ proteins from point‐centromere fungi possess a conserved central domain
Saccharomyces cerevisiae Hrr25 contains an N‐terminal kinase domain (amino acids ~1–290), a “central domain” (amino acids ~290–394) that we previously reported is found only in S. cerevisiae and its close relatives (Corbett & Harrison, 2012), and a C‐terminal proline/glutamine‐rich domain that is predicted to be largely disordered in solution (Fig 1A). To more clearly determine the species distribution and conservation of the Hrr25 central domain, we generated a phylogenetic tree from the sequences of 93 fungal Hrr25 orthologs (Figs 1B and EV1A). We found that Hrr25 orthologs from Saccharomycotina yeast split into two major groups, based on whether they possess the central domain or not. The set of species whose Hrr25 orthologs possess a central domain correlates with those species that possess a MAM1 gene, and with species known or suspected to possess point centromeres, short DNA sequence‐defined centromeres whose kinetochores bind a single microtubule as in S. cerevisiae (Meraldi et al, 2006; Westermann et al, 2007; Gordon et al, 2011). This striking correlation supports the idea that the Hrr25 central domain is involved in a monopolin complex‐specific role, likely through an interaction (physical or functional) with Mam1.
Figure 1. Hrr25 orthologs from point‐centromere fungi form a distinct group.
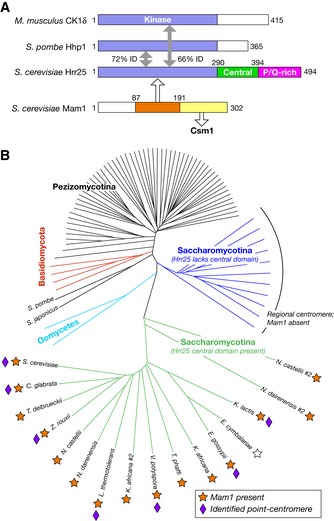
- Domain diagram of CK1δ orthologs from Saccharomyces cerevisiae, S. pombe, and M. musculus, with percent sequence identity (% ID) noted and of S. cerevisiae Mam1. Hrr25 orthologs from S. cerevisiae and its close relatives possess a “central domain” (green) and a C‐terminal proline/glutamine‐rich region predicted to be disordered in solution (magenta).
- Phylogenetic tree constructed from an alignment of 93 fungal Hrr25 orthologs. Branches in green represent Hrr25 proteins from Saccharomycotina yeast with the conserved central domain (see Fig EV1A for sequence alignment). Branches in blue represent Saccharomycotina Hrr25 proteins lacking this domain. Orange stars indicate the presence of a Mam1 ortholog in each species' genome. E. cymbalariae (white star) possesses an unannotated Mam1 ortholog on chromosome VII (region ˜313,000–315,000, 32% identity to S. cerevisiae Mam1 residues 83–242, identified by TBLASTN (NCBI)). Purple diamonds denote organisms in which point‐centromere sequences have been identified (Meraldi et al, 2006; Gordon et al, 2011). Point centromeres and Mam1 orthologs are both exclusive to the group of yeast whose Hrr25 orthologs possess the central domain.
Figure EV1. Sequence alignments of the Hrr25 central domain and the Hrr25‐binding region of Mam1.
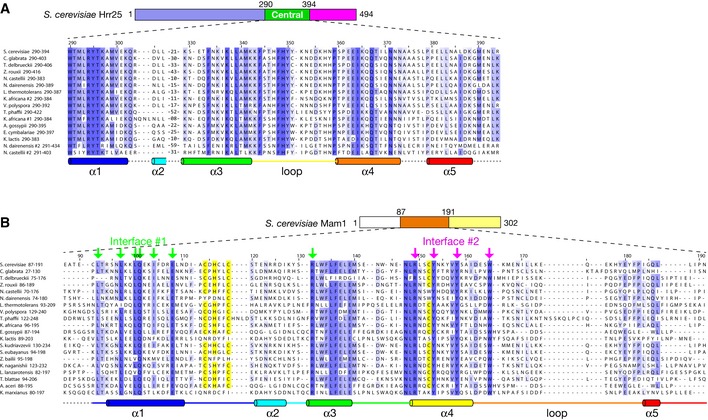
- Sequence alignment of the Hrr25 central domain from the Saccharomycotina fungi (green branches in Fig 1B).
Structure of the Hrr25 kinase domain
To explore the structural basis for Hrr25 function within the monopolin complex, we sought to determine the structure of the protein from point‐centromere fungi. We previously showed that an S. cerevisiae Hrr25 construct consisting of the kinase and central domains (residues 1–394 of 494) can be expressed in Escherichia coli (Corbett & Harrison, 2012). Because this protein is extensively and heterogeneously autophosphorylated when purified from E. coli, we purified a kinase‐dead version of the protein (lysine 38 to arginine; K38R) for crystallization trials. We obtained crystals of Hrr251–394 K38R in the presence of the CK1 inhibitor CK1‐7, which likely constrained the conformational freedom of the kinase domain and contributed to the formation of well‐ordered crystals (Chijiwa et al, 1989; Xu et al, 1996). We also cloned the equivalent construct of C. glabrata Hrr25 (residues 1–403 of 495) and obtained crystals both in the absence of bound nucleotide (Apo) and in the presence of ADP. We used single‐wavelength anomalous diffraction (SAD) methods to determine the structure of C. glabrata Hrr251–403 K38R bound to ADP at a resolution of 2.0 Å, then used this model to determine the structures of C. glabrata Hrr251–403 K38R in the Apo form at 2.9 Å and of CK1‐7‐bound S. cerevisiae Hrr251–394 K38R at 3.0 Å (Figs 2 and EV2, and Table EV1).
Figure 2. Structure and nucleotide/inhibitor binding by budding yeast Hrr25.
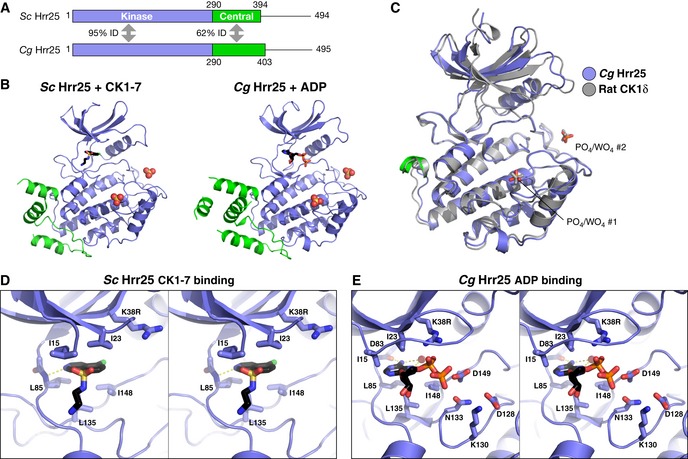
- Domain diagram of crystallized Hrr25 constructs from Saccharomyces cerevisiae and Candida glabrata, missing the P/Q‐rich C‐terminal domain.
- Structures of S. cerevisiae Hrr251–394 K38R bound to CK1‐7 (Chijiwa et al, 1989; Xu et al, 1996) and C. glabrata Hrr251–403 K38R bound to ADP, with domains colored as in (A). Bound / ions are shown as spheres (see Fig EV2A for additional C. glabrata Hrr25 structures and Fig EV3A for ion electron density).
- Stereo view of CK1‐7 binding to S. cerevisiae Hrr25. Bound drug is positioned identically to a previous structure of S. pombe Cki1 bound to CK1‐7 (Xu et al, 1996). All active‐site residues shown are conserved between S. cerevisiae and C. glabrata Hrr25.
- Stereo view of ADP binding to C. glabrata Hrr25 (formate structure; structure is equivalent, but the GxGxxG motif is disordered in that structure). All active‐site residues shown are conserved between S. cerevisiae and Candida glabrata Hrr25.
Figure EV2. binding sites in Hrr25.
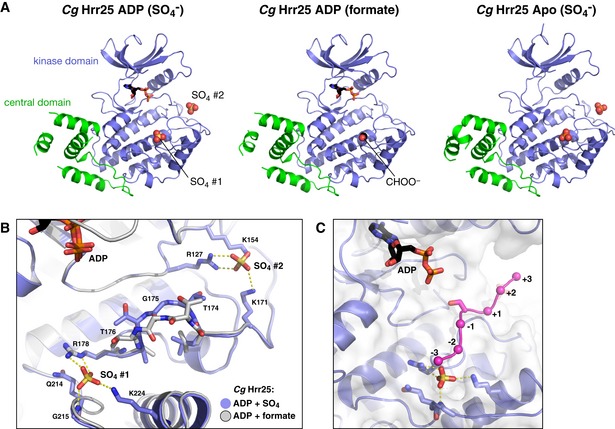
- Aligned structures of Candida glabrata Hrr251–403 crystallized without nucleotide (Apo, left), or in the presence of ADP in crystallization buffer with formate (center) or sulfate (, right). / ions bound to sites 1 and 2 are shown as spheres. In the ADP (formate) structure, there is no ion bound to site 2, but a formate ion (CHOO−) is observed at site 1.
- Close‐up view of binding sites S1 and S2 in C. glabrata Hrr25 (blue), overlaid with the formate‐bound structure (white).
- View of the C. glabrata Hrr25 active site and at S1, with modeled substrate peptide from a Cdk2:cyclin A:peptide complex (PDB ID 1GY3) (Cook et al, 2002).
The structures of S. cerevisiae and C. glabrata Hrr25 are nearly identical, showing between 0.3 and 0.7 Å overall Cα r.m.s.d. (Fig 2A and B). The structures are also highly similar to those of mammalian CK1δ and S. pombe Cki1, showing 0.6–0.7 Å Cα r.m.s.d. with both enzymes (over 215–226 Cα atom pairs) (Fig 2C). The juxtaposition of the N‐ and C‐lobes, and the position of important active‐site residues, is also nearly identical between Apo, ADP, and CK1‐7 bound Hrr25 structures. As observed in prior CK1 structures, Hrr25 bears the structural hallmarks of an active kinase in both the Apo and nucleotide‐bound structures. First, the DFG motif (residues 149–151) containing the catalytic aspartic acid residue is in its active conformation. Also, the αC helix bordering the nucleotide‐binding site is in the “in” position. CK1 enzymes do not require phosphorylation in the “activation loop” for full activity; however, a conserved binding site in Hrr25 does align structurally with other kinases' activation loop phosphothreonine residue (see below). Finally, both nucleotide and CK1‐7 bind Hrr25 in an identical manner to prior structures (Xu et al, 1996).
Hrr25 shares conserved binding sites with other CK1 enzymes
Prior structures of S. pombe Cki1 and mammalian CK1δ revealed two conserved sites that bind phosphate () or tungstate () ions in the crystals (Xu et al, 1995; Longenecker et al, 1996). Both S. cerevisiae and C. glabrata Hrr25 were crystallized in the presence of 0.2 M or higher or salts (see Materials and Methods), and we observed ions bound to both previously identified sites in all structures (Figs 2C and EV2). The first site, referred to here as S1, is likely involved in substrate recognition. CK1 enzymes are phosphate‐directed, preferring substrates with a “priming phosphate” in the −2 or −3 position (Flotow & Roach, 1991; Knippschild et al, 2005). S1 is positioned close to the active site, and modeling a substrate onto the Hrr25 structure shows that a phosphorylated residue in the −2 or −3 position could be accommodated at this site (Fig EV2C).
The second ion‐binding site, S2, is positioned behind the active site between the N‐ and C‐lobes, but is coordinated solely by C‐lobe residues in both Hrr25 and other CK1δ enzymes (Figs 2C and EV2). This site overlaps almost perfectly with the location of the phosphorylated activation loop threonine in other kinase families and has been proposed to recognize phosphorylated residues in the autoregulatory C‐terminal domain of CK1 enzymes (Longenecker et al, 1996). To determine whether ion occupancy at S2 affects the conformation of Hrr25, we attempted to crystallize the protein in the absence of or . We determined the structure of C. glabrata Hrr251–403 K38R bound to ADP in the presence of sodium formate instead of lithium sulfate and found that while S1 was bound to a formate ion, S2 was unoccupied. In this structure, the overall protein conformation is unchanged but residues 174–176, positioned directly between S1 and S2, shift position slightly and are less well defined in electron density maps in the formate‐bound structure than in the ‐bound structure (Fig EV2B). As this region makes up part of the substrate‐binding groove, local perturbations in structure induced by binding or release of ions at these positions may have an effect on substrate recognition or kinase activity.
Structure of the Hrr25 central domain
While the structure of its kinase domain is nearly identical to that of other CK1 enzymes, Hrr25 differs from these enzymes in possessing a conserved, well‐ordered “central domain” between the kinase domain and C‐terminal autoregulatory region (Figs 1A, 3 and EV1A). This domain adopts the same structure in both S. cerevisiae and C. glabrata Hrr25 and folds into a cluster of five short α‐helices and one extended loop, all packed tightly against the C‐lobe of the kinase domain (Fig 3B). Helices α2 and α5 are poorly ordered (as judged by B‐factor, a statistical measure of order in a refined crystal structure) in all structures, and helix α2 is visible in electron density maps in only one structure, that of C. glabrata Hrr251–403 K38R in the Apo state. Structural similarity searches using DALI reveal no similar folds, either as independently folded proteins or associated with kinases.
Figure 3. Structure of the Hrr25 central domain.
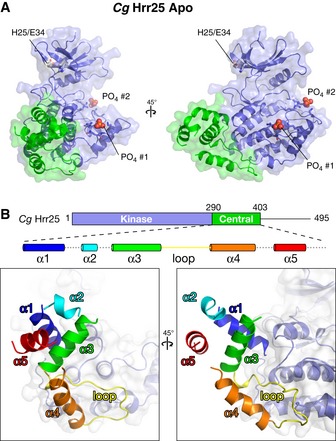
- Two views, roughly equivalent to the views in (A), of the Hrr25 central domain, colored as a rainbow according to the schematic at top. Dotted lines in the schematic indicate disordered loops (see Fig EV1A for sequence alignment of this domain).
Structure of the Hrr25:Mam1 complex
We previously reported that a construct of S. cerevisiae Mam1 containing residues 87–191 forms a stable complex with Hrr251–394 K38R when co‐expressed in E. coli (Corbett & Harrison, 2012). After initial crystallization trials with this complex (Hrr251–394 K38R:Mam187–191) were unsuccessful, we performed reductive methylation of surface‐exposed lysine residues (Walter et al, 2006; Kim et al, 2008) and identified conditions for crystallization of the modified complex. We determined the structure of the Hrr251–394 K38R:Mam187–191 complex by molecular replacement in two different crystal forms, one with one copy of the 1:1 complex per asymmetric unit (form 1, 1.84 Å resolution) and one with two copies (form 2, 2.89 Å resolution) (Table EV1). In both crystal forms, key crystal packing interactions are mediated by methylated surface lysine residues (Fig EV3). We have so far been unable to obtain crystals of the complex in the presence of nucleotides or inhibitors.
Figure EV3. Representative electron density maps.
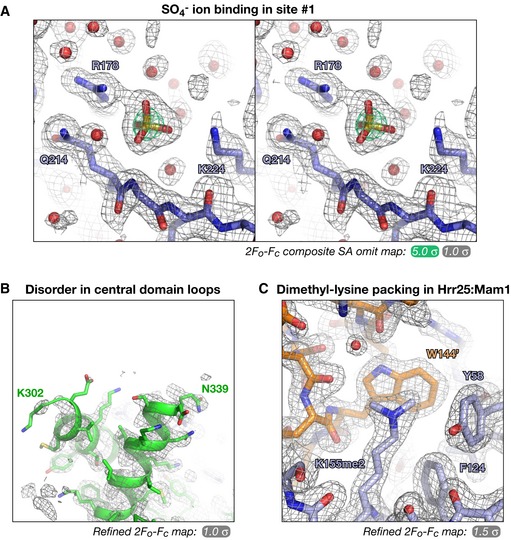
- Stereo view of a 2F o –F c simulated‐annealing composite all‐omit map from the Candida glabrata Hrr251–403·ADP complex (2.0 Å resolution), showing the sulfate ion bound to site #1.
- Refined 2F o –F c electron density for the α1–α3 segment (α2 is disordered in this structure) Hrr25 central domain in the C. glabrata Hrr251–403·ADP complex (2.0 Å resolution).
- Example crystal packing interaction mediated by dimethylated surface lysine residues in the structures of S. cerevisiae Hrr251–394 K38R:Mam187–191. Here, dimethylated Hrr25 Lys155 (foreground; blue) forms a hydrophobic packing interaction with a symmetry‐related Mam1 Trp144 (background; orange). Refined 2F o –F c electron density is shown at 1.5 σ.
Mam187–191 adopts a fold with five α‐helices and one extended loop (residues 162–191) that packs tightly against Hrr25. The protein possesses only a minimal buried hydrophobic core and is likely dependent on Hrr25 binding to fold properly. Mam187–191 unexpectedly contains a zinc‐binding motif that resembles a “zinc knuckle” but with a previously unobserved pattern of zinc‐coordinating residues. Three of these residues (Cys114, His116, and Cys119) are located in the short loop between α‐helices 1 and 2, with the fourth (Cys152) on α‐helix 4 (Fig 4C and D). These residues are highly conserved among budding yeast Mam1 orthologs, but are not universally conserved; several species have lost one or more of the zinc‐liganding residues (Fig EV1B), suggesting that a bound zinc is not absolutely required for Mam1's structure or activity. We confirmed that the coordinated atom in our structure is in fact zinc using a diffraction dataset collected at zinc's anomalous absorption edge; this map contains large anomalous peaks (~15 σ) at the expected locations (Fig 4D). As with the Hrr25 central domain, structural similarity searches using DALI reveal no similar folds to that of Mam187–191.
Figure 4. Structure of the Hrr25–Mam1 complex.
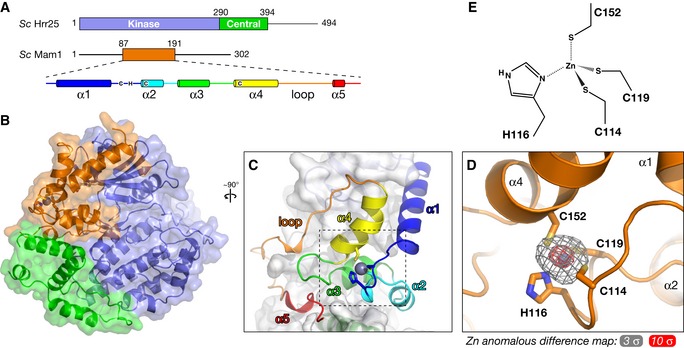
- Domain diagram of Saccharomyces cerevisiae Hrr25 (top) and Mam1 (bottom), with a schematic of Mam1 secondary structure. Zinc‐coordinating cysteine and histidine residues (Cys114, His116, Cys119, and Cys152) are shown in their approximate locations in the secondary structure (see Fig EV1B for Mam1 sequence alignment).
- Overall structure of the S. cerevisiae Hrr251–394 K38R:Mam187–191 complex, colored as in (A). View is equivalent to Fig 2B.
- Close‐up view of Mam187–191, colored as in the schematic in (A).
- Close‐up view of the variant zinc knuckle motif of Mam1, with zinc anomalous difference electron density shown at 3 σ (gray) and 10 σ (red).
- Geometry of zinc binding in Mam1. Zn‐S bonds and the Zn‐N bond were restrained to ˜2.3 Å and ˜2.0 Å, respectively, during refinement.
Mam1 forms an extensive interface with the Hrr25 kinase and central domains, burying over 2,700 Å2 of surface area per interacting partner. The main interface, between Mam1 and the Hrr25 kinase domain N‐lobe, can be broken into two parts, termed interfaces #1 and #2 (Fig 5A). Interface #1 involves hydrophobic residues in Mam1 α‐helix 1 inserting into a hydrophobic cavity on the top surface of the Hrr25 kinase domain N‐lobe. Mam1 binding imparts significant order to the N‐terminal ~5 residues of Hrr25, which are disordered in our other structures. The hydrophobic cavity on Hrr25, which comprises residues Leu3, Val5, Ile11, Tyr24, Leu39, and Tyr77, is conserved and solvent‐exposed in other CK1δ proteins and comprises part of a crystallographic dimer interface observed in several CK1δ crystal structures (Longenecker et al, 1996, 1998; Zeringo et al, 2013) (Fig EV4; see Discussion). Interface #1 also involves two conserved polar residues from Mam1, Gln101 and Glu108, forming a hydrogen‐bond network with several residues immediately preceding the Hrr25 GxGxxG motif (residues 16–21), including Gly12, Arg13, and Lys14. Mam1 residue Arg131 also interacts directly with Hrr25 residues Asp91 and Ile15, bridging the N‐ and C‐lobes of the kinase very near the ATP‐binding site. Together, these interactions likely impart additional order onto the Hrr25 N‐lobe and particularly the GxGxxG motif that drapes over a bound nucleotide, potentially affecting the kinetics of nucleotide binding and/or release by Hrr25. Mam1:Hrr25 interface #2 borders interface #1 and is anchored by a buried hydrogen‐bond network involving two residues in Hrr25, His25 and Glu34, that were previously shown to be involved in Mam1 binding (Petronczki et al, 2006). These residues form a buried hydrogen‐bond network with Mam1 residues Arg149 and Tyr158 that is surrounded by conserved hydrophobic residues in both proteins (Fig 5C).
Figure 5. Two interfaces in the Hrr25:Mam1 complex.
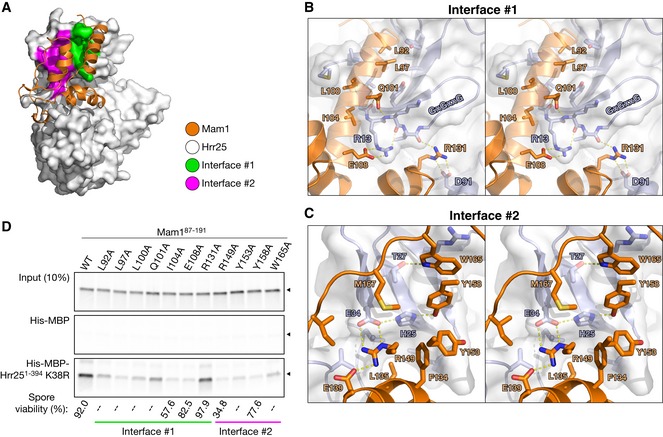
- The Hrr25:Mam1 complex with Mam1 shown as orange ribbons, and Hrr25 shown as a white surface with Mam1‐binding interface #1 shown in green and #2 in magenta.
- Stereo view of interface #1, which involves hydrophobic interactions by Mam1 L92, L97, L100, and I104, and a hydrogen‐bond network between Mam1 Q101/E108/R131 and Hrr25 residues 12–15.
- Stereo view of interface #2. Hrr25 residues H25 and E34 participate in a buried hydrogen‐bond network with Mam1 residues R149 and Y158 and are surrounded by hydrophobic residues on both proteins. For clarity, Mam1 helix α4, on which R149, Y153, and Y158 are located, is not shown.
- Pull‐down assay with in vitro‐translated Mam187–191 (wild‐type and alanine mutants) and Hrr251–394 K38R. Mam1 mutations also disrupt Hrr25 binding when the two are co‐expressed in Escherichia coli (Fig EV6B). Bottom: Spore viability of MAM1 mutant strains (see Table EV2 for strains; strains tested are KC549, KC560, KC566, KC552, KC554, KC556, and KC558; 46–48 tetrads were dissected for each strain). “–” indicates that this mutation was not tested for spore viability.
Figure EV4. Conservation of N‐lobe hydrophobic surface in CK1‐family kinases.
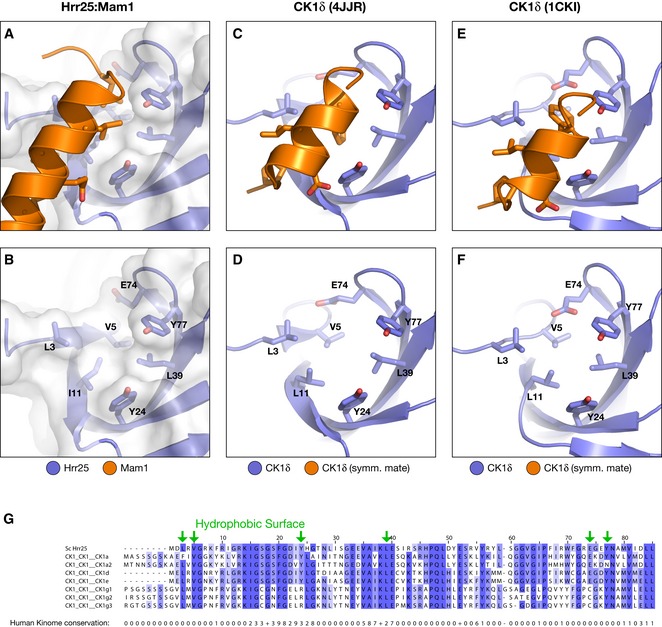
-
A–FViews of the kinase domain N‐lobes of Hrr25 (A, B), human CK1δ in PDB 4JJR (C, D) (Zeringo et al, 2013), and human CK1δ in PDB 1CKI (E, F) (Longenecker et al, 1996). Upper panels show kinase domain (blue) with conserved hydrophobic surface residues in stick view and bound α‐helix from Mam1 (A) or a symmetry‐related copy of CK1δ (C, E) in orange. Lower panels are missing bound partner for clarity, with conserved hydrophobic surface residues labeled.
-
GSequence alignment of the Hrr25 N‐lobe with all human CK1‐family kinases. At bottom is shown the conservation score (1–10, “+” indicates a score of 10) calculated by Jalview from an alignment of the entire human kinome (downloaded from http://kinase.com) plus Hrr25. Hydrophobic surface residues are shown with green arrows; most of these residues are highly conserved within CK1‐family kinases but poorly conserved in kinases as a whole.
To determine the importance of each interface for Mam1–Hrr25 binding, we generated a series of alanine mutants in Mam1 and examined their ability to bind Hrr25 in a pull‐down assay (Fig 5D). In interface #1, mutation of the hydrophobic amino acids Leu92, Leu97, Leu100, and Ile104 all disrupted Hrr25 binding. Mutation of the polar amino acids in interface #1 had a more variable effect: mutation of Glu108 strongly disrupted Hrr25 binding, while mutation of Gln101 weakly disrupted binding and mutation of Arg131 had no effect. In interface #2, mutation of either Mam1 Arg149 or Tyr158 strongly disrupted Hrr25 binding, consistent with the known role of Hrr25 residues His25 and Glu34 (Petronczki et al, 2006). Mutation of the surrounding Mam1 hydrophobic residues Tyr153 and Trp165 also affected Hrr25 binding, although not as strongly as Arg149 and Tyr158 (Fig 5C). To confirm that disrupting the Mam1:Hrr25 interface results in meiotic chromosome segregation defects, we tested spore viability for Mam1 point mutations in interfaces #1 and #2. While the effects were less severe than deleting MAM1 (8.5% viable for mam1Δ compared to 92% for wild‐type; Fig 5D), several mutations showed strong spore viability effects, most notably I104A in interface #1 (58% viable) and R149A in interface #2 (35% viable; Fig 5D). These results suggest that the point mutants tested do not completely disrupt Mam1–Hrr25 binding in vivo, but rather lower the affinity of the interaction by varying degrees. Nonetheless, these data strongly support the in vivo relevance of our structural findings.
Kinase activity of Hrr25 and Hrr25:Mam1
Our structures showed that Mam1 packs tightly against the N‐lobe and central domain of Hrr25, but does not significantly alter Hrr25's conformation. Mam1 binding does, however, significantly rigidify the Hrr25 N‐lobe, as judged by crystallographic B‐factors in this domain (Figs 6 and EV5). The GxGxxG motif in particular is well ordered in all three crystallographically unique views of the Hrr25:Mam1 structure, despite the lack of bound nucleotide. This loop is disordered in three of our four structures of Hrr25 alone and is only visible in one of the two structures of ADP‐bound C. glabrata Hrr251–403 K38R. These data support the idea that Mam1 binding imparts significant additional order onto Hrr25 and particularly the ATP‐binding site, suggesting that Mam1 may directly affect the kinase's activity and/or substrate specificity. Also, we previously showed that co‐expression of Mam1 with Hrr25 in E. coli leads to a reduction in its non‐specific autophosphorylation activity, suggesting that Mam1 binding directly affects the activity and/or specificity of Hrr25 (Corbett & Harrison, 2012). To more quantitatively examine the effects of Mam1 binding on Hrr25 activity, we next examined Hrr25's kinase activity directly.
Figure 6. Mam1 binding restricts Hrr25 N‐lobe mobility.
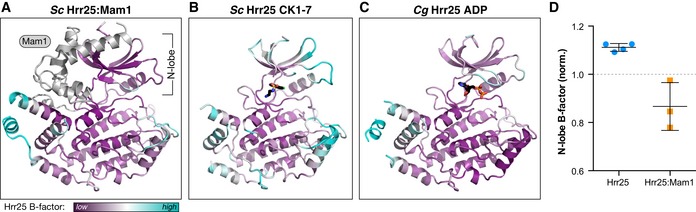
-
A–CRibbon views of the Saccharomyces cerevisiae Hrr251–394:Mam187–191 structure (form 1), CK1‐7 bound S. cerevisiae Hrr251–394, and ADP‐bound C. glabrata Hrr251–403 (formate condition), with Hrr25 colored according to main‐chain B‐factor from low (purple) to cyan (high). For each, coloring is normalized to correspond to the average of the 20 lowest (purple) or highest (cyan) main‐chain B‐factors in Hrr25. Mam1 (panel A) is colored in light gray.
-
DOverall main‐chain B‐factors for the N‐lobe (residues 1–85) of four structures of Hrr25 (both C. glabrata and S. cerevisiae) and the three crystallographically unique views of the S. cerevisiae Hrr25:Mam1 complex. Values are normalized to the average main‐chain B‐factor for the entire Hrr25 chain (dotted line at 1.0). P‐value = 0.004 (Student's t‐test) (see Fig EV5 for residue‐by‐residue B‐factor plots for each structure). Error bars represent standard deviation.
Figure EV5. N‐lobe B‐factors for Hrr25 and Hrr25:Mam1 structures.
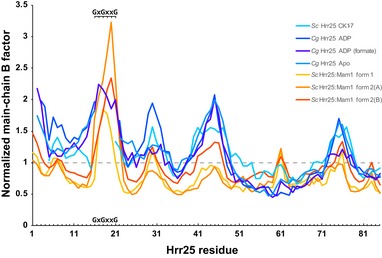
Residue‐by‐residue plot of main‐chain B‐factors from four structures of Hrr25 and three crystallographically unique views of Hrr25:Mam1. Values are normalized as in Fig 6D, with the average B‐factor for the entire Hrr25 chain adjusted to 1.0. Values calculated with BAVERAGE (Winn et al, 2011). GxGxxG indicates the glycine‐rich loop (residues 16–21 in both Saccharomyces cerevisiae and Candida glabrata Hrr25) that drapes over a bound nucleotide. This motif is well ordered in all three views of Hrr25:Mam1, but only one of the four structures of Hrr25. Because the GxGxxG motif was disordered in three of four Hrr25‐alone structures and was therefore not included in the analysis in Fig 6D, that analysis likely under‐estimates the stabilizing effect of Mam1 on Hrr25 structure.
To quantitatively study Hrr25 kinase activity in vitro, we used a high‐throughput ATP hydrolysis assay (ADP‐Glo; Promega). Both S. cerevisiae Hrr251–394 and the Hrr251–394:Mam187–191 complex showed low basal ATPase activity that was strongly stimulated by a non‐specific substrate protein, bovine casein (Fig 7A). We were unable to purify isolated full‐length Hrr25 due to poor expression and solubility, but we could purify full‐length Hrr25 in complex with Mam187–191. This complex showed similar activity to the truncated complex in all assays, indicating that the Hrr25 C‐terminal region does not strongly affect in vitro activity (Fig 7). Hrr251–394 showed a roughly twofold higher V max than Hrr251–394:Mam187–191 and also showed a roughly threefold tighter K m for casein (Fig 7A and D). In a second assay, we found that Hrr251–394:Mam187–191 shows a roughly twofold tighter K m for ATP than Hrr251–394 alone (Fig 7B). Together, these data suggest that Mam1 can alter several aspects of Hrr25's catalytic activity, including its nucleotide binding affinity. The Hrr251–394:Mam187–191 complex's weaker K m for casein suggests that Mam1 may weaken interactions with non‐specific substrates. This idea is supported by our observation (not shown) that Hrr251–394 ATPase activity increases with protein concentration in the absence of casein, suggesting that the kinase can act as a non‐specific substrate for itself. This behavior was not observed in the presence of casein and was also not observed for Hrr251–394:Mam187–191 in either the presence or absence of casein.
Figure 7. Mam1 binding regulates Hrr25 kinase activity.
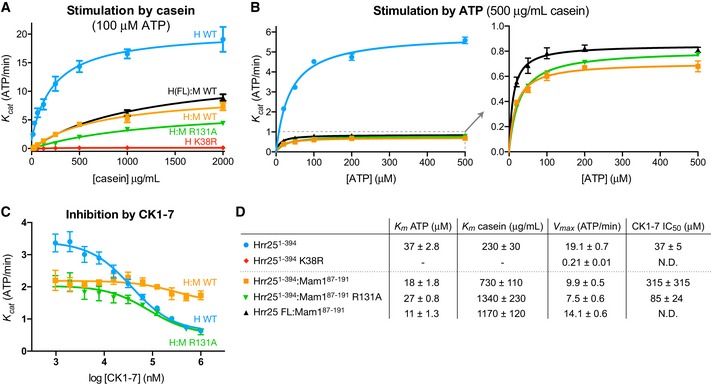
- ADP‐Glo ATPase assay showing stimulation of Saccharomyces cerevisiae Hrr251–394 (blue circles), Hrr251–394 K38R (red diamonds), Hrr25 full‐length:Mam187–191 (black triangles), Hrr251–394:Mam187–191 (orange squares), and Hrr251–394:Mam187–191 R131A (green triangles) by bovine casein (see Fig EV6A for SDS–PAGE analysis of purified proteins). Error bars represent standard deviation from triplicate measurements.
- Stimulation of Hrr251–394, Hrr251–394:Mam187–191, and Hrr251–394:Mam187–191 R131A by ATP, measured using an enzyme‐coupled ATPase assay. Error bars represent standard deviation from triplicate measurements.
- ADP‐Glo assay showing the effect of added CK1‐7 on Hrr251–394, Hrr251–394:Mam187–191, and Hrr251–394:Mam187–191 R131A. Error bars represent standard deviation from triplicate measurements.
- K m, V max, and IC50 values for tested enzymes. V max was calculated from curves in (A) at 100 μM ATP.
Mam1 residue Arg131 is highly conserved in Mam1 proteins and positioned close to the Hrr25 active site, but its mutation does not affect Mam1–Hrr25 binding in vitro or cause spore lethality (Fig 5D). This residue's proximity to the Hrr25 active site suggested a potential direct role in regulating ATP binding or hydrolysis, but we found that the catalytic parameters of Hrr251–394:Mam187–191 R131A were nearly identical to the wild‐type complex (Fig 7A and D). We did, however, notice a strong effect on the efficacy of the inhibitor CK1‐7. CK1‐7 inhibits Hrr251–394 with an IC50 of 37 ± 5 μM at 40 μM ATP (K i ~19 μM as calculated using the Cheng–Prusoff approximation; see Materials and Methods), but only very weakly inhibits the Hrr251–394:Mam187–191 complex (IC50 > 300 μM; Fig 7C and D). Structural modeling suggests that the aminoethyl group of CK1‐7, which extends out of the Hrr25 active site, would clash with Mam1 Arg131 (Fig EV6C). In support of this idea, the Mam1 R131A mutation rescued inhibition by CK1‐7, with the mutated complex showing an IC50 of 85 ± 24 μM (K i ~ 27 μM) (Fig 7C and D).
Figure EV6. Purification of Hrr25 and Hrr25:Mam1 for ATPase assays and modeling of Mam1 Arg131 interaction with nucleotide and CK1‐7.
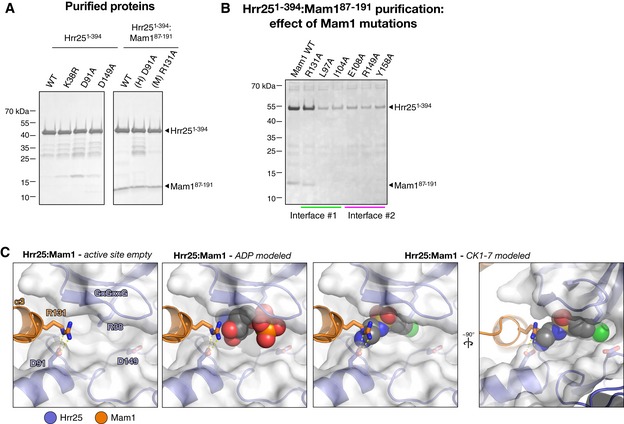
- SDS–PAGE analysis of purified Saccharomyces cerevisiae Hrr251–394 and Hrr251–394:Mam187–191 complexes used for ATPase assays.
- Ni2+ affinity pull‐down of co‐expressed His6‐Hrr251–394 K38R:Mam187–191 complexes with mutations to Mam1 in interfaces #1 or #2. Except for R131A, all mutations result in a loss of co‐purification of Mam1, indicating a loss of binding, and a significant reduction in the level of soluble Hrr251–394 K38R.
- Close‐up views of the Hrr25 active site in the structure of S. cerevisiae Hrr251–394:Mam187–191. Mam1 R131 extends toward the ATP‐binding pocket, and while it would not clash with a bound nucleotide (center panel), it would clash with the aminoethyl group of bound CK1‐7 (right panels).
Hrr25 phosphorylates Dsn1, the kinetochore receptor for monopolin
Hrr25 has been previously shown to phosphorylate Mam1, and this activity was proposed to regulate monopolin–kinetochore interactions (Petronczki et al, 2006; Corbett & Harrison, 2012). To further investigate the potential regulatory roles of Hrr25 in monopolin–kinetochore binding, we tested the activity of Hrr251–394 and Hrr251–394:Mam187–191 on the purified S. cerevisiae Mtw1 complex, which contains the kinetochore receptor for the monopolin complex, Dsn1. By examining band shifts on a Phos‐tag SDS–PAGE gel (Kinoshita et al, 2006), we observed that both Hrr251–394 and Hrr251–394:Mam187–191 phosphorylate Dsn1, but not the other subunits of this complex (Fig 8A). In contrast to our findings with bovine casein, the Hrr251–394:Mam187–191 complex was roughly as active as Hrr251–394 alone when phosphorylating Dsn1. This supports the idea that instead of globally suppressing Hrr25's kinase activity, Mam1 rather alters the kinase's substrate specificity. The observed band shifts are due specifically to phosphorylation by Hrr25, as the shift was eliminated by the addition of CK1‐7 (Fig 8A) or alkaline phosphatase (not shown).
Figure 8. Hrr25 phosphorylates Dsn1.
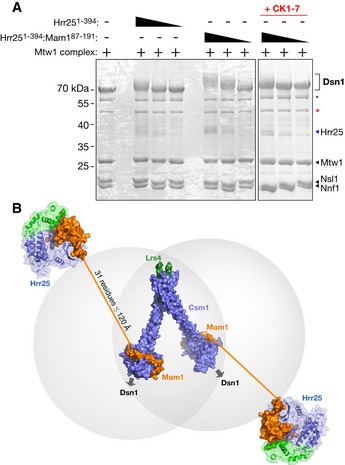
- Phos‐tag gels showing phosphorylation of purified S. cerevisiae Mtw1 complex by Hrr251–394 and Hrr251–394:Mam187–191. Asterisks denote N‐terminal proteolytic cleavage products of Dsn1, with the red asterisk denoting a previously characterized product lacking the first 171 residues (Hornung et al, 2011).
- Model of the intact monopolin complex. Four copies of Csm1 (blue) and two of Lrs4 (green) form the core “V”‐shaped kinetochore‐binding complex (Corbett et al, 2010). Each dimer of Csm1 binds one Mam1 (orange) (Corbett & Harrison, 2012), which in turn recruits one copy of Hrr25 (blue/green). Thirty‐one residues of Mam1 (192–222) separate the protein's Hrr25‐binding and Csm1‐binding domains, which when fully extended could reach as far as ˜120 Å.
Dsn1 contains 576 residues, with a proteolytically sensitive ~220 residue N‐terminal domain that is predicted to be mostly disordered (Hornung et al, 2011). This region also contains the Csm1‐binding site, which spans approximately residues 70–110 (Sarkar et al, 2013). Our preparation of the Mtw1 complex contained two major proteolytic degradation products of Dsn1, the smaller of which has previously been shown to lack the N‐terminal 171 residues of Dsn1 (Hornung et al, 2011). In contrast to the full‐length protein, neither proteolytic fragment of Dsn1 was detectably phosphorylated by Hrr25, indicating that the bulk of Hrr25‐mediated Dsn1 phosphorylation likely occurs in the disordered N‐terminal region (Fig 8A). Due to limited yield and purity of the Mtw1 complex, we have so far been unable to test its ability to stimulate Hrr25 ATPase activity, or determine the sites on Dsn1 that are phosphorylated by Hrr25.
Discussion
Here, we report the structure of budding yeast Hrr25 and its complex with the monopolin subunit Mam1. These reveal close structural similarity to other CK1δ‐family enzymes, including the conservation of two binding sites previously observed in both mammalian CK1δ and S. pombe Cki1. Unique to budding yeast Hrr25, however, is a “central domain” with a novel fold that packs tightly against the kinase domain's C‐lobe. The species distribution of this domain suggests that its main function is to cooperate with Mam1 in a monopolin‐specific role, likely in regulation and/or localization of Hrr25's kinase activity. A recent report has implicated this domain in its localization to P‐bodies, however, suggesting that the domain may also play a role in Hrr25 localization outside the context of monopolin (Zhang et al, 2016).
Mam1 adopts a novel fold anchored by a variant zinc knuckle motif, and forms an extensive interface with the Hrr25 N‐lobe, C‐lobe, and central domain. The most significant effect of Mam1 on Hrr25 structure is the evident suppression of conformational dynamics in the Hrr25 N‐lobe and especially the GxGxxG motif that participates in nucleotide binding. Our biochemical analysis shows that Mam1 binding reduces the K cat of Hrr25 by about half and also induces a tighter K m for ATP, both effects that are consistent with suppressed dynamics in the Hrr25 N‐lobe. The Hrr25:Mam1 complex's lowered affinity for a non‐specific substrate (bovine casein) suggests a role for Mam1 in regulating substrate recognition, but confirmation of this idea will require quantitative characterization of Hrr25's activity on a bona fide monopolin‐specific substrate. As Hrr25 is involved in a large number of distinct signaling pathways in budding yeast, it will also be important to determine whether Mam1's mode of Hrr25 recruitment and regulation is unique, or rather represents a mechanism shared by other pathways in budding yeast.
The Hrr25:Mam1 interface further provides a clue to how CK1δ‐family kinases may be regulated more globally. Hrr25:Mam1 interface #1 involves an α‐helix from Mam1 docking into a hydrophobic cavity on the Hrr25 N‐lobe. The residues within this cavity are highly conserved and consistently hydrophobic throughout the CK1 kinase family, but are typically hydrophilic or charged in other kinase families (Fig EV4). In addition, the same surface is involved in formation of a crystallographic dimer that has been observed in three different crystal forms of mammalian CK1δ and has been proposed to represent an autoinhibited state (Longenecker et al, 1996, 1998; Zeringo et al, 2013). In this dimer, an α‐helix from the CK1δ C‐lobe docks onto the hydrophobic cavity in a neighboring CK1δ N‐lobe, in a manner analogous to Mam1 α‐helix 1 in our structure (Fig EV4). This common interface suggests one of two possibilities, both of which implicate this surface as an important regulatory element: first, CK1 kinases may share a mechanism for autoinhibition through dimerization. Second, and more intriguingly, the hydrophobic cavity on the CK1δ N‐lobe may be a conserved site of recruiter/regulator binding. In this context, the CK1δ dimer structures may represent fortuitous interactions that utilize a conserved, solvent‐exposed hydrophobic surface.
The structure of the Hrr25:Mam1 complex represents the final element necessary for a complete structural model of the budding yeast monopolin complex (Fig 8B). Our earlier structures showed that Csm1 and Lrs4 form a “V”‐shaped complex with at least two binding sites for Dsn1 (Corbett et al, 2010). The C‐terminal domain of Mam1 (residues 223‐263 of 302) wraps around the Csm1 C‐terminal domains on each head of the “V”, close to the Dsn1‐binding surface (Corbett & Harrison, 2012). Our current structure shows that Mam1 residues 87–191 form a folded domain that tethers Hrr25, and sequence alignments/structure predictions indicate that the 31 residues of Mam1 between its Csm1‐ and Hrr25‐binding regions likely constitute a disordered linker. While the sequence of this region is highly variable, its length is remarkably consistent among Mam1 orthologs (not shown). If completely extended (3.8 Å per residue), this disordered linker region could stretch only to ~120 Å, implying that Hrr25 kinase activity is tightly constrained around the monopolin complex's kinetochore‐binding site. The consistent length of this region in different Mam1 orthologs also suggests that Hrr25's “range” when bound to kinetochores is functionally important.
How could highly localized Hrr25 kinase activity contribute to sister kinetochore cross‐linking specificity? A yeast kinetochore contains between 8 and 20 copies of Dsn1 (Joglekar et al, 2006; Lawrimore et al, 2011), and each Dsn1 contains a binding site for Csm1 in its disordered N‐terminal region (Sarkar et al, 2013). In order to cross‐link two kinetochores, a single monopolin complex must bind at least one Dsn1 from each of two separate kinetochores, rather than binding multiple copies of Dsn1 within a single kinetochore. Given the structure of the complex, with two Csm1 dimer “heads” separated by ~10 nm (Corbett et al, 2010), the most likely scenario is that each head binds Dsn1 from a different kinetochore. We propose that Hrr25's kinase activity is important to suppress binding of the second Csm1 dimer head to the same kinetochore as the first head is bound. This suppression is likely achieved through phosphorylation of Mam1 and/or Dsn1, which could modulate, either positively or negatively, their affinity for Csm1 (Corbett & Harrison, 2012). Because sister chromosomes are physically linked through cohesin complexes, pairs of kinetochores that approach one another closely enough to be bound by the two heads of monopolin are overwhelmingly likely to be sisters, meaning that even non‐specific kinetochore cross‐linking would mostly occur between sisters. A role for Hrr25 in suppressing intrakinetochore binding of the two monopolin heads is consistent with evidence implicating Hrr25 in negative regulation of monopolin–kinetochore binding (Petronczki et al, 2006; Corbett & Harrison, 2012) and could also explain the very low stoichiometry of the monopolin complex in purified meiotic kinetochore samples (Sarangapani et al, 2014). A more detailed analysis of the interactions between Csm1 and Dsn1, and the roles of Mam1 and Hrr25 in modulating these interactions, will be required to fully understand the mechanism of sister kinetochore co‐orientation by the monopolin complex.
Materials and Methods
Sequence alignments
For sequence alignments, fungal orthologs of S. cerevisiae Hrr25 and Mam1 were identified by PSI‐BLAST, then aligned with MAFFT (Katoh & Standley, 2013) in JalView (Waterhouse et al, 2009). An unrooted phylogenetic tree for Hrr25 was calculated in JalView and visualized using the Drawtree module of the PHYLIP package (Felsenstein, 2013).
Cloning and protein purification
Saccharomyces cerevisiae Hrr251–394 and C. glabrata Hrr251–403 proteins were cloned from genomic DNA into pET‐based vectors with N‐terminal, TEV protease‐cleavable His6 tags; then, kinase‐dead K38R variants were cloned using site‐directed mutagenesis. For co‐expression of Hrr251–394 (wild type or K38R) with Mam187–191, a polycistronic expression cassette was assembled by PCR, with Hrr25 tagged with an N‐terminal, TEV protease‐cleavable His6 tag. Proteins were expressed in E. coli strain Rosetta2(DE3)pLysS (EMD Millipore) transformed with a pCDFDuet vector encoding λ‐phosphatase (kindly provided by A. Motamedi); expression was induced with IPTG for 16–20 h at 20°C. For S. cerevisiae Mtw1 complex, the four proteins (Mtw1, Nnf1, Nsl1 and Dsn1) were sequentially cloned into a polycistronic expression plasmid (Tan, 2001) with Dsn1 fused to an N‐terminal, TEV protease‐cleavable His6 tag and expressed in E. coli Rosetta2(DE3)pLysS using autoinducing media (Studier, 2005). Proteins were purified by Ni2+‐affinity (Ni‐NTA; Qiagen) and ion‐exchange (Hitrap SP HPor Hitrap Q HP; GE Healthcare) chromatography; then, His6‐tags were cleaved by incubation with TEV protease (Kapust et al, 2001) at 4°C overnight, and the proteins were further purified by gel filtration (Superdex 200; GE Healthcare). Proteins were concentrated and stored at 4°C for crystallization, or at −80°C for biochemical assays.
Crystallization and structure determination
For crystallization, S. cerevisiae Hrr251–394 K38R was concentrated to 10 mg/ml and exchanged into a buffer containing 20 mM HEPES pH 7.5, 0.3 M NaCl, 1 mM DTT, 5 mM MgCl2, and 1 mM EDTA. The protein was mixed 1:1 in hanging‐drop format with well solution containing 0.1 M CAPS pH 11, 0.2 M lithium sulfate, and 1.5–1.6 M ammonium sulfate. Football‐shaped crystals (~100 × 100 × 200 μm) were cryoprotected by the addition of 25% glycerol and flash‐frozen in liquid nitrogen. Candida glabrata Hrr251–403 K38R was concentrated to 12 mg/ml and exchanged into a buffer containing 20 mM HEPES pH 7.5, 0.3 M NaCl, 1 mM DTT, 5 mM MgCl2, and 1 mM EDTA. For crystals of the ADP‐bound form, the protein was mixed 1:1 in hanging‐drop format with well solution containing 0.1 M Bis‐Tris pH 6.5, 0.2 M lithium sulfate, 19% PEG 3350, and 5 mM TCEP. Crystals (oval‐shaped plates ~300 × 700 × 40 μm) were cryoprotected by the addition of 10% glycerol and flash‐frozen in liquid nitrogen. For crystals grown with formate, lithium sulfate was replaced with 0.2 M sodium formate. For crystals of the Apo form, the protein was mixed 1:1 in hanging‐drop format with well solution containing 0.1 M CAPS pH 10.5, 0.2 M lithium sulfate, 0.56 M NaH2PO4, and 0.66 M K2HPO4. Crystals (triangular plates ~30 × 50 × 5 μm) were cryoprotected by addition of 25% glycerol and flash‐frozen in liquid nitrogen.
For crystallization of S. cerevisiae Hrr251–394 K38R:Mam187–191, purified protein at 0.5 mg/ml was subjected to reductive dimethylation of surface lysine residues by incubation (2 h, 4°C) with 50 mM borane dimethylamine complex and 0.1 M formaldehyde (Sigma‐Aldrich), followed by quenching with 25 mM glycine. Methylated protein was exchanged into a buffer containing 20 mM HEPES pH 7.5, 0.3 M NaCl, 1 mM DTT, 5 mM MgCl2, and 1 mM EDTA, concentrated to 10 mg/ml, and crystallized in hanging‐drop format by mixing 1:1 with well solution. There are two crystal forms. For form 1 crystals, well solution contained 0.1 M CHES pH 8.9, 5% Tacsimate, 25 mM YCl3, and 19% PEG 3350. Needle‐shaped crystals (~300 × 40 × 40 μm) were cryoprotected with the addition of 15% glycerol and flash‐frozen in liquid nitrogen. For form 2 crystals, well solution contained 0.1 M imidazole pH 8.0, 14% PEG 3350, and 6% glycerol. Crystals (arrow‐shaped plates, ~300 × 100 × 10 μm) were cryoprotected with the addition of 15% glycerol and flash‐frozen in liquid nitrogen.
Datasets for S. cerevisiae Hrr251–394 K38R and C. glabrata Hrr251–403 K38R were collected on beamline 24‐ID‐E at the Advanced Photon Source at Argonne National Laboratory, datasets for S. cerevisiae Hrr251–394 K38R:Mam187–191 form 1 crystals were collected on NE‐CAT beamline 24‐ID‐C at the Advanced Photon Source at Argonne National Laboratory, and datasets for S. cerevisiae Hrr251–394 K38R:Mam187–191 form 2 crystals were collected on beamline 12.3.1 at the Advanced Light Source at Lawrence Berkeley National Laboratory. The structure of C. glabrata Hrr251–403 ADP was determined by single‐wavelength anomalous diffraction (SAD) phasing using a 2.5 Å dataset collected from a crystal grown from selenomethionine‐derivatized protein. Phasing was carried out using an automated protocol (NE‐CAT RAPD; Frank Murphy) incorporating SHELXC/D/E (Sheldrick, 2010) for site identification followed by PHASER (McCoy et al, 2007) for SAD phasing. An initial model built by RESOLVE allowed us to dock a model of S. pombe CK1 (PDB ID 1CSN) (Xu et al, 1995) into the density. This model was iteratively rebuilt in COOT (Emsley et al, 2010) and refined against a 2.0 Å native dataset using phenix.refine (Adams et al, 2010), then used as a starting point for rebuilding and refinement of the C. glabrata Hrr251–403 K38R Apo model. A consistent free‐R set was used for refinement of the two structures of C. glabrata Hrr251–403 K38R. The C. glabrata model was then used for molecular replacement in PHASER to determine the S. cerevisiae Hrr251–394 K38R·CK1‐7 structure. All Hrr25 models were refined in phenix.refine using positional, B‐factor, and TLS refinement and have good R‐factors and stereochemical parameters (Table EV1). For S. cerevisiae Hrr251–394 K38R:Mam187–191, the structure of S. cerevisiae Hrr251–394 K38R was used as a molecular‐replacement model to generate initial phases. The Mam1 model was iteratively rebuilt in COOT and refined using phenix.refine. The location and identity of the Mam1‐bound Zn2+ ion was confirmed with an anomalous difference map generated from a form 2 dataset collected just above the zinc K‐edge (9964 eV/1.2829 Å) (Fig 4D). Original diffraction data for all structures are deposited at the SBGrid Data Bank, and refined structure factors and coordinates are deposited at the RCSB Protein Data Bank (see Table EV1 for accession numbers).
Kinase assays
Kinase assays were performed using the ADP‐Glo kinase assay kit (Promega), or an enzyme‐coupled assay, as indicated. All reactions were performed in kinase buffer (20 mM HEPES pH 7.5, 200 mM NaCl, 5% glycerol, 2 mM MgCl2, and 1 mM DTT) supplemented with ATP and dephosphorylated bovine casein (Sigma) as indicated. For casein titration, [ATP] was 100 μM; for ATP titration, [casein] was 500 μg/ml; and for inhibition by CK1‐7, [ATP] was 40 μM and [casein] was 100 μg/ml. For ADP‐Glo assays, reactions were performed for 60 min at 30°C. Enzyme‐coupled assays were performed essentially as described (Ye et al, 2015), in kinase buffer supplemented with 3 mM phosphoenolpyruvate, 20 U/ml lactate dehydrogenase (Sigma), 20 U/ml pyruvate kinase (Sigma), and 0.3 mM NADH. Enzyme‐coupled assays were performed at 28°C. Both luminescence and absorbance measurements were taken with a TECAN Infinite M1000 spectrophotometer (Mannedorf) in 384‐well microplates. All data analysis was performed with PRISM v. 6 (GraphPad Software). K i values were calculated using the Cheng–Prusoff equation: K i = IC50/(([S]/K m)+1) (Cheng & Prusoff, 1973).
For tests with purified S. cerevisiae Mtw1 complex, 9.2 μg (84 pM) of Mtw1 complex (purified as previously described) (Corbett et al, 2010) was incubated with Hrr251–394 or Hrr251–394:Mam187–191 (16.8, 8.4, or 4.2 pM; 5×, 10×, or 20× less kinase than substrate) in 20 μl reactions in kinase buffer (20 mM Tris–HCl pH 7.5, 300 mM NaCl, 10% glycerol, 1 mM DTT, 2 mM EDTA, 10 mM MgCl2, and 5 mM ATP) for 30 min at 20°C. Samples were run on a 10% polyacrylamide Phos‐tag gel (Kinoshita et al, 2006) and visualized by Coomassie staining. For inhibition by CK1‐7, 1 mM CK1‐7 was added at the start of the incubation.
In vitro translation and pull‐down assays
In vitro translation of MBP‐fused S. cerevisiae Mam187–191 fragments and pull‐down assays with His‐MBP‐Hrr251–394 K38R were performed as previously described (Corbett & Harrison, 2012). Prey constructs (Mam187–191 and mutants) were cloned into a plasmid with a T7 promoter, leading Kozak sequence, and an N‐terminal MBP‐fusion, and translated using a TNT T7 Transcription/Translation kit (Promega) with 35S‐methionine. Ten micrograms of purified bait protein (His‐MBP‐Hrr251–394 K38R) was incubated with 10 μl of the translation reaction in binding buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 20 mM imidazole, 5% glycerol, 1 mM dithiothreitol (DTT), and 0.1% NP‐40) for 90 min at 4°C, then 15 μl Ni‐NTA beads were added, and the mixture was incubated for further 45 min. Beads were washed three times with 0.5 ml buffer, then eluted with 25 μl elution buffer (2× SDS–PAGE loading dye plus 400 mM imidazole) and boiled. Samples were run on SDS–PAGE; then, the gel was dried and scanned with a phosphorimager.
Yeast strains and sporulation
Wild‐type S. cerevisiae SK1 strains (A4841, A4842) were generously provided by A. Amon. Mutant strains were generated with PCR‐based methods as described (Longtine et al, 1998). The natMX4 selection marker was derived from plasmid pAG25 (Goldstein & McCusker, 1999). For spore viability, cells were grown on YPD agar and then patched onto SPO medium (1% KOAc) for 48–72 h. About 46–48 tetrads were dissected for each strain. Yeast strain genotypes can be found in Table EV2.
Synchrotron support statements
Portions of this work were conducted at the Northeastern Collaborative Access Team beamlines at the Advanced Photon Source at Argonne National Laboratory, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24‐ID‐C beamline is funded by a NIH‐ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE‐AC02‐06CH11357.
Portions of this work were conducted at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by DOE Office of Biological and Environmental Research. Additional support for beamline 12.3.1 comes from the National Institute of Health project MINOS (R01GM105404).
Author contributions
QY designed the experiments, purified all proteins, determined all crystal structures, and performed quantitative ATPase assays; SNU performed spore viability measurements; TYS measured Hrr25 phosphorylation of the S. cerevisiae Mtw1 complex; KDC designed the experiments, helped with structure determination, and performed ATPase assays. KDC and QY wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Acknowledgements
The authors thank A. Shiau and A. Motamedi for providing the E. coli strain expressing λ‐phosphatase, A. Amon for providing yeast strains, J. Liang and R. Suhandynata for advice on yeast strains and cloning, and the staffs of the NE‐CAT beamlines at the Advanced Photon Source, Argonne National Laboratory; and beamline 12.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory (synchrotron support statements in Materials and Methods). We thank C. Asbury, S. Biggins, A. Marston, and members of the Corbett laboratory for helpful discussions and comments. This work was supported by the Ludwig Institute for Cancer Research and the National Institutes of Health R01‐GM104141.
The EMBO Journal (2016) 35: 2139–2151
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D, Zhang J, Menon S, Lord C, Chen S, Helm JR, Thorsen K, Corbett KD, Hay JC, Ferro‐Novick S (2013) Sit4p/PP6 regulates ER‐to‐Golgi traffic by controlling the dephosphorylation of COPII coat subunits. Mol Biol Cell 24: 2727–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito IL, Monje‐Casas F, Amon A (2010) The Lrs4‐Csm1 monopolin complex associates with kinetochores during anaphase and is required for accurate chromosome segregation. Cell Cycle 9: 3611–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Applen Clancey SE, Chacón JM, Gardner MK, Berman J (2013) Monopolin recruits condensin to organize centromere DNA and repetitive DNA sequences. Mol Biol Cell 24: 2807–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22: 3099–3108 [DOI] [PubMed] [Google Scholar]
- Chijiwa T, Hagiwara M, Hidaka H (1989) A newly synthesized selective casein kinase I inhibitor, N‐(2‐aminoethyl)‐5‐chloroisoquinoline‐8‐sulfonamide, and affinity purification of casein kinase I from bovine testis. J Biol Chem 264: 4924–4927 [PubMed] [Google Scholar]
- Cook A, Lowe ED, Chrysina ED, Skamnaki VT, Oikonomakos NG, Johnson LN (2002) Structural studies on phospho‐CDK2/cyclin A bound to nitrate, a transition state analogue: implications for the protein kinase mechanism. Biochemistry 41: 7301–7311 [DOI] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee L‐S, Walz T, Amon A, Harrison SC (2010) The monopolin complex crosslinks kinetochore components to regulate chromosome‐microtubule attachments. Cell 142: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Harrison SC (2012) Molecular architecture of the yeast monopolin complex. Cell Rep 1: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (2013) PHYLIP (Phylogeny Inference Package) version 3.695
- Flotow H, Roach PJ (1991) Role of acidic residues as substrate determinants for casein kinase I. J Biol Chem 266: 3724–3727 [PubMed] [Google Scholar]
- Gallego M, Virshup DM (2007) Post‐translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–148 [DOI] [PubMed] [Google Scholar]
- Ghalei H, Schaub FX, Doherty JR, Noguchi Y, Roush WR, Cleveland JL, Stroupe ME, Karbstein K (2015) Hrr25/CK1δ‐directed release of Ltv1 from pre‐40S ribosomes is necessary for ribosome assembly and cell growth. J Cell Biol 208: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae . Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH (2011) Mechanisms of chromosome number evolution in yeast. PLoS Genet 7: e1002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, Kearsey SE, Shirahige K, Allshire RC, Nasmyth K (2007) The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol 17: 1190–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra MF, Liskay RM, Ou AC, DeMaggio AJ, Burbee DG, Heffron F (1991) HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science 253: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Hornung P, Maier M, Alushin GM, Lander GC, Nogales E, Westermann S (2011) Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J Mol Biol 405: 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y (2010) Shugoshin‐PP2A counteracts casein‐kinase‐1‐dependent cleavage of Rec8 by separase. Nat Cell Biol 12: 500–506 [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom K, Salmon ED (2006) Molecular architecture of a kinetochore–microtubule attachment site. Nat Cell Biol 8: 581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild‐type catalytic proficiency. Protein Eng 14: 993–1000 [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W (2010) Rec8 phosphorylation by casein kinase 1 and Cdc7‐Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell 18: 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Quartey P, Li H, Volkart L, Hatzos C, Chang C, Nocek B, Cuff M, Osipiuk J, Tan K, Fan Y, Bigelow L, Maltseva N, Wu R, Borovilos M, Duggan E, Zhou M, Binkowski TA, Zhang R‐G, Joachimiak A (2008) Large‐scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods 5: 853–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita‐Kikuta E, Takiyama K, Koike T (2006) Phosphate‐binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5: 749–757 [DOI] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Löhler J, Stöter M (2005) The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal 17: 675–689 [DOI] [PubMed] [Google Scholar]
- Lawrimore J, Bloom K, Salmon ED (2011) Point centromeres contain more than a single centromere‐specific Cse4 (CENP‐A) nucleosome. J Cell Biol 195: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longenecker KL, Roach PJ, Hurley TD (1996) Three‐dimensional structure of mammalian casein kinase I: molecular basis for phosphate recognition. J Mol Biol 257: 618–631 [DOI] [PubMed] [Google Scholar]
- Longenecker KL, Roach PJ, Hurley TD (1998) Crystallographic studies of casein kinase I delta toward a structural understanding of auto‐inhibition. Acta Crystallogr D Biol Crystallogr 54: 473–475 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR‐based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lord C, Bhandari D, Menon S, Ghassemian M, Nycz D, Hay J, Ghosh P, Ferro‐Novick S (2011) Sequential interactions with Sec23 control the direction of vesicle traffic. Nature 473: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7: R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Ohsumi Y, Nakatogawa H (2014) Hrr25 phosphorylates the autophagic receptor Atg34 to promote vacuolar transport of α‐mannosidase under nitrogen starvation conditions. FEBS Lett 588: 3862–3869 [DOI] [PubMed] [Google Scholar]
- Peng Y, Grassart A, Lu R, Wong CCL, Yates J, Barnes G, Drubin DG (2015a) Casein kinase 1 promotes initiation of clathrin‐mediated endocytosis. Dev Cell 32: 231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Moritz M, Han X, Giddings TH, Lyon A, Kollman J, Winey M, Yates J, Agard DA, Drubin DG, Barnes G (2015b) Interaction of CK1δ with γTuSC ensures proper microtubule assembly and spindle positioning. Mol Biol Cell 26: 2505–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K (2006) Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell 126: 1049–1064 [DOI] [PubMed] [Google Scholar]
- Pfaffenwimmer T, Reiter W, Brach T, Nogellova V, Papinski D, Schuschnig M, Abert C, Ammerer G, Martens S, Kraft C (2014) Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep 15: 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Cipak L, Polakova S, Hyppa RW, Cipakova I, Anrather D, Karvaiova L, Mechtler K, Smith GR, Gregan J (2015) Casein kinase 1 and phosphorylation of cohesin subunit Rec11 (SA3) promote meiotic recombination through linear element formation. PLoS Genet 11: e1005225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MA (2006) CKI, there's more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev 20: 399–410 [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K (2003) Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 4: 535–548 [DOI] [PubMed] [Google Scholar]
- Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, Ammerer G, Mechtler K, Gregan J (2010a) Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle 9: 2657–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpf C, Cipak L, Schleiffer A, Pidoux A, Mechtler K, Tolić‐Nørrelykke IM, Gregan J (2010b) Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle 9: 3997–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T, Watanabe Y (2015) Phosphorylation of cohesin Rec11/SA3 by casein kinase 1 promotes homologous recombination by assembling the meiotic chromosome axis. Dev Cell 32: 220–230 [DOI] [PubMed] [Google Scholar]
- Sarangapani KK, Duro E, Deng Y, Alves Fde L, Ye Q, Opoku KN, Ceto S, Rappsilber J, Corbett KD, Biggins S, Marston AL, Asbury CL (2014) Sister kinetochores are mechanically fused during meiosis I in yeast. Science 346: 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Shenoy RT, Dalgaard JZ, Newnham L, Hoffmann E, Millar JBA, Arumugam P (2013) Monopolin subunit Csm1 associates with MIND complex to establish monopolar attachment of sister kinetochores at meiosis I. PLoS Genet 9: e1003610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T, Maco B, Petfalski E, Tollervey D, Böttcher B, Aebi U, Hurt E (2006) Hrr25‐dependent phosphorylation state regulates organization of the pre‐40S subunit. Nature 441: 651–655 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr 66: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW (2005) Protein production by auto‐induction in high density shaking cultures. Protein Expr Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T, Watanabe Y (2011) Condensin association with histone H2A shapes mitotic chromosomes. Nature 474: 477–483 [DOI] [PubMed] [Google Scholar]
- Tan S (2001) A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli . Protein Expr Purif 21: 224–234 [DOI] [PubMed] [Google Scholar]
- Tanaka C, Tan L‐J, Mochida K, Kirisako H, Koizumi M, Asai E, Sakoh‐Nakatogawa M, Ohsumi Y, Nakatogawa H (2014) Hrr25 triggers selective autophagy‐related pathways by phosphorylating receptor proteins. J Cell Biol 207: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SB, Nasmyth K (2000) Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103: 1155–1168 [DOI] [PubMed] [Google Scholar]
- Vancura A, Sessler A, Leichus B, Kuret J (1994) A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J Biol Chem 269: 19271–19278 [PubMed] [Google Scholar]
- Walter TS, Meier C, Assenberg R, Au K‐F, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM (2006) Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14: 1617–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hoekstra MF, DeMaggio AJ, Dhillon N, Vancura A, Kuret J, Johnston GC, Singer RA (1996) Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol Cell Biol 16: 5375–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Davis S, Menon S, Zhang J, Ding J, Cervantes S, Miller E, Jiang Y, Ferro‐Novick S (2015) Ypt1/Rab1 regulates Hrr25/CK1δ kinase activity in ER‐Golgi traffic and macroautophagy. J Cell Biol 210: 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G (2007) Structures and functions of yeast kinetochore complexes. Annu Rev Biochem 76: 563–591 [DOI] [PubMed] [Google Scholar]
- Winey M, Morgan GP, Straight PD, Giddings TH, Mastronarde DN (2005) Three‐dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol Biol Cell 16: 1178–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RM, Carmel G, Sweet RM, Kuret J, Cheng X (1995) Crystal structure of casein kinase‐1, a phosphate‐directed protein kinase. EMBO J 14: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RM, Carmel G, Kuret J, Cheng X (1996) Structural basis for selectivity of the isoquinoline sulfonamide family of protein kinase inhibitors. Proc Natl Acad Sci USA 93: 6308–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Rosenberg SC, Moeller A, Speir JA, Su TY, Corbett KD (2015) TRIP13 is a protein‐remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. Elife 4: 213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeringo NA, Murphy L, McCloskey EA, Rohal L, Bellizzi JJ (2013) A monoclinic crystal form of casein kinase 1|δ. Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Shi Q, Varia SN, Xing S, Klett BM, Cook LA, Herman PK (2016) The activity‐dependent regulation of protein kinase stability by the localization to P‐bodies. Genetics 203: 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File


