Abstract
To cope with DNA damage, proliferating cells have evolved sophisticated mechanisms including cell cycle arrest and activation of DNA repair. Paradoxically, various DNA damage response pathways are promoted by cyclin‐dependent kinase (CDK) activity, while cell cycle remains arrested. New work in The EMBO Journal shows that plant cells have evolved intricate ways to resolve this dilemma, by utilizing distinct and specialized CDKs for cell cycle progression and homologous recombination.
Subject Categories: Cell Cycle; DNA Replication, Repair & Recombination; Plant Biology
The cell division cycle consists of a unidirectional series of highly coordinated events that end up in the production of two daughter cells. Its main driving force is the activity of various cyclin/CDK complexes, allowing cells to progress from G1 into S phase, through S phase, and in G2 phase enabling chromosome segregation during mitosis. Unidirectional progression is achieved by fluctuations in CDK activity, which results from a sophisticated regulatory balance involving primarily cyclical accumulation and timely degradation of their cyclin subunits, the presence of CDK inhibitors, and the action of CDK‐activating or CDK‐inhibiting kinases and phosphatases. In addition to normal cell cycle progression, the unavoidable occurrence of various types of DNA damage either resulting from normal cellular physiology or from attack by a plethora of exogenous DNA‐damaging agents requires a fast and precise response of the proliferating cell.
The cellular response to the presence of DNA damage is of fundamental importance to prevent the accumulation of DNA lesions, the occurrence of chromosomal replication and segregation errors, and the transmission of mutated genomes to daughter cells. Briefly, such cellular responses include (i) various complementary mechanisms for immediate cell cycle arrest to prevent cell division with damaged DNA, (ii) activation of appropriate DNA repair pathways to fix the DNA damage in a highly lesion‐specific manner, and (iii) activation of DNA damage tolerance pathways.
Analyses of the pathways contributing to damage‐induced cell cycle arrest in yeast, animal, and plant cells have revealed that they all use very similar overall strategies, usually converging on transient inhibition of cyclin/CDK activity (Ciccia & Elledge, 2010; Aguilera & Garcia‐Muse, 2013; Hu et al, 2016). Strikingly, however, some of the main players involved are nevertheless distinct and unique in plants (Fig 1). Notably, while yeast and animal cells share most of the basic components, plant cells lack checkpoint and response factors such as Chk1, Chk2, p53, Mdm2, as well as the CDK inhibitor p21 and the checkpoint‐targeted CDK activator Cdc25 (Hu et al, 2016). Furthermore, although plant cells encode a homologue of the CDK inhibitory kinase Wee1, it does not participate in normal cell cycle regulation but only in the DNA damage response (Hu et al, 2016).
Figure 1. Summary of the major DNA damage response pathways in plant cells.
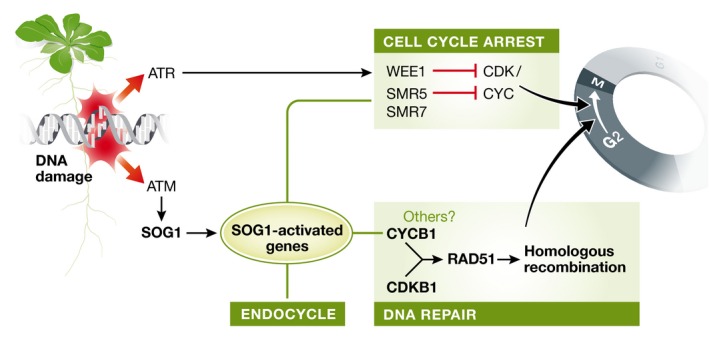
After sensing damage such as DSBs, an ATR‐dependent pathway contributes to cell cycle arrest by inhibiting CDK/cyclin complexes needed for the G2/M transition. Concomitantly, an ATM‐dependent pathway upregulates the plant‐specific transcription factor SOG1 that activates expression of hundreds of genes, including the CDK inhibitors SMR5 and SMR7, genes required for entering and progressing through the endocycle program, and CYCB1. CYCB1 together with the plant‐specific CDKB1 phosphorylates RAD51 to promote HR repair. Meanwhile, the key cell cycle CDK activity remains suppressed solving the dilemma of conflicting CDK regulation requirements in response to DNA damage.
Among the cellular DNA repair mechanisms, non‐homologous end joining (NHEJ) and homologous recombination (HR) pathways are crucial for repairing double‐strand breaks (DSBs), one of the most harmful lesions. HR is a highly accurate mechanism that relies on the use of a homologous template for DNA repair and can therefore only operate once the two daughter strands have been generated by bulk DNA replication. HR initiates with the recognition and subsequent resection of the DSB, resulting in formation of a 3′ DNA overhang covered by the single‐stranded DNA binding protein RPA, and finally proceeds to the exchange of RPA by members of the RAD51 recombinase family. All this occurs entirely while cells remain arrested after inactivation of CDK/cyclin complexes. This leads to a very important caveat, because CDK activity is also required to promote HR (Zhang et al, 2009), even though the underlying mechanisms are not yet fully understood.
How do cells solve this conundrum? As shown now by Weimer and coworkers, plants have devised a unique way to make cell cycle arrest by CDK inhibition and HR activation by CDK activation compatible (Weimer et al, 2016). Based on a combination of molecular, cellular, and genetic approaches at the organismal level to study the response of Arabidopsis thaliana to the induction of DSBs, the authors found that plant cells have solved the problem by using different CDK/cyclin complexes to drive G2/M transition and to trigger HR, respectively (Fig 1). Taking into account the high DNA damage tolerance of plants and their efficient DNA repair pathways, this makes the study of primary relevance not only from the basic science point of view, but also with regard to potential biotechnological benefits that can be derived from understanding the processes involved.
The study was footed on the somewhat paradoxical observation that in Arabidopsis, expression of the typical G2/M cyclin CYCB1;1 is strongly upregulated in response to DNA‐damaging treatments or DNA replication stress (Chen et al, 2003; Culligan et al, 2006; Ramirez‐Parra & Gutierrez, 2007). To test the possible implication of the CYCB1 gene family in the DNA damage response, the authors tested single and double mutants of the four Arabidopsis CYCB1 genes and found that all of them showed retarded root growth after cisplatin‐induced DNA interstrand crosslinking, which can only be repaired by HR, but not after treatment with bleomycin or hydroxyurea, which induces lesions that are also subject to NHEJ repair. Hypersensitivity to cisplatin seems to be specific to cycb1 mutants, because a triple CYCA mutant did not show exaggerated root growth defects after treatment with any of these drugs. As a consequence of DSB accumulation, a significant increase in γ‐H2AX foci was observed in the cycb1;1,cycb1;3 double mutant after cisplatin treatment. Furthermore, the authors found that cycb1 mutants were markedly impaired in HR both in the absence and presence of cisplatin. Together, these initial analyses suggested that CYCB1‐type cyclins were required for HR.
In Arabidopsis, CDKA;1 is functionally homologous to metazoan CDK1/2 and likewise used to control G1/S and G2/M transitions. However, Arabidopsis additionally encodes two CDKB1 and two CDKB2 kinases, all of which are plant specific. Experiments using single and double mutants in the CDKB genes clearly showed that CDKB1;1 and CDKB1;2, but not the CDKB2 members, were required for proper DNA damage responses similar to CYCB1 cyclins. In particular, the cdkb1;1,cdkb1;2 double mutant is hypersensitive to cisplatin and exhibits highly reduced root growth, high levels of DNA damage, and accumulation of γ‐H2AX foci. Finally, combining mutations in the CYCB1;1, CDKB1;1, and CDKB1;2 genes did not exacerbate the hypersensitivity phenotype, strongly suggesting that these CDKs and cyclins function in a common pathway.
These data together support the conclusion that CDKB/CYCB1 kinase activity is required for efficient HR‐mediated DNA repair. The use of a distinct, non‐CDKA kinase in DNA repair appears to be an evolutionary bypass solution for cellular situations where the major CDKA;1 activity is blocked in order to arrest the cell cycle as a primary response to DNA damage. This posed two obvious questions: How are CDKB1 and CYCB1 made available after DNA damage, and what are the downstream targets of the CDKB1/CYCB1 complex?
Using fluorescent translational fusion constructs for each of the four B1‐type cyclins and of the two CDKB1 kinases, Weimer and colleagues showed that only the promoters of CYCB1;1 and CDKB1;1 were directly activated in response to DNA damage (Weimer et al, 2016). Furthermore, ChIP experiments demonstrated that the plant‐specific transcriptional regulator SOG1, which is required for CYCB1;1 upregulation in an ATM‐dependent manner (Culligan et al, 2006; Yoshiyama et al, 2013), binds to the 5′ and 3′ regions of the CYCB1;1 gene. Of note, based on its combination of transcriptional activity and other properties, plant SOG1 is a potential functional homolog of animal p53, despite lacking sequence/structural similarity (Hu et al, 2016).
A candidate approach was used to identify downstream targets of CDKB1;1/CYCB1;1. Mutants in genes encoding members of the RAD51 recombinase family were hypersensitive to cisplatin and exhibited reduced root growth and γ‐H2AX foci accumulation in response to the drug. Moreover, damage‐induced RAD51 foci were highly reduced in the CYCB1 and CDKB1 double mutants, strongly pointing to RAD51 as a potential CDKB1;1/CYCB1;1 target. In vitro kinase assays using recombinant proteins demonstrated that RAD51 could be efficiently phosphorylated by CYCB1;1‐containing CDK complexes. Interestingly, recent work in human cells also pointed to a phosphorylation‐mediated Rad51 activation mechanism. Here, a yet‐to‐be‐identified CDK phosphorylates Rad51‐bound BRCA2, facilitating recruitment of the kinase Plk1 that, in turn, phosphorylates Rad51 to promote HR (Yata et al, 2014).
Although current results support the idea that NHEJ is the predominant pathway for DSB repair in plants, the current study clearly points to the relevance of HR in proliferating cells—the expression domain of SOG1—where CYCB1 and CDKB1 are expressed in a SOG1‐dependent manner after DNA damage (Weimer et al, 2016). These new concepts explain some of the discrepancies in the mode that plant cells cope with DNA damage. They will also serve as the basis for future work to understand the choice of DNA repair pathways depending on the type of DNA lesion, on the proliferative state of the cells and on their developmental stage. The use of Arabidopsis offers unique opportunities to carry out these studies at adult stages and at the genetic, molecular, cellular, and organismal levels.
Acknowledgements
The authors want to thank J.A. Tercero for insightful comments on the DNA response in yeast and animal cells. The CG laboratory is supported by grants BFU2015‐68396‐R (MINECO‐FEDER) and BIO2013‐50098‐EXP (MINECO). The CBMSO receives an institutional grant from Fundación Ramón Areces. We apologize to colleagues whose work was not cited or cited indirectly due to space limitations.
See also: AK Weimer et al (October 2016)
References
- Aguilera A, Garcia‐Muse T (2013) Causes of genome instability. Annu Rev Genet 47: 1–32 [DOI] [PubMed] [Google Scholar]
- Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H (2003) The transcriptional response of Arabidopsis to genotoxic stress – a high‐density colony array study (HDCA). Plant J 35: 771–786 [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB (2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Hu Z, Cools T, De Veylder L (2016) Mechanisms used by plants to cope with DNA damage. Annu Rev Plant Biol 67: 439–462 [DOI] [PubMed] [Google Scholar]
- Ramirez‐Parra E, Gutierrez C (2007) E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol 144: 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, Van Damme D, Sugimoto K, Koncz C, Doerner P, Umeda M, Schnittger A (2016) The plant‐specific CDKB1‐CYCB1 complex mediates homologous recombination repair in Arabidopsis . EMBO J 35: 2068–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yata K, Bleuyard JY, Nakato R, Ralf C, Katou Y, Schwab RA, Niedzwiedz W, Shirahige K, Esashi F (2014) BRCA2 coordinates the activities of cell‐cycle kinases to promote genome stability. Cell Rep 7: 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M (2013) ATM‐mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis . EMBO Rep 14: 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shim EY, Davis M, Lee SE (2009) Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks. DNA Repair 8: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]


