Abstract
The pervasive view of bacteria as strictly pathogenic has given way to an appreciation of the widespread prevalence of beneficial microbes within the human body1–3. Given this milieu, it is perhaps inevitable that some bacteria would evolve to preferentially grow in environments that harbor disease and thus provide a natural platform for the development of engineered therapies4–6. Such therapies could benefit from bacteria that are programmed to limit bacterial growth while continually producing and releasing cytotoxic agents in situ7–10. Here, we engineer a clinically relevant bacterium to lyse synchronously at a threshold population density and to release genetically encoded cargo. Following quorum lysis, a small number of surviving bacteria reseed the growing population, thus leading to pulsatile delivery cycles. We use microfluidic devices to characterize the engineered lysis strain and we demonstrate its potential as a drug delivery platform via co-culture with human cancer cells in vitro. As a proof of principle, we track the bacterial population dynamics in ectopic syngeneic colorectal tumors in mice. The lysis strain exhibits pulsatile population dynamics in vivo, with mean bacterial luminescence that remained two orders of magnitude lower than an unmodified strain. Finally, guided by previous findings that certain bacteria can enhance the efficacy of standard therapies11, we orally administer the lysis strain, alone or in combination with a clinical chemotherapeutic, to a syngeneic transplantation model of hepatic colorectal metastases. We find that the combination of both circuit-engineered bacteria and chemotherapy leads to a notable reduction of tumor activity along with a marked survival benefit over either therapy alone. Our approach establishes a methodology for leveraging the tools of synthetic biology to exploit the natural propensity for certain bacteria to colonize disease sites.
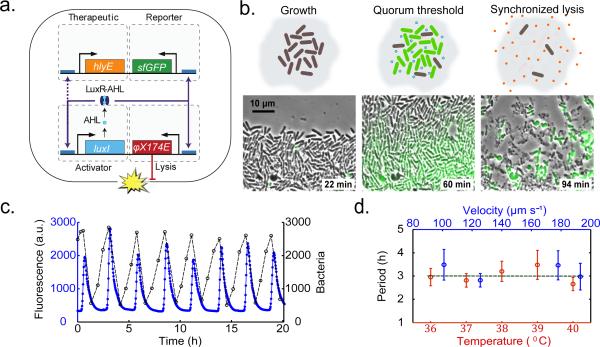
In order to control population levels and facilitate drug delivery using bacteria, we engineered a synchronized lysis circuit (SLC) using coupled positive and negative feedback loops that have previously been used to generate robust oscillatory dynamics12,13. The circuit (Fig. 1a) consists of a common promoter that drives expression of both its own activator (positive feedback) and a lysis gene (negative feedback). Specifically, the luxI promoter regulates production of autoinducer (AHL), which binds LuxR and enables it to transcriptionally activate the promoter. Negative feedback arises from cell death that is triggered by a bacteriophage lysis gene (ϕX174 E) which is also under control of the luxI promoter13–15. Importantly, AHL can diffuse to neighboring cells and thus provides an intercellular synchronization mechanism.
Fig. 1.
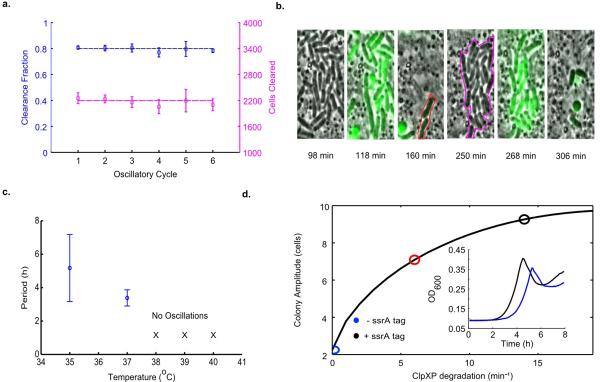
Construction and characterization of the SLC. (a) The circuit contains an activator13 and lysis plasmid. When the population reaches the quorum threshold at a critical AHL concentration, the luxI promoter drives the transcription of gene E for lysis, LuxI, and sfGFP or luxCDABE as the reporter module. The luxI or the ptac promoter also drives the transcription of the therapeutic gene for the stabilized circuit used in vivo. LuxR in this system is driven by the native pLuxR promoter. (b) A schematic that illustrates the main stages of each lysis cycle from seeding to quorum ‘firing’. Shown below are typical time series images of the circuit-harboring cells undergoing the three main stages of quorum firing in a microfluidic growth chamber12. (c) Fluorescence profile of a typical microfludic experiment. The estimated cell population trajectory reveals that lysis events correspond to peaks of sfGFP fluorescence. (d) Period as a function of estimated flow velocity in the media channel of the microfluidic device and environmental temperature. Error bars indicate ±1 standard deviation for 13 - 50 peaks. The above experiments were performed with Strain 1, see Supplementary Information for complete strain information.
The bacterial population dynamics arising from the synchronized lysis circuit can be conceptualized as a slow buildup of the signaling molecule (AHL) to a threshold level, followed by a lysis event that rapidly prunes the population and enables the release of bacterial contents (Fig. 1b). After lysis, a small number of remaining bacteria begin to produce AHL anew, allowing the “integrate and fire” process to be repeated in a cyclical fashion. We used microfluidic devices to observe growth and lysis with a fluorescent protein (sfGFP) as a proxy for circuit dynamics in attenuated S. typhimurium (Supplementary Videos 1 and 2). We observed periodic lysis events characterized by peaks in the fluorescent reporter expression that correspond to population lysis (Fig. 1c). The fraction of lysed cells remains consistent across subsequent cycles, suggesting that lysis and survival occur in a stochastic manner (Extended Data Fig. 1a-b). Given the ultimate goal of implementation in an in vivo microenvironment characterized by variable growth conditions, we tested a range of incubation temperatures (36°C to 40°C) and perfusion rates (100μm/s to 200μm/s), measuring an average period of 3 hours across all conditions (Fig. 1d). These findings demonstrate that the SLC has the capacity to generate robust cycles of bacterial lysis in our microfluidic devices across a spectrum of environmental fluctuations that is likely to exist in an in vivo context.
The emergence of bacterial therapies in synthetic biology has accentuated the need for predictive modeling. This need stems from a bottleneck created by a difference in the timescales for bacterial cloning versus animal experiments; the circuits required for candidate therapies can be created much faster than they can be tested in vivo. Therefore, in order to quantitatively characterize the SLC concept before testing in animal models, we developed a computational model (Fig. 2a and and Supplementary Information) in order to define an optimal strategy for subsequent testing in a lower-throughput animal model setting. We found that high production and degradation rates of the feedback-controlling proteins resulted in a wider domain of oscillatory dynamics in the parameter space (Fig. 2b). This model is consistent with our observations that oscillations in S. typhimurium were more robust than in E. coli, where rates of protein production and degradation were previously found to be lower16 (Extended Data Fig. 1c and Supplementary Video 3). Since the ability to manipulate circuit behavior enhances the versatility of the system, we explored the tunability of the lysis period by adding an ssrA degradation tagging sequence on the LuxI protein. Consistent with model predictions, we observed an increased period and colony firing amplitude when tracking bacterial population dynamics (Fig. 2c-d and Extended Data Fig. 1d). The SLC thus enables tuning of the period and magnitude of delivery, which will be necessary for eventual application of this platform in the complex and fluctuating conditions present in vivo.
Fig. 2.
Computational modeling and tunability. (a) The model consists of intracellular variables (lysis gene E and LuxI concentrations) and extracellular variables (colony size and AHL concentrations). A time series of colony size (black line), colony AHL (blue line), intracellular LuxI (green line) and lysis protein concentrations (red line) are shown on the right. (b) The region in the model parameter space for clpXP mediated degradation (see Supplementary information) and flow where the model output is oscillatory increases with higher production and degradation terms. (c) Results from the computational model showing the ability to tune the oscillatory period by varying ClpXP mediated degradation of LuxI. (d) Fluorescence profiles showing lysis oscillations for LuxI ssrA (black line, Strain 2) and non-ssrA (blue line, Strain 1) tagged versions of the circuit. See Supplementary Information for complete model information.
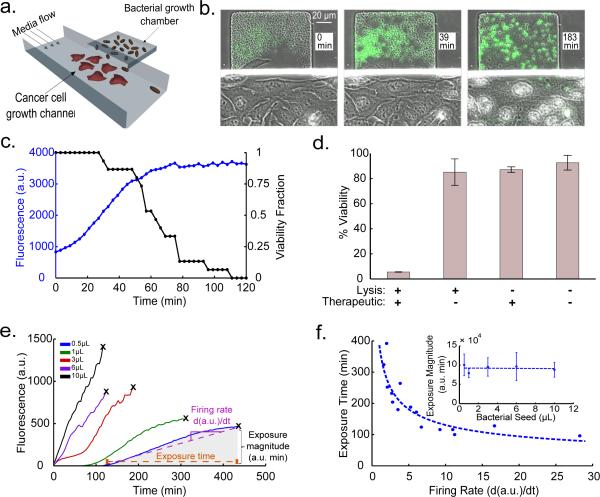
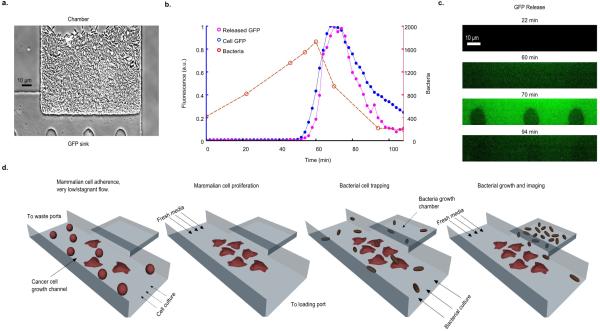
To incorporate a cytotoxic payload into the SLC strain, we added expression of Hemolysin E, encoded by hlyE of E. coli, which has been tested as a pore-forming anti-tumor toxin17. We initially confirmed the capability of the circuit to release intracellular contents by visualizing released sfGFP with a small microfluidic sink located beneath the growth chamber (Extended Data Fig. 2a-c). Then, in order to visualize bacterial lysis and killing of cancer cells in vitro via HlyE, we engineered a microfluidic device so that cancer cells adhere inside a growth channel that is flanked by smaller bacterial growth chambers, which permits simultaneous single-cell visualization of bacterial lysis and cancer cell death (Extended Data Fig. 2d). After co-culturing human cervical cancer HeLa cells with S. typhimurium harboring the SLC circuit, we observed HeLa cell death upon the onset of bacterial lysis, indicating efficient toxin release (Fig. 3a-b, Supplementary Videos 4 and 5). Complete cell death occurred in the growth channel within ~111 min of initial sfGFP fluorescence (Fig. 3c). Thus, the SLC bacteria were capable of releasing HlyE at levels necessary to kill tumor-derived cells.
Fig. 3.
In vitro co-culture. (a) Schematic of the microfluidic co-culture with cancer cells and bacteria. Fluidic resistance was modified in this chip to achieve stable near-stagnant flow reduction to allow for cancer cell adherence and for diffusion of released therapeutic from the trap to the channel (methods in Supplementary Information). (b) Frames from the co-culture time series sequentially visualizing S. typhimurium (Strain 3) ‘firing’, lysis, and HeLa cell death. (c) Fluorescent profile of the bacteria and HeLa cell viability fraction (# live cells / # dead cells in image frames) from (b) with time. (d) % viability of HeLa cells co-cultured with supernatant from S. typhimurium culture harboring the SLC + HlyE (Strain 4), the SLC only (Strain 5), constitutive hlyE only (Strain 6), or no plasmid (Strain 7). Error bars indicate ±1 standard error averaged over three measurements. (e) Fluorescence profile of the SLC + HlyE (Strain 4) co-cultured with HeLa cells at various initial seeding densities. The black ‘x’ marks the point of complete HeLa cell death. (f) The toxin exposure time, measured from the initial presence of fluorescence to HeLa cell death, as a function of the sfGFP production rate (see example in (e)). Although the time to death depends on seeding, the total magnitude of exposure remains conserved (inset). Error bars indicate ±1 standard error for three measurements. See Supplementary Information for ELH1301 host strain information.
We assessed the toxicity of released SLC or control bacterial contents in batch culture. As anticipated, we found that HeLa cells exposed to supernatant from a culture of the SLC bacteria bearing the hlyE module exhibited almost complete loss of viability (Fig. 3d), while the viability of HeLa cells exposed to supernatants of and equivalent dose of non-payload bearing SLC bacteria were only slightly impacted (~15%). We concluded that bacterial lysis allowed for efficient HlyE release in vitro and that natural intracellular bacterial contents do not significantly affect HeLa cell viability. We further investigated the delivery characteristics of the SLC bacteria with hlyE by seeding variable amounts of circuit-harboring bacteria with HeLa cultures in well plates. We observed that the time to HeLa cell death following initial seeding increased with lower bacterial seeding volumes, presumably resulting from the extended time needed for bacteria to reach the quorum threshold (Fig. 3e and Supplementary Video 6). Initial seeding with a larger volume of bacteria resulted in increased firing rates which corresponded to shorter HlyE exposure times until cell death, consistent with a greater magnitude of lysis and payload release, although the cumulative toxicity threshold appears to be similar in all cases (Fig. 3f). Based on these observations, one can adjust the seeding size of the bacterial population to determine the initial timing and release characteristics of the circuit.
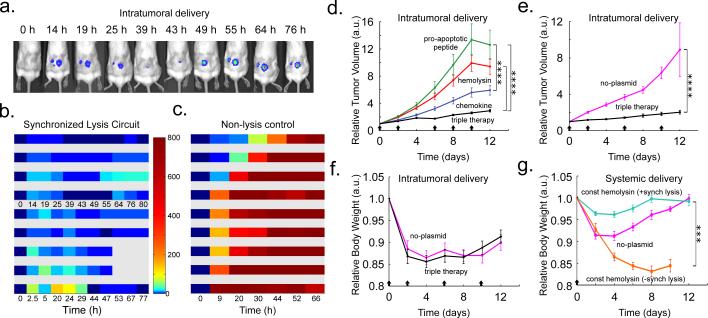
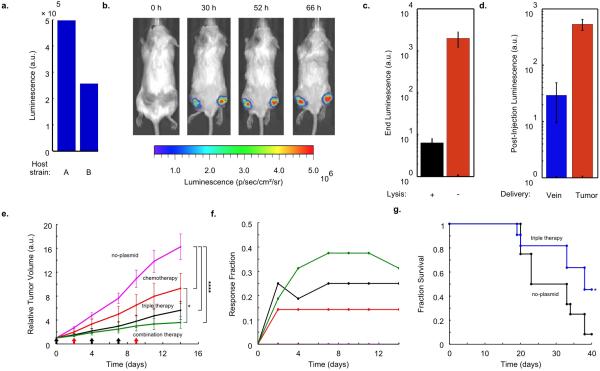
We next used a luciferase reporter to monitor bacterial population dynamics in grafted syngeneic colorectal tumors in mice. To minimize the extent of plasmid loss in the absence of antibiotic selection in vivo, we incorporated previously described stabilizing elements for plasmid retention and segregation into the SLC strain18–22. Additionally, we placed both the payload and luxCDABE genes (the in vivo reporter module) under the luxI promoter as an indicator of hlyE production and quorum firing via bacterial luminescence (Fig. 1a). Using a subcutaneous model of colorectal cancer (MC26 cell line) in immunocompetent mice, we intratumorally injected a strain of SLC bacteria (SLC-hly). We observed pulsatile bacterial population dynamics within the tumor (Fig. 4a-c, Extended Data Fig. 3a-b) using In-Vivo Imaging (IVIS) technology23, consistent with the design and in vitro characterization (Fig. 2). Importantly, the end luminescence intensity was on average ~300-fold lower than the constitutive control strain, indicating a significant decrease in bacterial population levels within the tumor (Extended Data Fig. 3c).
Fig. 4.
In vivo bacterial dynamics, tumor impact, and tolerability in a subcutaneous tumor model. (a) IVIS imaging over time of a mouse bearing two hind flank tumors injected once with the stabilized SLC-hly strain (Strain 8). (b) Single tumor density map trajectories of bacterial luminescence (relative to luminescence at 0h) for the SLC-hly strain (Strain 8). Data for each axis represents separate experiments. (c) Single tumor density map trajectories of bacterial luminescence for the genomically integrated constitutively luminescent strain (Strain 9). Intratumoral injection resulted in over 35-fold higher post-injection luminescence compared to intravenous injection (Extended Data Fig. 3d). (d) Average relative tumor volume over time for subcutaneous tumor bearing mice injected with SLC-hly (red line, Strain 10), SLC-cdd (green line, Strain 14), SLC-ccl21 (blue line, Strain 15), and all together (SLC-3) (black line). Bacteria were injected intratumorally on days 0, 2, 6, 8, and 10 (black arrows) (****P < 0.0001, two-way ANOVA with Bonferroni post test, n = 14 - 17 tumors, s.e.). (e) Average relative tumor volume over time for mice with subcutaneous tumors injected with the SLC-3 strains (black line, Strain 10, 14, and 15) and the no-plasmid control (magenta line, Strain 7). Bacteria were injected intratumorally on days 0, 2, 6, and 10 (black arrows) (****P < 0.0001, two-way ANOVA with Bonferroni post test, n = 18 - 19 tumors, s.e.). (f) Average relative body weight over time for mice with subcutaneous tumors injected with the SLC-3 strains (black line, Strain 10, 14, and 15) and the no-plasmid control (magenta line, Strain 7). Bacteria were injected intratumorally on days 0, 2, 6, and 10 (black arrows) (n=10 mice for both cases, s.e.). (g) Average relative body weight over time for subcutaneous tumor-bearing mice with a single intravenous injection of the SLC + constitutive hlyE (turquoise line, n=9 mice, Strain 11), a non-SLC strain with constitutive hlyE (orange line, n=5 mice, Strain 12), or the no-plasmid control strain (magenta line, n=9 mice, Strain 7) (***P < 0.001, two-way ANOVA with Bonferroni post test, s.e.).
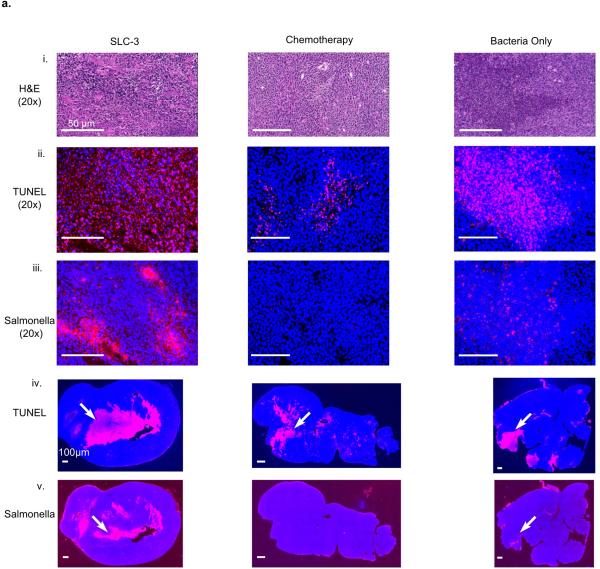
Given the ability to engineer bacterial population dynamics in tumor grafts, we next leveraged the versatility of the SLC bacteria as a delivery system to compare different classes of previously developed payloads. In addition to the hemolysin strain that was characterized in microfluidic devices, we created two additional SLC strains expressing genes to activate a host immune response (via T-cell and dendritic cell recruitment, using mCCL21) or trigger tumor cell apoptosis (using CDD-iRGD)24,25. Upon intratumoral injection, the immune recruitment strain elicited the strongest effect on tumor growth when compared to the hemolysis or apoptotic strains (Fig. 4d). We observed that an equal mixture of the three strains generated a stronger response than any single strain (Fig. 4d and Extended Data Fig.3 e-g), and on this basis we elected to pursue the “triple-strain” dose for further testing in order to minimize animal usage. In a side by side comparison, we observed that the tumor response to SLC triple-strain (SLC-3) injections was significantly larger than the response to unmodified bacteria (Fig. 4e). Additionally, upon necropsy, histopathological analysis of remnant tumors was performed for mice treated with the SLC-3 strains, chemotherapy, or unmodified bacteria. In mice treated with SLC-3 and non-circuit bacterial strains, robust staining of bacteria was observed by anti-Salmonella antibodies, showing localization of Salmonella within tumors. TUNEL staining indicated higher levels of apoptosis and cell death in SLC-3 treated tumors (Extended Data Fig. 4).
As a first step towards monitoring the effect of bacterial injections on the host, we compared how the triple-strain system affected body weight when administered intratumorally and intravenously, since the administration route affects bacterial localization (Extended Data Fig. 3d). We found that treatment with the SLC strains generated the same weight change as unmodified bacteria when administered intratumorally (Fig. 4f). However, intravenous administration of the SLC conferred a greater health benefit based on observations that SLC strains producing constitutive therapy were better tolerated than unmodified bacteria or non-SLC strains producing constitutive therapy (Fig. 4g). While further targeted studies are required to systematically explore the impact of these bacteria on host health, these preliminary experiments suggest that the SLC design can reduce the burden of bacterial injections.
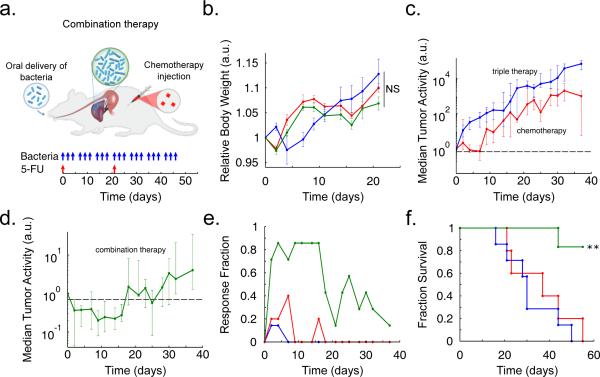
To explore a proof-of-principle for the application of our circuit in the context of in vivo tumors, we examined the efficacy of our system in an experimental syngeneic transplantation model of colorectal metastases within the liver. We had previously established that oral delivery of these bacterial strains led to safe and efficient colonization of hepatic colorectal metastases (see Methods), and that mice tolerated repeated dosing without overt adverse effects (Fig. 5a-b)22. In the context of bacteria-based therapeutic candidates, previous studies have shown that anaerobic bacteria can occupy avascular tumor compartments where chemotherapy is thought to be ineffective due to poor drug delivery11. Thus a synergistic effect may arise when bacteria are used to deliver drugs to the necrotic core of a tumor, while standard chemotherapy is used for the vascularized regions11,26. Inspired by this paradigm, we tested the combination of SLC-3 bacteria with a common clinical chemotherapy (5-fluorouracil, 5-FU). Tumors exhibited similar growth trajectories in response to repeated oral delivery of either the bacterial therapy alone, or two i.v. doses of 5-FU on Days 0 and 21 (Fig. 5c). In contrast, combination of these two applications led to a marked decrease in tumor activity over a period of 18 days, followed by a return to growth (Fig. 5d). During the initial 18-day period, a large fraction of the tumors were scored as eliciting at least a 30% reduction in tumor activity (Fig. 5e). The overall response led to roughly a 50% increase in the mean survival time for animals harboring incurable colorectal metastases (Fig. 5f). Improvements may arise from strategies for long term circuit stability or the utilization of additional therapeutic cargo.
Fig. 5.
In vivo testing in an experimental model of colorectal metastases in the liver via oral delivery of bacteria. (a) Schematic of the experimental syngeneic transplantation model of hepatic colorectal metastases in a mouse, with the dosing schedule of either engineered bacteria (SLC-3) or a common cytotoxic chemotherapeutic, the antimetabolite 5-FU. The SLC-3 strains were delivered orally while 5-FU was delivered via intraperitoneal injection. (b) Relative body weight over time for the mice with with hepatic colorectal metastases fed with the SLC-3 strains (blue line), injected with 5-FU chemotherapy (red line), or a combination of the two (green line). Error bars indicate ±1 standard error for 5 - 7 mice. (c) Median relative tumor activity, measured via tumor cell luminescence using IVIS imaging, for the chemotherapy and SLC-3 cases from (b). (d) Median relative tumor activity for the combination therapy case from (b). Error bars for (c) and (d) indicate the interquartile ranges for 5 - 7 mice. The dashed line marks relative tumor activity of 0.70. (e) Fraction of mice from the cases in (b) which respond with 30% reduction of tumor activity over time. (f) Fraction survival over time for the mice in (b) (**P < 0.01, log rank test; n = 5 - 7 mice).
The synchronized lysis circuit exemplifies a methodology for leveraging the tools of synthetic biology to exploit the ability of certain bacteria to colonize disease sites. In contrast to most drug delivery strategies, the synchronized lysis paradigm does not require pre-loading of a drug or the engineering of additional secretion machinery. In addition, it has the potential to decrease the likelihood of a systemic inflammatory response through population control; since the bacterial colony is pruned after each oscillatory lysis event, the design could mitigate an undesirable host response. The circuit may enable new bacterial drug delivery strategies through modulation of the frequency and amplitude of the population cycles over time. Along these lines, cyclic drug release may have broader implications, given recent insights into the effects of circadian rhythms on host-microbial interactions and metabolic disorders27,28. Such engineering strategies may allow for the development of therapeutic communities within in vivo environments, where population dynamics are driven by interacting viruses, bacteria, and host immune cells29.
Methods
Strains and Plasmids
Our circuit strains were cultured in LB media with 50 μg ml−1 and 34 μg ml−1 of Kanamycin and Chloramphenicol respectively, along with 0.2% glucose, in a 37°C incubator. Mammalian cells (HeLa CCL-2 from ATCC, verified by third party cell line authentication services using an STR multiplex system) were cultured in DMEM media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (CellGro 30-002-CI), placed inside a tissue culture incubator at 37°C maintained at 5% CO2. Plasmids were constructed using the CPEC method of cloning or using standard restriction digest/ligation cloning. The activator plasmid (Kan, ColE1) was used in previous work from our group, while the lysis plasmid was constructed by taking the lysis gene, E, from the ePop plasmid via PCR and cloning it into a vector (Chlor, p15A) under the control of the LuxI promoter13,15. The hlyE gene was taken via PCR from the genomic DNA of MG1655, while mCCL21 (mouse CCL21) and CDD-iRGD were synthesized. These genes were cloned into the lysis plasmid, under the control of either the ptac or pLuxI promoters. Co-culturing was performed with HeLa cells and either motile or non-motile S. typhimurium, SL1344. For full strain and plasmid information, please refer to the Supplementary Information.
Microfluidics and Microscopy
The microfluidic devices and experiment preparation protocols used in this study are similar to those previously reported from our group13. The bacteria growth chambers were 100×100 μm in area and approximately 1.4 μm in height. For co-culture experiments on the chip, we first loaded a suspended culture of HeLa cells in the device media channels at very low flow rates, to allow for adherence, and then incubated the device in a tissue culture incubator for 0.5 - 2 days to allow for proliferation. On the day of the experiment, the device was transferred to the microscope and circuit-containing bacteria were loaded in the growth chambers before imaging. Acquisition of images was performed with a Nikon TI2 using a Photometrics CoolSnap cooled CCD camera. The scope and accessories were programmed using the Nikon Elements software. Additional details on microfluidics and microscopy can be found in the Supplementary Information.
In vivo Experiments
All animal work was approved by the committee on animal care (MIT, protocol 0414-022-17). The cell line (MC26-LucF, Tanabe lab, Massachusetts General Hospital) was obtained from, and authenticated by, the Tanabe lab, MGH. The cell line was tested several times to be mycoplasma free before implantation in mice. Sample sizes for mice were determined by expected effect size to produce a power of 0.8-0.9. Mice were blindly randomized into various groups using a random number generator.
Subcutaneous Tumor Model
Animal experiments were performed on 6 week old female BALB/c mice (Taconic Biosciences, Inc.) with bilateral subcutaneous hind flank tumors from an implanted mouse colon cancer cell line. The concentration for implantation of the tumor cells was 1e8 cells ml−1 in DMEM (no phenol red). Cells were then implanted subcutaneously at a volume of 100 μL per flank, with each implant consisting of 1e7 cells. Tumors were typically grown to an average of 300 mm3 before experiments.
Experimental Liver Metastasis Model
The experimental metastasis model was generated by injecting luciferase-producing mouse cancer cells into surgically externalized spleens of immuno-competent mice. Tumor cells seeded the liver during 90 seconds after which the spleen was removed to prevent ectopic tumor growth30. The MC26-LucF cell line was used (Tanabe Lab, MGH) and injected at 5e4 cells/100 uL PBS into the spleens of female Balb/c mice 6 weeks of age (Taconic Biosciences, Inc.). For the liver metastasis model, tumors were grown for 5-7 days to an average total tumor burden of 143 mm3 before experiments.
Bacterial Growth and Administration
Bacterial strains were grown overnight in LB media containing appropriate antibiotics and 0.2% glucose as for the in vitro experiments. A 1/100x dilution in fresh media with antibiotics was started the day of injection and grown until an OD < 0.1 to prevent bacteria from reaching the quorum threshold (for SLC specifically). Bacteria were spun down and washed 2-3x with sterile PBS before injection into mice. Intratumoral injections of bacteria were performed at a concentration of 5e7 cells ml−1 in PBS with a total volume of 10-20 μL injected per tumor, while intravenous injections were given at a total volume of 100 μL. For the SLC-3 strains injection, this final volume was equally divided between the three strains at the indicated density. For liver metastasis experiments, bacteria were grown in LB media containing appropriate antibiotics and 0.2% glucose until they reached an OD of 0.05, after which they were concentrated to 1-5e9 bacteria/mL and delivered via oral gavage.
Post-Administration Monitoring for Subcutaneous Liver Metastasis Models
Luminescent signal was measured with the IVIS Spectrum in vivo imaging system following bacterial injection. Measurements were compared relative to pre-injection values to follow dynamics. Subcutaneous tumor volume was quantified using calipers to measure the length, width, and height of each tumor throughout the imaging course (V=L×W×H). Volumes were compared to pre-injection values to follow physical tumor growth. Survival of mice was measured as the time from the beginning of the experiment up to the day when mice were moribund and euthenized.
Statistical Analysis
Statistical tests were calculated either in Microsoft Excel (Student's t-test) or GraphPad Prism 5.0 (ANOVA with Bonferroni post test, Log-rank test). The details of the statistical tests carried out are indicated in respective figure legends. Where data were approximately normally distributed, values were compared using either a Student's t-test or one-way ANOVA for single variable, or a two-way ANOVA for two variables. Mice were randomized in different groups before experiments.
Extended Data
Extended Data Fig. 1.
Various properties of the SLC. (a) The fraction and number of bacterial cells cleared per consecutive oscillatory cycle in the growth chamber for a typical microfluidic experiment for S. typhimurium, including the effects of lysis and flow of cells outside of the trap (Strain 1). (b) Subset of time series images from the experiment in (a) showing a portion of the growth chamber where survivors of the initial lysis event (160 min frame, red outline) produce progeny (250 min frame, magenta outline) which are lysis sensitive. (c) Period as a function of the environmental temperature for E. coli (Strain 13). The circuit does not oscillate for temperatures above 37°C in E. coli. Error bars indicate ±1 standard deviation for 12 - 19 peaks. (d) Colony amplitude at quorum firing for increasing degradation on the LuxI activator protein in the computational model. These simulation results are supported by batch well-plate experiments of the LuxI ssrA (black line, Strain 2) and non-ssrA (blue line, Strain 1) tagged versions of the circuit in S. typhimurium (inset).
Extended Data Fig. 2.
Investigating lysis mediated intracellular release. (a) A bacterial growth chamber with a 0.4μm sink for sfGFP visualization after release. (b) Number of bacteria (red line), bacterial fluorescence (blue line), sink fluorescence (pink line) for a typical oscillatory cycle (Strain 1). (c) Fluorescence time series images of the microfluidic sink from (b). (d) General procedure for performing bacterial and cancer cell co-culture experiments in a microfluidic device (also see Supplementary Information).
Extended Data Fig. 3.
In vivo expression and therapy testing. (a) End-point in vitro luminescence intensity for SLC strains after ~20 h of growth. Host strains A and B are the host bacteria for Strains 8 and 10. They are ELH1301 and ELH 430, respectively. Host A exhibits ~2-fold higher luminescence with the same circuit than Host B. (b) IVIS imaging over time of a mouse bearing subcutaneous tumors injected with a genomically integrated constitutively luminescent strain (Strain 9). (c) End-point in vivo bacterial luminescence of the SLC-hly strain and the constitutively luminescent strain from the experiments presented in Fig. 4. Error bars represent the standard error of the mean bacterial luminescence from 9 tumors. (d) Post-injection in vivo bacterial luminescence for the constitutively luminescent strain administered intravenously (vein) or intratumorally (tumor). Luminescence was measured ~20 h post-injection. Error bars represent the standard error of the mean bacterial luminescence from 6 and 9 tumors for the intravenous and intratumoral cases, respectively. (e) Average relative tumor volume over time for subcutaneous tumor bearing mice injected with the no-plasmid bacterium (Strain 7), 5-FU chemotherapy, the SLC-3 strains, and the combination of SLC-3 with chemotherapy. Bacteria were injected intratu-morally on days 0, 4, and 7 (black arrows), and chemotherapy was administered on days 2 and 9 (red arrows) (*P < 0.05, ****P < 0.0001, two-way ANOVA with Bonferroni post test, n = 12 - 16 tumors, s.e.). (f) Fraction of mice from the cases in (e) which respond with 30% reduction of tumor volume over time. (g) Fraction survival over time for mice with hepatic colorectal metastases fed with either the SLC-3 strains (blue line) or the no-plasmid control (black line) (*P < 0.05, log rank test; n = 11 - 12 mice).
Extended Data Fig. 4.
(a) Histology of tumor sections taken from mice with different treatments 3 days post administration: (i) H and E staining for tissue sections intravenously injected with a combination of therapeutic bacteria (SLC-3), chemotherapy (5-FU), or a bacteria control with no therapeutic (Strain 7). (ii) TUNEL staining (red) in the same sections indicating cell apoptosis. (iii) Salmonella immunohistochemistry (red) in the same sections confirming presence of bacteria in tumors. Scale bars for (i), (ii), and (iii) denote 50μm. (iv) and (v) TUNEL and Salmonella staining (red) in the entire tumor sections (examples indicated by arrows). Scale bars for (iv) and (v) denote 100μm. DAPI staining (blue) was used to obtain a measure of live and dead cells in (ii) - (iv). Histology slices (n=6) from 20x images were compared across the groups and mean intensity of TUNEL staining, normalized by sample area, was demonstrated to be significantly higher for SLC-3 compared to the other two groups (P<0.0001, one-way ANOVA), and not significantly different between the chemotherapy and bacteria only cases.
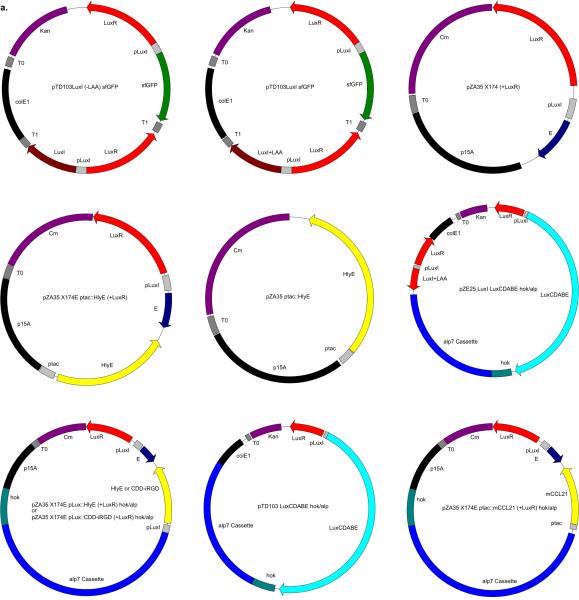
Extended Data Fig. 5.
Shown are the main plasmids used in this study (see Supplementary Information for more details).
Extended Data Table 1.
A list of strains and respective plasmids used in this study (see Supplementary Information for more details).
| Strain # | Strain Name | Host Bacterium | Plasmid(s) |
|---|---|---|---|
| 1 | MOD47 | SL1344, M913 | pTD103 luxI (−LAA) sfGFP + pZA35 X714E (+LuxR) |
| 2 | MOD46a | SL1344, M913 | pTD103 luxI sfGFP + pZA35 X714E (+LuxR) |
| 3 | MOD67 | SL1344, M913 | pTD103 luxI (−LAA) sfGFP + pZA35 X714E (+LuxR) ptac::HlyE |
| 4 | MOD61 | SL1344, ELH1301 | pTD103 luxI sfGFP + pZA35 X714E (+LuxR) ptac::HlyE |
| 5 | MOD64 | SL1344, ELH1301 | pTD103 luxI sfGFP + pZA35 X714E (+LuxR) |
| 6 | MOD65 | SL1344, ELH1301 | pZA35 ptac::HlyE |
| 7 | ELH1301 | SL1344, ELH1301 | N/A |
| 8 | MOD105 | SL1344, ELH430 | pZE25 luxI luxCDABE hok/alp + pZA35 X714E (+LuxR) pLux::HlyE hok/alp |
| 9 | EcN-luxCDABE | Nissle 1917 | N/A |
| 10 | MOD101 | SL1344, ELH1301 | pZE25 luxI luxCDABE hok/alp + pZA35 X714E (+LuxR) pLux::HlyE hok/alp |
| 11 | MOD102 | SL1344, ELH1301 | pZE25 luxI luxCDABE hok/alp + pZA35 X714E (+LuxR) ptac::HlyE hok/alp |
| 12 | MOD69 | SL1344, ELH1301 | pTD103 LuxCDABE hok/alp + pZA35 X714E (+LuxR) ptac::HlyE hok/alp |
| 13 | MOD29 | JS006, BW25113 | pTD103 luxI sfGFP + pZA35 X714E (+LuxR) |
| 14 | MOD110 | SL1344, ELH1301 | pZE25 luxI luxCDABE hok/alp + pZA35 X714E (+LuxR) pLux::CDD-iRGD hok/alp |
| 15 | MOD112 | SL1344, ELH1301 | pZE25 luxI luxCDABE hok/alp + pZA35 X714E (+LuxR) ptac::mCCL21 hok/alp |
Supplementary Material
Acknowledgements
This work was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health (GM069811), the San Diego Center for Systems Biology (P50 GM085764), a Koch Institute Support Grant (P30-CA14051) from the National Cancer Institute (Swanson Biotechnology Center), a Core Center Grant (P30-ES002109) from the National Institute of Environmental Health Sciences, and the NIH Pathway to Independence Award NIH (K99 CA197649-01). TD was supported by the Misrock Postdoctoral fellowship. AP was supported by the Department of Defense National Defense Science and Engineering Graduate Fellowship. SNB is an HHMI Investigator. We would like to thank Ryan Johnson for help with constructing microfluidic devices, Heather Fleming for help with editing the manuscript, and H. Ding of The Barbara K. Ostrom (1978) Bioinformatics and Computing Facility in the Swanson Biotechnology Center for help with the statistical tests carried out in this study. We would also like to thank Lingchong You for kindly providing the lysis gene used in this study.
Footnotes
Author Contributions
All authors (MOD, TD, AP, MS, JS, KA, EJ, EA, LT, SB, and JH) contributed extensively to the work presented in this paper.
Author Information
The authors declare no competing financial interests.
Supplementary Information
Supplementary information, including methods, supplementary figures and tables, is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature Reviews Genetics. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xuan C, et al. Microbial dysbiosis is associated with human breast cancer. PloS one. 2014;9:e83744. doi: 10.1371/journal.pone.0083744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Science translational medicine. 2013;5:179ps7. doi: 10.1126/scitranslmed.3005568. 179ps7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawelek JM, Low KB, Bermudes D. Tumor-targeted Salmonella as a novel anticancer vector. Cancer research. 1997;57:4537–4544. [PubMed] [Google Scholar]
- 5.Ruder WC, Lu T, Collins JJ. Synthetic biology moving into the clinic. Science. 2011;333:1248–1252. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 6.Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nature Reviews Genetics. 2011;13:21–35. doi: 10.1038/nrg3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baban CK, Cronin M, O'Hanlon D, O'Sullivan GC, Tangney M. Bacteria as vectors for gene therapy of cancer. Bioeng Bugs. 2010;1:385–394. doi: 10.4161/bbug.1.6.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cann SH, Van Netten J, Van Netten C. Dr William Coley and tumour regression: a place in history or in the future. Postgraduate medical journal. 2003;79:672–680. [PMC free article] [PubMed] [Google Scholar]
- 9.Davila ML, et al. Efficacy and toxicity management of 19-28z car T cell therapy in B cell acute lymphoblastic leukemia. Science translational medicine. 2014;6:224ra25–224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang LH, Bettegowda C, Huso DL, Kinzler KW, Vogelstein B. Combination bacteriolytic therapy for the treatment of experimental tumors. Proceedings of the National Academy of Sciences. 2001;98:15155–15160. doi: 10.1073/pnas.251543698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danino T, Mondragón-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prindle A, et al. A sensing array of radically coupled genetic ‘biopixels’. Nature. 2012;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young KD, Young R. Lytic action of cloned φX174 gene E. Journal of virology. 1982;44:993–1002. doi: 10.1128/jvi.44.3.993-1002.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marguet P, Tanouchi Y, Spitz E, Smith C, You L. Oscillations by minimal bacterial suicide circuits reveal hidden facets of host-circuit physiology. PloS one. 2010;5:e11909. doi: 10.1371/journal.pone.0011909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prindle A, et al. Genetic circuits in Salmonella typhimurium. ACS synthetic biology. 2012;1:458–464. doi: 10.1021/sb300060e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan R, et al. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene therapy. 2009;16:329–339. doi: 10.1038/gt.2008.188. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes K. The parB (hok/sok) locus of plasmid R1: a general purpose plasmid stabilization system. Nature Biotechnology. 1988;6:1402–1405. [Google Scholar]
- 19.Wood T, Kuhn R, Peretti S. Enhanced plasmid stability through post-segregational killing of plasmid-free cells. Biotechnology techniques. 1990;4:39–44. [Google Scholar]
- 20.Derman AI, et al. Phylogenetic analysis identifies many uncharacterized actin-like proteins (alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in alp7a. Molecular microbiology. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danino T, Lo J, Prindle A, Hasty J, Bhatia SN. In vivo gene expression dynamics of tumor-targeted bacteria. ACS synthetic biology. 2012;1:465–470. doi: 10.1021/sb3000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danino T, et al. Programmable probiotics for detection of cancer in urine. Science translational medicine. 2015;7:289ra84–289ra84. doi: 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danino T, Prindle A, Hasty J, Bhatia S. Measuring growth and gene expression dynamics of tumor-targeted S. typhimurium bacteria. JoVE (Journal of Visualized Experiments) 2013:e50540–e50540. doi: 10.3791/50540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R, et al. Application of a proapoptotic peptide to intratumorally spreading cancer therapy. Cancer research. 2013;73:1352–1361. doi: 10.1158/0008-5472.CAN-12-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeffler M, Le'Negrate G, Krajewska M, Reed JC. Salmonella typhimurium engineered to produce CCL21 inhibit tumor growth. Cancer immunology, immunotherapy. 2009;58:769–775. doi: 10.1007/s00262-008-0555-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nature Reviews Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leone V, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell host & microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thaiss CA, Levy M, Elinav E. Chronobiomics: The biological clock as a new principle in host–microbial interactions. PLoS Pathog. 2015;11:e1005113. doi: 10.1371/journal.ppat.1005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheong I, et al. A bacterial protein enhances the release and efficacy of liposomal cancer drugs. Science. 2006;314:1308–1311. doi: 10.1126/science.1130651. [DOI] [PubMed] [Google Scholar]
- 30.Soares KC, et al. A preclinical murine model of hepatic metastases. JoVE (Journal of Visualized Experiments) 2014:e51677–e51677. doi: 10.3791/51677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.