Abstract
As an inevitable by-product of the biofuel industry, glycerol is becoming an attractive feedstock for biorefinery due to its abundance, low price and high degree of reduction. Converting crude glycerol into value-added products is important to increase the economic viability of the biofuel industry. Metabolic engineering of industrial strains to improve its performance and to enlarge the product spectrum of glycerol biotransformation process is highly important toward glycerol biorefinery. This review focuses on recent metabolic engineering efforts as well as challenges involved in the utilization of glycerol as feedstock for the production of fuels and chemicals, especially for the production of diols, organic acids and biofuels.
Keywords: Glycerol, Metabolic engineering, Biorefinery, Biofuel, Biochemical
Background
Glycerol is a traditionally valuable compound which is used in a variety of areas, such as foods and beverages, pharmaceuticals, and cosmetics. In recent years, the fast development of biofuel industry has generated a large amount of crude glycerol as a by-product [1]. Approximately, 10 kg of crude glycerol is generated for every 100 kg of biodiesel produced [2]. The bioethanol process also generates glycerol up to 10 % of the total sugar consumed. The excess of crude glycerol produced in the biofuel industry is leading to a dramatic decrease in glycerol price, making it a waste with a disposal cost for many biodiesel plants. Converting crude glycerol into value-added products is an urgent need and also a good opportunity to improve the viability of biofuel economy. So far, both chemical and biological approaches have been explored to convert glycerol into more valuable products [3–5]. Compared to chemical transformation, biological routes bear several advantages, such as higher specificity, higher tolerance to impurity, being more environmentally friendly (e.g., low temperature/pressure) [6]. Compared to other carbon sources (e.g., glucose and xylose), glycerol has higher degree of reduction and thus can generate more reducing equivalents, conferring the ability to produce fuels and reduced chemicals at higher yields [7]. High abundance, low price and high degree of reduction make glycerol an attractive feedstock for biorefinery [8].
There are a number of microorganisms which can metabolize glycerol. However, direct utilization of natural organisms at the industrial level is often limited by the low production rates, titers, or yields. Thus, metabolic engineering is often needed to improve strain performance, including: (a) elimination of transcriptional repression and enzyme feedback inhibition; (b) reduction of undesired by-products; (c) enhancement of building blocks, reducing power or energy supply; (d) expansion of substrate and product portfolio; and (e) enhancement of strain tolerance to inhibitors and environmental stress [9–14]. More recently, the integration of protein engineering, systems biology and synthetic biology into metabolic engineering has extended strain engineering from local modification to system-wide optimization. Powerful omics technologies, such as genomics, transcriptomics, proteomics and fluxomics, have been combined for in-depth understanding of glycerol metabolism and regulation of microorganism at the system level [15–18].
In general, glycerol is channeled into glycolysis via two metabolic routes (Fig. 1) [7]. In one route, glycerol is dehydrogenated by NAD-dependent glycerol dehydrogenase (gldA or dhaD) to dihydroxyacetone (DHA), which is then phosphorylated by phosphoenolpyruvate (PEP)- or ATP-dependent dihydroxyacetone kinases (DHAK, dhaKLM) to dihydroxyacetone phosphate (DHAP). In the other route, glycerol is first phosphorylated by ATP-dependent glycerol kinase (glpK) to glycerol 3-phosphate, and the latter is further reduced by NAD-dependent glycerol 3-phosphate dehydrogenase (glpD) to DHAP. The catabolism of glycerol suffers from various genetic and enzymatic regulations. Metabolic control analysis showed that the glycolytic flux during glycerol anaerobic fermentation was almost exclusively controlled by glycerol dehydrogenase and dihydroxyacetone kinase in Escherichia coli [19]. Large efforts have been devoted to engineer glycerol catabolism and downstream metabolic pathways for production of various valuable products, including fuels, chemicals, and biomaterials (Table 1). In this review, recent efforts of metabolic engineering to develop industrial strains for glycerol biorefinery are described. Especially, we systematically summarize and discuss different strategies of metabolic engineering for the production of diols, organic acids and biofuels, which represent the most important categories of bulk chemicals in the current biotechnology industry.
Fig. 1.

Metabolic pathways of glycerol for the production of various products. Production of diols, amino acids, organic acids, and biofuels via the catabolic pathway of glycerol are illustrated with different colors: diols, blue; amino acids, green; organic acids, yellow; biofuels, orange. In some microorganisms, glycerol can be dehydrated to 3-hydroxypropionaldehyde (3-HPA) by glycerol dehydratase. 3-HPA can be converted to 1,3-propanediol by alcohol dehydrogenase or 3-hydroxypropionic acid by aldehyde dehydrogenase. Glycerol can also enter the central metabolic pathway via two routes: (1) glycerol kinase and glycerol 3-phosphate dehydrogenase; (2) glycerol dehydrogenase and dihydroxyacetone kinase
Table 1.
Fermentative production of diols, organic acids, and biofuels from glycerol by metabolic engineered strains
| Product and strain | Genetic modification | Titer [g/L] | Yield [g/g] | Culture | References |
|---|---|---|---|---|---|
| 1,3-Propanediol | |||||
| Klebsiella pneumoniae | ∆ldhA | 102.06 | 0.43 | Fed-batch | [27] |
| Klebsiella oxytoca | ∆aldA | 70.5 | 0.58 | Fed-batch | [26] |
| Clostridium acetobutylicum | Overexpression of dhaB1B2 from C. butyricum and yqhD from E. coli | 84 | 0.51 | Fed-batch | [24] |
| Lactobacillus reuteri | Overexpression of yqhD from E. coli | 9.1 | 0.42 | Batch | [22] |
| Citrobacter werkmanii | ∆dhaD∆ldhA∆adhE | 30.7 | 0.59 | Batch | [23] |
| Escherichia coli | Overexpression of dhaB1B2 and dhaT from Clostridium butyricum | 104.4 | 0.9 | Two-stage fed-batch | [34] |
| 1,2-Propanediol | |||||
| Escherichia coli | ∆ackA-pta∆ldhA∆dhaK, overexpression gldA, mgsA, yqhD from E. coli and dhaKL from C. freundii | 5.6 | 0.21 | Batch | [36] |
| Escherichia coli | Overexpression of 1,2-propanediol synthesis enzymes with targeting sequences for inclusion into microcompartments | – | – | Shake flask | [39] |
| Saccharomyces cerevisiae | Overexpression gldA, mgsA from E. coli and GUP1, GUT1, GUT2, gdh from S. cerevisiae | 2.19 | 0.27 | Batch | [37] |
| 2,3-Butanediol | |||||
| Klebsiella oxytoca | ∆pduC∆ldhA | 131.5 | 0.44 | Fed-batch | [43] |
| Bacillus amyloliquefaciens | Overexpression of dhaD, acr and alsR | 102.3 | 0.44 | Fed-batch | [45] |
| Escherichia coli | Overexpression of budA and budC | 6.9 | 0.21 | Batch | [44] |
| l-Lactate | |||||
| Escherichia coli | ∆pta∆adhE∆frdA∆mgsA∆ldhA∆lldD, overexpression of glpK, glpD and ldh | 50 | 0.89 | Shake flask | [50] |
| Enterococcus faecalis | ΔpflB | 27 | 0.98 | Shake flask | [51] |
| d-Lactate | |||||
| Klebsiella pneumoniae | ΔdhaTΔyqhD, overexpression of ldhA | 142.1 | 0.82 | Fed-batch | [46] |
| Escherichia coli | ΔackA-ptaΔppsΔpflBΔdldΔpoxBΔadhEΔfrdA, overexpression of ldhA | 100.3 | 0.75 | Fed-batch | [47] |
| Succinate | |||||
| Escherichia coli | ΔackA-ptaΔppcΔldhAΔpoxBΔadhE, overexpression of pyc | 14 | 0.69 | Shake flask | [53] |
| Escherichia coli | ΔptsIΔpflB, overexpression of pck | 9.4 | 1.03 | Shake flask | [55] |
| Corynebacterium glutamicum | ΔackA-ptaΔsdhCABΔactΔpqo, overexpression of glpFKD | 9.3 | 0.27 | Shake flask | [56] |
| 3-Hydroxypropionic acid | |||||
| Klebsiella pneumoniae | ΔdhaTΔyqhD, overexpression of puuC | 28 | 0.39 | Fed-batch | [59] |
| Escherichia coli | ΔackA-ptaΔyqhDΔglpR, overexpression of dhaB123,gdrAB and mutant gabD4 | 71.9 | – | Fed-batch | [65] |
| Pseudomonas denitrificans is | Δ3hpdhΔ3hibdhIV, overexpression of dhaB123, gdrAB and aldH | 3 | 0.76 | Shake flask | [68] |
| Glutamate | |||||
| Corynebacterium glutamicum | Overexpression of glpF, glpK, glpD | 2.2 | 0.11 | Shake flask | [70] |
| Lysine | |||||
| Corynebacterium glutamicum | lysC P458S, hom V59A, pyc T311I, Δpck, overexpression of glpF, glpK, glpD | 3.8 | 0.19 | Shake flask | [70] |
| Ornitine | |||||
| Corynebacterium glutamicum | ΔargR, ΔargF, overexpression of glpF, glpK, glpD | 2.2 | 0.11 | Shake flask | [71] |
| Arginine | |||||
| Corynebacterium glutamicum | ΔargR, overexpression of argB A26VM31V , glpF, glpK, glpD | 0.8 | 0.04 | Shake flask | [71] |
| Putrescine | |||||
| Corynebacterium glutamicum | ΔargR, overexpression of speC, argF, glpF, glpK, glpD | 0.4 | 0.02 | Shake flask | [71] |
| Ethanol | |||||
| Escherichia coli | ΔfrdABCDΔackA-ptaΔldhAΔpoxB, overexpression of dhaKLM, gldA and adhE | 40.8 | 0.44 | Fed-batch | [76] |
| Butanol | |||||
| Clostridium pasteurianum | ΔdhaT | 8.6 | 0.26 | Shake flask | [78] |
| Fatty acid | |||||
| Escherichia coli | ΔfadDΔfadR, overexpression of pntAB, nadK, TE and fabZ | 4.82 | 0.3 | Fed-batch | [90] |
Metabolic engineering for the production of diols from glycerol
Diols are compounds with two hydroxyl groups which have a wide range of important applications as chemicals and fuels. The C2–C4 diols are the most important and have been widely used as monomers in the polymer industry. In recent years, microbial production of 1,3-propanediol (1,3-PDO), 1,2-propanediol (1,2-PDO), and 2,3-butanediol (2,3-BDO) from glycerol have been widely investigated (Fig. 1). The production of 1,3-PDO and 2,3-BDO has reached commercial scales.
1,3-Propanediol
1,3-PDO is an important diol which has received broad interest in recent years [20, 21]. Of particular interest is its use as a monomer for the synthesis of polyethers, polyurethanes and polyesters such as polytrimethylene terephthalate (PTT). Several species of microorganisms, including Klebsiella (K. pneumoniae and K. oxytoca), Clostridia (C. butyricum and C. pasteurianum), Enterobacter (E. agglomerans), Citrobacter (C. freundii) and Lactobacilli (L. brevis and L. buchneri), possess the ability to convert glycerol into 1,3-PDO [22–25]. Among the natural producers, K. pneumoniae and C. butyricum are the most promising species due to their high tolerance to glycerol inhibition and high production of 1,3-PDO. DuPont has developed a recombinant E. coli which can directly utilize glucose for 1,3-PDO production with high titer and yield [21]. This process has been commercialized and considered as a milestone of metabolic engineering. With the dramatic decrease in glycerol price over the past few years, the direct conversion of glycerol to 1,3-PDO is becoming economically competitive to the glucose-based process.
The production of 1,3-PDO from glycerol is a reduction process which consumes 1 mol NADH per mol of 1,3-PDO. In the reductive pathway, glycerol is first dehydrated into 3-hydroxypropionaldehyde (3-HPA) by B12-dependent or B12-independent glycerol dehydratase, and the latter is reduced to 1,3-PDO by alcohol dehydrogenase (Fig. 1). The regeneration of reducing equivalent should be achieved via a glycerol oxidative pathway. Glycerol can be converted into different oxidation products with different yields of reducing equivalent. Under anaerobic condition, oxidation of glycerol to acetate (with formate) is the most effective route for NADH generation with a yield of 2 mol NADH/mol glycerol. Oxidation of glycerol to 2,3-BDO and lactate also generate reducing equivalent with yields of 1.5 mol NADH/mol glycerol and 1 mol NADH/mol glycerol, respectively. Conversion of glycerol to succinate or ethanol, however, does not generate NADH. Thus, coproduction of acetate with 1,3-PDO would give the highest yield of 1,3-PDO under anaerobic condition (0.67 mol/mol). To this end, reduction of succinate and ethanol synthesis could be the first step to channel metabolic flux to more efficient NADH generation pathways. This is proved by Zhang et al. who successfully increased the production of 1,3-PDO by 32.8 % only by knocking out the aldA gene to block the synthesis of ethanol by K. pneumoniae [26]. Xu et al. attempted to enhance 1,3-PDO production by K. pneumoniae by reducing lactate accumulation with the single deletion of d-lactate dehydrogenase gene ldhA [27]. Although the formation of lactate was decreased by 95 %, most of the reduced flux in the lactate pathway was not channeled to 1,3-PDO but to the branch of 2,3-BDO, indicating that the flux distribution in the pyruvate node is more flexible than in the glycerol node. Since 2,3-BDO is less toxic to K. pneumoniae and synthesis of 2,3-BDO gives higher yield of NADH than lactate, the altered fluxes in the oxidative pathway also benefited 1,3-PDO synthesis, resulting in an increase of 1,3-PDO production by 7 % [27]. Surprisingly, reduction of both lactate and 2,3-BDO formation was reported to be detrimental for glycerol consumption and 1,3-PDO production [28, 29]. Metabolic flux was not channeled to 1,3-PDO or acetate pathway, while high accumulation of pyruvate was observed in the double mutant strain (ΔldhAΔbudO), indicating a metabolic imbalance at the pyruvate node [28]. High accumulation of glyceraldehyde 3-phosphate and pyruvate was postulated to be detrimental for 1,3-PDO production, since glyceraldehyde 3-phosphate was strongly inhibitory to glycerol dehydratase [28]. A potential strategy to remove the metabolic imbalance at the PEP/pyruvate node is fine-tuning of downstream enzymes. For example, overexpression of phosphoenolpyruvate carboxylase (ppc) and deletion of arcA gene, a transcriptional regulator mediating tricarboxylic acid (TCA) cycle flux, were successfully implemented by Dupont to channel the flux from PEP/pyruvate to TCA cycle for by-product’s reduction and NADH regeneration for 1,3-PDO overproduction [21]. Metabolic pathway analysis suggested a maximum yield of 0.84 mol 1,3-PDO/mol glycerol in which only TCA cycle was activated for NADH generation under microaerobic condition [30]. In this case, inactivation of NADH dehydrogenase to block the transfer of electrons from NADH to the quinone pool would be important to increase the NADH pool for 1,3-PDO production.
Minimization of the accumulation of toxic intermediates is another important issue for the high production of 1,3-PDO. 3-HPA is a toxic intermediate during 1,3-PDO synthesis which may cause cell death and fermentation cessation. Reduction of 3-HPA accumulation can be achieved by accelerating the conversion of 3-HPA to 1,3-PDO either by overexpression of 1,3-PDO dehydrogenase (NADH-dependent enzyme encoded by dhaT gene or NADPH-dependent enzyme encoded by yqhD gene) or by increasing the NADH pool via the modification of fluxes in the oxidation pathway as previously discussed or via the introduction of formate dehydrogenase (fdh) [31–33]. Formate dehydrogenase catalyzes the oxidation of formate to carbon dioxide, providing an additional NADH for 3-HPA reduction.
Non-natural producers have also been engineered to produce 1,3-PDO from glycerol. Tang et al. reported the introduction of B12-independent glycerol dehydratase and its activator from C. butyricum into E. coli in addition to the overexpression of the native alcohol dehydrogenase YqhD [34]. Based on two-stage fermentation (aerobic growth stage and anaerobic production stage), the overall 1,3-PDO yield and productivity reached 104.4 g/L and 2.61 g/L/h at the production stage. Similarly, a recombinant C. acetobutylicum was constructed by overexpression of B12-independent glycerol dehydratase and 1,3-PDO dehydrogenase from C. butyricum, which can produce 84 g/L of 1,3-PDO with a yield of 0.65 mol/mol [24]. The utilization of B12-independent glycerol dehydratase enhances the economic viability of the designed processes since no expensive B12 is required during the fermentation.
1,2-Propanediol
1,2-PDO is another important diol with a global market of about 1.5 million tons/year. It is used in a wide spectrum of areas such as: (a) production of polyester resins; (b) antifreezing; (c) detergents; and (d) cosmetics [35]. Several attempts have been made to optimize 1,2-PDO production from glycerol [36, 37]. Similar to 1,3-PDO, the conversion of glycerol to 1,2-PDO is a reduction process which consumes 1 mol NADH per mol of 1,2-PDO (Fig. 2). In the 1,2-PDO synthesis pathway, DHAP is first converted to methylglyoxal (MG) via methylglyoxal synthase (mgsA). The conversion of MG into 1,2-PDO takes place through two alternative pathways, one with acetol as an intermediate through the action of alcohol dehydrogenase (yqhD) and glycerol dehydrogenase (gldA), and the other with lactaldehyde as an intermediate mediated by glycerol dehydrogenase (gldA) and 1,2-PDO reductase (fucO). Overexpression of either pathway is technologically feasible for 1,2-PDO production [36]. However, to overproduce 1,2-PDO, several issues should be considered. First, glycerol assimilation is coupled with PEP synthesis in E. coli since DHAK utilizes PEP as the phosphate donor in the phosphorylation of DHA. Thus, a high flux from DHAP to PEP is required during glycerol assimilation which significantly reduces the precursor availability for 1,2-PDO synthesis. This problem could be overcome by substitution of PEP-dependent DHAK with ATP-dependent DHAK. This was proved to be highly efficient for enhancing 1,2-PDO yield and glycerol consumption in E. coli [36]. Second, since 1,2-PDO synthesis is both redox and ATP consuming (with ATP-dependent DHAK), the coproduction of another oxidation product is necessary in the anaerobic condition. As discussed in the previous session, the conversion of glycerol to acetate or lactate generates NADH, while the conversion of glycerol to ethanol (with formate) or succinate is redox neutral. Coupling the production of acetate with 1,2-PDO should give the highest yield of 1,2-PDO. However, this maximum yield is never quite reached because there is no net gain of ATP for cell growth and maintenance. Thus, coproduction of ethanol to provide ATP is considered to be necessary for 1,2-PDO production under anaerobic conditions [36]. Interestingly, it was found that blocking the synthesis of acetate (ΔackA-pta) and/or lactate (ΔldhA) increased the production of 1,2-PDO, while disrupting the synthesis of ethanol (ΔadhE) or succinate (ΔfrdA) reduced both glycerol consumption and 1,2-PDO production by E. coli under anaerobic conditions [36]. Deletion of ethanol or succinate channeled most of the flux toward acetate or lactate formation, which would cause significant redox imbalance and growth deficiency. On the other hand, it was observed that there was an increase of ethanol production for the mutants with the disruption of acetate and/or lactate production pathways [36]. The conversion of glycerol to ethanol (and formate) is a redox-neutral process with the generation of 1 mol ATP. Thus, the enhanced ethanol production could provide more ATP for cell growth without affecting the redox balance. However, since the ethanol (and formate) pathway does not generate NADH, the accumulation of pyruvate as oxidized product and the activation of pyruvate dehydrogenase and/or formate hydrogen lyase (FHL) were necessary for redox balance in the acetate- (ΔackA-pta) and/or lactate-deficient (ΔldhA) mutants [36]. With the deletion of ackA-pta, ldhA, and PEP-dependent dhaK in addition to overexpression of gldA, mgsA, yqhD, and ATP-dependent DHAK (C. freundii dhaKL), the mutant E. coli can produce 5.6 g/L of 1,2-PDO with a yield of 0.213 g/g glycerol. Similarly, S. cerevisiae strains have been engineered to produce 1,2-PDO by modifying both glycerol dissimilation and 1,2-PDO synthesis pathways [37]. Overexpression of glycerol kinase (GUT1), glycerol 3-phosphate dehydrogenase (GUT2), glycerol dehydrogenase (gdh), and a glycerol transporter gene (GUP1) increased glycerol utilization and cell growth rate. Further introduction of mgsA and gldA from E. coli enabled the production of 2.19 g/L of 1,2-PDO by the metabolically engineered S. cerevisiae.
Fig. 2.
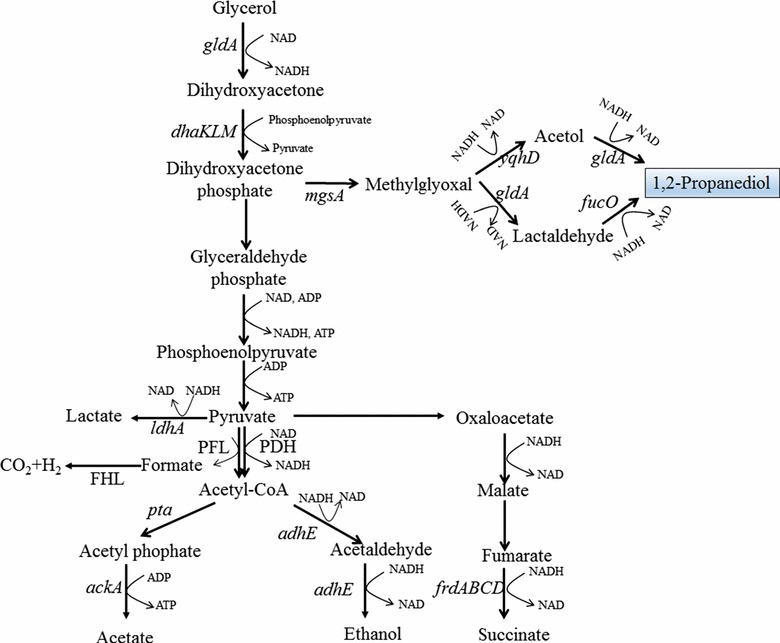
The metabolic pathway for the production of 1,2-propanediol (1,2-PDO). Glycerol is converted to 1,2-PDO via two alternative pathways: one with acetol as an intermediate and the other with lactaldehyde as an intermediate. ackA acetate kinase; adhE acetaldehyde/alcohol dehydrogenase; dhaKLM dihydroxyacetone kinase; FHL fomate hydrogen lyase complex; frdABCD fumarate reductase; fucO 1,2-PDO reductase; gldA glycerol dehydrogenase; ldhA lactate dehydrogenase; mgsA methylglyoxal synthase; PDH pyruvate dehydrogenase; PFL pyruvate formate-lyase; pta phosphate acetyltransferase; yqhD alcohol dehydrogenase
The conversion of glycerol to 1,2-PDO generates several toxic intermediates such as MG and lactaldehyde. To minimize the accumulation of toxic intermediates, assembly of pathway enzymes by DNA scaffolding and bacterial microcompartment (BMC) have been tried. Conrado et al. utilized fused zinc-finger domains which can specifically bind the DNA scaffold to assemble methylglyoxal synthase, 2,5-diketo-d-gluconic acid reductase and glycerol dehydrogenase in E. coli [38]. Under optimized enzyme/scaffold ratio, the production of 1,2-PDO was improved by ~3.5 times. In another attempt to construct artificial BMC for 1,2-PDO synthesis, N-terminal targeting sequences derived from Pdu BMC were added to glycerol dehydrogenase, dihydroxyacetone kinase, methylglyoxal synthase and 1,2-PDO oxidoreductase, resulting in the formation of protein inclusions within the recombinant E. coli cell [39]. Interestingly, irrespective of the presence of BMCs, a strain containing the fused enzymes exhibited a 245 % increase of 1,2-PDO production in comparison to the strain with free enzymes. It was observed that the enzymes with targeting peptides formed dense protein aggregates, which may result in increased channeling of substrates and products between proteins due to proximity effects.
2,3-Butanediol
2,3-BDO is a potential platform chemical which can be used as biofuel as well as building blocks for the synthesis of 2-butanone [40], 2-butanol [41] and 1,3-butadiene [42]. As discussed before, the conversion of glycerol to 2,3-BDO is an oxidation process with a theoretical yield of 0.5 mol/mol. The NADH generated during the production of 2,3-BDO should be consumed by coupling it with the generation of a reduced product or with oxidative phosphorylation. Thus, aerobic condition is often employed for 2,3-BDO production in which oxygen can be used as electron acceptor. One well-studied case is K. oxytoca which can efficiently synthesize both 1,3-PDO and 2,3-BDO from glycerol. Aeration and pH are two critical factors affecting the distribution between 1,3-PDO and 2,3-BDO [43–45]. High aeration and low pH (pH6) are beneficial for 2,3-BDO production, while low aeration and higher pH (pH7) enhance the accumulate of 1,3-PDO. With optimal pH and aeration, a mutant K. oxytoca strain blocking the synthesis of 1,3-PDO and lactate (ΔpduCΔldhA) can accumulate 131.5 g/L of 2,3-BDO from crude glycerol, with a productivity and a yield of 0.84 g/L/h and 0.44 g/g. In another study, overexpression of glycerol dehydrogenase, acetoin reductase, and a transcriptional regulator ALsR were combined to enhance the metabolic flux toward 2,3-BDO synthesis in Bacillus amyloliquefaciens. The engineered strain showed reduced accumulation of acetoin and other by-products. Similarly, a combination of three-stage dissolved oxygen control strategy (350 rpm for the first 5 h; 400 rpm from 5 to 22 h; and 350 rpm thereafter) and two-stage pH control strategy (without pH control for the first 16 h and with constant pH at 6.5 thereafter) was applied for this mutant during the fermentation, which allowed a high titer (102.3 g/L) and yield (0.44 g/g) of 2,3-BDO production [45]. The high titer and yield in these examples demonstrated that the biological production of 2,3-BDO could be both technologically and economically competitive.
Metabolic engineering for the production of organic acids from glycerol
Beyond traditional uses in the feed and food industry, recent interest in bioplastics and bulk chemicals has pushed the biological production of organic acids from renewable raw materials. Glycerol has been utilized to produce various organic acids, including lactate, succinate, fumarate, 3-hydroxypropionic acid (3-HP), and amino acids (Fig. 1).
Lactate
Lactate is an important organic acid with many applications in the food, pharmaceutical, and chemical industries. In particular, the synthesis of biodegradable polylactate (PLA) from d- or l-lactate holds a great potential to substitute for the synthetic plastics produced by the petrochemical industry. Biological production of lactate from glycerol is a promising process with a theoretical yield of 1.0 mol/mol. Similar to 2,3-BDO, biological production of lactate from glycerol is an oxidation process which generates net NADH and ATP. Thus, microaerobic condition is preferred for lactate production from glycerol, while anaerobic fermentation is preferred when glucose is used as substrate. Several strains can natively produce a large amount of lactate under anaerobic and microaerobic condition. For example, d-lactate is produced as a main by-product by K. pneumoniae and E. coli during glycerol assimilation. With the deletion of dhaT and yqhD gene to block the synthesis of 1,3-PDO and overexpression of the native d-lactate dehydrogenase gene ldhA, K. pneumoniae was able to accumulate up to 142.1 g/L of d-lactate with a yield of 0.82 g/g glycerol under microaerobic condition [46]. More systematical strategies have also been applied to E. coli for d-lactate production [47–49]. A homolactic strain for d-lactate production from glycerol was constructed by systematic elimination of all by-products synthetic routes, including acetate (ΔackA-ptaΔpoxB), ethanol (ΔadhE), succinate (ΔppsΔfrdA), formate (ΔpflB), and d-lactate assimilation (Δdld). The yield of d-lactate was further improved from 0.67 to 0.75 g/g by overexpression of d-lactate dehydrogenase from a low-copy plasmid [47]. Similar strategies can be adopted for l-lactate production [50, 51]. Mazumdar et al. developed a highly efficient l-lactate producer by four steps: (1) replacing the native d-lactate dehydrogenase (ΔldhA) with l-lactate dehydrogenase from Streptococcus bovis; (2) inactivating the methylglyoxal pathways (ΔmgsA) to avoid the synthesis of lactate racemic mixture; (3) inactivating l-lactate assimilation pathway (ΔlldD); and (4) blocking the synthesis of acetate (Δpta), ethanol (ΔadhE), and succinate (ΔfrdA) [50]. The resulting E. coli strain produced l-lactate from crude glycerol with high yield (0.89 g/g) and optical purity (99.9 %). Thus, it is highly competitive to produce both d-lactate and l-lactate from cheap crude glycerol as compared to the glucose-based routes.
Succinate
Succinate is considered as one of the top 12 building block chemicals [52, 53]. It can be used as solvents, pharmaceutical products, or monomer for the synthesis of biodegradable plastics. Transformation of glycerol to succinate is a redox-neutral process. Theoretically, glycerol can be converted into succinate with a yield of 1 mol/mol with the addition of supplementary bicarbonate in the medium for anaplerotic reaction [52]. This yield is much higher than the one observed when utilizing glucose as carbon source. Enhancing anaplerotic flux is considered to be the most important factor for high yield production of succinate. There are three enzymes involved in anaplerotic reactions: phosphoenolpyruvate carboxylase (PEPC), phosphoenolpyruvate carboxykinase (PEPCK), and pyruvate carboxylase (PYC). Overexpression of both PEPC and PYC are shown to be beneficial for succinate production. In most bacteria, PEPC and PYC catalyze the carboxylation reactions, while PEPCK catalyzes the decarboxylation reaction. Reversing the direction of the gluconeogenic PEPCK reaction could generate net ATP during CO2 fixation and thus can be an efficient way to enhance succinate production under anaerobic condition. Overexpression of PEPCK together with the increase of the PEP pool is important to reverse the direction of PEPCK. For example, overexpression of the native PEPCK was combined with the deletion of pyruvate formate-lyase (ΔpflB) to increase the yield and production of succinate in E. coli. Similarly, overexpression of PEPCK from Actinobacillus succinogenes, the main CO2-fixing enzyme in A. succinogenes, increased succinate production 6.5-fold in an E. coli mutant when combined with the deletion of PEPC [54]. Overexpression of PEPCK in E. coli, together with the disruption of pyruvate formate-lyase (ΔpflB) and phosphotransferase system (ΔptsI) to increase the pool of PEP, allowed the conversion of glycerol to succinate with a yield of 0.80 mol/mol under anaerobic condition [55]. This yield is much higher than the strain which overexpressed PYC and suppressed all by-products synthesis (acetate, ethanol, and lactate) [53]. Aerobic succinic acid production from glycerol was realized in C. glutamicum by disruption of the TCA cycle downstream of succinate (ΔsdhCAB) and suppression of acetate formation (ΔackA-ptaΔactΔpqo) in addition to overexpression of PYC, PEPC, and glp regulon for glycerol assimilation (glpFKD) [56]. Under optimized fermentation conditions, the strain exhibited a yield of 0.21 mol/mol and a titer of 9.3 g/L [56]. From a practical point of view, aerobic production of succinate is not economically reasonable, since it has lower theoretical yield and higher energy requirement as compared to anaerobic condition.
3-Hydroxypropionic acid
3-Hydroxypropionic acid (3-HP) is a versatile platform chemical which can be used for the production of acrylic acid, acrylonitrile, malonic acid, and 1,3-PDO [57]. Different metabolic pathways have been predicted and tried for the production of 3-HP from several intermediates including glycerol, lactate, malonyl-CoA, and β-alanine [57]. Direct conversion of glycerol to 3-HP is considered as one of the most promising routes for 3-HP production. Unlike the production of 1,3-PDO, the formation of 3-HP from glycerol is an oxidation process generating net NADH. Thus, it is feasible to coproduce 3-HP with 1,3-PDO to recycle the reducing equivalent. This concept was proved in recombinant K. pneumoniae which produced 48.9 g/L of 3-HP and 25.3 g/L of 1,3-PDO by overexpressing an E. coli aldehyde dehydrogenase (aldH) [58]. On the other hand, 3-HP can be produced as the sole product if an electron acceptor is provided [59]. Under anaerobic condition, nitrate was used as an electron acceptor for 3-HP production by a mutant K. pneumoniae with double deletion of dhaT and glpK to reduce the flux toward 1,3-PDO synthesis and the glycerol oxidation pathway [60]. Under aerobic condition, oxygen can be utilized as an electron acceptor. However, control of the aeration is important for the production of 3-HP. Increasing dissolved oxygen is beneficial for NAD recycling; however, high oxygen concentration also inhibits the synthesis of coenzyme B12 and reduces the activity of glycerol dehydratase. Thus, it is important to combine both metabolic engineering (reduction of 1,3-PDO and other by-products formation) and process optimization for the high production of 3-HP by K. pneumoniae.
Escherichia coli as a heterologous host has also been engineered to produce 3-HP from glycerol. Similarly, reduction of 1,3-PDO synthesis by yqhD deletion is important for increasing the production of 3-HP [61]. Knockout of glpK to block glycerol oxidation pathway, however, significantly reduced cell growth and glycerol consumption [62]. Thus, L-arabinose-inducible promoter was used for fine-tuning of the expression of glpK, and deletion of glpR, a repressor of glp regulon, was also combined to enhance glycerol metabolism [62]. Another smart strategy using toggle switch to conditionally repress gene expression has also been tried. By controlling the adding time of IPTG, a toggle switch to simultaneously induce the 3-HP synthesis pathway (glycerol dehydratase and aldehyde dehydrogenase) and repress glyceraldehyde 3-phosphate dehydrogenase could increase 3-HP production compared to the control strain [63]. In silico metabolic simulation has been utilized to identify new targets for increasing the yield of 3-HP. A genome-scale modeling identified triosephosphate isomerase (tpiA) and glucose 6-phosphate dehydrogenase (zwf) as candidates for improving 3HP production [64]. The 3-HP yield of a triple E. coli mutant (ΔtpiAΔzwfΔyqhD) was increased by 7.4-fold in comparison to the parental strain. Compared to K. pneumoniae, the key challenge of using E. coli as host for 3-HP production is the need to add expensive coenzyme B12 during fermentation.
Metabolic balance between glycerol dehydratase and aldehyde dehydrogenase to reduce 3-HPA accumulation is also important for 3-HP production. This could be achieved using more active aldehyde dehydrogenase or controlling the expression of glycerol dehydratase [65, 66]. Chu et al. screened a highly active aldehyde dehydrogenase from Cupriavidus necator and used site-directed mutagenesis to improve its activity [65]. The most active mutant bears two point mutations: E209Q/E269Q. Escherichia coli transformed with this mutant produced up to 71.9 g/L of 3-HP with a productivity of 1.8 g/L/h. Lim et al. used UTR (untranslated region) engineering to precisely control the expression of glycerol dehydrogenase (dhaB1) for optimal metabolic balance [67]. It was observed that high expression of glycerol dehydrogenase resulted in low cell growth and high accumulation of 3-HPA. Under optimal expression level of glycerol dehydratase, the best strain produced 40.51 g/L of 3-HP with a yield of 0.97 g/g glycerol (using glycerol and glucose as co-substrates).
Since it is necessary to add the expensive coenzyme B12 during 3-HP production by E. coli, Pseudomonas denitrificans which can naturally produce coenzyme B12 under aerobic conditions was engineered to produce 3-HP. When glycerol dehydratase and its activator and an aldehyde dehydrogenase from K. pneumoniae (puuC) were overexpressed, the recombinant strain could produce 4.9 g/L of 3-HP using glycerol and glucose as co-substrates [68]. However, P. denitrificans can degrade 3-HP and utilize it as carbon source. Thus, the 3-HP degradation pathway should be identified and deleted [69].
Amino acids
l-Amino acids are very important fermentation products which have been widely used in industry. Due to the high reduction degree of glycerol, it is very promising to utilize glycerol as substrate for amino acid production, which often consumes a large amount of reducing equivalents. The dominant strain for l-amino acid production, C. glutamicum, has been engineered to utilize different substrates including glycerol. Although C. glutamicum possesses putative homologs of glycerol kinase (glpK) and glycerol-3-phosphate dehydrogenase (glpD), wild-type C. glutamicum cannot grow with glycerol as the sole carbon source. Glutamate production from glycerol was enabled by plasmid-borne expression of E. coli glpF, glpK, and glpD in C. glutamicum with the titer of 2.2 g/L [70]. Introduction of glpFKD into C. glutamicum strains producing other amino acids led to the synthesis of lysine, ornithine, arginine, phenylalanine, and putrescine from crude glycerol (Table 1) [71, 72]. The titers (0.4–3.8 g/L) and yields (0.02–0.2 g/g), however, are much lower compared to cultures using glucose as carbon source. One of the potential reasons is the low yield of NADPH during glycerol catabolism. Further enhancement could be achieved by introducing novel NADPH generation pathways such as transhydrogenase, NADPH-dependent glycerol dehydrogenase or NADPH-dependent glyceraldehyde phosphate dehydrogenase [73].
Metabolic engineering for the production of biofuels from glycerol
Due to its low price and high reduction degree, glycerol is a very promising feedstock for biofuel production in comparison to glucose and other sugars. Production of ethanol from glycerol has bene intensively studied, while the research on the transformation of glycerol to butanol and lipids/fatty acids has just started in recent years.
Ethanol
As discussed before, the transformation of glycerol to ethanol (with formate) is a redox-neutral process with net ATP generation; thus, it is possible to eliminate most by-products formation in anaerobic condition. Specifically, co-production of ethanol-hydrogen or ethanol-formate can be obtained with yield close to 1.0 mol/mol [74]. For example, double mutation of fumarate reductase (ΔfrdA) and phosphotransacetylase (Δpta) to eliminate succinate and acetate synthesis is enough to engineer an E. coli strain producing ethanol and hydrogen at maximum theoretical yield (1.0 mol/mol) at pH 6.3 under anaerobic condition [75]. The final titer and productivity, however, were relatively low although glycerol consumption could be partially strengthened by overexpression of glycerol dehydrogenase (gldA) and dihydroxyacetone kinase (dhaKLM). Wong et al. recently constructed an inducer- and antibiotic-free system to increase the titer, yield and specific productivity of ethanol under microaerobic condition [76]. To achieve such a goal, the pathways to by-products (succinate, acetate, and lactate) were first inactivated (ΔfrdABCDΔackA-ptaΔpoxBΔldhA). The glycerol uptake genes dhaKLM, gldA, and glpK and the ethanol pathway gene adhE were then overexpressed under native E. coli promoters in a plasmid. The chromosomal gldA and glpK were deleted to inactivate the native glycerol consumption pathway. Thus, only the strains bearing a plasmid-borne gldA can utilize glycerol as the sole carbon source. This strategy eliminates the need for antibiotics to retain the plasmid during fermentation. The final engineered strain can produce ethanol with high titer and yield (40.8 g/L and 0.44 g/g) without using expensive inducers and antibiotics. This process is very promising for industrial ethanol production.
Butanol
Butanol has received renewed interest recently as a potential green biofuel [77]. Compared to ethanol, butanol has higher energy density and lower hygroscopicity, and can be directly blended with gasoline without the need for modifying the current vehicle system. Production of butanol is a reducing equivalent-intensive process. Thus, glycerol can be utilized as a good substrate for butanol production. Three isoforms of butanol (n-butanol, 2-butanol, and isobutanol) can be used as biofuel and can be directly produced from glycerol. Clostridium pasteurianum can utilize crude glycerol to produce n-butanol and 1,3-PDO. To enhance the production of n-butanol, the dhaT gene was knocked out, resulting in an 83 % reduction of 1,3-PDO production [78]. The yield and selectivity (reported as g n-butanol produced per g of total solvents) of butanol was increased by 24 and 63 %. The n-butanol synthesis pathway has also been heterogeneously expressed in E. coli and K. pneumoniae (Fig. 3) [79, 80]. However, the currently reported titers were much lower in comparison to those using glucose as substrate [77]. More systematic strategies to optimize heterologous pathways and to reduce by-products formation should be applied to increase both the titer and yield. Isobutanol production from glycerol has also been achieved by expressing acetolactate synthase (ilvIH), keto-acid reducto-isomerase (ilvC), α-ketoisovalerate decarboxylase (kivd), and alcohol dehydrogenase (adhA) in K. pneumoniae (Fig. 3) [81]. By eliminating the lactate and 2,3-butanediol pathway (ΔldhAΔadc), the resulting strain can produce about 320 mg/L of isobutanol. 2-Butanol production from glycerol could be achieved via the extension of 2,3-BDO synthesis pathway with diol dehydratase (pduCDE) and alcohol dehydrogenase (adh) (Fig. 3) [41]. However, the activity of diol dehydratase toward meso-2,3-BDO dehydration should be significantly improved by protein engineering to get a higher titer of 2-butanol.
Fig. 3.

Metabolic pathway for the production of n-butanol, 2-butanol, and isobutanol. adc acetolactate decarboxylase; adhA alcohol dehydrogenase; ilvC keto-acid reducto-isomerase; ilvIH acetolactate synthase; kivd α-ketoisovalerate decarboxylase; ldhA lactate dehydrogenase; pduCDE: diol dehydratase
Lipids and fatty acids
Microbial lipids and free fatty acids (FFAs) are important sources for the production of biodiesel and other chemicals. A broad spectrum of microbes including microalgae, yeasts and fungi can efficiently utilize crude glycerol for the production of lipids [82–84]. Overexpression of glycerol utilization pathway such as glycerol kinase and glycerol 3-phosphate dehydrogenase was shown to be important for increasing lipid productivity in Fistulifera solaris [85] and Synechocystis sp. [86]. The theoretical yield of FFAs on glycerol is higher than that on glucose due to the higher reduction degree of glycerol [87]. To produce FFAs, effective acyl–acyl carrier protein (ACP) thioesterase (TE) should be introduced to release FFAs from acyl-ACP generated in fatty acid elongation cycle. Other important factors to increase the yield of FFAs include (1) enhancement of precursor supply (malonyl-CoA) by overexpression of acetyl-CoA carboxylase; (2) repression of FFAs consumption via the deletion of acyl-CoA synthetase (ΔfadD); (3) enhancement of fatty acid elongation cycle; (4) enhancement of NADPH generation (Fig. 4). The first two strategies were combined with the overexpression of a TE from Cinnamomum camphora by Lu et al. who constructed a mutant E. coli strain that can accumulate FFAs up to 2.5 g/L from glycerol [88]. Similar strategies were adopted by Lenne et al. who further optimized the copy number of TE from Umbellularia californica [89]. A lower copy of TE was shown to be more beneficial for FFAs production. To enhance the supply of NADPH for FFAs production, overexpression of both NAD kinase (nadK) and membrane-bound transhydrogenase (pntAB) was shown to be an effective strategy to enhance the production and yields of FFAs (4.82 g/L and 0.3 g/g) [90]. Production of FFAs with varied chain length from glycerol by the engineered reversal of β-oxidation cycle was also reported [91]. The system contained thiolase (fadA), 3-hydroxyacyl-CoA dehydrogenase (fadB), enoyl-CoA hydratase (fadB), enoyl-CoA reductase (yidO), and thioesterases (TE) (Fig. 4). Thioesterases could be used to control the chain length of FFAs.
Fig. 4.

Metabolic pathway for the production of free fatty acids (FFAs) via fatty acid elongation cycle and reversed β-oxidation cycle. Strategies used to increase the yield of FFAs include (1) enhancement of precursor supply by overexpression of acetyl-CoA carboxylase (ACC); (2) repression of FFAs consumption via the deletion of acyl-CoA synthetase (fadD); (3) enhancement of fatty acid elongation cycle via the overexpression of (3R)-hydroxymyristoyl-ACP dehydrase (fabZ); (4) enhancement of NADPH generation via the overexpression of NAD kinase (nadK) and membrane-bound transhydrogenase (pntAB). ACC acetyl-CoA carboxylase; fabZ: (3R)-hydroxymyristoyl-ACP dehydrase; fadA, thiolase; fadB, functional hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase; fadD: acyl-CoA synthetase; nadK NAD kinase; pntAB membrane-bound transhydrogenase; TE thioesterase; ydiO, enoyl-CoA reductase
Challenges and future prospects
Biorefineries are based on the integration of various conversion processes to fully utilize biomass for the production of energy, fuels, and chemicals. In this context, the utilization of crude glycerol generated in the biodiesel and bioethanol production processes to produce valuable chemicals and fuels offers an excellent opportunity to improve resource efficiency and economic viability of the biofuel industry. So far, most of the glycerol transformation processes are based on the fermentation of refined glycerol. The performance of the engineered strains on crude glycerol should be further examined. The composition of crude glycerol varies significantly, depending on the feedstock, the process used for biodiesel production, and the post-treatment methods. The impurities of crude glycerol include water, methanol, salts, soap, fatty acids, and other unknown substances [2, 8]. Some of the impurities may significantly affect the performance of microorganisms. For example, it was reported that only three of the seven tested samples can be utilized by a metabolic engineered C. glutamicum for amino acids production [71]. Thus, it is important to elucidate the inhibition mechanism and enhance the tolerance of engineered strains to inhibitory substances. To this end, metabolic engineered strains can be further evolved to adapt to different glycerol lots by adaptive laboratory evolution and systems biology can be utilized to understand how specific mutations affect cellular metabolism. Moreover, systems biology can be utilized to identify novel targets which cannot be found by rational analysis for designing more efficient strains [15, 17, 18].
Recent development of synthetic biology can also be utilized to develop strains for the production of natural and non-natural products from glycerol. For example, novel metabolic pathways have recently been developed to produce ethylene glycol [13] and 1,4-butanediol [92], two most widely used diols in the polymer industry. Production of highly valuable pharmaceuticals such as artemisinin can also be implemented in glycerol metabolic pathway, providing new potentials for glycerol biorefinery [5, 93].
Conclusions
The use of metabolic engineering to improve the performance of industrial strain for converting abundant and low-priced crude glycerol into higher value products represents a promising route toward glycerol biorefinery, which will significantly increase the economic viability of the biofuels industry. Current development of systems biology, synthetic biology, and new tools of metabolic engineering need to be integrated to improve the process efficiency (titer, yield and productivity) and to produce new products such as non-natural chemicals. Efforts also need to be paid to engineer and adapt strains for industrial implementation, especially for enhanced tolerance to toxic substances of crude glycerol and variable industrial environments.
Authors’ contributions
ZC and DHL wrote the manuscript. Both authors read and approved the final manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant no. 21406131), the Science and Technology Planning Project of Guangdong Province (Grant no. 2015A020215001), and the Tsinghua University’s Initiative Scientific Research Program (Grant no. 20151080362).
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
All data generated or analyzed during this study are included in this published article.
Abbreviations
- DHA
dihydroxyacetone
- 1,3-PDO
1,3-propanediol
- 1,2-PDO
1,2-propanediol
- ACP
acyl–acyl carrier protein
- TE
thioesterase
- PEPC
phosphoenolpyruvate carboxylase
- PEPCK
phosphoenolpyruvate carboxykinase
- PYC
pyruvate carboxylase
- 3-HP
3-hydroxypropionic acid
Contributor Information
Zhen Chen, Phone: +86-10-62772130, Email: zhenchen2013@mail.tsinghua.edu.cn.
Dehua Liu, Email: dhliu@tsinghua.edu.cn.
References
- 1.Jose A, James M. Fermentation of glycerol and production of valuable chemical and biofuel molecules. Biotechnol Lett. 2013;35:831–842. doi: 10.1007/s10529-013-1240-4. [DOI] [PubMed] [Google Scholar]
- 2.Garlapati VK, Shankar U, Budhiraja A. Bioconversion technologies of crude glycerol to value added industrial products. Biotechnol Reports. 2016;9:9–14. doi: 10.1016/j.btre.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida JRM, Fávaro LCL, Quirino BF. Biodiesel biorefinery : opportunities and challenges for microbial production of fuels and chemicals from glycerol waste. Biotechnol Biofuels. 2012;5:48. doi: 10.1186/1754-6834-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quispe CAG, Coronado CJR, Carvalho JA. Glycerol : production, consumption, prices, characterization and new trends in combustion. Renew Sustain Energy Rev. 2013;27:475–493. doi: 10.1016/j.rser.2013.06.017. [DOI] [Google Scholar]
- 5.Tan HW, Aziz ARA, Aroua MK. Glycerol production and its applications as a raw material: a review. Renew Sustain Energy Rev. 2013;27:118–127. doi: 10.1016/j.rser.2013.06.035. [DOI] [Google Scholar]
- 6.Clomburg JM, Gonzalez R. Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends Biotechnol. 2013;31:20–28. doi: 10.1016/j.tibtech.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Yazdani SS, Gonzalez R. Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr Opin Biotechnol. 2007;18:213–219. doi: 10.1016/j.copbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Luo X, Ge X, Cui S, Li Y. Value-added processing of crude glycerol into chemicals and polymers. Bioresour Technol. 2016;215:144–154. doi: 10.1016/j.biortech.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Bommareddy RR, Frank D, Rappert S, Zeng A-P. Deregulation of feedback inhibition of phosphoenolpyruvate carboxylase for improved lysine production in Corynebacterium glutamicum. Appl Environ Microbiol. 2014;80:1388–1393. doi: 10.1128/AEM.03535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng F, Chen Z, Zheng P, Sun J, Zeng A-P. Exploring the allosteric mechanism of dihydrodipicolinate synthase by reverse engineering of the allosteric inhibitor binding sites and its application for lysine production. Appl Microbiol Biotechnol. 2013;97:1963–1971. doi: 10.1007/s00253-012-4062-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Rappert S, Zeng A. Rational design of allosteric regulation of homoserine dehydrogenase by a nonnatural inhibitor. ACS Synth Biol. 2015;4:126–131. doi: 10.1021/sb400133g. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Chen Z, Zheng P, Sun J, Zeng A-P. Study and reengineering of the binding sites and allosteric regulation of biosynthetic threonine deaminase by isoleucine and valine in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:2939–2949. doi: 10.1007/s00253-012-4176-z. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Huang J, Wu Y, Liu D. Metabolic engineering of Corynebacterium glutamicum for the de novo production of ethylene glycol from glucose. Metab Eng. 2016;33:12–18. doi: 10.1016/j.ymben.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, Geng F, Zeng A-P. Protein design and engineering of a de novo pathway for microbial production of 1,3-propanediol from glucose. Biotechnol J. 2015;10:284–289. doi: 10.1002/biot.201400235. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Sun J, Hartlep M, Deckwer W-D, Zeng A-P. Combined use of proteomic analysis and enzyme activity assays for metabolic pathway analysis of glycerol fermentation by Klebsiella pneumoniae. Biotechnol Bioeng. 2003;83:525–536. doi: 10.1002/bit.10701. [DOI] [PubMed] [Google Scholar]
- 16.Liao Y-C, Huang T-W, Chen F-C, Charusanti P, Hong JSJ, Chang H-Y, Tsai S-F, Palsson BO, Hsiung CA. An experimentally validated genome-scale metabolic reconstruction of Klebsiella pneumoniae MGH 78578, iYL1228. J Bacteriol. 2011;193:1710–1717. doi: 10.1128/JB.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beckers V, Castro IP, Tomasch J, Wittmann C. Integrated analysis of gene expression and metabolic fluxes in PHA—producing Pseudomonas putida grown on glycerol. Microb Cell Fact. 2016;15:73. doi: 10.1186/s12934-016-0470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salazar M, Vongsangnak W, Panagiotou G, Andersen MR, Nielsen J. Uncovering transcriptional regulation of glycerol metabolism in Aspergilli through genome-wide gene expression data analysis. Mol Genet Genomics. 2009;282:571–586. doi: 10.1007/s00438-009-0486-y. [DOI] [PubMed] [Google Scholar]
- 19.Cintolesi A, Clomburg JM, Rigou V, Zygourakis K, Gonzalez R. Quantitative analysis of the fermentative metabolism of glycerol in Escherichia coli. Biotechnol Bioeng. 2012;109:187–198. doi: 10.1002/bit.23309. [DOI] [PubMed] [Google Scholar]
- 20.Kaur G, Srivastava AK, Chand S. Advances in biotechnological production of 1, 3-propanediol. Biochem Eng J. 2012;64:106–118. doi: 10.1016/j.bej.2012.03.002. [DOI] [Google Scholar]
- 21.Celińska E. Debottlenecking the 1,3-propanediol pathway by metabolic engineering. Biotechnol Adv. 2010;28:519–530. doi: 10.1016/j.biotechadv.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Vaidyanathan H, Kandasamy V, Ramakrishnan GG, Ramachandran KB. Glycerol conversion to 1, 3-propanediol is enhanced by the expression of a heterologous alcohol dehydrogenase gene in Lactobacillus reuteri. AMB Express. 2011;1:37. doi: 10.1186/2191-0855-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maervoet VET, De Maeseneire SL, Avci FG, Beauprez J, Soetaert WK. High yield 1,3-propanediol production by rational engineering of the 3-hydroxypropionaldehyde bottleneck in Citrobacter werkmanii. Microb Cell Fact. 2016;15:23. doi: 10.1186/s12934-016-0421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meynial-salles I, Mendes F, Andrade C, Vasconcelos I, Soucaille P. Metabolic engineering of Clostridium acetobutylicum for the industrial production of 1, 3-propanediol from glycerol. Metab Eng. 2005;7:329–336. doi: 10.1016/j.ymben.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Sun J, Hartlep M, Deckwer W, Zeng A. Combined use of proteomic analysis and enzyme activity assays for metabolic pathway analysis of glycerol fermentation by Klebsiella pneumoniae. Biotechnol Bioeng. 2003;5:525–536. doi: 10.1002/bit.10701. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Li Y, Du C, Liu M. Inactivation of aldehyde dehydrogenase: a key factor for engineering 1, 3-propanediol production by Klebsiella pneumoniae. Metab Eng. 2006;8:578–586. doi: 10.1016/j.ymben.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Xu YZ, Guo NN, Zheng ZM, Ou XJ, Liu HJ, Liu DH. Metabolism in 1,3-propanediol fed-batch fermentation by a d-lactate deficient mutant of Klebsiella pneumoniae. Biotechnol Bioeng. 2009;104:965–972. doi: 10.1002/bit.22455. [DOI] [PubMed] [Google Scholar]
- 28.Rathnasingh C, Song H, Seung D, Park S. Effects of mutation of 2,3-butanediol formation pathway on glycerol metabolism and 1,3-propanediol production by Klebsiella pneumoniae J2B. Bioresour Technol. 2016;214:432–440. doi: 10.1016/j.biortech.2016.04.032. [DOI] [PubMed] [Google Scholar]
- 29.Oh B-R, Seo J-W, Heo S-Y, Hong W-K, Luo LH, Kim S, Park D-H, Kim CH. Optimization of culture conditions for 1,3-propanediol production from glycerol using a mutant strain of Klebsiella pneumoniae. Appl Biochem Biotechnol. 2012;166:127–137. doi: 10.1007/s12010-011-9409-6. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Xiu Z. Metabolic pathway analysis of glycerol metabolism in Klebsiella pneumoniae incorporating oxygen regulatory system. Biotechnol Prog. 2009;25:103–115. doi: 10.1002/btpr.70. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z, Liu H, Liu D. Regulation of 3-hydroxypropionaldehyde accumulation in Klebsiella pneumoniae by overexpression of dhaT and dhaD genes. Enzyme Microb Technol. 2009;45:305–309. doi: 10.1016/j.enzmictec.2009.04.005. [DOI] [Google Scholar]
- 32.Chen Z, Liu H, Liu D. Metabolic pathway analysis of 1,3-propanediol production with a genetically modified Klebsiella pneumoniae by overexpressing an endogenous NADPH-dependent alcohol dehydrogenase. Biochem Eng J. 2011;54:151–157. doi: 10.1016/j.bej.2011.02.005. [DOI] [Google Scholar]
- 33.Zhang Y, Huang Z, Du C, Li Y, Cao Z. Introduction of an NADH regeneration system into Klebsiella oxytoca leads to an enhanced oxidative and reductive metabolism of glycerol. Metab Eng. 2009;11:101–106. doi: 10.1016/j.ymben.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Tang X, Tan Y, Zhu H, Zhao K, Shen W. Microbial conversion of glycerol to 1, 3-propanediol by an engineered strain of Escherichia coli. Appl Environ Microbiol. 2009;75:1628–1634. doi: 10.1128/AEM.02376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxena RK, Anand P, Saran S, Isar J, Agarwal L. Microbial production and applications of 1,2-propanediol. Indian J Microbiol. 2010;50:2–11. doi: 10.1007/s12088-010-0017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clomburg JM, Gonzalez R. Metabolic engineering of Escherichia coli for the production of 1, 2-propanediol from glycerol. Biotechnol Bioeng. 2011;108:867–879. doi: 10.1002/bit.22993. [DOI] [PubMed] [Google Scholar]
- 37.Joon-Young J, Yun HS, Lee J, Oh M. Production of 1,2-propanediol from glycerol in Saccharomyces cerevisiae. J Microbiol Biotechnol. 2011;21:846–853. doi: 10.4014/jmb.1103.03009. [DOI] [PubMed] [Google Scholar]
- 38.Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, Lebar T, Turnšek J, Tomšič N, Avbelj M, Gaber R, Koprivnjak T, Mori J, Glavnik V, Vovk I, Benčina M, Hodnik V, Anderluh G, Dueber JE, Jerala R, DeLisa MP. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 2012;40:1879–1889. doi: 10.1093/nar/gkr888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MJ, Brown IR, Juodeikis R, Frank S, Warren MJ. Employing bacterial microcompartment technology to engineer a shell-free enzyme-aggregate for enhanced 1,2-propanediol production in Escherichia coli. Metab Eng. 2016;36:48–56. doi: 10.1016/j.ymben.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z, Sun H, Huang J, Wu Y, Liu D. Metabolic engineering of Klebsiella pneumoniae for the production of 2-butanone from glucose. PLoS One. 2015;10:e0140508. doi: 10.1371/journal.pone.0140508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, Wu Y, Huang J, Liu D. Metabolic engineering of Klebsiella pneumoniae for the de novo production of 2-butanol as a potential biofuel. Bioresour Technol. 2015;197:260–265. doi: 10.1016/j.biortech.2015.08.086. [DOI] [PubMed] [Google Scholar]
- 42.Duan H, Yamada Y, Sato S. Efficient production of 1, 3-butadiene in the catalytic dehydration of 2, 3-butanediol. Appl Catal A Gen. 2015;491:163–169. doi: 10.1016/j.apcata.2014.12.006. [DOI] [Google Scholar]
- 43.Cho S, Kim T, Woo HM, Kim Y, Lee J, Um Y. High production of 2,3-butanediol from biodiesel-derived crude glycerol by metabolically engineered Klebsiella oxytoca M1. Biotechnol Biofuels. 2015;8:146. doi: 10.1186/s13068-015-0336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Kim B, Park K, Um Y, Lee J. Synthesis of pure meso-2,3-butanediol from crude glycerol using an engineered metabolic pathway in Escherichia coli. Appl Biochem Biotechnol. 2012;166:1801–1813. doi: 10.1007/s12010-012-9593-z. [DOI] [PubMed] [Google Scholar]
- 45.Yang T, Rao Z, Zhang X, Xu M, Xu Z, Yang ST. Enhanced 2,3-butanediol production from biodiesel-derived-glycerol by engineering of cofactor regeneration and manipulating carbon flux in Bacillus amyloliquefaciens. Microb Cell Fact. 2015;14:122. doi: 10.1186/s12934-015-0317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng X, Ding Y, Xian M, Xu X, Zhang R, Zhao G. Production of optically pure d-lactate from glycerol by engineered Klebsiella pneumoniae strain. Bioresour Technol. 2013;172:269–275. doi: 10.1016/j.biortech.2014.09.074. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Tian K, Niu D, Shen W, Algasan G, Singh S, Wang Z. Efficient bioconversion of crude glycerol from biodiesel to optically pure d-lactate by metabolically engineered Escherichia coli. Green Chem. 2014;16:342. doi: 10.1039/C3GC41769G. [DOI] [Google Scholar]
- 48.Wang ZW, Saini M, Lin L, Chiang C, Chao Y. Systematic engineering of Escherichia coli for d-lactate production from crude glycerol. J Agric Food Chem. 2015;63:9583–9589. doi: 10.1021/acs.jafc.5b04162. [DOI] [PubMed] [Google Scholar]
- 49.Mazumdar S, Clomburg JM, Gonzalez R. Escherichia coli strains engineered for homofermentative production of d-lactic acid from glycerol. Appl Environ Microbiol. 2010;76:4327–4336. doi: 10.1128/AEM.00664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazumdar S, Blankschien MD, Clomburg JM, Gonzalez R. Efficient synthesis of l-lactic acid from glycerol by metabolically engineered Escherichia coli. Microb Cell Fact. 2013;12:7. doi: 10.1186/1475-2859-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doi Y. Production from biodiesel-derived crude glycerol by metabolically engineered Enterococcus faecalis: cytotoxic evaluation of biodiesel waste and development of a glycerol-inducible gene expression system. Appl Environ Microbiol. 2015;81:2082–2089. doi: 10.1128/AEM.03418-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, Liu H, Zhang J, Liu D. Elementary mode analysis for the rational design of efficient succinate conversion from glycerol by Escherichia coli. J Biomed Biotechnol. 2010;2010:518743. doi: 10.1155/2010/518743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blankschien MD, Clomburg JM, Gonzalez R. Metabolic engineering of Escherichia coli for the production of succinate from glycerol. Metab Eng. 2010;12:409–419. doi: 10.1016/j.ymben.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Kim P, Laivenieks M, Vieille C, Zeikus JG. Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl Environ Microbiol. 2004;70:1238–1241. doi: 10.1128/AEM.70.2.1238-1241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X, Shanmugam KT, Ingram LO. Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol. 2010;76:2397–2401. doi: 10.1128/AEM.02902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Litsanov B, Brocker M, Bott M. Glycerol as a substrate for aerobic succinate production in minimal medium with Corynebacterium glutamicum. Microb Biotechnol. 2013;6:189–195. doi: 10.1111/j.1751-7915.2012.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valdehuesa KNG, Liu H, Nisola GM, Chung W-J, Lee SH, Park SJ. Recent advances in the metabolic engineering of microorganisms for the production of 3-hydroxypropionic acid as C3 platform chemical. Appl Microbiol Biotechnol. 2013;97:3309–3321. doi: 10.1007/s00253-013-4802-4. [DOI] [PubMed] [Google Scholar]
- 58.Huang Y, Li Z, Shimizu K, Ye Q. Co-production of 3-hydroxypropionic acid and 1,3-propanediol by Klebseilla pneumoniae expressing aldH under microaerobic conditions. Bioresour Technol. 2013;128:505–512. doi: 10.1016/j.biortech.2012.10.143. [DOI] [PubMed] [Google Scholar]
- 59.Ashok S, Sankaranarayanan M, Ko Y, Jae K-E, Ainala SK, Kumar V, Park S. Production of 3-hydroxypropionic acid from glycerol by recombinant Klebsiella pneumoniae ΔdhaTΔyqhD which can produce vitamin B12 naturally. Biotechnol Bioeng. 2013;110:511–524. doi: 10.1002/bit.24726. [DOI] [PubMed] [Google Scholar]
- 60.Ashok S, Mohan Raj S, Ko Y, Sankaranarayanan M, Zhou S, Kumar V, Park S. Effect of puuC overexpression and nitrate addition on glycerol metabolism and anaerobic 3-hydroxypropionic acid production in recombinant Klebsiella pneumoniae ΔglpKΔdhaT. Metab Eng. 2013;15:10–24. doi: 10.1016/j.ymben.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 61.Kim K, Kim S-K, Park Y-C, Seo J-H. Enhanced production of 3-hydroxypropionic acid from glycerol by modulation of glycerol metabolism in recombinant Escherichia coli. Bioresour Technol. 2014;156:170–175. doi: 10.1016/j.biortech.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 62.Jung WS, Kang JH, Chu HS, Choi IS, Myung K. Elevated production of 3-hydroxypropionic acid by metabolic engineering of the glycerol metabolism in Escherichia coli. Metab Eng. 2014;23:116–122. doi: 10.1016/j.ymben.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Tsuruno K, Honjo H, Hanai T. Enhancement of 3-hydroxypropionic acid production from glycerol by using a metabolic toggle switch. Microb Cell Fact. 2015;14:155. doi: 10.1186/s12934-015-0342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tokuyama K, Ohno S, Yoshikawa K, Hirasawa T, Tanaka S, Furusawa C, Shimizu H. Increased 3-hydroxypropionic acid production from glycerol by modification of central metabolism in Escherichia coli. Microb Cell Fact. 2014;13:64. doi: 10.1186/1475-2859-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu HS, Kim YS, Lee CM, Lee JH, Jung WS, Ahn J-H, Song SH, Choi IS, Cho KM. Metabolic engineering of 3-hydroxypropionic acid biosynthesis in Escherichia coli. Biotechnol Bioeng. 2015;112:356–364. doi: 10.1002/bit.25444. [DOI] [PubMed] [Google Scholar]
- 66.Rathnasingh C, Raj SM, Jo J-E, Park S. Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioeng. 2009;104:729–739. doi: 10.1002/bit.22429. [DOI] [PubMed] [Google Scholar]
- 67.Lim HG, Noh MH, Jeong JH, Park S, Jung GY. Optimum rebalancing of the 3-hydroxypropionic acid production pathway from glycerol in Escherichia coli. ACS Synth Biol. 2016 doi: 10.1021/acssynbio.5b00303. [DOI] [PubMed] [Google Scholar]
- 68.Zhou S, Catherine C, Rathnasingh C, Somasundar A, Park S. Production of 3-hydroxypropionic acid from glycerol by recombinant Pseudomonas denitrificans. Biotechnol Bioeng. 2013;110:3177–3187. doi: 10.1002/bit.24980. [DOI] [PubMed] [Google Scholar]
- 69.Zhou S, Ashok S, Ko Y, Kim D-M, Park S. Development of a deletion mutant of Pseudomonas denitrificans that does not degrade 3-hydroxypropionic acid. Appl Microbiol Biotechnol. 2014;98:4389–4398. doi: 10.1007/s00253-014-5562-5. [DOI] [PubMed] [Google Scholar]
- 70.Rittmann D, Lindner SN, Wendisch VF. Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl Environ Microbiol. 2008;74:6216–6222. doi: 10.1128/AEM.00963-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meiswinkel TM, Rittmann D, Lindner SN, Wendisch VF. Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum. Bioresour Technol. 2013;145:254–258. doi: 10.1016/j.biortech.2013.02.053. [DOI] [PubMed] [Google Scholar]
- 72.Gottlieb K, Albermann C, Sprenger GA. Improvement of l-phenylalanine production from glycerol by recombinant Escherichia coli strains: the role of extra copies of glpK, glpX, and tktA genes. Microb Cell Fact. 2014;13:96. doi: 10.1186/s12934-014-0096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bommareddy RR, Chen Z, Rappert S, Zeng A-P. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase. Metab Eng. 2014;25:30–37. doi: 10.1016/j.ymben.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Valle A, Cabrera G, Muhamadali H, Trivedi DK, Ratray NJW. A systematic analysis of TCA Escherichia coli mutants reveals suitable genetic backgrounds for enhanced hydrogen and ethanol production using glycerol as main carbon source. Biotechnol J. 2015;10(11):1750–1761. doi: 10.1002/biot.201500005. [DOI] [PubMed] [Google Scholar]
- 75.Shams Yazdani S, Gonzalez R. Engineering Escherichia coli for the efficient conversion of glycerol to ethanol and co-products. Metab Eng. 2008;10:340–351. doi: 10.1016/j.ymben.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 76.Wong MS, Li M, Black RW, Le TQ, Puthli S, Campbell P, Monticello DJ. Microaerobic conversion of glycerol to ethanol in Escherichia coli. Appl Environ Microbiol. 2014;80:3276–3282. doi: 10.1128/AEM.03863-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lan EI, Liao JC. Microbial synthesis of n-butanol, isobutanol, and other higher alcohols from diverse resources. Bioresour Technol. 2013;135:339–349. doi: 10.1016/j.biortech.2012.09.104. [DOI] [PubMed] [Google Scholar]
- 78.Pyne M, Sokolenko S, Liu X, Srirangan K, Bruder M, Aucoin M, Moo-Yong M, Chung D, Chou C. Disruption of the reductive 1,3-propanediol pathway triggers production of 1,2-propanediol for sustained glycerol fermentation by Clostridium pasteurianum. Appl Environ Microbiol. 2016 doi: 10.1128/AEM.01354-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang M, Fan L, Tan T. 1-Butanol production from glycerol by engineered Klebsiella pneumoniae. RSC Adv. 2014;4:57791–57798. doi: 10.1039/C4RA09016K. [DOI] [Google Scholar]
- 80.Zhou P, Zhang Y, Wang P, Xie J, Ye Q. Butanol production from glycerol by recombinant Escherichia coli. Ann Microbiol. 2014;64:219–227. doi: 10.1007/s13213-013-0654-5. [DOI] [Google Scholar]
- 81.Oh B-R, Heo S-Y, Lee S-M, Hong W-K, Park JM, Jung YR, Kim D-H, Sohn J-H, Seo J-W, Kim CH. Production of 2-butanol from crude glycerol by a genetically-engineered Klebsiella pneumoniae strain. Biotechnol Lett. 2014;36:57–62. doi: 10.1007/s10529-013-1333-0. [DOI] [PubMed] [Google Scholar]
- 82.Tchakouteu S, Kalantzi O, Gardeli C, Koutinas A, Aggelis G, Papanikolaou S. Lipid production by yeasts growing on biodiesel-derived crude glycerol: strain selection and impact of substrate concentration on the fermentation efficiency. J Appl Microbiol. 2015;118:911–927. doi: 10.1111/jam.12736. [DOI] [PubMed] [Google Scholar]
- 83.Polburee P, Yongmanitchai W, Honda K, Ohashi T. Lipid production from biodiesel-derived crude glycerol by Rhodosporidium fluviale DMKU-RK253 using temperature shift with high cell density. Biochem Eng J. 2016;112:208–218. doi: 10.1016/j.bej.2016.04.024. [DOI] [Google Scholar]
- 84.Karamerou EE, Theodoropoulos C, Webb C. A biorefinery approach to microbial oil production from glycerol by Rhodotorula glutinis. Biomass Bioenergy. 2016;89:113–122. doi: 10.1016/j.biombioe.2016.01.007. [DOI] [Google Scholar]
- 85.Muto M, Tanaka M, Liang Y, Yoshino T, Matsumoto M, Tanaka T. Enhancement of glycerol metabolism in the oleaginous marine diatom Fistulifera solaris JPCC DA0580 to improve triacylglycerol productivity. Biotechnol Biofuels. 2015;8:4. doi: 10.1186/s13068-014-0184-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X, Xiong X, Sa N, Roje S, Chen S. Metabolic engineering of enhanced glycerol-3-phosphate synthesis to increase lipid production in Synechocystis sp. PCC 6803. Appl Microbiol Biotechnol. 2016:6091–6101. [DOI] [PubMed]
- 87.Bommareddy RR, Sabra W, Maheshwari G, Zeng A-P. Metabolic network analysis and experimental study of lipid production in Rhodosporidium toruloides grown on single and mixed substrates. Microb Cell Fact. 2015;14:36. doi: 10.1186/s12934-015-0217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu X, Vora H, Khosla C. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab Eng. 2008;10:333–339. doi: 10.1016/j.ymben.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 89.Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol Bioeng. 2010;106:193–202. doi: 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu H, Karanjikar M, San K-Y. Metabolic engineering of Escherichia coli for efficient free fatty acid production from glycerol. Metab Eng. 2014;25:82–91. doi: 10.1016/j.ymben.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 91.Clomburg JM, Vick JE, Blankschien MD, Rodríguez-Moyá, Gonzalez R. A synthetic biology approach to engineer a functional reversal of the beta-oxidation cycle. ACS Synth Biol. 2012;1:541–554. doi: 10.1021/sb3000782. [DOI] [PubMed] [Google Scholar]
- 92.Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, Estadilla J, Teisan S, Schreyer HB, Andrae S, Yang TH, Lee SY, Burk MJ, Van Dien S. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 93.Chen Z, Zeng A-P. Protein engineering approaches to chemical biotechnology. Curr Opin Biotechnol. 2016;42:198–205. doi: 10.1016/j.copbio.2016.07.007. [DOI] [PubMed] [Google Scholar]


