Abstract
Translesion DNA synthesis (TLS) operates when replicative polymerases are blocked by DNA lesions. To investigate the mechanism of mammalian TLS, we employed a plasmid bearing a single 7-(deoxyadenosine-N6-yl)-aristolactam I (dA-AL-I) adduct, which is generated by the human carcinogen, aristolochic acid I, and genetically engineered mouse embryonic fibroblasts. This lesion induces A to T transversions at a high frequency. The simultaneous knockouts of the Polh, Poli and Polk genes did not influence the TLS efficiency or the coding property of dA-AL-I, indicating that an unknown DNA polymerase(s) can efficiently catalyze the insertion of a nucleotide opposite the adduct and subsequent extension. Similarly, knockout of the Rev1 gene did not significantly affect TLS. However, knockout of the Rev3l gene, coding for the catalytic subunit of polζ, drastically suppressed TLS and abolished dA-AL-I to T transversions. The results support the idea that Rev1 is not essential for the cellular TLS functions of polζ in mammalian cells. Furthermore, the frequency of dA-AL-I to T transversion was affected by a sequence context, suggesting that TLS, at least in part, contributes to the formation of mutational hot and cold spots observed in aristolochic acid-induced cancers.
Keywords: Aristolochic acid, translesion DNA synthesis, Y-family polymerases, Rev1, polζ, mutational hotspot
1. Introduction
DNA damage, generated by endogenous and environmental agents, often blocks DNA synthesis catalyzed by replicative DNA polymerases (1). Under this situation, translesion DNA synthesis (TLS) catalyzed by specialized DNA polymerases operates across a lesion, often resulting in a mutation. Among mammalian DNA polymerases, the Y-family polymerases, polη, polι, polκ and Rev1, play important roles in TLS (1). A defect in human polη, the XPV gene product, is responsible for the xeroderma pigmentosum variant syndrome, an inherited disorder in individuals highly predisposed to sunlight-induced skin cancer (2, 3). Polη catalyzes accurate TLS across UV-induced cyclobutane pyrimidine dimers (2, 3) to avoid mutation induction. Polι, together with polη, suppresses the development of skin cancer in mice (4, 5). It also plays a role in protecting human cells against oxidative damages (6). Polκ protects mouse cells against genotoxicity of benzo[a]pyrene dihydrodiol epoxide-derived lesion (7). It also plays a role in the bypass of cholesterol-induced guanine lesions in mice (8). Rev1 has deoxycytidyl transferase activity (9, 10) and catalyzes TLS across a certain class of lesions (11, 12). Rev1 also plays a non-catalytic role in TLS (13) by physically interacting with other Y-family polymerases (14, 15) and the Rev7 subunit of polζ (16, 17). Polζ, consisting of Rev3, Rev7, Pold2 and Pold3 subunits (Pold2 and Pold3 subunits are shared with DNA polymerase δ) (18–21), belongs to the B family and also plays an important role in TLS (1). This pol is especially competent for extending a primer from a 3’-terminal nucleotide pairing to a template DNA lesion (1). Although Rev1 plays a critical non-catalytic role in the polζ activity in yeast (22), this role is questioned in mammalian cells: Rev1 is critical for the activity of Y-family polymerases, but not polζ (23).
Although many recently discovered specialized polymerases can catalyze TLS in vitro, Y-family polymerases likely play a major role. If a recruited polymerase cannot extend a primer following nucleotide insertion, a second polymerase such as polζ and polκ extends from the newly formed primer terminus (1). In this case, TLS is accomplished by two specialized polymerases, often called two-step TLS. However, our previous study questioned the essential role for the Y-family polymerases in TLS: neither the TLS efficiency nor the coding properties was greatly affected in the polη/polι/polκ triple-gene knockout (TKO) mouse embryonic fibroblasts (MEFs) when TLS across a single benzo[a]pyrene-derived dG was studied (24). To further explore the mechanism of mammalian TLS, we employed another environmental human carcinogen (aristolochic acid)-derived bulky adenine adduct in this study.
Aristolochic acid (AA), a nephrotoxin and human carcinogen, is found in Aristolochia plants and associated with both chronic kidney disease and urothelial carcinomas of the upper urinary tract (25, 26). Following metabolic activation, a metabolite(s) reacts with DNA to form covalent aristolactam-DNA adducts (27, 28). The aristolactam-dA adducts persist in the renal cortex for many years and are also found in urothelial tissues, where they initiate cancers bearing characteristic mutations in oncogenes and tumor suppressor genes (25, 26, 29, 30). The mutational spectrum in the urothelial carcinomas associated with AA exposure is dominated by A to T transversions (73% of single-base substitutions) of a non-transcribed strand (29, 30). The A to T transversions are rare in other cancers (4.4%) (31). A sequence preference has also been observed for 5’pyrimidineAG, which coincides with the splicing acceptor sequence of a non-transcribed strand (29, 30). In this study, we have again observed that the Y family polymerases, including Rev1, are not essential for the efficient TLS across this adduct, but Polζ is.
2. Materials and Methods
2.1. Cell lines
Rev1−/− MEFs (32), Rev3l−/− Trp53−/− MEFs (33) and Polh−/− Poli−/− Polk−/− TKO MEFs (24, 34) have been described. The genomic reconfirmation of these knockouts is presented in Supplementary Fig. S1. Figure S2 of reference 24 shows the UV sensitivity of TKO MEFs.
2.2. Construction of gapped, site-specifically modified plasmid containing 7-(deoxyadenosin-N6-yl)-aristolactam I (dA-AL-I, A)
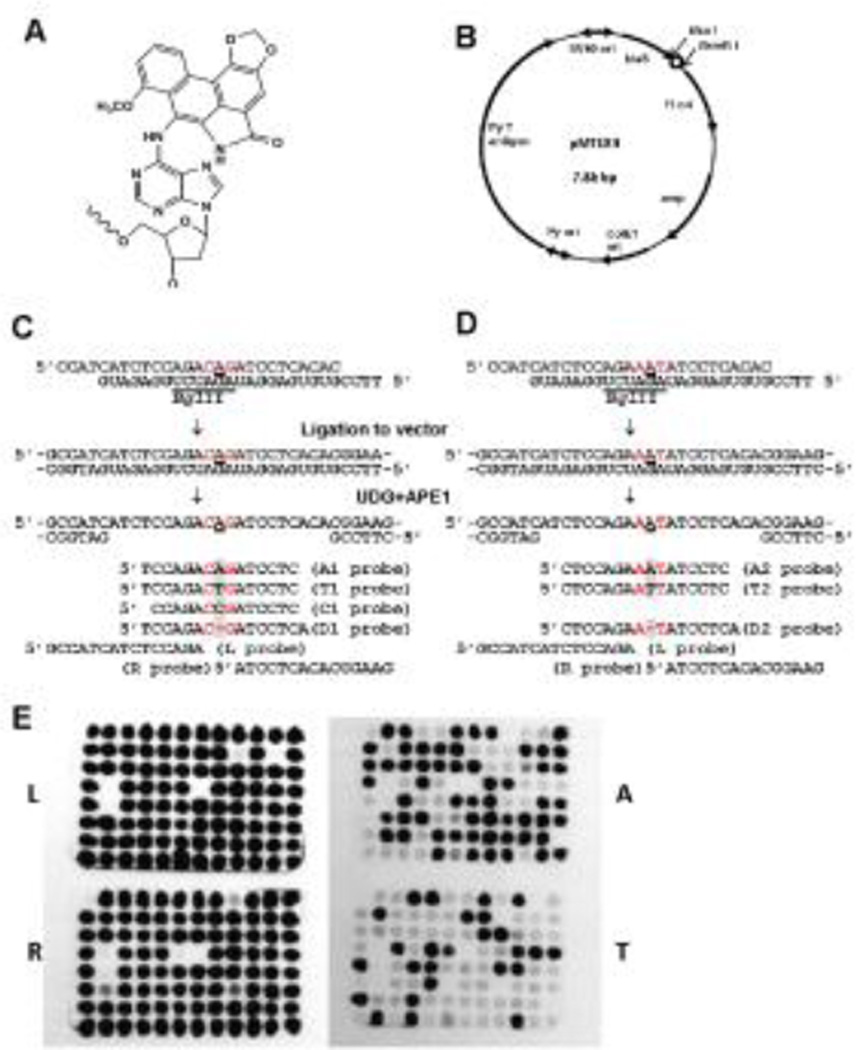
The 27-mer oligonucleotides containing dA-AL-I (Fig. 1A) were synthesized as described previously (35, 36). The oligonucleotide, 5’CCATCATCTCCAGACAGATCCTCACAC (Fig. 1C) or 5’CCATCATCTCCAGAAATATCCTCACAC (Fig. 1D), was annealed to a complementary, uracil-containing 27-mer, 5’TTCCGUGUGAGGAUAGAUCUGGAGAUG. The annealing resulted in the formation of four-nucleotide overhangs on both ends with two or three base mismatches opposite and adjacent to the adduct site (Figs. 1C and 1D). The annealed oligonucleotides were incorporated into pMTEX4 by ligating to the BsaI and BsmBI sites of the vector (Fig. 1B). Closed circular DNA was separated by ultracentrifugation in a cesium chloride-ethidium bromide continuous density gradient. To generate a gap opposite dA-AL-I, 200 ng of a modified construct was incubated, prior to transfection, with 2.5 units of uracil-DNA glycosylase (NEB) for 30 min at 37°C, followed by treatment with 25 units of apurinic/apyrimidinic endonuclease I (NEB) for 30 min at 37°C (Figs. 1C and 1D). These treatments rendered dA-AL-I resistant to nucleotide excision repair.
Fig. 1. Preparation of dA-AL-I-bearing plasmid and oligonucleotide probes.
(A) Chemical structure of dA-AL-I; (B) Structure of a MEF–E. coli shuttle vector: Py, mouse polyoma virus; ori, replication origin; amp, ampicillin resistance gene; blaS, blasticidin S-resistance gene; the dA-AL-I insertion site (open circle) is located between BsaI and BsmBI.; (C) and (D) Preparation of two gapped plasmids and probes employed: they differ only in the immediate neighboring bases (shown in red) flanking the adduct. A represents dA-AL-I. Note three mismatches at 5’CAG/5’AGA and two mismatches at 5’AA/5’GA. A1, A2, T1, T2, C1, D1, and D2 probes detect a coding event at dA-AL-I, and L and R probes confirm the presence of the inserted modified oligonucleotide; and (E) Examples of colony hybridization with oligonucleotide probes. Colonies showing positive hybridization signals of both L and R probes are considered to be derived from TLS. E. coli transformants that did not hybridize to L and R probe were excluded from analysis. Probes A and T detect A→A correct TLS events and A→T transversions, respectively.
2.3. Transfection of MEFs with a modified construct and recovery of plasmid
Cells were cultured under 5% (v/v) CO2 at 37°C in Dulbecco’s modified Eagle’s medium supplemented with fetal bovine serum (10%, v/v), penicillin (100 units/ml), and streptomycin (100 µg/ml). Cells (1×106) were plated in a 25-cm2 flask, cultured overnight, and then transfected overnight with 200 ng of a freshly prepared, gapped construct together with 400 ng of internal control plasmid, pMTKm, using the X-tremeGene 9 DNA transfection reagent (Roche). pMTKm was constructed by replacing the blasticidin S and ampicillin resistance genes in pMTEX4 with the kanamycin resistance gene (24). The following day, cells were detached by treating with trypsin/EDTA, transferred to a 150-cm2 flask and cultured for 3 days. Progeny plasmids were recovered from cells by the method of Hirt (37).
2.4. Determination of TLS efficiencies
Progeny plasmids were analyzed for TLS events. Recovered plasmids were treated with DpnI (10 units) and BglII for 1 h to remove unreplicated input DNA and progeny derived from the residual complementary strand, respectively. NEB 10-beta electro-competent E. coli (Δ(ara-leu)7697 araD139 fhuA ΔlacX74 galK16 galE15 e14-ϕ80dlacZΔM15 recA1 relA1 endA1 nupG rpsL (StrR) rph spoT1 Δ(mrr-hsdRMS-mcrBC)) (NEB) was transformed with progeny plasmid and plated on YT (1×) agar plates containing both ampicillin (100 µg/ml) and blasticidin S (50 µg/ml) for the detection of progeny derived from the modified construct or kanamycin (50 µg/ml) alone for progeny of the internal control, pMTKm. Because the adduct incorporation site is located very close to the blasticidin resistance gene (Fig. 1B), E. coli transformants carrying a progeny plasmid with deletions around the adduct site do not grow on a blasticidin S-containing plate and are therefore excluded from the analysis. The ratio of the number of ampicillin/blasticidin S-resistant colonies (TLS products) to the number of kanamycin-resistant colonies (internal control) was determined for each MEF line, and the relative TLS efficiency was determined by setting the ratio obtained in experiments with wild-type MEFs to 100% (24).
2.5. Analysis of TLS events
E. coli colonies on plates containing ampicillin and blasticidin S were picked up individually and analyzed for a sequence of the adducted region by oligonucleotide hybridization using probes shown in Figs. 1C and 1D. Probes L and R were used to confirm the presence of the oligonucleotide insert and to detect untargeted mutations and small deletions around the adduct site. These mutants were also excluded from the analysis. Probes A1, A2, T1, T2, and C1 detect targeted base substitutions. Probes D1 and D2 detect targeted one-base deletions. An example of oligonucleotide hybridization was presented in Fig. 1E. DNA sequencing was performed when none of these probes hybridized or when the confirmation of hybridization results was necessary.
3. Results
3.1. Efficient TLS in the absence of the three Y-family polymerases, polη, polι, and polκ
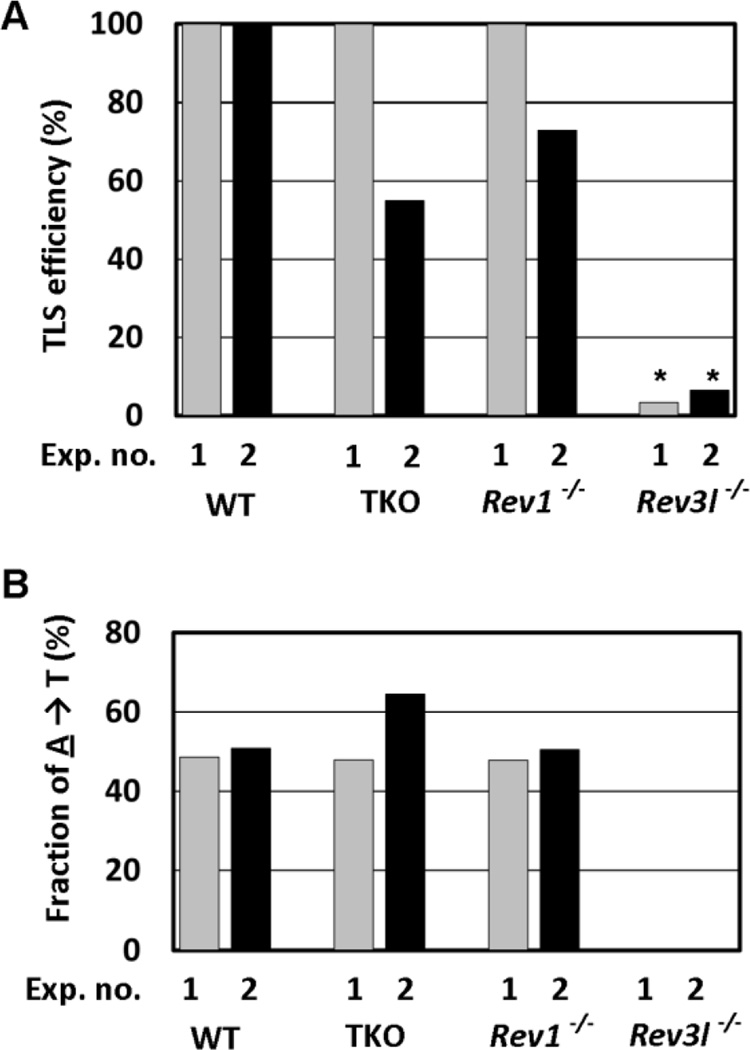
When considering the structure and size of dA-AL-I, Y-family polymerases were anticipated to be involved in TLS across this lesion. Therefore, we analyzed for TLS in TKO MEFs, using the 5’CAG sequence context. Unexpectedly, neither TLS efficiency (Fig. 2A) nor coding specificity (Table 1) was markedly affected in TKO MEFs. The TLS efficiency remained greater than 50% when compared with that in the wild-type cells in two independent experiments. The major coding events were A→A and A→T in both wild-type and TKO MEFs (Table 1): the former non-miscoding event and the latter transversion accounted for 30–40% and 50–60% of the coding events, respectively. Thus, the coding specificity did not change in the absence of the three Y-family polymerases (Fig. 2B). No clear difference in the TLS efficiency and coding specificity between wild-type and TKO MEFs was also confirmed when the 5’AAT context was employed (Fig. S2, Table S1). These results indicate that a yet undesignated polymerase(s) must insert a nucleotide across from dA-AL-I and extend from the newly formed abnormal pairs of dA-AL-I:dA and dA-AL-I:dT in the absence of the three Y-family polymerases.
Fig. 2. TLS efficiency (A) and the fraction of A→T transversion (B) in 5’CAG context in gene knockout MEFs.
TLS efficiency in wild-type MEFs was set to 100 %. Data in (B) were extracted from Table 1.
*p<0.001 when compared with a value for wild-type (WT) MEFs.
Table 1.
Targeted coding specificity of translesion DNA synthesis across dA-AL-I in various MEFs
| Host cells | Exp. | Total no. analysed |
[dA-AL-I] → |
|||||
|---|---|---|---|---|---|---|---|---|
| A | T | G | C | Δ | Others | |||
| Wild-type | 1 | 181 | 71 (39)a |
88 (49) |
0 (0) |
4 (2) |
5 (3) |
13 (7) |
| 2 | 189 | 76 (40) |
96 (51) |
2 (1) |
1 (0.5) |
6 (3) |
8 (4) |
|
| TKO | 1 | 94 | 33 (35) |
45 (48) |
0 (0) |
2 (2) |
1 (1) |
13 (14) |
| 2 | 93 | 27 (29) |
60 (65) |
1 (1) |
2 (2) |
0 (0) |
3 (3) |
|
| Rev1−/− | 1 | 92 | 34 (37) |
44 (48) |
0 (0) |
0 (0) |
5 (5) |
9 (10) |
| 2 | 91 | 36 (40) |
46 (51) |
0 (0) |
5 (6) |
0 (0) |
4 (4) |
|
| Rev3l−/− | 1 | 90 | 49 (54) |
0 (0) |
0 (0) |
3 (3) |
36 (40) |
2 (2) |
| 2 | 70 | 18 (26) |
0 (0) |
0 (0) |
30 (43) |
20 (29) |
2 (3) |
|
Sequence context is 5’CAG.
%. ‘Others’ includes untargeted and multiple mutations.
3.2. Deficiency in Rev3l, but not in Rev1, greatly affects TLS
We then explored the potential involvement of Rev1 and polζ in the TLS. Inactivation of the Rev3l polζ catalytic subunit gene drastically reduced the TLS efficiency to 10% or less (Fig. 2A) and completely abolished A→T transversions (Fig. 2B), indicating that polζ is essential for the TLS and the induction of A→T transversion mutations. The same was true for the other sequence context of 5’AAT (Fig. S2, Table S1). In contrast, a defect in the Rev1 gene affected neither the TLS efficiency (Figs. 2A and S2) nor the coding specificity (Tables 1 and S1) in the two sequence contexts of 5’CAG and 5’AAT: the relative TLS efficiency remained high, and A→A and A→T events were prominent in Rev1−/− MEFs. The fraction of the miscoding A→T transversion did not change in the absence of Rev1 (Fig. 2B, Table S1). These results demonstrate that Rev1 is dispensable for the TLS. Accordingly, Rev1 is not epistatic to Rev3 in mouse cells, suggesting that the physical interaction of Rev1 with the Rev7 regulatory subunit of polζ is not essential for the polζ function.
In the experiments using Rev3l−/− MEFs, the number of progeny was small due to the low TLS efficiency, causing a fluctuation in coding events: a large fraction of A→C transversions in one experiment and A→Δ targeted single-base deletions in the other (Table 1 and S1).
3.3. 5’CAG is more miscoding than 5’AAT
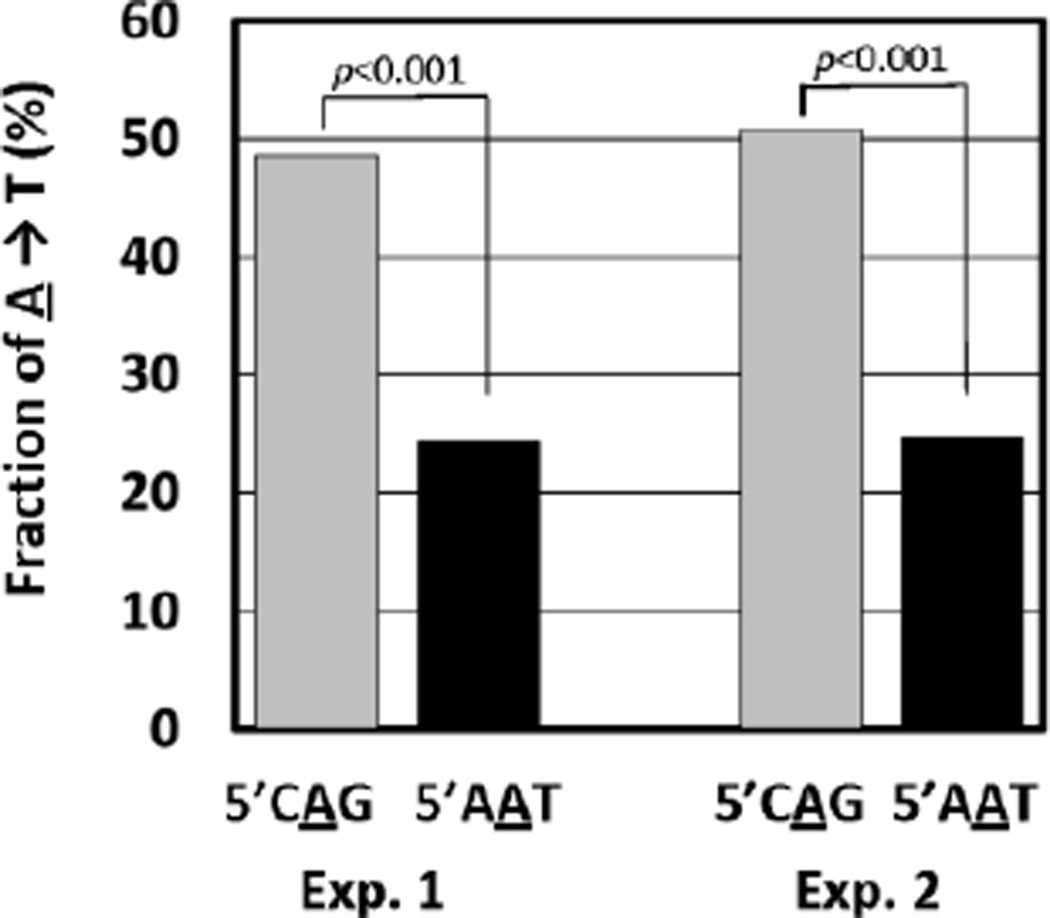
Most mutations found in tumors associated with AA exposure involve A→T transversions occurring principally on the non-transcribed strand (29, 30). Furthermore, these transversions display a strong preference for deoxyadenosine within the consensus sequence of 5’T/CAG (29, 30). In contrast, the transversions are rarely observed in 5’AAT (29). To investigate the role for TLS in the formation of mutational hotspots, we examined the coding property in the 5’CAG and 5’AAT sequences. The fractions of A→T transversion were about 50% and 25% of TLS events in 5’CAG and 5’AAT, respectively (Fig. 3). The difference is statistically significant and reproducible. Furthermore, A→T transversions observed in TKO and Rev1 knockout MEFs (Table S1) showed a similar preference for 5’CAG. These results demonstrate that TLS, at least in part, can contribute to the formation of mutational hotspots in AA-induced mutagenesis.
Fig. 3. Fraction of A→T transversion in wild-type MEFs.
Data were extracted from Supplementary Table S1.
4. Discussion
4.1 TLS taking place at a replication fork and a single-stranded gap
Recent studies have indicated that TLS is conducted at a replication fork and also a single-stranded gap (38–40). The mechanisms of the two TLS pathways may be different (40). Sale’s group indicated that Rev1 and K164-monoubiquitinated proliferating cell nuclear antigen (PCNA) act independently to facilitate polζ-dependent TLS across T-T (6–4) photoproducts at a fork and during gap filling, respectively (40, 41). Our experimental design is well suited for the study of the gap-filling mechanism. Polζ is proposed to be essential for gap-filling TLS (38), suggesting its role for extension from a synthesis-blocking abnormal pair although it may also function in the nucleotide insertion opposite a lesion (42, 43). Our results obtained using the single-stranded gap substrate with dA-AL-I and benzo[a]pyrene-dG are consistent with the idea of its essential role in gap-filling TLS.
4.2. A DNA polymerase(s) other than Y-family polymerases can efficiently insert a nucleotide across from dA-AL-I
TLS consists of two steps: insertion of a nucleotide opposite a lesion and extension from the newly formed primer terminus (1). It is likely that the Y-family polymerases, polη, polι, polκ and Rev1, play a principal role in the nucleotide insertion (1), facilitated by a large catalytic site that can accommodate an unusual base pair formed between the incoming nucleotide and an adducted template nucleotide (44, 45). Because Rev1 is a dCMP insertase, it does not insert dAMP or dTMP opposite dA-AL-I. Surprisingly, our results have revealed that none of polη, polι, and polκ is required for the TLS across bulky dA-AL-I: neither the efficiency nor the coding property of TLS was significantly affected in TKO MEFs. These results are in a full agreement with the results of our previous study employing a benzo[a]pyrene dihydrodiol epoxide-derived dG adduct (24). Thus, a question has been raised regarding nucleotide insertion opposite these bulky adducts. Candidate polymerases include repair polymerases such as polβ, polλ, polμ, polν and polθ (1), the recently identified PrimPol, which has both primase and polymerase activities (46, 47), and polζ four subunits holoenzyme (Rev3-Rev7-Pold2-Pold3), which is more versatile than polζ two subunits enzyme (Rev3-Rev7) (20, 21). Another possibility is that replicative polymerases, such as polδ and polε, insert a nucleotide, as observed with several DNA lesions in vitro (42, 48–53), before dissociating from a replication complex. In conclusion, it is surprising that a DNA polymerase other than Y-family members can efficiently insert a nucleotide opposite a template with such bulky DNA adducts.
4.3. Polζ is essential for TLS across dA-AL-I
Once a nucleotide is inserted opposite a lesion, the same polymerase may extend from an abnormal base pair formed. It is well established that polη catalyzes insertion and extension across T-T cyclobutane dimers (2, 3). However, not all abnormal pairs can be extended by the same polymerase. It has been shown that polζ is very competent for the extension role (42, 54): it efficiently catalyzes extension in vitro from the unusual primer terminus formed by an incoming nucleotide and a DNA lesion (1). Our results show that knockout of the Rev3l gene drastically reduces TLS efficiency and completely abolishes A→T transversions. Two scenarios are imaginable: one is that polζ catalyzes both insertion of dAMP or dTMP opposite dA-AL-I and subsequent extension. The other scenario is that an unidentified polymerase catalyzes the insertion and polζ performs the subsequent extension.
In contrast, the lack of Rev1 did not affect the TLS events: TLS efficiency was not significantly reduced when compared to that in wild-type MEFs, nor was the coding property. In yeast, Rev1 is required for the function of polζ, possibly for the recruitment of polζ to a stalled site (22), but our results indicate that this is not the case in mammalian cells and are consistent with the results of Yoon et al (23), who claim that Rev1 plays a critical role in the TLS function, possibly recruitment, of Y-family polymerases but not polζ. The notion that mammalian polζ may act independently of Rev1 was also proposed by others (23, 55). Our previous study (24) also revealed that the effect of a Rev1 defect was much milder than that of a polζ defect in the gap-filling TLS across a benzo[a]pyrene-dG adduct. These results suggest that an unknown mechanism exists to recruit polζ for gap-filling TLS in mammalian cells. Because polζ shares the two subunits (Pold2 and Pold3) of polδ holoenzyme (18–21), a Pold2-Pold3 heterodimer might be involved in the recruitment of polζ. Pold2 and Pold3 interact with polζ (Rev3) (18–21) and PCNA (a sliding clamp) (56–59), respectively.
4.4. Preference for the 5’CAG sequence in the induction of A→T transversions
Previous studies (25, 26, 29) have revealed that the molecular signature of mutations in AA-associated cancers involves A→T transversions predominantly located in the non-transcribed strand and a strong preference for 5’T/CAG. This trinucleotide overlaps the canonical splice acceptor site, facilitating inappropriate splicing in the messages of tumor suppressor genes such as TP53 (29). Our results show that the frequency of A→T transversions was two-fold higher in 5’CAG (~50%) than in 5’AAT (~25%). This result suggests that the fidelity of TLS can also contribute to the sequence preference for mutation induction as well as the ease of adduct formation and the resistance to nucleotide excision repair. Another study has also reported that sequence contexts contribute to the efficiency and coding property of TLS across a T-T (6–4) photoproduct (60). The mechanism by which neighboring bases influence the property of TLS remains to be determined
Supplementary Material
Highlights.
The dA adduct of aristolochic acid I (dA-AL-I) causes A to T mutations at a high frequency.
Y-family DNA polymerases are not essential for TLS across dA-AL-I.
Polζ, but not Rev1, is indispensable for TLS and A to T transversions.
TLS contributes in part to the formation of dA-AL-I-induced mutational hotspots.
Acknowledgments
We thank Drs. Haruo Ohmori (Gakushuin University, Japan) for polη/polι/polκ triple-gene knockout MEFs and Niels de Wind (Leiden University Medical Center, The Netherlands) for Rev1 and Rev3l knockout MEFs. This work was supported by the grants from National Institutes of Health (ES018833 to MM) and Henry and Marsha Laufer (to APG).
Abbreviations
- AA
Aristolochic acid
- dA-AL-I
7-(deoxyadenosine-N6-yl)-aristolactam I
- MEF
mouse embryonic fibroblast
- pol
DNA polymerase
- TKO
triple-gene knockout
- TLS
translesion DNA synthesis
- PCNA
proliferating cell nuclear antigen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 3.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 4.Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, Kanao R, Higashi Y, Kondoh H, Tatematsu M, Masutani C, Hanaoka F. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA Polymerase ι. Mol. Cell. Biol. 2006;26:7696–7706. doi: 10.1128/MCB.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA. Participation of mouse DNA polymerase ι in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petta TB, Nakajima S, Zlatanou A, Despras E, Couve-Privat S, Ishchenko A, Sarasin A, Yasui A, Kannouche P. Human DNA polymerase iota protects cells against oxidative stress. EMBO J. 2008;27:2883–2895. doi: 10.1038/emboj.2008.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo C, Kosarek-Stancel JN, Tang T-S, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell. Mol. Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singer WD, Osimiri LC, Friedberg EC. DNA Repair (Amst.) 2013;12:817–823. doi: 10.1016/j.dnarep.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 10.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Wang J, Zhang Y, Wang Z. The catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis. Nucleic Acids Res. 2010;38:5036–5046. doi: 10.1093/nar/gkq225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiltrout ME, Walker GC. The DNA polymerase activity of Saccharomyces cerevisiae Rev1 is biologically significant. Genetics. 2011;187:21–35. doi: 10.1534/genetics.110.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross AL, Simpson LJ, Sale JE. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 16.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J. Biol. Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 17.Masuda Y, Ohmae M, Masuda K, Kamiya K. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J. Biol. Chem. 2003;278:12356–12360. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J. Biol. Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y-S, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence CW. Cellular functions of DNA polymerase ζ and Rev1 protein. Adv. Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 23.Yoon J-H, Park J, Conde J, Wakamiya M, Prakash L, Prakash S. Rev1 promotes replication through UV lesions in conjunction with DNA polymerases η, ι, and κ but not DNA polymerase ζ. Genes Dev. 2015;29:2588–2602. doi: 10.1101/gad.272229.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto K, Cho Y, Yang I-Y, Akagi J-i, Ohashi E, Tateishi S, de Wind N, Hanaoka F, Ohmori H, Moriya M. The vital role of polymerase ζ and REV1 in mutagenic, but not correct, DNA synthesis across benzo[a]pyrene-dG and recruitment of polymerase ζ by REV1 to replication-stalled site. J. Biol. Chem. 2012;287:9613–9622. doi: 10.1074/jbc.M111.331728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, Jakovina K, Brdar B, Slade N, Turesky RJ, Goodenough AK, Rieger R, Vukelic M, Jelakovic B. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C-H, Dickman KG, Moriya M, Zavadil J, Sidorenko VS, Edwards KL, Gnatenko DV, Wu L, Turesky RJ, Wu X-R, Pu Y-S, Grollman AP. Aristolochic acid-associated urothelial cancer in Taiwan. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidorenko VS, Attaluri S, Zaitseva I, Iden CR, Dickman KG, Johnson F, Grollman AP. Bioactivation of the human carcinogen aristolochic acid. Carcinogenesis. 2014;35:1814–1822. doi: 10.1093/carcin/bgu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto K, Zaitseva IN, Bonala R, Attaluri S, Ozga K, Iden CR, Johnson F, Moriya M, Grollman AP, Sidorenko VS. Sulfotransferase-dependent bioactivation of aristolochic acid I and N-hydroxyaristolactam I in human cells. Carcinogenesis. 2016;37:647–655. doi: 10.1093/carcin/bgw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoang ML, Chen C-H, Sidorenko VS, He J, Dickman KG, Yun BH, Moriya M, Niknafs N, Douville C, Karchin R, Turesky RJ, Pu Y-S, Vogelstein B, Papadopoulos N, Grollman AP, Kinzler KW, Rosenquist TA. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci. Transl. Med. 2013;5:197ra102. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenquist TA, Grollman AP. Mutational signature of aristolochic acid: clue to the recognition of a global disease. DNA Repair (Amst.) 2016;44:205–211. doi: 10.1016/j.dnarep.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 31.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U, Campbell PJ. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. http://cancer.sanger.ac.uk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Verspuy J, Gali H, Haracska L, de Wind N. Mammalian polymerase ζ is essential for post-replication repair of UV-induced DNA lesions. DNA Repair (Amst.) 2009;8:1444–1451. doi: 10.1016/j.dnarep.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Ito W, Yokoi M, Sakayoshi N, Sakurai Y, Akagi J-i, Mitani H, Hanaoka F. Stalled Polη at its cognate substrate initiates an alternative translesion synthesis pathway via interaction with REV1. Genes Cells. 2012;17:98–108. doi: 10.1111/j.1365-2443.2011.01576.x. [DOI] [PubMed] [Google Scholar]
- 35.Attaluri S, Bonala RR, Yang I-Y, Lukin MA, Wen Y, Grollman AP, Moriya M, Iden CR, Johnson F. DNA adducts of aristolochic acid II: total synthesis and site-specific mutagenesis studies in mammalian cells. Nucleic Acids Res. 2010;38:339–352. doi: 10.1093/nar/gkp815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Attaluri S, Iden CR, Bonala RR, Johnson F. Total synthesis of the aristolochic acids, their major metabolites, and related compounds. Chem. Res. Toxicol. 2014;27:1236–1242. doi: 10.1021/tx500122x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 38.Quinet A, Martins DJ, Vessoni AT, Biard D, Sarasin A, Stary A, Menck CFM. Translesion synthesis mechanisms depend on the nature of DNA damage in UV-irradiated human cells. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinet A, Vessoni AT, Rocha CRR, Gottifredi V, Biard D, Sarasin A, Menck CFM, Stary A. Gap-filling and bypass at the replication fork are both active mechanisms for tolerance of low-dose ultraviolet-induced DNA damage in the human genome. DNA Repair (Amst.) 2014;14:27–38. doi: 10.1016/j.dnarep.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 41.Szuts D, Marcus AP, Himoto M, Iwai S, Sale JE. REV1 restrains DNA polymerase ζ to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res. 2008;36:6767–6780. doi: 10.1093/nar/gkn651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PMJ, Prakash S, Prakash L. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbs PEM, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Polζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase η. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biertümpfel C, Zhao Y, Kondo Y, Ramón-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase η. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, Taylor E, Stevanovic I, Green AJ, Stracker TH, Lindsay HD, Doherty AJ. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell. 2013;52:566–573. doi: 10.1016/j.molcel.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mouron S, Rodriguez-Acebes S, Martinez-Jimenez MI, Garcia-Gomez S, Chocron S, Blanco L, Mendez J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 2013;20:1383–1389. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 48.Hirota K, Yoshikiyo K, Guilbaud G, Tsurimoto T, Murai J, Tsuda M, Phillips LG, Narita T, Nishihara K, Kobayashi K, Yamada K, Nakamura J, Pommier Y, Lehmann A, Sale JE, Takeda S. The POLD3 subunit of DNA polymerase δ can promote translesion synthesis independently of DNA polymerase ζ. Nucleic Acids Res. 2015;43:1671–1683. doi: 10.1093/nar/gkv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narita T, Tsurimoto T, Yamamoto J, Nishihara K, Ogawa K, Ohashi E, Evans T, Iwai S, Takeda S, Hirota K. Human replicative DNA polymerase δ can bypass T-T (6-4) ultraviolet photoproducts on template strands. Genes to Cells. 2010;15:1228–1239. doi: 10.1111/j.1365-2443.2010.01457.x. [DOI] [PubMed] [Google Scholar]
- 50.Meng X, Zhou Y, Zhang S, Lee EYC, Frick DN, Lee MYWT. DNA damage alters DNA polymerase δ to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 2009;37:647–657. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase δ. Biochimie. 2009;91:1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan K, Resnick MA, Gordenin DA. The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst.) 2013;12:878–889. doi: 10.1016/j.dnarep.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin Y-C, Li L, Makarova AV, Burgers PM, Stone MP, Lloyd RS. Error-prone replication bypass of the primary aflatoxin B1 DNA adduct, AFB1-N7-Gua. J. Biol. Chem. 2014;289:18497–18506. doi: 10.1074/jbc.M114.561563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 55.Temviriyanukul P, van Hees-Stuivenberg S, Delbos F, Jacobs H, de Wind N, Jansen JG. Temporally distinct translesion synthesis pathways for ultraviolet light-induced photoproducts in the mammalian genome. DNA Repair (Amst.) 2012;11:550–558. doi: 10.1016/j.dnarep.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Johansson E, Garg P, Burgers PMJ. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 57.Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17927–17932. doi: 10.1073/pnas.1109981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ducoux M, Urbach S, Baldacci G, Hubscher U, Koundrioukoff S, Christensen J, Hughes P. Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21Cip1-like PCNA-binding motif present in the third subunit of human DNA polymerase δ. J. Biol. Chem. 2001;276:49258–49266. doi: 10.1074/jbc.M106990200. [DOI] [PubMed] [Google Scholar]
- 59.Pohler JRG, Otterlei M, Warbrick E. An in vivo analysis of the localisation and interactions of human p66 DNA polymerase δ subunit. BMC Mol. Biol. 2005;6:17. doi: 10.1186/1471-2199-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shriber P, Leitner-Dagan Y, Geacintov N, Paz-Elizur T, Livneh Z. DNA sequence context greatly affects the accuracy of bypass across anultraviolet light 6-4 photoproduct in mammalian cells. Mutat. Res. 2015;780:71–76. doi: 10.1016/j.mrfmmm.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.