Abstract
Retinoblastoma is a rare childhood cancer of the developing retina, and studies on this orphan disease have led to fundamental discoveries in cancer biology. Retinoblastoma has also emerged as a model for translational research for pediatric solid tumors, which is particularly important as personalized medicine expands in oncology. Research on retinoblastomas has been combined with the exploration of retinal development and retinal degeneration to advance a new model of cell type–specific disease susceptibility termed ‘cellular pliancy’. The concept can even be extended to species-specific regeneration. This review discusses the remarkable path of retinoblastoma research and how it has shaped the most current efforts in basic, translational, and clinical research in oncology and beyond.
Keywords: retinoblastoma, pediatric solid tumors, orthotopic patient-derived xenografts, cellular pliancy, retinal regeneration
Early Landmark Discoveries in Retinoblastoma Cancer Biology
Retinoblastoma is a rare childhood cancer of the developing retina, begins during fetal development and is diagnosed at birth or during early childhood [1]. The first sign is often an abnormal white reflection in the eye called leukocoria. In developed countries, most children survive retinoblastoma because it is detected before it metastasizes and the eye can be removed if the tumor is not responding to treatment. The goal of retinoblastoma treatment in developed countries is to save eyes and functional vision. In developing countries, approximately half of the children who are diagnosed with retinoblastoma still die of the disease. In those regions, the goal of therapy is to diagnose the tumor early enough to save the child’s life save eyes and vision in children with early stage disease. With only 300 cases in the United States (US) and 5–10,000 cases worldwide reported annually [2], prospective clinical trials for patients with retinoblastoma pose a challenge. Despite the rarity of retinoblastoma cases, many landmark discoveries have been made by studying this cancer because of several reasons. First, retinoblastoma exhibits little molecular or cellular heterogeneity across patients and is thus ideal for studying fundamental principles of human cancer genetics and biology[3–5]. Second, retinoblastoma is easy to detect, and long before researchers had access to sophisticated diagnostic imaging tools, they could identify patients with retinoblastoma and monitor disease progression. Third, retinoblastoma is one of the earliest diagnosed cancers, making it ideal for studying cancer genetics, because the inheritance pattern of disease susceptibility mutations can be established in early childhood [1]. Finally, there is little evidence of environmental factors associated with retinoblastoma, making it easier to identify its molecular and cellular origins.
The first human tumor suppressor gene RB1 was identified by studying the genomes of children with inherited retinoblastoma [6, 7]. These data provided genetic validation of Knudson’s two-hit hypothesis (see Glossary) for the initiation of cancer by inactivation of tumor suppressors [8]. Also, some initial attempts to model cancer in genetically engineered mouse models (GEMM) were focused on retinoblastoma, because nearly all retinoblastoma patients present biallelic inactivation of RB1 [9, 10]. Interestingly, mice with a germline mutation in Rb1 do not develop retinoblastoma, and even biallelic inactivation fails to produce retinal tumors [11–13]. Subsequent work has also demonstrated the intrinsic species-specific genetic compensation and redundancy of Rb family members p107 and p130 in preventing retinoblastoma formation [13–18].
In this review, I will present an update on what we have learned about the genomics of retinoblastoma since those early landmark discoveries. In addition, efforts to advance our understanding of retinoblastoma biology have led to the development of some of the first orthotopic patient derived xenografts (O-PDXs) and I discuss how preclinical testing using those models has improved outcomes for patients with retinoblastoma. This approach has now been extended to all pediatric solid tumors in a large-scale effort to validate ‘druggable’ mutations for personalized medicine in children with cancer. Finally, studies on the biology of retinal development, retinoblastoma and retinal degeneration have led to new insights into cell-type specific susceptibility to malignant transformation and degeneration. I outline a new model to explain why some cells are intrinsically more susceptible to malignant transformation and others are more susceptible to degeneration.
GEMMS, Chromothripsis and Orthotopic Patient-derived Xenografts
Since these early discoveries, retinoblastoma has continued to provide new insights into cancer biology. Retinoblastoma was the first pediatric solid tumor grown as an orthotopic patient-derived xenografts (O-PDXs), retaining molecular, cellular, and genetic features of a patient’s tumor [3]. As shown in Figure 1, retinoblastoma O-PDX tumors grow as a disorganized mass with intercellular regions of neuronal plexus reflecting their retinal origins. This finding was important because, contrary to established dogma in the field, studies using O-PDX showed that retinoblastomas have remarkably stable genomes [3]. Another important recent advance in cancer genetics has been the discovery of chromothripsis contributing to retinoblastoma initiation through inactivation of RB1 in humans [5]. Chromothripsis was first described by Stratton’s group as the shattering of a chromosome [19], and recently, Pellman’s group suggested that its underlying mechanism involved uncoupled DNA replication in micronuclei [20]. Inactivation of RB1 by chromothripsis in retinoblastoma was the first example of this process contributing to tumor initiation [5]. Subsequent studies demonstrated that epigenetic deregulation of genes such as SYK could contribute to tumor progression after RB1 inactivation [3]. Specifically, in human retina, the SYK gene is not normally expressed but following RB1 inactivation it is epigenetically deregulated and expressed at high levels [3]. SYK protein expression is required in retinoblastoma to prevent programmed cell death through MCL1 [3]. This discovery provided an important mechanistic explanation as to why retinoblastomas progress very quickly after RB1 inactivation. In particular, there are widespread epigenomic changes that lead to changes in cancer gene expression such as SYK that result from the loss of the RB1 protein. Unlike other types of tumors that rely on sequential accumulation of genetic lesions that alter cancer gene expression, loss of RB1 in retinoblastoma leads to rapid expression changes because the underlying mechanism is epigenetic. It also provided the impetus for many other studies to begin determining how epigenetics contribute to tumor progression in pediatric cancers, as in the case of rhabdoid tumors and diffuse intrinsic pontine gliomas. Also, retinoblastoma was the first example of a cross-species comparison of O-PDXs and GEMMs reporting fundamental species-specific differences in their epigenomes [4]. For example, contrary to human tumors, SYK was not found to be epigenetically deregulated in murine retinoblastomas [4]. These findings were instrumental in shifting away from GEMMs for preclinical testing of pediatric solid tumors. Although GEMMs are still useful for testing genetic hypotheses and elucidating fundamental biological processes, O-PDX models are better suited by providing translational relevance for new putative therapies to treat pediatric solid tumors [21].
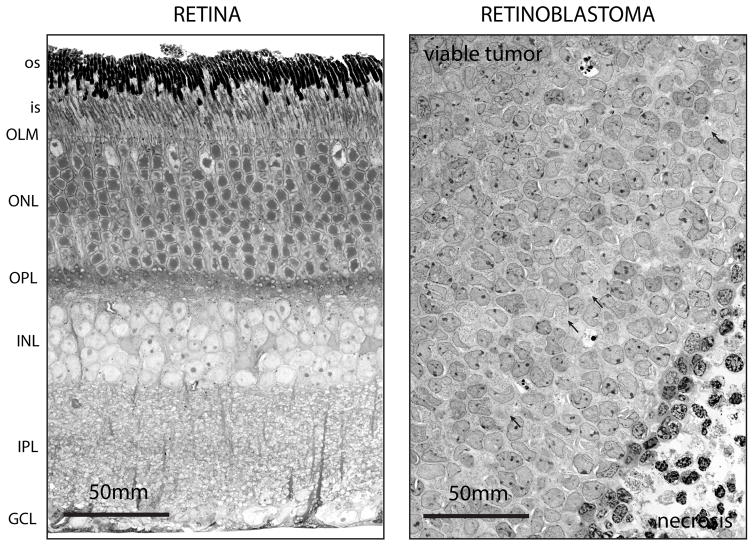
Figure 1. Disruption of Retinal Lamination in Murine Retinoblastoma.
Representative electron micrographs of normal mouse retinal tissue (left) showing all 3 cellular layers and a highly organized retinal lamination adjacent to retinoblastoma tissue (right) with disrupted lamination. The tumor is an orthotopic patient derived xenograft (O-PDX) in the eye of an immunocompromised mouse. The normal retinal lamination is disorganized in the tumors and most retinoblastomas have regions of necrosis that result from oxygen depletion. Some neuronal plexus (arrows) is retained surrounding the tumor cells reflecting partial differentiation of retinoblastoma along the neuronal lineages. Abbreviations: os, outer segments; is, inner segments; OLM, outer limiting membrane; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Scale bar, 50 mm.
Retinoblastoma: A Model for Translational Research
In the US, most prospective clinical trials on retinoblastoma are conducted by St. Jude Children’s Research Hospital (St. Jude) or by Children’s Oncology Group (COG). A typical phase II clinical trial for retinoblastoma takes 3–4 years, depending on the trial’s objectives [22–24]. Therefore, the treatment regimen that is selected to move into clinical trials must first be carefully vetted in the laboratory to increase the likelihood of improving patient outcome [25]. Figure 2 provides an example of the workflow for preclinical studies to support a clinical trial. Moreover, careful monitoring during treatment and long-term follow up is essential to identifying therapy-related late effects [26, 27]. Comprehensive preclinical testing and validation necessitate a strong investment in preclinical studies not just for retinoblastoma, but also for other childhood cancers, as well as orphan diseases in adults [28].
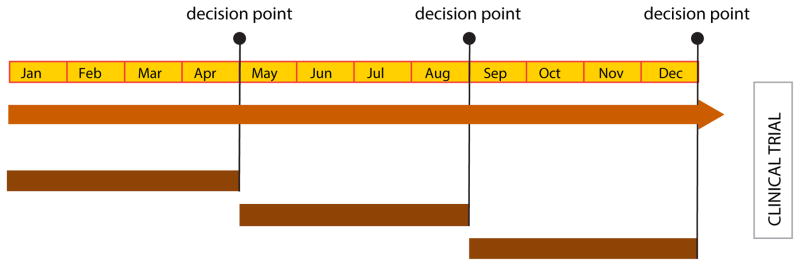
Figure 2. Workflow for Translational Studies in Support of Clinical Trials.
Diagram of the timeline and sequence for development and characterization of O-PDX models as well as drug screening efforts to identify unique tumor cell vulnerabilities that can be exploited therapeutically. Over the course of a year, it is feasible to complete the necessary preclinical studies to move a new treatment regimen into clinical trials. After careful review of those pharmacokinetic data, a decision is made about whether to move into preclinical phase I/II studies based on the penetration of the drug into the tumor. If sufficient drug penetration is not achieved at a clinically relevant dose, then the treatment regimen is not pursued further. The next key decision point follows the preclinical phase I and II studies to determine tolerability (phase I) and the pilot study to identify any indication of efficacy (phase II). If the treatment regimen is not tolerated or has no indication of efficacy, it is abandoned. Finally, after completion of the preclinical phase III (randomized, double blind, placebo controlled study) the decision is made whether to move into a clinical trial based on efficacy and tolerability.
Clearly, there is limited investment from pharmaceutical companies to develop drugs for rare childhood cancers. Instead, therapies developed for large markets are repurposed for orphan diseases, including childhood cancer. Although there are currently incentives for pharmaceutical companies to identify pediatric indications for their cancer drugs, they often lack the expertise or infrastructure to properly test preclinical efficacy. Another major limitation is the lack of validated and predictive preclinical models of pediatric cancer.
With the stakes so high for pediatric cancer in general and retinoblastoma in particular, it is critical to establish the scientific justification and translational relevance for a regimen before moving it into the clinic [28]. To establish scientific justification, investigators need to demonstrate that the therapeutic regimen targets a bona fide tumor vulnerability necessary for tumorigenesis. Investigators are often misled by recurrent pathway perturbations or biomarkers that play little or no role in tumorigenesis. Importantly, tumor vulnerabilities are not necessarily genetic; there are few ‘druggable’ mutations that could be targeted in pediatric solid tumors, and none in retinoblastoma [28]. More often, tumor vulnerabilities result from a combination of intrinsic factors and the tumor microenvironment. Therefore, if a mutation or pathway perturbation is shown through genetic and other methods to be required for tumorigenesis, it is considered scientifically justified [28].
For the few scientifically justified tumor vulnerabilities identified in retinoblastoma or other pediatric solid tumors, clinical trials should be initiated only after establishing translational relevance [28]. In vivo tumor studies in preclinical models do not necessarily prove translational relevance. Many animal studies are performed at drug doses and schedules not tolerable by patients. Translational researchers should also consider the current standard of care, randomization, and statistical design. To standardize preclinical testing for pediatric solid tumors, we developed a comprehensive preclinical testing paradigm that incorporates preclinical phase I, II, and III trials (Figure 2) and provides sufficient translational relevance for scientifically justified therapeutic regimens [28, 29]. Importantly, the efficacy of new treatment regimens is compared to the corresponding standard of care as a baseline for response. This approach has been successful for retinoblastoma and is being extended to other pediatric solid tumors [29].
Improving Outcomes for Children with Retinoblastoma
In the past years, 2 major therapeutic trials for retinoblastoma have been conducted: administration of intra-arterial (IA) melphalan by the COG (Clinicaltrials.gov identifier: NCT02097134) and combined administration of systemic topotecan with local ocular delivery of carboplatin by St. Jude (NCT01783535, NCT00186888). The scientific justification and translational relevance of IA melphalan have not been established. The only preclinical studies on IA melphalan were performed in non-human primates, which revealed significant toxicities (e.g., ophthalmic artery thrombosis, choroidal atrophy, and retinal vasculopathy)[30–32]. Owing to conflicting reports from retinoblastoma treatment centers on the safety and efficacy of IA melphalan, the COG trial was designed to document and determine the safety of administering IA melphalan.
In contrast to IA melphalan, the carboplatin–topotecan combination exploits unique vulnerabilities of retinoblastoma in the intraocular environment. First, retinoblastomas grow in a hypoxic environment (Figure 1), and, combined with their metabolic demands, present high levels of oxidative stress[3, 4, 33–35]. This can lead to DNA lesions that can slow down DNA replication. Carboplatin causes intercalation of platinum into the DNA of retinoblastoma cells, further exacerbating the DNA replication defects leading to replicative stress [36]. In addition, the p53 pathway is intact in retinoblastoma but kept in check by increased levels of MDM4, a p53 antagonist [21, 37–40]. Importantly, suppression of the p53 pathway by MDM4 can be overcome by inducing DNA breaks in retinoblastoma cells, which leads to p53-mediated cell death [37, 41]. Although ionizing radiation is very effective for retinoblastoma, damage to surrounding tissues remains a major complication [1]. Therefore, topotecan, which induces DNA breaks and overcomes MDM4 suppression of p53, has been tested in combination with carboplatin. This combination efficiently kills retinoblastoma cells and has a synergistic action when combined with carboplatin[40]. Next, a comprehensive series of preclinical studies was performed to establish the translational relevance of topotecan–carboplatin combination therapy for retinoblastoma[40]. The pharmacokinetics, dose, route, and schedule of each drug were studied to increase the likelihood of clinical success [41, 42].
As the IA mephalan and topotecan–carboplatin trials have moved forward, many predictions from preclinical testing have been validated in patients. The topotecan–carboplatin combination appears to remarkably improve ocular survival and vision preservation in children with very advanced retinoblastoma, a significant step forward in recent years (Brennan et al.). Unfortunately, ocular toxicities seen in preclinical trials of IA melphalan in non-human primates have also been reported in human patients [30–32, 43, 44]. Further, the incidence of grade 3/4 neutropenia in patients receiving IA melphalan suggests that IA administration of mephalan might not be very beneficial in reducing systemic exposure to this potent alkylating agent [45].
Accelerating Discovery in Pediatric Solid Tumors
Over the past 5 years, large-scale efforts have been launched to replicate the success of the retinoblastoma program in establishing scientific justification and translational relevance for a novel therapeutic approach to other pediatric solid tumors[46, 47]. The outcomes of children with solid tumors have not significantly ameliorated over the past 2 decades [48]. The introduction of next-generation sequencing in cancer research has substantially improved our knowledge of the genomic landscapes of diverse pediatric cancers [49]. However, a gene mutation alone cannot provide scientific justification or translational relevance for a therapeutic regimen. Moreover, very few mutations in pediatric solid tumors are considered ‘druggable’, even by the most generous criteria used in clinical genomic studies [50, 51]. To overcome this major barrier in identifying novel therapeutic combinations for pediatric solid tumors, examples from basic research were drawn to validate preclinical models of retinoblastoma and provide scientific justification and translational relevance for other pediatric solid tumors.
To engage leading basic scientists worldwide in pediatric solid tumor research and improve the quality of preclinical models, we established a protocol (NCT01050296) to develop O-PDXs from pediatric solid tumor patients at St. Jude [47]. To date, more than 250 patients have consented to the protocol, and we have established O-PDXs from more than 80 tumors, including some very rare types. The O-PDXs have been extensively characterized by whole-genome and exome sequencing, RNA-sequencing, whole-genome bisulfite sequencing, and epigenetic profiling by ChIP-sequencing for histone marks and components of the transcriptional machinery. Histologic and subcellular features have been validated by electron microscopy, and clonal analysis of O-PDX models relative to patient tumors has been performed. Novel high-throughput screening methods for these tumors as primary cultures have allowed the exploration of their drug sensitivities in relation to genomic or cellular features. O-PDX models are ideal for in vivo translational studies, because they represent diverse tumor subtypes and tend to be aggressive and invasive. All models and data are shared freely with researchers, with no obligation to collaborate through the Childhood Solid Tumor Network (http://www.stjude.org/CSTN)[47].
With these models and data in hand, how do we identify novel therapeutic combinations to move into clinical trials? This is particularly challenging given that few mutations in pediatric solid tumors directly indicate deregulated pathways that can be interfered with by molecular targeted therapy. Another concern is how efficacy in preclinical models relates to that in patients. Does a small extension in survival in preclinical models translate to an improvement in patient outcomes? Even if there is a dramatic initial response with a novel therapy, tumor cells might acquire resistance to therapy. One approach is to identify vulnerable pathways and combine drugs that target that pathway to induce synthetic lethality. Apart from retinoblastoma, the best example of synthetic lethality in pediatric solid tumors is the use of poly ADP ribose polymerase (PARP) inhibitors in combination with irinotecan and temozolomide for Ewing sarcoma [29].
Ewing sarcoma cells express high levels of SLFN11 and PARP1, which make them sensitive to PARP inhibitors in combination with irinotecan and temozolomide. These cells have defects in DNA repair and might exhibit a cellular phenotype that resembles (breast cancer) BRCA deficiency [29], even though BRCA1/2 genes are wild type in Ewing sarcoma cells [52]. BRCA deficieny leads to a defect in DNA repair which can be exploited by combining PARP inhibitors with chemotherapeutic agents that cause DNA damage such as irinotecan and temozolomide. Importantly, preclinical phase I, II, and III studies have shown that these therapeutic combinations can be tolerated at clinically relevant doses and schedules to cure disease in O-PDXs [29]. Cure is defined as the absence of any detectable tumor at the end of treatment and several months after the completion of therapy in preclinical models [29]. Currently, 2 ongoing trials (NCT02392793, NCT01858168) are testing these novel regimens for Ewing sarcoma. This is one of the many examples of how the Childhood Solid Tumor Network (CSTN) has accelerated discovery in pediatric solid tumors and established a benchmark for moving novel combinations into the clinic. However, despite these advances in our understanding of retinoblastoma biology and treatment and the extension to other pediatric solid tumors, we still understand very little about why some cell types and some tissues are more susceptible to disease than others.
Cellular Pliancy: a Unified Framework for Disease Susceptibility
The impact of retinoblastoma research extends beyond cancer biology and translational research. A long-held belief in developmental neurobiology is that neurons cannot divide to produce more neurons once they have differentiated. While studying retinae from a series of mouse strains lacking different combinations of the Rb family of proteins (Rb, p107 and p130) [11, 53–55], mature horizontal neurons were discovered to re-enter the cell cycle and clonally divide while maintaining their differentiated features such as neurites and synaptic connections[54]. If left unchecked, the proliferating horizontal neurons formed aggressive, invasive retinoblastomas [54]. In addition, the tumor cells displayed all the features of differentiated horizontal neurons including neurites, synaptic vesicles and synaptic densities [54].
Not only did this discovery challenge the dogma in neuroscience that postmitotic differentiated neurons cannot divide, but it also created a new paradigm for understanding why some cell types are more susceptible to cell death and degeneration while others are more susceptible to cell cycle re-entry and malignant transformation. Importantly, these two cellular properties (susceptibility to cell death and susceptibility to cell cycle re-entry) are inversely correlated. For example, rod photoreceptors are highly prone to undergo cell death following stress but are virtually impossible to push back into the cell cycle. In contrast, horizontal neurons can be pushed back into the cell cycle but rarely undergo cell death (Figure 3) [56]. The differential susceptibility to cell death or cell-cycle re-entry is conserved across species but most of the experimental manipulations were performed using genetically engineered mouse models. While rods and horizontal neurons appear to be two extreme examples of the interplay between susceptibility to cell cycle re-entry and cell death, by studying such examples we hope to elucidate more generally the underlying biological mechanisms of cell type specific susceptibility to degeneration or malignant transformation.
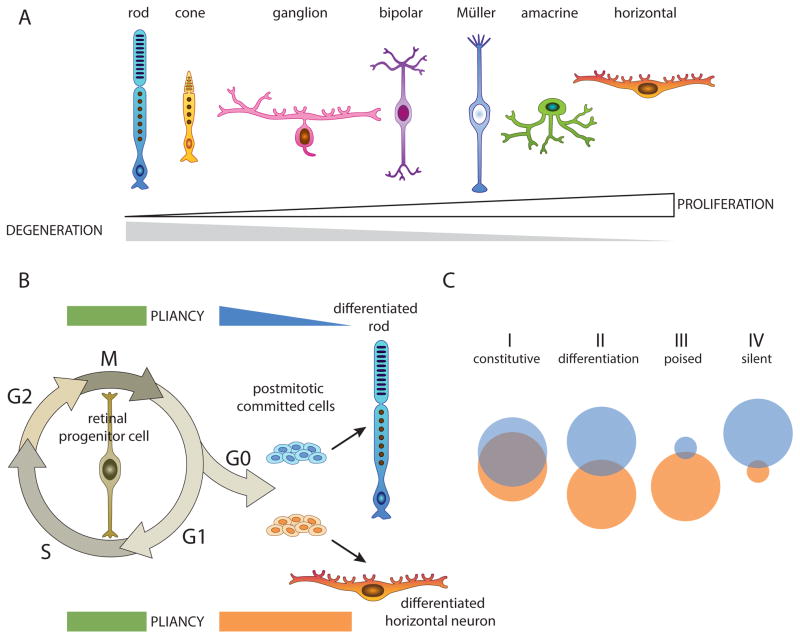
Figure 3. Cellular Pliancy Model.
A) The 7 major classes of cell types in the retina are lined up along a continuum based on their susceptibility to cell death and degeneration (gray) or proliferation and malignant transformation (white). B) Diagram of the cellular pliancy during development of rod photoreceptors and horizontal neurons: Proliferating retinal progenitor cells are highly pliant and their genomes are organized in a more open epigenetic configuration. As cells exit the cell cycle and differentiate, they activate a cell-type specific pliancy program that is cell type specific and interconnected with the differentiation program. C) Some genes are constitutively expressed (group I) in rods and horizontal cells and in an open chromatin configuration. Some genes are expressed only in rods (blue circle) or horizontal cells (orange circle) with little overlap (group II). The organization of genes that are not normally expressed into poised (group III) or epigenetically silent (group IV) domains of the genome contributes to cellular pliancy. If a stress response gene is in a poised state, it can be turned on and protect the cell from injury. If it is in a silent state, the cell will be more prone to cell death and degeneration.
The property of cells to confer susceptibility to either death or cell cycle re-entry has been termed cellular pliancy (Box 1) [56]. Cells that are more prone to degeneration, such as rods, have pliancy as they are less able to respond to extrinsic or intrinsic perturbations in homeostasis such as oxidative stress. In contrast, cells with high pliancy are more adaptable to such changes, making them more resistant to programmed cell death or necrosis, but this state may also confers susceptibility to acquiring oncogenic lesions because they can survive long enough to acquire all the hallmarks of cancer [57]. We propose that one underlying mechanism of cellular pliancy is the organization of the epigenome (Figure 3). As neurons in the retina undergo cell fate specification, they reorganize their epigenome to support retinal differentiation and cellular homeostasis [58, 59]. Clearly, the genes that are expressed in the differentiating neurons are in an active chromatin state. However, the genes that are not actively expressed are then organized into permissive or repressed epigenetic states. If a gene that is required for response to cellular stress or an oncogenic lesion is in a repressed epigenetic state, this may make a cell more prone to cell death or transformation, respectively.
Box 1. Cellular Pliancy Model.
Cellular pliancy refers to an intrinsic property of cells contributing to their specific susceptibility to malignant transformation or degeneration. One underlying mechanism of cellular pliancy is the organization of the epigenome: Some genes are organized into an accessible epigenetic state and others in an inaccessible epigenetic state in differentiated neurons. If a neuron encounters stress and a gene that is required to respond to that stress is sequestered in an inactive epigenetic state, it will be less likely to survive that stress. If a cell sustains an oncogenic mutation and a tumor suppressor that is needed to prevent malignant transformation is in an inaccessible epigenetic state, it may be more likely to become a tumor.
The simplest model to explain cellular pliancy is that as cells activate differentiation genes and turn off progenitor genes during development, they must also decide the fate of all other genes in the genome that are not directly required for differentiation or homeostasis. Some of these genes will be retained in an epigenetic state (H3K4me1,2,3;H3K36me3, H3K20Ac, H3K27Ac) and nuclear locale that make them accessible, whereas others will be sequestered in an inaccessible state (H3K27me3, H4K20me3, H3K9me3) [60]. In Figure 4, the 3 different chromatin domains (constitutive heterochromatin, facultative heterochromatin and euchromatin) of rod nuclei are visualized and measured using 3-dimensional electron microscopy (Figure 4A,B). The enrichment of particular histone modification to the 3 domains of the rod nuclei has been previously mapped [60] and individual genes can be localized within the nucleus using fluorescence in situ hybridization (Figure 4C,D). According to the pliancy model, the distribution of genes into accessible and inaccessible chromatin domains and nuclear regions are cell type-specific and ongoing studies are currently validating this model.
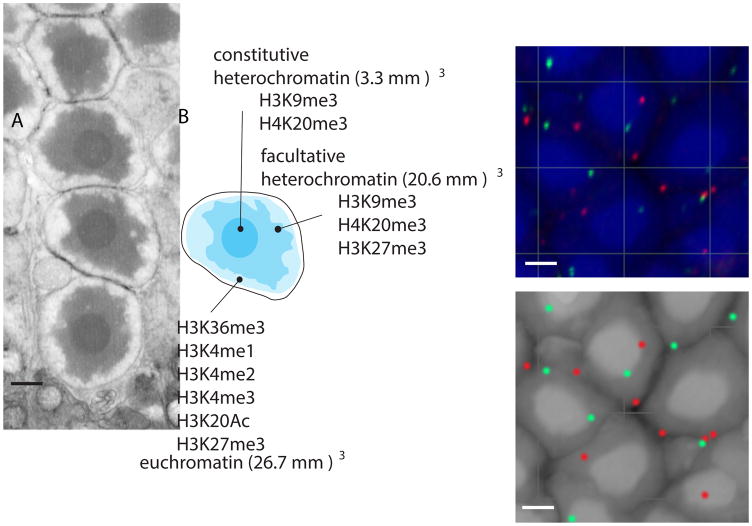
Figure 4. 3D Organization of Murine Rod Nuclei.
A) Representative electron micrograph of rod nuclei displaying the 3 stereotypical chromatin domains (constitutive heterochromatin, facultative heterochromatin and euchromatin). B) 3D electron microscopy allows a reconstruction of individual nuclei and a measurement of the volume of the central heterochromatin domain that contains the telomeres and centromeres, a more diffuse region of heterochromatin enriched in H3K27me3 histone acetylation marks, and, the euchromatin domain. Indicated in the diagram are histone modifications characteristic of these 3 chromatin domains C) Representative confocal microscopy image of DNA Fluorescence in situ hybridization (FISH) for individual genetic loci to precisely localize these within the different nuclear domains of rod cells. A BAC clone spanning the Sil1 gene is shown in red and another BAC clone spanning the Ezh2 gene is shown in green. D) When combined with ChIP-seq for histone marks, the 2D and 3D epigenetic maps can be overlaid to assess individual rods or other neuronal types. Scale bars: 1μm.
Although it is possible to describe the 2-dimensional epigenetic state of particular genes and their location within the 3-dimensional (3D) confines of the nucleus for individual neuronal classes, it is much more difficult to measure cellular pliancy. To compare cellular pliancy across neuronal classes at different stages of development, a system for cellular reprogramming was developed based on the rationale that the major barrier to cellular reprogramming involves epigenetic and organizational changes in the epigenome, which significantly impact cellular pliancy [61]. Therefore, the model is based on the premise that cells with low pliancy would be more difficult to reprogram than cells with high pliancy. For quantitative cellular reprogramming of individual neuronal classes, a series of genetically engineered mouse strains were developed which harbored a doxycycline-inducible cassette containing 4 reprogramming Yamanaka factors (Oct3/4, Myc, Klf4, and Sox2) and cell type–specific GFP reporter transgenes [61]. Individual retinal neuron types were subsequently purified by fluorescence activated cell sorting, and reprogramming was induced by adding doxycycline. Reprogramming was carried out in mosaic pellets (a mixture of wild type retinal cells and reprogrammable neurons) to ensure that neurons could survive for the 10–14 days when undergoing reprogramming, after which individual induced pluripotent stem cells (iPSCs) were cloned and characterized [61]. By performing the initial reprogramming and subsequent plating in limiting dilution assays, the efficiency of reprogramming could be quantitated. Pending further validation, data suggest that mature rods are more difficult to reprogram while immature rods are more efficiently reprogrammed. Rods also appear to be substantially more difficult to reprogram than horizontal neurons, consistent with our hypothesis of cellular pliancy; namely, that cells with low pliancy (rods) would be more difficult to reprogram than cells with high pliancy (horizontal neurons).
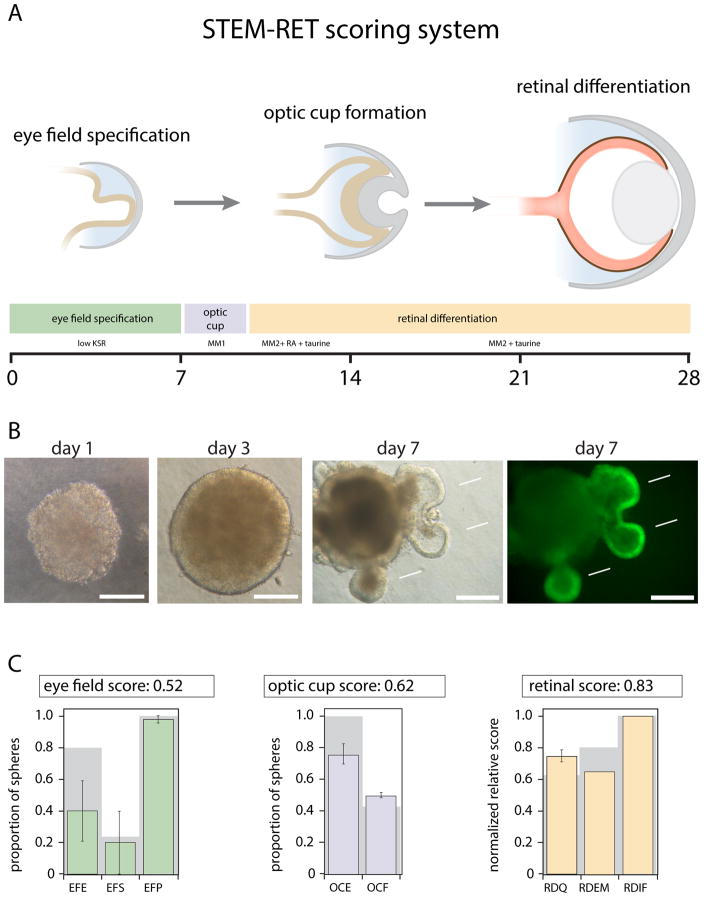
Interestingly, cells with low pliancy that were more difficult to reprogram were more likely to retain epigenetic memory as measured by their ability to produce retina [61]. As shown in Figure 5, we have developed a quantitative system called ‘Standard Transmission Electron Microscopy – Retinae’ (STEM-RET) for scoring retinal differentiation from stem cells (SC), which is critical for comparing across different stem cell lines [61]. Although pending further validation, preliminary data using the STEM-RET scoring method suggest that rod-derived iPSCs are more likely to retain epigenetic memory and produce retina more efficiently than horizontal neuron-derived iPSCs. This is encouraging because it might indicate that this type of cellular reprogramming assay can function as a reasonable measure of cellular pliancy. In addition, the underlying epigenetic factors that contribute to cellular pliancy may be elucidated by profiling the epigenome of iPSCs derived from differentiated neurons.
Figure 5. STEM-RET Scoring System of Retinal Differentiation from Stem Cells (SC).
A) Diagram of the three key stages of retinal development scored in the STEM-RET protocol (eye field specification, optic cup formation and retinal differentiation). The corresponding growth conditions (KSR: ;MM1: ;MM2: +RA: +taurine) and timing (days) are indicated below each stage for mouse iPSCs or ESCs. B) Representative micrographs of spheres from SC corresponding to days 1, 3 and 7 of STEM-RET using bright-field light microscopy. The outcroppings at day 7 (arrows) represent retinal tissue as indicated by the expression of GFP from the Rx-GFP transgenic EB5 mouse ESC line. The right panel for day 7 shows GFP expression alone. Scale bar, 200 mm. C) Representative examples of STEM-RET scoring of spheres of retinal differentiation for a single mouse SC line are shown. Eye field scoring: green; optic cup scoring: blue; and retinal differentiation: orange. Each bar represents the mean and standard deviation of integrate scores for each parameter on day 7 (eye field), day 10 (optic cup) and day 28 (retinal differentiation). Gray background bars: score for the positive control EB5 ESC line. All scores are normalized to normal mouse retina (score of 1.0). EFE, eye field efficiency; EFS, eye field specificity; EFP, eye field proliferation; OCE, optic cup efficiency; OCF, optic cup frequency; RD Q, retinal differentiation Q-PCR; RDEM, retinal differentiation electron microscopy; RDIF; retinal differentiation immunofluorescence.
Species-Specific Differences in Cellular Pliancy and Retinal Regeneration
During retinal development, retinal progenitor cells undergo unidirectional changes in their competence to produce the 7 major classes of cell types in an evolutionarily conserved birth order. Proliferation and differentiation during retinal development must be precisely coordinated to ensure that each cell type is produced at the appropriate stage and in the correct proportion. Subtle perturbations in the balance between proliferation and differentiation can lead to evolutionary change, an excellent example of which is the evolution of nocturnal vision in the owl monkey [62]. This species has a larger eye and retina adapted for low-light nighttime vision. Also, the composition of neurons is shifted to favor its nocturnal niche. The owl monkey evolved from diurnal ancestors by extension of the proliferation period during retinal development, which led to a larger retina as well as a disproportionate increase in late-born cell types that favor low-light vision. This remarkable mechanism of scaling the size of regions of the central nervous system (CNS) by changing the cell proliferation pattern across species has recently been extended to the cerebral cortex in macaques, chimpanzees and humans [63, 64]. The number and proportion of neurons in the cerebral cortex are much higher for humans than for other primates, which accounts for the increased cognitive ability in humans [65–67]. Recent studies using cortical organoids made from SC derived from humans, macaques, and chimpanzees have shown that human neural progenitor cells exhibit a longer period of neurogenic proliferation than those from other species [63, 64]. These findings are similar to those obtained for the owl monkey during retinal development [62].
Beyond the differences in coordination of proliferation and differentiation across CNS regions, there are opportunities to determine whether cellular pliancy is conserved across species. By comparing the stress response and epigenetic organization across diverse species, we can determine whether pliancy is further refined evolutionarily to accommodate differences in lifespan, neuronal physiology, and environmental stress. This is particularly important in the context of retinal regeneration. Lower-vertebrate species such as fish can regenerate retinal neurons after injury [68–71]. Radial glia of the retina, called Müller glia, can re-enter the cell cycle to produce retinal progenitor cells, which then undergo neurogenesis to produce retinal neurons [72]. Müller glia are also present in mammalian retinae and can re-enter the cell cycle after injury, but they undergo reactive gliosis rather than neurogenesis [73]. It is reasonable to suppose that species-specific differences in the cellular pliancy of Müller glia are responsible for the lack of retinal regeneration in mammalian retinae. It is thus essential to determine whether genes and cellular pathways that support retinal regeneration in fish are epigenetically silent in mammalian Müller glia, or whether there are other fundamental differences between species. This will be informative in that it may provide researchers a better molecular assessment of the regenerative potential of mammalian retinae. Advances in our understanding of the interplay between proliferation and differentiation processes in normal retinal progenitor and retinoblastoma cells, in conjunction with cellular pliancy, may ultimately allow us to promote retinal regeneration without causing deregulated proliferation and tumorigenesis.
Concluding Remarks
It is remarkable that a rare childhood cancer of the retina has—and continues to—provide numerous fundamental insights into diverse biological processes. Retinoblastoma has emerged as the model for translational research for different types of pediatric solid tumors. Retinoblastoma was a model for cancer genetics and genomics and one of the first cancers in which the role of epigenetics in disease progression was elucidated. Studies on retinoblastoma have provided a new paradigm for tackling a fundamental question, namely, why some cells are more susceptible to malignant transformation than others. We propose that cellular pliancy and corresponding epigenetic states and nuclear organization contribute to cell type–specific susceptibility in cancer. We extend this concept of cellular pliancy to cell type–specific degeneration and even species-specific regeneration. Much work remains to be done to understand the relationship between cell type–specific epigenetic profiles at individual genes, their location within the nucleus, and their ability to be acutely activated in response to stress or injury (see Outstanding Questions and Box 2). By using the retina as a model of tumorigenesis and degeneration, we hope to continue to advance the fundamental knowledge of biological processes with implications beyond the eye.
Outstanding Questions.
What is the relationship between the 2-dimensional and 3-dimensional organization of the epigenome in the nucleus to understand how cells are poised to respond to stress or oncogenic mutations?
How quickly can epigenetically inaccessible genes be turned on in response to oncogenic mutations or other types of stress, and how does this relate to susceptibility of particular retinal cell types to undergo malignant transformation?
How does the 2D and 3D organization of the epigenome contribute to epigenetic memory in stem cells and can this be used to predict which cell type will represent the best source to produce iPSCs for retinal development and retinoblastoma formation?
Can retinoblastoma tumor initiation be modeled using iPSCs derived from retinoblastoma patients that harbor germline RB1 mutations?
Box 2. The Clinician’s Corner.
The precise regulation of cell division during retinal development is essential to producing differentiated cell types (Müller glia, rods, cones, horizontal, bipolar, amacrine and ganglion cells) in the retina in the correct place and at the correct time.
Disruption in proliferation during retinogenesis can lead to defects in visual function, retinal degeneration or retinoblastoma.
As retinal progenitor cells stop dividing and form differentiated neurons, they must condense certain regions of their genome that are no longer needed and retain other regions in a more accessible state for gene transcription.
The cell type-specific distribution of genes into open (euchromatin) or condensed (heterochromatin) regions of the genome has been termed cellular pliancy and may have a profound impact on susceptibility to diseases such as cancer and retinal degeneration.
In addition, stem cells produced from differentiated cells may be more efficient at producing a particular cell type when retaining some of those open and condensed genomic regions.
In the future, it may be possible to exploit these features and customize the production of SC for cell-based therapies to more effectively treat diseases such as retinoblastoma and degenerative disorders, among others.
Trends Box.
Basic and translational research on retinoblastomas has led to improved outcomes in patients. However, there have not been significant improvements in outcomes for other pediatric solid tumor patients in the past 2 decades. The approach used to advance cures in retinoblastoma are now being applied more broadly across childhood solid tumors.
Preclinical studies have predicted toxicity of intra-arterial chemotherapy for retinoblastoma and this supports the value of using preclinical models to anticipate therapeutic toxicity and exclude ineffective therapies from clinical development.
Cellular pliancy is a new model of cell type-specific disease susceptibility and begins to explain why children with RB1 mutations develop retinoblastoma but not other tumors of the nervous system.
STEM-RET is a new method for scoring retinal differentiation from human and mouse stem cells. This can be used to directly measure the retinal epigenetic memory of iPSCs derived from different cell types. The cells with low pliancy are more likely to retain epigenetic memory of their cellular origins and this in turn relates to retinoblastoma susceptibility.
Acknowledgments
We thank Vani Shanker for editing the manuscript. This work was supported, in part, by Cancer Center Support (CA21765) from the NCI, grants to M.A.D from the NIH (EY014867 and EY018599 and CA168875), and the American Lebanese Syrian Associated Charities (ALSAC). M.A.D. was also supported by a grant from Alex’s Lemonade Stand Foundation for Childhood Cancer and Howard Hughes Medical Institute.
GLOSSARY
- Knudson’s two-hit hypothesis
Alfred Knudson was the first to propose that cancer can arise by inactivation of two copies of a tumor suppressor gene (biallelic inactivation).
- Orthotopic Patient Derived Xenograft
To establish new preclinical models of retinoblastoma and other pediatric solid tumors, cells are collected after surgery and orthtopically implanted in immunocompromised mice. For retinoblastoma, the tumor cells are injected into the vitreous of the eye. The tumors are then passaged from mouse to mouse and can be cryopreserved for long term storage.
- Chromothripsis
type of genetic lesion found in cancer and thought to involve local fragmentation of the genome and random reassembly. In some tumor types, it is a hallmark of genomic instability and asynchronous DNA replication but in retinoblastoma, chromothripsis can lead to RB1 inactivation and contribute to tumor initiation.
- Malignant Rhabdoid Tumor
aggressive cancer occurring primarily in children in diverse organ sites ranging from kidney to brain. The gene that encodes a component of the SWI/SNF complex called SMARCB1 undergoes biallelic inactivation in many rhabdoid tumors leading to changes in the epigenome, contributing to tumor progression.
- Diffuse Intrinsic Pontine Glioma
aggressive childhood glioma occurring in the brain stem and infiltrating the surrounding region. It is impossible to remove surgically and the 2-year survival rate is less than 10%.
- Grade 3/4 Neutropenia
abnormally low count of neurtrophils in the blood.
- Synthetic Lethality
related to the identification and perturbation of essential (and multiple) cellular pathways leading to a dramatic increase in cell death. In pediatric oncology, a good example is the treatment combination of PARP inhibitors with irinotecan and temozolomide.
- Irinotecan
prodrug hydrolyzed to an active chemotherapeutic agent (SN-38) in the body to inhibit topoisomerase I, and consequently, DNA replication and transcription by RNA polymerase. Treatment with irinotecan can lead to DNA breaks and kill cancer cells.
- Temozolomide
chemotherapeutic agent alkylating DNA, leading to DNA breaks and cancer cell death.
- Ewing sarcoma
cancer of the bone and soft tissue, most commonly in children, adolescents, and young adults. It usually occurs in the pelvis, long bones of the arms and legs and the ribs, and is more prevalent in males than females. The tumors initiate with a translocation creating a fusion oncoprotein between the EWS and FLI1 genes.
- Mature Horizontal Neurons
type of interneuron that forms synaptic connections with photoreceptors and bipolar neurons in the retina. Their processes extend laterally in the retina and they function by integrating light signals across large areas of the retina.
- Rod Photoreceptors
The light sensing neuron in the retina. Rods are used for low light night time vision. Rod photoreceptors are prone to die following injury or stress leading to retinal degeneration and blindness.
- Constitutive Heterochromatin (CH)
Regions of the genome composed primarily of high copy number tandem repeats that are highly condensed in the nucleus. CH has few genes and those that are present must overcome regional epigenetic silencing.
- Facultative Heterochromatin (FH)
Regions of the genome that are epigenetically silenced but do not comprise repeat sequences. Under certain circumstances, genes in FH can be derepressed and activated.
- Euchromatin
The least condensed form of chromatin in the nucleus comprising gene rich regions of the genome. Expressed genes are found, but it can also contain transcriptionally inactive genes.
- Yamanaka Factors
Four transcription factors, OCT3/4, MYC, KLF4 and SOX2, identified by Shinya Yamanaka as being sufficient to reprogram differentiated cells into pluripotent stem cells.
- iPSCs
Induced pluripotent stem cells are multipotent cells derived from differentiated cells by ectopic expression of OCT3/4, MYC, KLF4 and SOX2. These cells can produce all 3 germ layers (endoderm, mesoderm, ectoderm) and are immortal.
Footnotes
Resources
Clinicaltrials.gov identifiers: NCT02097134; NCT01783535; NCT00186888 -Retinoblastoma
Clinicaltrials.gov identifiers: NCT02392793, NCT01858168 – Ewing’s sarcoma
Clinicaltrials.gov identifiers: NCT01050296 – Orthotopic Patient Derived Xenografts
Childhood Solid Tumor Network (CSTN): http://www.stjude.org/CSTN
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rodriguez-Galindo C, et al. Retinoblastoma. In: Orkin S, editor. Hematology and Oncology of Infancy and Childhood. Elsevier; 2015. pp. 1747–1778. [Google Scholar]
- 2.Rodriguez-Galindo C, et al. Retinoblastoma: one world, one vision. Pediatrics. 2008;122:e763–770. doi: 10.1542/peds.2008-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benavente CA, et al. Cross-species genomic and epigenomic landscape of retinoblastoma. Oncotarget. 2013;4:844–859. doi: 10.18632/oncotarget.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McEvoy J, et al. RB1 gene inactivation by chromothripsis in human retinoblastoma. Oncotarget. 2014;5:438–450. doi: 10.18632/oncotarget.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dryja TP, et al. Genetic sequences that predispose to retinoblastoma and osteosarcoma. Symp Fundam Cancer Res. 1986;39:115–119. [PubMed] [Google Scholar]
- 7.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 8.Knudson A. Mutation and Cancer:statistical study of retinoblastoma. PNAS. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacks T, et al. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 10.Clarke AR, et al. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet. 2004;36:351–360. doi: 10.1038/ng1318. [DOI] [PubMed] [Google Scholar]
- 12.Donovan SL, Dyer MA. Developmental defects in Rb-deficient retinae. Vision Res. 2004;44:3323–3333. doi: 10.1016/j.visres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 13.MacPherson D, et al. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18:1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robanus-Maandag E, et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998;12:1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, et al. The First Knockout Mouse Model of Retinoblastoma. Cell Cycle. 2004;3:952–959. [PubMed] [Google Scholar]
- 16.Macpherson D, Dyer MA. Retinoblastoma: from the two-hit hypothesis to targeted chemotherapy. Cancer Res. 2007;67:7547–7550. doi: 10.1158/0008-5472.CAN-07-0276. [DOI] [PubMed] [Google Scholar]
- 17.MacPherson D, et al. Murine bilateral retinoblastoma exhibits rapid onset, metastasic progression and N-myc amplification. Embo J. 2007;26:784–794. doi: 10.1038/sj.emboj.7601515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, et al. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5:539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 19.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brennan RC, et al. Targeting the p53 pathway in retinoblastoma with subconjunctival Nutlin-3a. Cancer Res. 2011;71:4205–4213. doi: 10.1158/0008-5472.CAN-11-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen S, et al. Comparison of two methods for carboplatin dosing in children with retinoblastoma. Pediatr Blood Cancer. 2010;55:47–54. doi: 10.1002/pbc.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qaddoumi I, et al. Topotecan and vincristine combination is effective against advanced bilateral intraocular retinoblastoma and has manageable toxicity. Cancer. 2012;118:5663–5670. doi: 10.1002/cncr.27563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Galindo C, et al. Treatment of intraocular retinoblastoma with vincristine and carboplatin. J Clin Oncol. 2003;21:2019–2025. doi: 10.1200/JCO.2003.09.103. [DOI] [PubMed] [Google Scholar]
- 25.Dyer MA, et al. Use of preclinical models to improve treatment of retinoblastoma. PLoS Med. 2005;2:e332. doi: 10.1371/journal.pmed.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhagat SP, et al. Monitoring carboplatin ototoxicity with distortion-product otoacoustic emissions in children with retinoblastoma. Int J Pediatr Otorhinolaryngol. 2010;74:1156–1163. doi: 10.1016/j.ijporl.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qaddoumi I, et al. Carboplatin-associated ototoxicity in children with retinoblastoma. J Clin Oncol. 2012;30:1034–1041. doi: 10.1200/JCO.2011.36.9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langenau DM, et al. Preclinical Models Provide Scientific Justification and Translational Relevance for Moving Novel Therapeutics into Clinical Trials for Pediatric Cancer. Cancer Res. 2015;75:5176–5186. doi: 10.1158/0008-5472.CAN-15-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart E, et al. Targeting the DNA repair pathway in Ewing sarcoma. Cell Rep. 2014;9:829–841. doi: 10.1016/j.celrep.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse BC, et al. Enophthalmos and choroidal atrophy after intraophthalmic artery chemotherapy for retinoblastoma. Ophthalmology. 2015;122:435–437. doi: 10.1016/j.ophtha.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 31.Tse BC, et al. Superselective intraophthalmic artery chemotherapy in a nonhuman primate model: histopathologic findings. JAMA Ophthalmol. 2013;131:903–911. doi: 10.1001/jamaophthalmol.2013.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ditta LC, et al. Validating a nonhuman primate model of super-selective intraophthalmic artery chemotherapy: comparing ophthalmic artery diameters. Invest Ophthalmol Vis Sci. 2012;53:7791–7794. doi: 10.1167/iovs.12-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benavente CA, Dyer MA. Genetically engineered mouse and orthotopic human tumor xenograft models of retinoblastoma. Methods Mol Biol. 2015;1267:307–317. doi: 10.1007/978-1-4939-2297-0_15. [DOI] [PubMed] [Google Scholar]
- 34.Benavente CA, Dyer MA. Genetics and epigenetics of human retinoblastoma. Annu Rev Pathol. 2015;10:547–562. doi: 10.1146/annurev-pathol-012414-040259. [DOI] [PubMed] [Google Scholar]
- 35.Benavente CA, et al. Chromatin remodelers HELLS and UHRF1 mediate the epigenetic deregulation of genes that drive retinoblastoma tumor progression. Oncotarget. 2014;5:9594–9608. doi: 10.18632/oncotarget.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobbelstein M, Sorensen CS. Exploiting replicative stress to treat cancer. Nat Rev Drug Discov. 2015;14:405–423. doi: 10.1038/nrd4553. [DOI] [PubMed] [Google Scholar]
- 37.Laurie NA, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 38.Laurie NA, et al. Targeting MDM2 and MDMX in retinoblastoma. Curr Cancer Drug Targets. 2007;7:689–695. doi: 10.2174/156800907782418266. [DOI] [PubMed] [Google Scholar]
- 39.Marine JC, et al. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 40.Nemeth KM, et al. Subconjunctival carboplatin and systemic topotecan treatment in preclinical models of retinoblastoma. Cancer. 2011;117:421–434. doi: 10.1002/cncr.25574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurie NA, et al. Topotecan combination chemotherapy in two new rodent models of retinoblastoma. Clin Cancer Res. 2005;11:7569–7578. doi: 10.1158/1078-0432.CCR-05-0849. [DOI] [PubMed] [Google Scholar]
- 42.Brennan RC, et al. Targeting the p53 pathway in retinoblastoma with subconjunctival Nutlin-3a. Cancer research. 2011;71:4205–4213. doi: 10.1158/0008-5472.CAN-11-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tse DT, et al. Author reply: To PMID 23582989. Ophthalmology. 2014;121:e8–e10. doi: 10.1016/j.ophtha.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Steinle JJ, et al. Intra-ophthalmic artery chemotherapy triggers vascular toxicity through endothelial cell inflammation and leukostasis. Invest Ophthalmol Vis Sci. 2012;53:2439–2445. doi: 10.1167/iovs.12-9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunkel IJ, et al. Risk factors for severe neutropenia following intra-arterial chemotherapy for intra-ocular retinoblastoma. PLoS One. 2014;9:e108692. doi: 10.1371/journal.pone.0108692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langenau DM, et al. Preclinical Models Provide Scientific Justification and Translational Relevance for Moving Novel Therapeutics into Clinical Trials for Pediatric Cancer. Cancer Research. 2015 doi: 10.1158/0008-5472.CAN-15-1308. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart E, et al. The childhood solid tumor network: A new resource for the developmental biology and oncology research Communities. Dev Biol. 2015 doi: 10.1016/j.ydbio.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith MA, et al. Declining childhood and adolescent cancer mortality. Cancer. 2014;120:2497–2506. doi: 10.1002/cncr.28748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Downing JR, et al. The Pediatric Cancer Genome Project. Nature genetics. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris MH, et al. Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2015.5689. [DOI] [PubMed] [Google Scholar]
- 51.Janeway K. iCAT for Recurrent/Refractory/HR Solid Tumors. (2013) 2013 https://http://www.clinicaltrial.gov/ct2/show/NCT01853345?term=iCAT&rank=5.
- 52.Tirode F, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson DA, et al. Neuronal differentiation and synaptogenesis in retinoblastoma. Cancer Res. 2007;67:2701–2711. doi: 10.1158/0008-5472.CAN-06-3754. [DOI] [PubMed] [Google Scholar]
- 54.Ajioka I, et al. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell. 2007;131:378–390. doi: 10.1016/j.cell.2007.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donovan SL, et al. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006;4:14. doi: 10.1186/1741-7007-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, et al. Pediatric solid tumor genomics and developmental pliancy. Oncogene. 2014 doi: 10.1038/onc.2014.474. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 58.Mo A, et al. Epigenomic landscapes of retinal rods and cones. Elife. 2016:5. doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solovei I, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 60.Solovei I, Joffe B. Inverted nuclear architecture and its development during differentiation of mouse rod photoreceptor cells: a new model to study nuclear architecture. Genetika. 2010;46:1159–1163. [PubMed] [Google Scholar]
- 61.Hiler D, et al. Quantification of Retinogenesis in 3D Cultures Reveals Epigenetic Memory and Higher Efficiency in iPSCs Derived from Rod Photoreceptors. Cell Stem Cell. 2015;17:101–115. doi: 10.1016/j.stem.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dyer MA, et al. Developmental sources of conservation and variation in the evolution of the primate eye. Proc Natl Acad Sci U S A. 2009;106:8963–8968. doi: 10.1073/pnas.0901484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dyer MA. Stem Cells Expand Insights into Human Brain Evolution. Cell Stem Cell. 2016;18:425–426. doi: 10.1016/j.stem.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 64.Otani T, et al. 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell. 2016;18:467–480. doi: 10.1016/j.stem.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cahalane DJ, et al. Modeling local and cross-species neuron number variations in the cerebral cortex as arising from a common mechanism. Proc Natl Acad Sci U S A. 2014;111:17642–17647. doi: 10.1073/pnas.1409271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clancy B, et al. Translating developmental time across mammalian species. Neuroscience. 2001;105:7–17. doi: 10.1016/s0306-4522(01)00171-3. [DOI] [PubMed] [Google Scholar]
- 67.Clancy B, et al. Web-based method for translating neurodevelopment from laboratory species to humans. Neuroinformatics. 2007;5:79–94. doi: 10.1385/ni:5:1:79. [DOI] [PubMed] [Google Scholar]
- 68.Nagashima M, et al. A self-renewing division of zebrafish Muller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–4521. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qin Z, Raymond PA. Microarray-based gene profiling analysis of Muller glia-derived retinal stem cells in light-damaged retinas from adult zebrafish. Methods Mol Biol. 2012;884:255–261. doi: 10.1007/978-1-61779-848-1_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernardos RL, et al. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bernardos RL, et al. Notch-Delta signaling is required for spatial patterning and Muller glia differentiation in the zebrafish retina. Dev Biol. 2005;278:381–395. doi: 10.1016/j.ydbio.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 72.Goldman D. Muller glial cell reprogramming and retina regeneration. Nat Rev Neurosci. 2014;15:431–442. doi: 10.1038/nrn3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dyer MA, Cepko CL. Control of Muller glial cell proliferation and activation following retinal injury. Nat Neurosci. 2000;3:873–880. doi: 10.1038/78774. [DOI] [PubMed] [Google Scholar]