Abstract
Plants are unrivaled in the natural world in both the number and complexity of secondary metabolites they produce, and the ubiquitous phenylpropanoids and the lineage-specific glucosinolates represent two such large and chemically diverse groups. Advances in genome-enabled biochemistry and metabolomic technologies have greatly increased the understanding of their metabolic networks in diverse plant species. There also has been some progress in elucidating the gene regulatory networks that are key to their synthesis, accumulation and function. This review highlights what is currently known about the gene regulatory networks and the stable sub-networks of transcription factors at their cores that regulate the production of these plant secondary metabolites and the differentiation of specialized cell types that are equally important to their defensive function. Remarkably, some of these core components are evolutionarily conserved between secondary metabolism and specialized cell development and across distantly related plant species. These findings suggest that the more ancient gene regulatory networks for the differentiation of fundamental cell types may have been recruited and remodeled for the generation of the vast majority of plant secondary metabolites and their specialized tissues.
Graphical Abstract
This review highlights the recent developments in the investigation of gene regulatory networks controlling the production of plant secondary metabolites and the differentiation of specialized cell types that are important to their defensive function. Also discussed are their possible origins in the more ancient gene regulatory networks controlling the differentiation of fundamental cell types.
1. Introduction
Plant secondary metabolites (also called phytochemicals or natural products) constitute a large reservoir of natural chemical diversity that is constantly generated through genetic adaptations to the prevailing and ever-fluctuating abiotic and biotic environment. As a consequence, plant secondary metabolites have become essential to the survival and reproductive fitness of a plant species within its natural environment. Often derived from intermediates or end products of primary metabolic pathways, these secondary metabolites possess a variety of bioactivities, including cytotoxicity, and are biochemically expensive to synthesize. For these reasons, the formation of many plant secondary metabolites is regulated by external stresses (Patra 2013). The constant evolutionary pressure towards chemical innovation has also caused the majority of secondary metabolites to be restricted to narrow phylogenetic lineages, such as a family, genus or single species. Interestingly, despite this strong ecological and phylogenetic character of plant secondary metabolism, not all lineage-specific metabolites are distributed consistently across ancestral and sister taxa. While structural diversity is the rule for secondary metabolites, even in plants from related taxa, the metabolic responses of flowering plants to certain environmental stresses are for the most part functionally conserved. For example, the biosynthesis and accumulation of antimicrobial compounds is one of the evolutionary conserved plant responses to pathogenic microbes (Hammerschmidt 1999; Mansfield 2000; Dixon 2001). Thus, the regulatory principles behind plant secondary metabolism appear to be conserved and may have evolved earlier than many of the specific biosynthetic pathways (Wink 2003).
Plant secondary metabolites are often produced at high levels in specific tissues or cell types of individual species or across related species. Economically important examples of tissue-specific metabolites include alkaloids (e.g., morphine and codeine) and latex in laticifer cells in poppy and rubber trees, terpenes and saponins in epidermal cells of many plant families, and resins in trees (DellaPenna and Last, 2008). In addition, plant secondary metabolites have been shown to control physiological processes, such as the circadian clock and phototropism (Hasegawa et al., 2000; Yamada et al., 2003; Kerwin et al., 2011). Plant secondary metabolites, both constitutively and inducibly formed, may also be interlinked with plant immunity, controlling evolutionarily conserved immune responses, such as callose deposition and programmed cell death, in higher plants (Quidde et al., 1998; Bouarab et al., 2002; Clay et al., 2009; Ahmad et al., 2011). For example, lineage-specific defense-related alkaloids in the California poppy (Eschscholzia californica) and rose periwinkle (Catharanthus roseus) regulate their own production in a species-specific manner by using a negative feedback mechanism that blocks the generation of immune signals that initiate the induction of biosynthetic enzymes (Heinze et al., 2015); poppy-specific alkaloids do not regulate the production of periwinkle-specific alkaloids, and vice versa.
Recently, WD40 domain proteins and activator- and repressor-type transcription factors of the R2R3-type MYELOBLASTOSIS (MYB) (Figure 1), R3-type MYB, and basic helix-loop-helix (bHLH) families have been identified to regulate plant secondary metabolism and associated specialized cell development across diverse plant species. These stable regulatory components also control differentiation of fundamental plant tissues, such as single-celled trichomes, non-hair root epidermal cells, and stomata, which suggests that they form an ancient transcriptional component of gene regulatory networks in land plants (Oppenheimer 1991; Galway 1994; Lee 1999; Ramsay and Glover, 2005; Serna and Martin, 2006; Kliebenstein, 2013). Highlighted here are the biosynthetic pathways of phenylpropanoids and glucosinolates, their associated specialized cell types, and the transcriptional mechanisms that regulate the production of both.
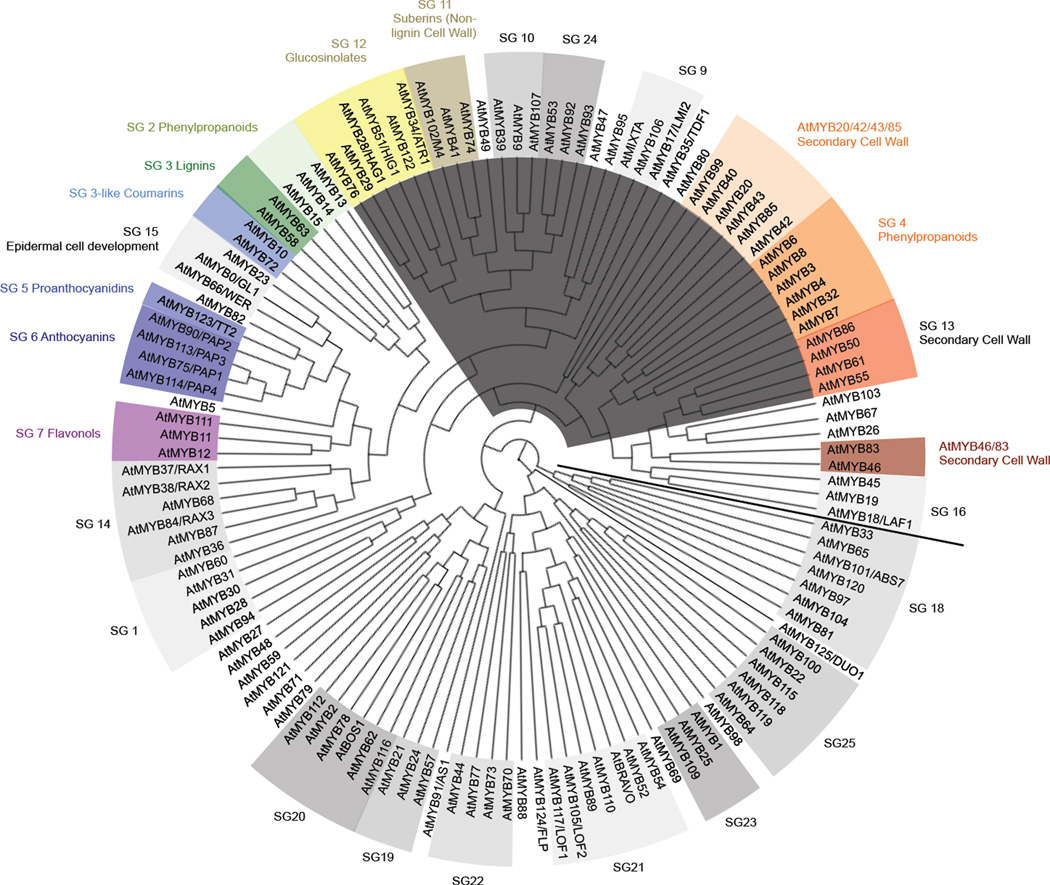
Figure 1. Putative evolutionary relationships among Arabidopsis R2R3-type MYB proteins in secondary metabolism and associated cell development.
Phylogenetic maximum likelihood tree of the complete Arabidopsis family of R2R3-type MYB proteins was generated using MUSCLE multiple sequence alignment, PhyML phylogeny, and TreeDyn tree viewer programs (http://phylogeny.lirmm.fr; Dereeper et al., 2008). Highlighted in color are MYB subgroups that regulate secondary metabolism. Highlighted in dark gray are MYB subgroups that contain the GIDPxxH motif of unknown function after the end of the MYB DNA-binding domain.
2. Secondary Metabolic Pathways
2.1. General phenylpropanoid pathway
Phenylpropanoids are a class of phenolic secondary metabolites that are mainly derived from the aromatic amino acid L-phenylalanine (1), and are ubiquitous in terrestrial plants and present in other eukaryotes. Lignins and the related lignans, flavonoids (e.g., flavonols, anthocyanins and proanthocyanidins), stilbenoids, coumarins, and hydroxycinnamic acid conjugates constitute the bulk of the known plant phenylpropanoids, and are synthesized by both general and specific pathways in phenylpropanoid metabolism. Phenylpropanoid metabolism often begins with three reactions catalyzed by phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H) and p-coumarate:coenzyme A ligase (4CL) enzymes, leading to the synthesis of the activated hydroxycinnamic acid p-coumaroyl-CoA (2) (Figures 2 and 3). This reaction sequence is known as the general phenylpropanoid pathway, and the remaining reactions of phenylpropanoid metabolism comprise the lignin, flavonoid, and many species- and clade-specific pathways (Vogt 2010).
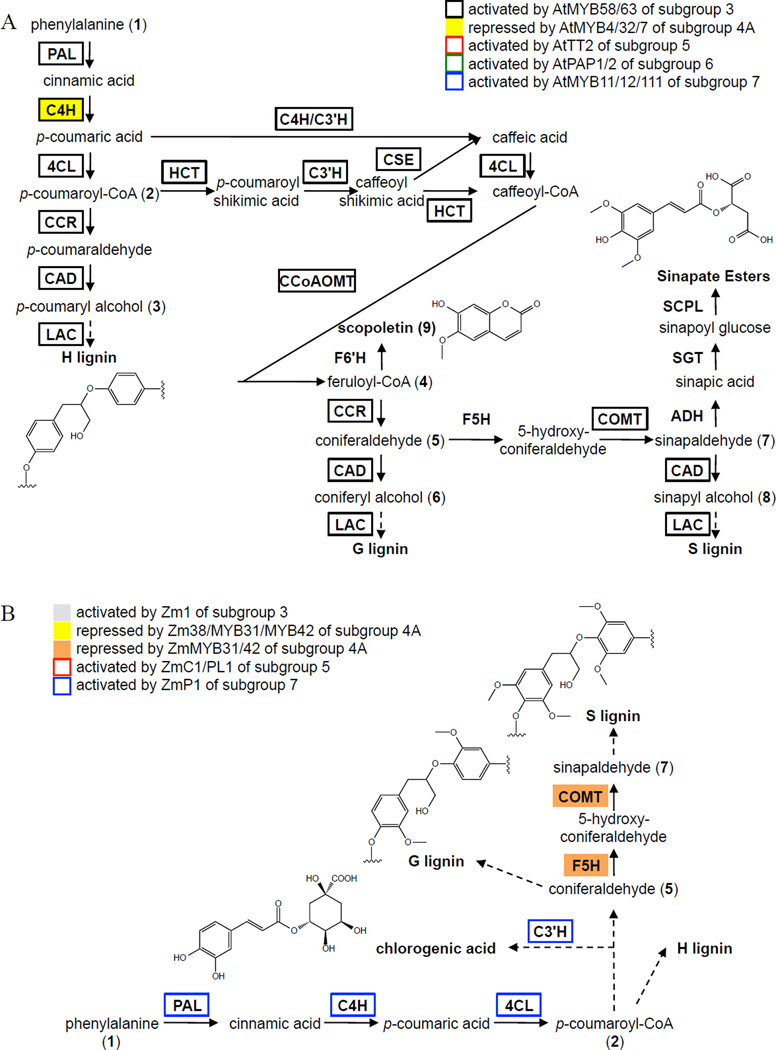
Figure 2. Multiple R2R3-type MYB Subgroups Regulate the Lignin Biosynthetic Pathways in Arabidopsis and Maize.
Unbroken arrows indicate single enzymatic conversions, and broken arrows indicate multiple enzymatic conversions. (A) Arabidopsis lignin pathways. Adapted from Vanholme et al. (2010, 2013). (B) Maize lignin pathways. Feruloyl-CoA (4) is at the intersection of the G/S lignin and coumarin biosynthetic pathways. Sinapaldehyde (16) is at the intersection of the sinapate ester and S lignin biosynthetic pathways. Monolignols, p-coumaryl (3), coniferyl (6), and sinapyl (8) alcohols, polymerize to form H, G and S lignin, respectively. ADH, alcohol dehydrogenase; C3’H, p-coumaroyl ester 3’-hydroxylase; C4H; cinnamate 4-hydroxylase; CAD, cinnamoyl-alcohol dehydrogenase; 4CL, 4-coumarate:CoA ligase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; COMT, caffeic acid O-methyltransferase; CSE, caffeoyl shikimate esterase; F5H, ferulate 5-hydroxylase; F6’H, feruloyl-CoA 6’-hydroxylase; HCT, hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyltransferase; LAC, laccase; PAL, phenylalanine ammonia-lyase; SCPL, serine carboxypeptidase-like; SGT, sinapate UDP-glucose sinapoyltransferase.
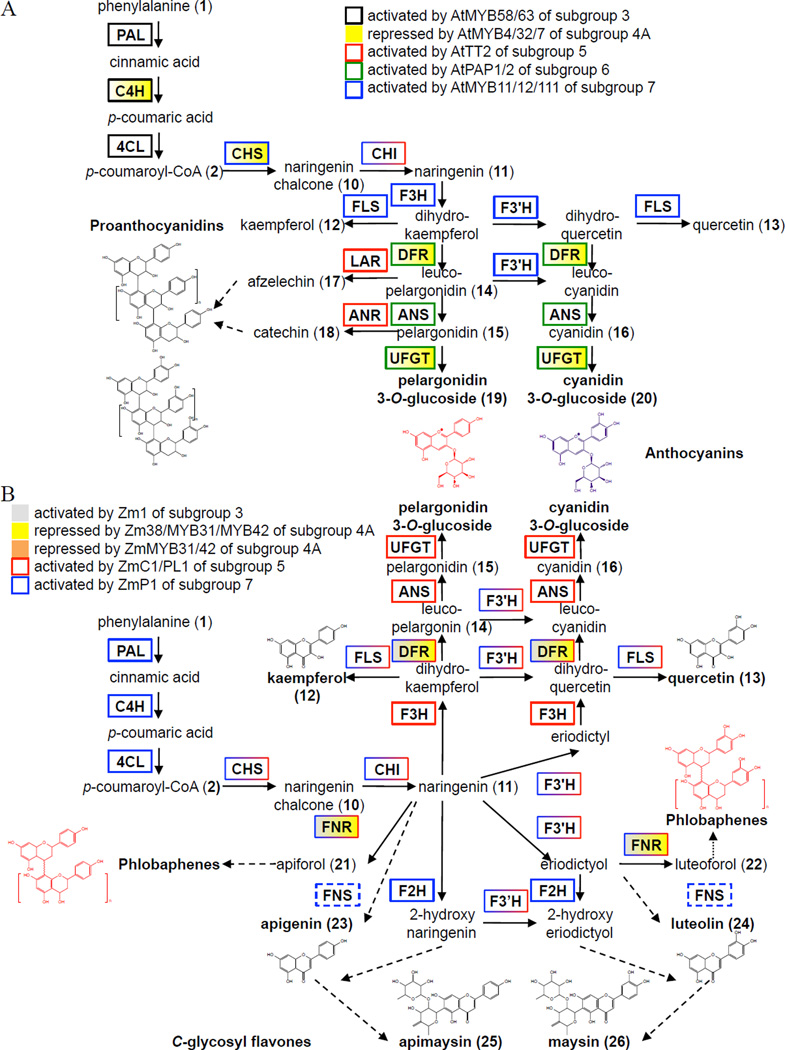
Figure 3. Multiple R2R3-type MYB Subgroups Regulate the Flavonoid Biosynthetic Pathways in Arabidopsis and Maize.
(A) Arabidopsis flavonoid pathway. (B) Maize flavonoid pathways. Adapted from Morohashi et al. (2012). Naringenin (11) is at the intersection between the 3-hydroxyflavonoid and 3-deoxyflavonoid pathways. Unbroken arrows indicate single enzymatic conversions, and broken arrows indicate multiple enzymatic conversions. Flavan-3-ols afzelechin (17) and catechin (16) polymerize to form proanthocyanidins. 3-hydroxyflavonoids include flavonols kaempferol (12) and quercetin (13), and anthocyanins pelargonin 3-glucoside (19) and cyanidin 3-glycoside (20). 3-deoxyflavonoids include the flavan-4-ols apiforol (21) and luteoforol (22), the flavones apigenin (23) and luteolin (24), and the C-glycosyl flavones apimaysin (25) and maysin (26). Flavan-4-ols polymerize to form the red phlobaphene pigments. Unbroken arrows indicate single enzymatic conversions, and broken arrows indicate multiple enzymatic conversions. ANR, anthocyanidin reductase; ANS, anthocyanidin synthase; C3’H, p-coumaroyl ester 3’-hydroxylase; C4H, cinnamate 4-hydroxylase; CHI, chalcone isomerase; CHS, chalcone synthase; 4CL, 4-coumarate-CoA ligase; COMT, caffeic acid O-methyltransferase; DFR, dihydroflavonol 4-reductase; F2H, flavanone 2-hydroxylase; F3H, flavanone 3-hydroxylase; F3’H, flavonoid 3’-hydroxylase; FLS, flavonol synthase; FNR, flavanone 4-reductase; FNS, Type I flavone synthase; LAR, leucoanthocyanidin reductase; PAL, phenylalanine ammonia-lyase; UFGT, UDP-glucose flavonoid 3-O-glucosyltransferase.
2.2. Lignin pathways
Lignin is an important structural and defensive component of plant secondary cell walls and the second most abundant biopolymer on earth. Lignin is composed primarily of three hydroxycinnamyl alcohol monomers, referred to as monolignols. These monolignols are oxidatively polymerized to form a variety of substructures that give the lignin polymer its complexity (See Davin et al., 2008 for a review on the structure and assembly of lignin). The lignin biosynthetic pathway is indispensable to plants for not only its role in the production of the monolignols, but also its role in the production of a host of other small molecules such as the UV-B-protecting sinapate esters and the iron-chelating coumarins (Ruegger et al., 1999; Kai et al., 2008; Fourcroy et al., 2013; Schmid et al., 2014). The lignin pathway is also involved in the production of the polyphenolic domains of suberin, which is a lipid and phenolic cell wall heteropolymer found in the roots and other organs of all vascular plants. Suberin’s polyphenolic domain is similar to lignin, but differs by containing a large proportion of hydroxycinnamic acids (especially ferulate) in addition to the monolignols normally found in lignin (Bernards et al., 1995; Yan and Stark, 2000).
In Arabidopsis, there are three iterations of the lignin pathway that result in the production of the three monolignols (p-coumaryl (3), coniferyl (6) and sinapyl (8) alcohols) from their respective precursors (p-coumaroyl-CoA (4), feruloyl-CoA (11) and sinapaldehyde (16)) by an enzyme cascade of cinnamoyl-CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD), or CAD alone (Figure 2; Vanholme et al., 2010). The three monolignols are then oxidatively polymerized to form their respective lignin polymers, H, G and S lignins (Figure 2). The monolignol precursor feruloyl-CoA (4) is synthesized from p-coumaryl-CoA (2) by an enzyme cascade of hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyltransferase (HCT), caffeoyl shikimate esterase (CSE), 4CL and caffeoyl-CoA O-methyltransferase (CCoAOMT) (Figure 2; Vanholme et al., 2013). The monolignol precursor sinapaldehyde (7) is synthesized from coniferaldehyde (5) by an enzyme cascade of ferulate 5-hydroxylase (F5H) and caffeic acid O-methyltransferase (COMT) (Figure 2). Although there are genomics-based descriptions of lignin pathway genes in monocots (Barriére et al., 2007; Lawrence and Walbot, 2007; Andersen et al., 2008), the corresponding enzymatic functions by many of the gene candidates have yet to be verified biochemically or genetically. In addition, it is not yet known whether monocot CSE homologues are involved in lignification.
2.3. Flavonoid pathways
Flavonoids are important for plant pigmentation and protection against biotic and abiotic stressors (e.g., herbivores, microbial pathogens, UV-B, free radicals). In Arabidopsis, there is only one flavonoid biosynthetic pathway that begins with the chalcone synthase (CHS) enzyme, a type-III polyketide synthase that catalyzes the formation of naringenin chalcone (10) from malonyl-CoA and p-coumaroyl-CoA (2). Downstream steps produce the commonly occurring flavonols kaempferol (12) and quercetin (13), the 3-hydroxyflavonoids pelargonidin (15) and cyanidin (16) as well as their derived plant pigments, the anthocyanins pelargonin 3-glucoside (19) and cyanidin 3-glycoside (20) (Figure 3A; Shirley et al., 1995; Bharti and Khurana, 1997). Pelargonidin (15) and its reduced precursor leucopelargonidin (14) are also precursors to the flavan-3-ol polymers, the proanthocyanidins (also called condensed tannins), which accumulate in the seed coat as antimicrobials and feeding deterrents (Figure 3A; Winkel-Shirley 1998). In maize, there are at least two flavonoid biosynthetic pathways that branch from the flavanone intermediate naringenin (11). One pathway results in the commonly occurring 3-hydroxyflavonoids pelargonidin (15) and cyanidin (16) and their derived pigments, the anthocyanins (Figure 3B; Dooner et al., 1991; van der Meer 1993). The other pathway produces the floral organ-specific 3-deoxyflavonoids, which include the flavan-4-ols apiforol (21) and luteoforol (22), their derived pigments the red phlobaphenes, the flavones apigenin (23) and luteolin (24), and their derived insecticidal and antifungal products, the C-glycosyl flavones apimaysin (25) and maysin (26) (Figure 3B; Styles and Ceska, 1975; Waiss et al., 1979; Elliger et al., 1980; Snyder and Nicholson, 1990; Reid et al., 1992; Byrne et al., 1996a, 1996b). Phlobaphenes and C-glycosyl flavones are lineage-restricted flavonoids found in a number of unrelated plant species (e.g., lotus, maize and redwoods). There are also lineage-specific flavonoids, such as the isoflavonoids, which are synthesized primarily in legumes from chalcone and flavanone precursors.
Closely related to CHS, the stilbene synthase (STS) enzymes, another of several specialized type-III polyketide synthases found in plants (reviewed in Yu et al., 2012), utilize the same precursors as CHS to instead form resveratrol, the simplest stilbene. Downstream steps produce the stilbenoids, which are hydroxylated derivatives of stilbenes. Unlike lignins and flavonoids, stilbenoids have been detected in a limited number of unrelated plant species and rapidly accumulate only in response to a wide range of biotic and abiotic stresses. Not present in Arabidopsis or maize, the stilbenoid biosynthetic pathway is best characterized in grapevine (Vitis vinifera), peanut (Arachis hypogea) and pine (Pinus spp.) (Jeandet et al., 2010).
2.4. Glucosinolate pathways
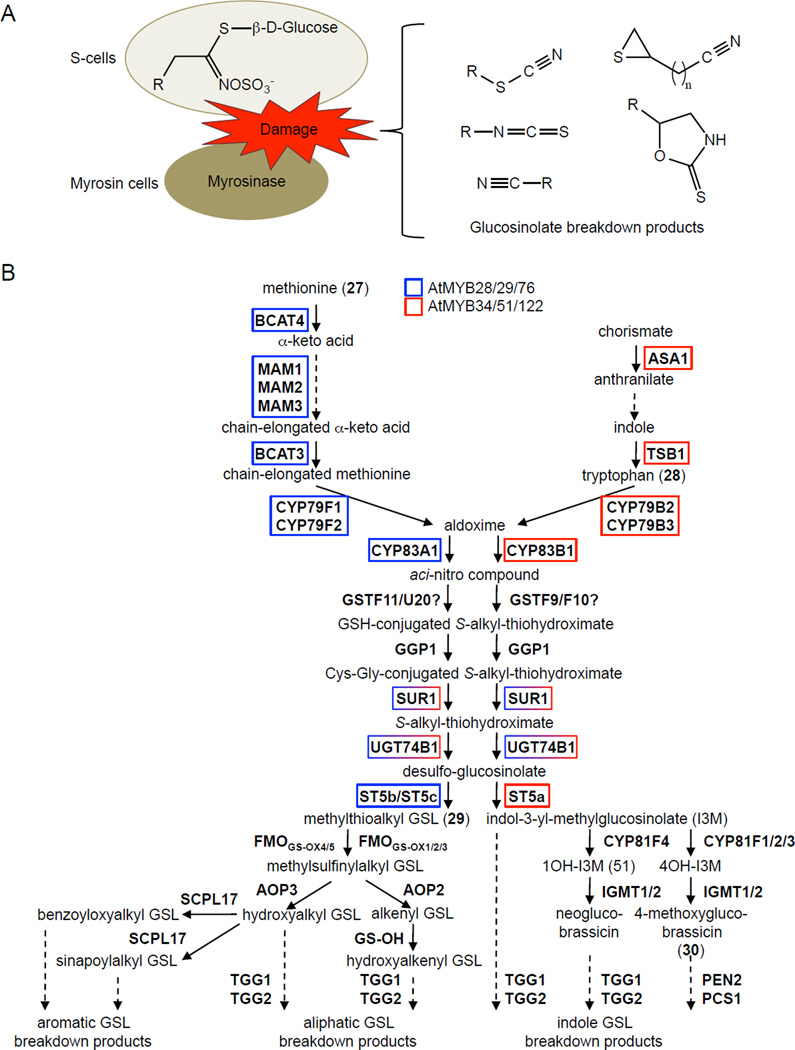
Glucosinolates are β-thioglucoside N-hydroxysulfates that are found principally in plants of the Brassicales order. Together with their hydrolytic enzymes, the myrosinases, they constitute the ‘mustard oil bomb’ (Matile, 1980; Luthy and Matile, 1984; Ratzka et al., 2002) that is involved in both constitutive and inducible plant defenses. Glucosinolates are characterized by a core sulfonated oxime group (also called a sulfated isothiocyanate group), which is conjugated to thioglucose and a side (R-) group (Figure 4). The chemical diversity of the more than 200 reported structures of GSLs lie in the elongations and modifications to the side-chain, and the esterification of the thioglucose moiety by benzoates and hydroxycinnamates (Clarke, 2010).
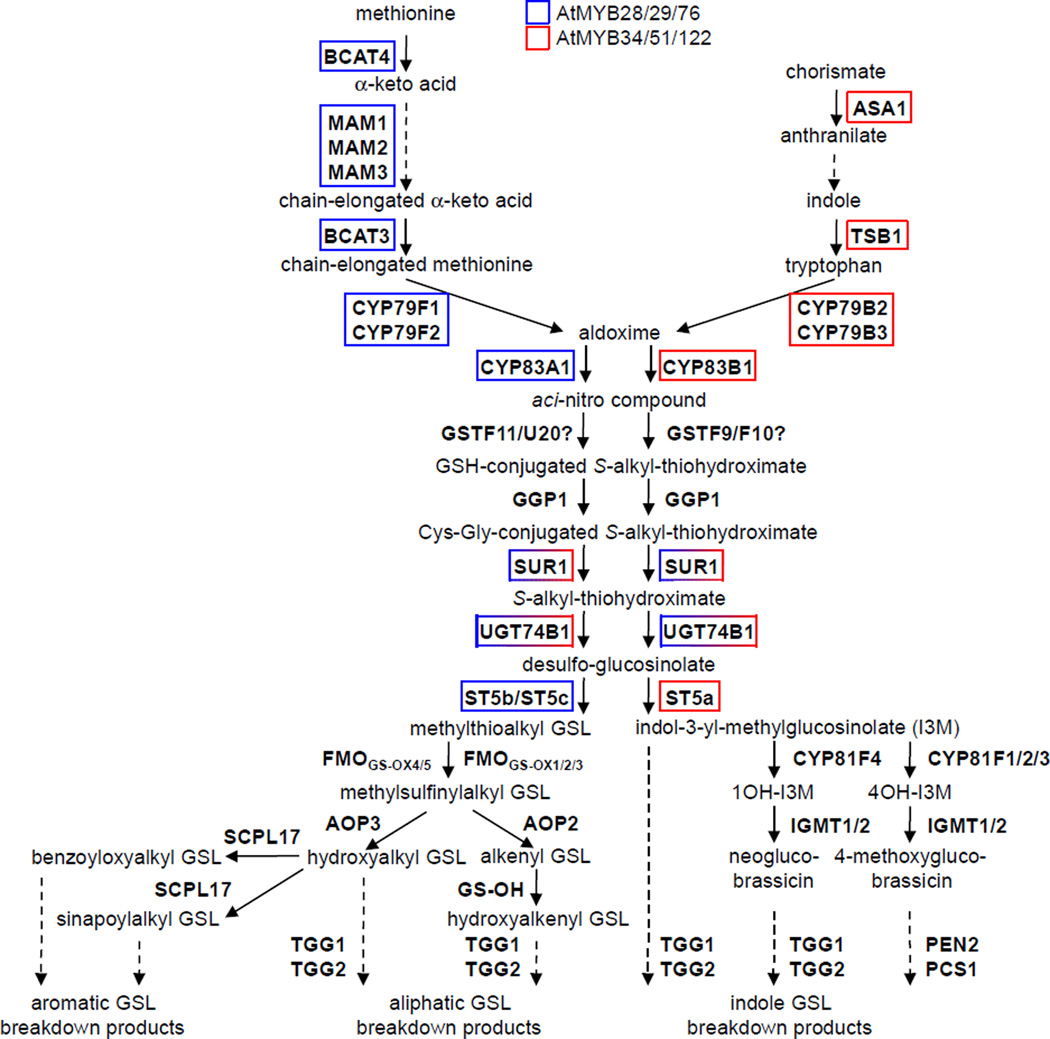
Figure 4. One R2R3-type MYB Subgroup Regulates the Glucosinolate Biosynthetic Pathways in Arabidopsis.
(A) Classical glucosinolate-myrosinase chemical defense system or “mustard oil bomb”. R is the side-chain of amino acids Trp, Tyr, Phe, Ile, Leu, Val, Ala or Met. Upon tissue damage, glucosinolates are hydrolyzed by myrosinase enzymes to glucose and unstable thiohydroximate-O-sulfate intermediates, which spontaneously eliminate the sulfate group and rearrange to form thiocyanates at pH >8; isothiocyanates at pH 5–8; nitriles and elemental sulfur if guided by epithiospecifier-like proteins, or at pH 2–5 and in the presence of Fe2+; epithionitriles if a terminal double bond captures the elemental sulfur released during nitrile formation; and oxazolidine-2-thiones if a hydroxyl function is present on carbon 3 of the glucosinolate. Adapted from Grubb and Abel et al. (2006). (B) Arabidopsis glucosinolate pathways. Adapted from Sønderby et al. (2010) and Maruyama-Nakashita et al. (2006). Unbroken arrows indicate single enzymatic conversions, and broken arrows indicate multiple enzymatic conversions. AOP2, alkenyl glucosinolate-producing 2-oxoglutarate-dependent dioxygenase; AOP3, hydroxyalkyl glucosinolate-producing 2-oxoglutarate-dependent dioxygenase; ASA1, anthranilate synthase alpha subunit; BCAT3, branched-chain aminotransferase; CYP79B2/3, tryptophan N-hydroxylases; CYP81F2, indol-3-ylmethylglucosinolate 4-hydroxylase; CYP83B1, indole-3-acetaldoxime N-hydroxylase; FMOGS-OX, flavin-monooxygenase GSL S-oxygenase; GST, glutathione transferase; GGP1, gamma-glutamyl peptidase 1; GS-OH, hydroxyalkenyl glucosinolate-producing 2-oxo acid-dependent dioxygenase; MAM, ; IGMT2 and IGMT3, indole glucosinolate O-methyltransferases; PEN2, 4-methoxyindol-3ylmethylglucosinolate thioglucosidase; PCS1, phytochelatin synthase 1; SCPL17, serine carboxypeptidase-like acyltransferase; ST5, desulfoglucosinolate sulfotransferase; SUR1, S-alkyl-thiohydroximate lyase; TGG, thioglucosidase; TSB1, tryptophan synthase β-subunit 1; UGT74B1, UDP-glucose:thiohydroximate S-glucosyltransferase.
Glucosinolates (GSLs) are among the best studied secondary metabolites of Arabidopsis, which contains at least 37 different GSLs (Hogge et al., 1988; Kliebenstein et al., 2001a; Reichelt et al., 2002; Kliebenstein et al., 2007). The two major GSL groups in Arabidopsis are the methionine (27) (Met)-derived aliphatic GSLs and the tryptophan (28) (Trp)-derived indole GSLs. Extensive functional genomics, metabolomics and systems biology approaches have led to the identification and functional characterization of nearly all of the biosynthetic enzymes of aliphatic and indole GSL metabolism (Wittstock and Halkier, 2002; Halkier and Gershenzon, 2006; Sønderby et al., 2010). GSL biosynthesis proceeds through three independent stages: chain elongation (for aliphatic GSLs), core structure formation and side-chain modification (Figure 4). Short-chain Met-derived aliphatic GSLs undergo one to four cycles of chain elongation, whereas long-chain aliphatic GSLs undergo five to six cycles of chain elongation. Minor GSL groups are the Phe- and Met-derived benzoyloxyalkyl GSLs and sinapoylalkyl GSLs, which are synthesized from the short-chain Met-derived methylthioalkyl GSL (29) by the enzyme cascade of flavin-monoxygenase glucosinolate S-oxygenases FMOGS-OX4/5, 2-oxoglutarate-dependent dioxygenase AOP3 and serine carboxypeptidase-like acyltransferase SCPL17 (Figure 4; Kliebenstein et al., 2001b; Lee et al., 2012).
3. Regulatory Components in Plant Secondary Metabolism
3.1. bHLH-bHLH, MYB-bHLH and MYB-bHLH-WD40 regulatory complexes
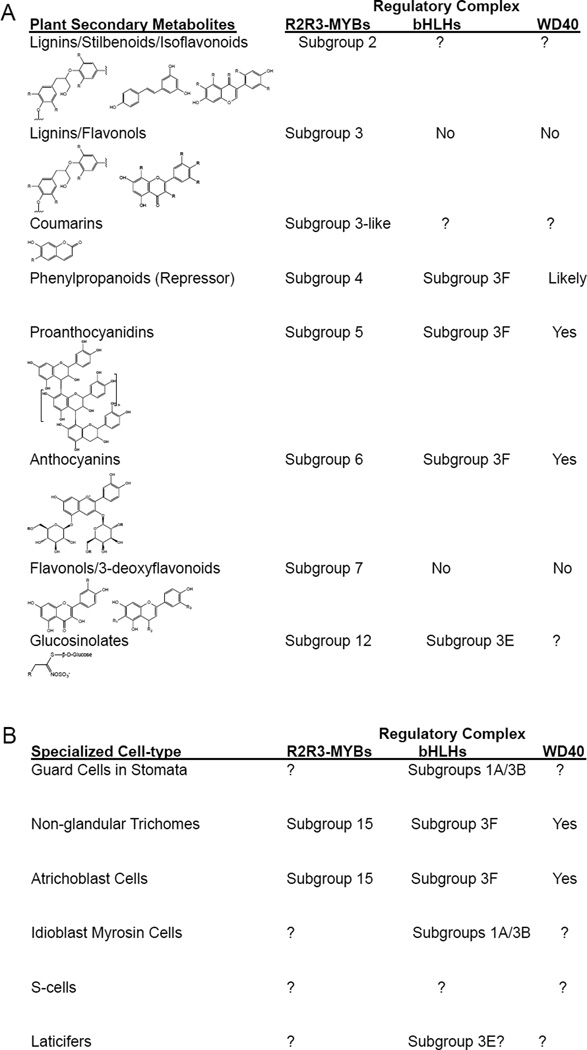
Gene regulation of plant secondary metabolism typically involves the formation of multiprotein complexes through combinatorial interactions between MYB and bHLH family of proteins, or between two different subgroups of bHLH proteins. Furthermore, similar transcription factor complexes have been shown to regulate cell differentiation and cell fate specification in eukaryotes. For example, the interactions between MYB and bHLH family proteins have been shown to activate or repress different gene modules in glucosinolate biosynthesis in Arabidopsis (Figures 4 and 5A; Schweizer et al., 2013; Frerigmann et al., 2014a), and in skeletal muscle differentiation in animals (Kaspar et al., 2005). In addition, depending on the plant lineage, the interactions between MYB, bHLH and WD40 domain proteins have been shown to activate all or a set of biosynthetic genes in anthocyanin and proanthocyanidin biosynthesis (Figure 3, 5A; Goff et al., 1992; Grotewold et al., 2000; Aharoni et al., 2001; Zimmermann et al., 2004; Quattrocchio et al., 2006), as well as the cell differentiation of single-celled trichomes, non-hair root epidermal cells (atrichoblasts) in the embryonic stem and mucilage secretory cells in the outer seed coat (Figure 5B; Payne et al., 2000; Bernhardt et al., 2003; Esch et al., 2003; Zhang et al., 2003; Gonzalez et al., 2009; Li et al., 2009). Of the plant MYB-bHLH-WD40 ternary complexes, there is considerable redundancy for the bHLH cofactors, while the MYB protein typically provides regulatory specificity for a given target pathway in secondary metabolism and cell differentiation by binding to specific cis-regulatory elements in gene promoters (Figure 5; Broun 2005; Feller et al., 2011). In addition, the conserved WD40 domain proteins are shared by the plant MYB-bHLH-WD40 complexes between secondary metabolism and cell differentiation (Broun 2005; Feller et al., 2011).
Figure 5. Regulatory Components in Plant Secondary Metabolism and Associated Specialized Cell Development.
Summary of the R2R3-type MYB subgroups, bHLH subgroups and WD40 domain proteins that regulate (A) the production of plant secondary metabolites and (B) the differentiation of specialized cell types.
Based on the phylogenetic tree of the MYB proteins, Serna and Martin (2006) suggested that the most ancient function of the MYB-bHLH-WD40 complex is in the regulation of anthocyanin and proanthocyanin production, and that the additional role of the MYB-bHLH-WD40 complex in single-celled trichome formation has been adopted after the Asterid-Rosid division (Serna and Martin, 2006). However, similar analyses have not been made for the other MYB-bHLH-WD40 complexes in epidermal cell differentiation, and thus it remains an open question whether the ancestral function of the MYB-bHLH-WD40 complex is in secondary metabolism or in differentiation of fundamental tissues.
Recently, interactions between bHLHs of different subfamilies in Arabidopsis have been shown to regulate the development of two different cell types that synthesize and accumulate the myrosinase enzymes that activate glucosinolate metabolites (Figure 5B; Shirakawa et al., 2014; Li and Sack, 2014). It is not yet known whether MYBs interact with one or more bHLH subgroups to regulate the development of specialized cell-types for secondary metabolism.
3.2. Activator-type R2R3-type MYBs
MYB transcription factors are exclusive to eukaryotes (Yang et al., 2003; Jiang et al., 2004), and the plant-specific double-repeat R2R3-type MYBs constitute one of the largest families of transcription factors in angiosperms, with 126 members in Arabidopsis (Stracke et al., 2001; Dubos et al., 2010) and at least 125 and 173 in rice and maize, respectively (Yilmaz et al., 2009). The R2R3 domain is responsible for DNA binding, for promoter specificity, and for interaction with other co-factors (e.g., bHLHs). The majority of the Arabidopsis R2R3-type MYBs are classified into 22 subgroups, based on the presence of distinctive motifs outside of the conserved MYB domains (Kranz et al., 1998; Stracke et al., 2001), while the remainder are grouped based on overall amino acid sequence alignments (e.g., the related MYBs AtMYB20, AtMYB42, AtMYB43 and AtMYB85 as well as general activators of lignin biosynthesis, AtMYB46 and AtMYB83) (Figures 1 and 6A).
Figure 6. Motifs defining MYB and bHLH subgroups in pathway regulation of secondary metabolism and associated specialized cell development.
The numbers shown at the left and right of each sequence are the positions of the first and last amino acid relative to the translational start site. (A) Activator-type R2R3-MYBs. Asterisk indicates that the reported cDNA sequence may contain an extra nucleotide that when removed produces the full conserved subgroup 2 motif. (B) Repressor-type R2R3-MYBs. Double asterisks indicate that the reported cDNA sequence is incomplete. (C) Activator-type bHLHs in subgroups as defined by Heim et al. (2003) and Pires and Dolan (2010). At, Arabidopsis thaliana; Eg, Eucalyptus gunnii; Ej, Eriobotrya japonica (loquat); Ph, Petunia x hybrida; Pm, Physcomitrella patens (moss); Ps, Pisum sativum (pea); Sm, Selaginella moellendorffii (spikemoss); Vv, Vitis vinifera (grapevine); Zm, Zea mays (maize). F denotes aliphatic amino acid; W denotes aromatic amino acid; uppercase letters represent strictly conserved amino acids; lowercase letters represent amino acids that are conserved in the majority of the sequences.
While MYB function in animals is restricted to the control of cell division and differentiation (Weston 1998), the plant-specific R2R3-type MYBs are involved in many diverse such as epidermal cell differentiation (Oppenheimer et al., 1991; Nesi et al., 2001; Ramsay and Glover, 2005), seed development (Penfield et al., 2001), abiotic and biotic stress regulation (Abe et al., 1997; Lee et al., 2001; Vailleau et al., 2002; Mengiste et al., 2003; Zhu et al., 2005; Agarwal et al., 2006), light and sugar regulation (Ballesteros et al., 2001; Teng et al., 2005), stomatal movements (Cominelli et al., 2005; Liang et al., 2005), and secondary metabolism (Martin and Paz-Ares, 1997). Of these functions, nearly half of the Arabidopsis R2R3-type MYBs activate genes in epidermal cell differentiation or secondary metabolism (Figures 1 and 5). Among those involved in secondary metabolism, only one MYB subgroup (subgroup 12) thus far is known to activate the lineage-specific glucosinolate pathway (Figures 1 and 5A), whereas at least 8 MYB subgroups have been shown to activate one or more branches of phenylpropanoid metabolism, including subgroups 2, 3, 5, 6, 7, 13, AtMYB20/42/43/85, and AtMYB46/83 (Figures 1 and 5; Grotewold et al., 1994; Sablowski et al., 1994; Tamagnone et al., 1998; Borevitz et al., 2000; Preston et al., 2004; Deluc et al., 2006; Patzlaff et al., 2003a, 2003b; Goicoechea et al., 2005; Demura and Fukuda, 2007; Zhong and Ye, 2007).
3.3. Repressor-type R2R3-MYBs and R3-MYBs
While the majority of R2R3-type MYBs in Arabidopsis activate transcription, some operate as transcriptional repressors. Subgroup 4 MYBs all contain a repressor motif known as the ethylene response factor (ERF)-associated amphiphilic repressor (EAR) motif (Kranz et al.,1998) involved in the repression of transcription (Figure 6B; Jin et al., 2000). These repressor-type MYBs transcriptionally repress different branches of phenylpropanoid metabolism, including the lignin and flavonoid pathways. Subgroup 4 is further divided into 4 clades (A–D) according to the presence of additional motifs (Figure 6B; Cavallini et al., 2015). A number of subgroup 4A/B MYBs have been shown to down-regulate the lignin pathway (and in the case of Arabidopsis, the derived sinapate ester pathway). They include the snapdragon (Antirrhinum majus) AmMYB308 and AmMYB330, Arabidopsis AtMYB4 and AtMYB32, Eucalyptus gunnii EgMYB1 and maize ZmMYB31 and ZmMYB42 proteins (Figure 6B; Tamagnone et al., 1998; Jin et al., 2000; Preston et al., 2004; Legay et al., 2007; Sonbol et al., 2009; Fornalé et al., 2010; Colquhoun et al., 2011). Subgroup 4A MYBs have also been shown to down-regulate one or more genes in the flavonoid pathway, such as Arabidopsis AtMYB4 and AtMYB7 and maize ZmMYB31 and Zm38 (Franken et al.,1994; Fornalé et al., 2000; Fornalé et al., 2014). Grapevine and a few other plant species additionally contain the subgroup 4D2 MYBs, which down-regulate the anthocyanin pathway (Figure 6B; Cavallini et al., 2015).
The EAR-repressor motif is also present in a small family of single-repeat R3-type MYBs. These repressor-type MYBs are thought to fine-tune metabolic and cell differentiation pathways to achieve different metabolic outputs and cell fates, respectively, in accordance with the tissue-type or environmental stress. Five of the 8 Arabidopsis R3-type MYBs (i.e., TRY, CPC, ETC1, ETC2, ETC3/CPL3) are functionally similar to one another and counteract the activity of the MYB-bHLH-WD40 ternary complexes in epidermal cell patterning in plants by sequestering their bHLH components (Esch et al., 2003; Zimmermann et al., 2004; Kirik et al., 2004; Simon et al., 2007; Tominaga et al., 2007). In addition, the R3-type MYB TCL1 may act to inhibit the expression of the MYB component of the MYB-bHLH-WD40 complex, through direct binding to its promoter (Wang et al., 2007). The R3-type MYB TCL2 functions redundantly with AtTCL1 in controlling trichome formation on inflorescences (Gan et al., 2011). Interestingly, the R3-type MYB MYBL2 not only inhibits trichome development but also flavonoid biosynthesis (Sawa 2002; Dubos et al., 2008; Matsui et al., 2008), presumably by sequestering the bHLH components in both active MYB-bHLH-WD40 complexes. Similarly, its petunia ortholog, the R3-type MYB PhMYBx, acts as an inhibitor of anthocyanin biosynthesis by sequestering the bHLH protein PhAN1 in active complexes (Koes et al., 2005).
Overexpression of subgroup 15 MYBs (AtGL1 and AtMYB0) promoted the formation of MYB homodimers or heterodimers at the MYB domains and counteracted the activity of the MYB-bHLH-WD40 complexes by preventing the formation of MYB-bHLH interactions (Larken et al., 1994; Liang et al., 2014).
3.4. Cis-regulatory elements of R2R3-type MYBs in phenylpropanoid metabolism
The tissue specificity and metabolic output of a particular biosynthetic pathway are dependent on not only the expression profiles and properties of the regulatory genes, but also the promoter regions of the target genes. In vitro promoter binding studies to date indicate that the distinct subgroups of R2R3-type MYBs from different subfamilies appear to bind to the same or very similar cis-regulatory elements, which are described generally as “AC-like” sequences. For example, systematic evolution of ligands by exponential enrichment (SELEX) experiments demonstrated that both the maize subgroup 4 phenylpropanoid-repressing MYB ZmMYB31 and the maize subgroup 7 flavonoid-activating MYB ZmP1 preferentially bind to ACC(T/A)ACC consensus sequence in vitro (Fornalé et al., 2010; Grotewold et al., 1994; Williams and Grotewold, 1997). In addition, electrophoretic mobility shift assay (EMSA) experiments have shown that this motif is similar in sequence to the ACCCGCC sequence that is bound by the Arabidopsis subgroup 4 phenylpropanoid-repressing MYB AtMYB4 (Zhao et al., 2007), and identical in sequence to two of the three AC elements that are bound by the Arabidopsis subgroup 3 lignin-activating MYBs AtMYB58 and AtMYB63 (Raes et al., 2003; Zhou et al., 2009). Similar to AtMYB58/63, the Arabidopsis subgroup 2 MYB AtMYB15 has been shown by EMSA to bind to all three AC elements (Romero et al., 1998), which are enriched in the promoters of lignin biosynthetic genes (Raes et al., 2003). Consistent with this finding, the loquat (Eriobotrya japonica) subgroup 2 MYB EjMYB1 has recently been shown to transactivate the promoters of both Arabidopsis and loquat lignin biosynthetic genes (Xu et al., 2014), presumably by binding to similar cis-regulatory elements.
AC-like sequences are also present in the promoters of target R2R3-type MYB genes to generate regulatory feedback and feedforward loops. For example, the Arabidopsis subgroup 4 MYB AtMYB4 directly represses its own transcription by binding to its promoter (Zhao et al., 2007). In addition, the related MYBs AtMYB46 and AtMYB83 proteins bind to AC-like sequences in the promoters of the lignin-activating AtMYB58/63 regulatory genes and the lignin biosynthetic genes, activating both and amplifying the lignin induction signal in the process (Zhong and Ye, 2012).
However, in vitro-defined consensus motifs for these R2R3-type MYBs may not be sufficient and/or present in the majority of their in vivo target promoters, as was shown for several animal transcription factors (Carr and Biggin, 1999; Yang et al., 2006; Rabinovich et al., 2008). For example, chromatin immunoprecipitation (ChIP)-PCR experiments demonstrated that the maize subgroup 4 MYB ZmMYB31 binds only a small subset of the maize lignin gene promoters containing the ACC(T/A)ACC consensus sequence (Fornalé et al., 2010). In addition, ChIP-Seq experiments indicated that the maize subgroup 7 MYB ZmP1 preferentially binds to sequences containing 6 to 8 repeats of CxxC (where X corresponds to any nucleotide) with no preference for A or T between the C base pairs in vivo (Morohashi et al., 2012). It is not known whether ZmP1 directly binds the (CxxC)6–8 motif in vivo or whether this motif reflects a tethering of ZmP1 thorough an unknown regulatory cofactor.
3.5. MYB-interacting bHLHs
bHLHs are characterized by a conserved DNA-binding domain and diverse sequence motifs that participate in transcriptional regulation (Heim et al., 2003; Pires and Dolan, 2010). The majority of the approximately 160 Arabidopsis bHLHs contain His (H), Glu (E), and Arg (R) residues in the core bHLH region, and these residues have been shown to facilitate interaction with DNA at G-box (CACGTG) elements (Martínez-Garcia et al., 2000; Toledo-Ortiz et al., 2003; Qian et al., 2007). The G-box cis-element, alone or in tandem, is found in the promoters of genes that control a variety of developmental and homeostatic processes (Menkens et al.,1995; Chakravarthy et al., 2003; Chandrasekharan et al., 2003). Recently, bHLHs have been shown to play key roles in several cell fate decision points in the differentiation of two different myrosin cell types (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007; Li and Sack, 2014; Shirakawa et al., 2014).
Activator-type MYBs controlling flavonoid biosynthesis and epidermal cell development have a signature motif ([DE]Lxx[RK]xxxLxxxxxxLxxxR) located in their R3 repeats (Zimmerman et al., 2004; Serna and Martin, 2006). This motif is responsible for their interaction with the subgroup 3F bHLHs (Zimmermann et al., 2004). This bHLH-interacting motif is also shared by repressor-type MYBs controlling phenylpropanoid metabolism and single-celled trichome development, including all characterized subgroup 4 R2R3-type MYBs in plants and six of the eight Arabidopsis R3-type MYBs (Zimmermann et al., 2004; Serna and Martin, 2006).
4. Metabolic Pathway Regulation
4.1. Regulation of lignin pathways in the context of secondary cell wall biosynthesis
At least four subgroups of Arabidopsis R2R3-MYBs have been demonstrated to transactivate the promoters of general phenylpropanoid and lignin-specific pathway genes, including subgroup 3 MYBs AtMYB58 and AtMYB63, subgroup 13 MYB AtMYB61, related MYBs AtMYB46 and AtMYB83, and related MYBs AtMYB20, AtMYB42, AtMYB43, and AtMYB85 (Newman et al., 2004; Zhong et al., 2007a; Zhong et al., 2008; McCarthy et al., 2009; Zhou et al., 2009). Of these, only AtMYB58 and AtMYB63 have been shown to directly activate nearly all of the structural genes for lignin biosynthesis (Zhong and Ye, 2009). In Arabidopsis, lignin biosynthesis is one of a number of metabolic and developmental pathways involved in secondary cell wall biosynthesis, which is regulated by a three-tiered hierarchical network of transcription factors. AtMYB46 and AtMYB83 as well as AtMYB58 and AtMYB63 occupy the bottom two tiers of this gene regulatory network, with AtMYB46 and AtMYB83 directly activating AtMYB58 and AtMYB63.
AtMYB46 and AtMYB83 together with their regulators, the secondary wall NAC transcription factors (SWNs), are functionally conserved in other angiosperm and gymnosperm species, including the SWN orthologs in Medicago, poplar, Eucalyptus, rice, maize and Brachypodium (Zhong et al., 2010a; Zhao et al., 2010; Zhong et al., 2011a; Zhong et al., 2011b; Valdivia et al., 2013), as well as the AtMYB46/83 orthologs in poplar, Eucalyptus and pine (Patzlaff et al., 2003a; Goichoechea et al., 2005; McCarthy et al., 2010; Zhong et al., 2010b; Zhong et al., 2013). By contrast, the AtMYB58/63 orthologs are present only in species within the rosid eudicot clade (Plant Genome Duplication Database; chibba.agtec.uga.edu/duplication/). In addition, even though the Arabidopsis AtMYB58 and AtMYB63 proteins are lignin-specific, their orthologs in other plant lineages may have different regulatory roles. For example, the AtMYB58/63 ortholog in maize, Zm1, has been shown to transactivate the maize DFR/FNS gene A1, which is involved in flavonoid biosynthesis and not lignin biosynthesis (Franken et al., 1994).
4.2. Regulation of flavonoid pathways
In Arabidopsis, the regulation of the single flavonoid biosynthetic pathway is mediated by three regulatory complexes, each responsible for regulating either the early biosynthetic genes that are common to the different flavonoid subpathways or the late genes that are specific for the anthocyanin or proanthocyanidin subpathways. In Arabidopsis, the MYB-bHLH-WD40 complex that activates genes specific for proanthocyanidin biosynthesis consists of the subgroup 5 MYB AtTT2, subgroup 3F bHLH EGL3, GL3, or TT8, and the WD40 domain protein AtTTG1 (Figure 5A; Nesi et al., 2000, 2001; Baudry et al., 2004; Zimmermann et al., 2004). Similarly, the MYB-bHLH-WD40 complex that activates genes specific for anthocyanin biosynthesis consists of subgroup 6 MYB PAP1 or PAP2, subgroup 3F bHLH EGL3, GL3 or TT8, and the WD40 protein AtTTG1 (Figure 5A; Borevitz et al., 2000; Gonzalez et al., 2008; Teng et al., 2005; Zhang et al., 2003; Zimmermann et al., 2004). In both cases, the activator-type MYBs are responsible for pathway specificity. Finally, the subgroup 7 MYBs AtMYB11, AtMYB12, and AtMYB111 control flavonol biosynthesis, independently of any bHLH and WD40 cofactors (Figure 5A; Mehrtens et al., 2005; Stracke et al., 2007; Zimmermann et al., 2004).
The Arabidopsis subgroup 3F bHLHs EGL3, GL3, and TT8 also interact with another class of transcription factors, the JA-ZIM domain (JAZ) proteins, which can interact with MYBs and bHLHs to repress jasmonic acid (JA)-regulated signaling (Chini et al., 2007; Seo et al., 2011; Song et al., 2011). Here, the JAZ proteins inhibit the MYB-bHLH interaction between the subgroup 3F bHLHs and the subgroup 6 MYB PAP1 to repress JA-regulated anthocyanin accumulation, as well as inhibit the MYB-bHLH interaction between the subgroup 3F bHLHs and the R2R3-type MYB AtGL1 to repress JA-regulated trichome initiation (Qi et al., 2011).
Similar to Arabidopsis, the anthocyanin and 3-deoxyflavonoid pathways in maize are controlled by two different regulatory complexes. A MYB-bHLH-WD40 complex consisting of subgroup 5 MYB ZmC1 or ZmPL1, subgroup 3F bHLH ZmR or ZmB, and the WD40 protein ZmPAC1 activates early biosynthetic genes that are shared by all of the flavonoid pathways (e.g., CHS, chalcone isomerase [CHI], flavanone 3-hydroxylase [F3H] and dihydroflavonol 4-reductase [DFR] genes) as well as genes that are specific for the anthocyanin pathway (e.g., anthocyanidin synthase [ANS] and UDP-glucose flavonoid 3-O-glucosyltransferase [UFGT] genes) (Figure 3; Goff et al., 1990; Roth et al., 1991; Tuerck and Fromm, 1994; Grotewold et al., 1998; Selinger and Chandler, 1999; Walker et al., 1999; Carey 2004). By contrast, the subgroup 7 MYB ZmP1 activates the production of the floral organ-specific 3-deoxyflavonoids, independently of any bHLH and WD40 cofactors (Grotewold et al., 1994). Additionally, ZmP1 also activates general phenylpropanoid pathway genes (Figures 2 and 3). Contrary to previous expectations that ZmP1 is a bottom-tiered transcription factor in a hierarchical gene regulatory network, recent ChIP-Seq results suggest that ZmP1 functions both as a transcriptional activator and a repressor with a wider regulatory role that includes not only the PAL, C4H and 4CL genes of the general phenylpropanoid pathway but also the p-coumaroyl ester 3’-hydroxylase (C3’H) gene of the lignin pathway (Morohashi et al., 2012).
In petunia petal cells, anthocyanin synthesis and vacuole acidification are regulated by a MYB-bHLH-WD40 complex, consisting of subgroup 6 MYB PhAN2 or PhAN4, subgroup 3F bHLH PhJAF13 or PhAN1, and the WD40 protein PhAN11 (Koes et al., 2005; Quattrocchio et al., 2006; Gerats and Strommer, 2009). In addition, subgroup 6 MYBs PhAN2 and PhAN4 regulate overlapping sets of target genes, providing gene specificity to the pathway (Quattrocchio et al., 2006). Finally, the R3-type MYB PhMYBx downregulates anthocyanin synthesis (Koes et al., 2005; Quattrocchio et al., 2006), presumably by sequestering the bHLH component of the MYB-bHLH-WD40 complex.
Similar to Arabidopsis, the proanthocyanidin and anthocyanin pathways in grapevine are activated by the subgroup 5 MYB VvMYBPA2 and the subgroup 6 MYBs VvMYBA1 and VvMYBA2, respectively. MYBs from subgroups 5 and 6 typically interact with subgroup 3F bHLHs and a conserved WD40 protein for their activity. However, as yet, no bHLH or WD40 cofactors have been identified for these grapevine flavonoid-activating MYBs, raising questions over the conservation of the MYB-bHLH-WD40 regulatory model in flavonoid biosynthesis for different plant species (Walker et al., 2007; Kobayashi et al., 2005).
In addition to activating complexes, flavonoid regulation is also controlled by multiple repressive transcription factors to fine-tune phenylpropanoid metabolism or serve as a regulatory loop. The subgroup 4 MYBs, which include AtMYB4, AtMYB6, AtMYB7, and AtMYB32 (Jin et al., 2000; Fornalé et al., 2014; Preston et al., 2004) have been thought to also form MYB-bHLH-WD40 complexes (Zimmermann et al., 2004). There appears to be distinctions between the gene targets of these repressors. AtMYB4 primarily targets the main phenylpropanoid pathway enzyme gene C4H and the flavonoid enzyme gene CHS to promote sinapate ester production (Jin et al., 2000), In contrast, the atmyb7 mutant has increased transcription of early steps of phenylpropanoid biosynthesis as well as enzymes in flavonol and anthocyanin biosynthesis (Fornalé et al., 2014), and the atmyb32 mutant has no change in core phenylpropanoid gene expression and has reduced transcription of anthocyanin biosynthetic genes (Preston et al., 2004). In spite of their transcriptional phenotypes, the atmyb4 mutant produces greater levels of flavonols and reduced anthocyanin content, while the atmyb7 mutant has the opposite metabolic phenotype, and the atmyb4/myb7 double mutant has substantial increases in both flavonols and anthocyanins (Fornalé et al., 2014). These results suggest a more complex system of regulation of flavonoids.
These repressors are themselves activated by other MYBs, It was recently shown that AtMYB4, AtMYB7, and AtMYB32 genes are direct targets of the secondary cell wall regulators AtMYB46 and AtMYB83 (Zhong and Ye, 2012), which suggested their involvement in fine-tuning of lignin biosynthesis. However, more recently MYB112, a subgroup 20 MYB, was found to directly activate MYB4/6/7/32 expression in response to abiotic stresses such as salinity and UV light (Lotkowska et al., 2015) to accumulate anthocyanins. These contrasting results suggest that the particular role of these regulatory proteins and their complexes can vary depending upon tissue or cell type and can be stress-dependent.
4.3. Regulation of other phenylpropanoid pathways
In Arabidopsis, the phenylpropanoid biosynthetic pathway is indispensable to plants for not only its role in the production of the monolignols, but also its role in the production of the phenolic moieties of suberin and a host of other small molecules such as the UV-protecting sinapate esters and the iron-chelating coumarins (Ruegger et al., 1999; Kai et al., 2008; Fourcroy et al., 2013; Schmid et al., 2014).
A recent study identified AtMYB41, a subgroup 11 MYB, as a regulator of suberin and cutin production, including the activation of phenylpropanoid biosynthesis and fatty acid-ferulate transferases such as ASFT (Kosma et al., 2014). Analysis of AtMYB41 expression by GUS staining found expression solely in the root following treatment with abscisic acid or salt, consistent with its role in suberin metabolism. Overexpression of MYB41 results in increased suberin production including ferulate-conjugated fatty acids in both Arabidopsis and Nicotiana benthamiana. In addition, overexpression of MYB41 increases total lignin content in both species, identifying MYB41 as a novel regulator of lignin metabolism.
The Arabidopsis subgroup 3-like MYB AtMYB72 has recently been shown to modulate iron deficiency responses in roots by not only regulating the lignin biosynthetic genes involved in the production of feruloyl-CoA (4), the biosynthetic precursor of the coumarin scopoletin (9), but also activating genes involved in the secretion of the iron-chelating coumarins into the soil (Zamioudis et al., 2014), presumably by binding to AC-like elements in the target gene promoters.
The subgroup 2 MYBs have particularly interesting roles in phenylpropanoid metabolism. They are commonly known to be induced by both biotic and abiotic stresses including elicitor treatment, cold stress, and drought (Sugimoto et al., 2000; Maeda et al., 2005; Agarwal et al., 2006; Ding et al., 2009), and have been characterized as regulators of diverse phenylpropanoid metabolites. Recently, subgroup 2 MYBs have been found to upregulate lignin biosynthesis in loquat (e.g., EjMYB1), stilbene biosynthesis in grapevine (e.g, VvMYB14/15), and isoflavonoid biosynthesis in Lotus japonicus in response to abiotic and biotic stresses (Shelton et al., 2012; Höll et al., 2013; Xu et al., 2014). These results are in agreement with earlier studies that had identified subgroup 2 MYBs in tobacco and carrot (Sugimoto et al., 2000; Maeda et al., 2005) that activate early steps of phenylpropanoid biosynthesis. Phylogenetically, the subgroup 2 MYBs are closely related to the subgroup 3 MYBs; however, the subgroup 2 MYBs are more evolutionarily ancient, and have been identified in conifers (Bedon et al., 2007), although gymnosperm subgroup 2 MYBs have not yet been characterized. These findings suggest that subgroup 2 MYBs in other plant lineages may regulate both lignin and clade-specific pathways under certain stress conditions, and may make attractive targets for engineering phenylpropanoid metabolism and exploring plant metabolic diversity.
4.4. Regulation of glucosinolate pathways
The direct regulatory network controlling GSL synthesis in Arabidopsis involves activator-type MYBs from subgroup 12 (Figure 2A). The subgroup 12 MYBs AtMYB34, AtMYB51, and AtMYB122 regulate the tryptophan-derived indolic GSLs (Celenza et al., 2005; Gigolashvili et al., 2007a; Malitsky et al., 2008), of which AtMYB34/51 are the main regulators (Frerigmann et al., 2014b). By contrast, the subgroup 12 MYBs AtMYB28, AtMYB29, and AtMYB76 regulate the methionine-derived aliphatic GSLs (Gigolashvili et al., 2007b, 2008; Hirai et al., 2007; Sønderby et al., 2007; Beekwilder et al., 2008; Malitsky et al., 2008). In addition, all six subgroup 12 MYBs interact directly with the subgroup 3E bHLHs, including AtMYC2 (also known as AtbHLH06), AtMYC3 (also known as AtbHLH05), AtMYC4 (also known as AtbHLH04) and AtbHLH28 (Schweizer et al., 2013; Frerigmann et al., 2014). Moreover, certain MYB-bHLH protein pairs are preferred over others, with strong interactions found between the pairing of AtMYB28/51/76 with AtMYC4, AtMYB29 with AtMYC2, and AtMYB34/122 with AtMYC3/4 (Frerigmann et al., 2014).
Similar to the subgroup 3F bHLHs in anthocyanin biosynthesis and trichome development, the Arabidopsis subgroup 3E bHLHs AtMYC2/3/4 also interact with the JAZ proteins. The JAZ proteins inhibit the MYB-bHLH interaction to repress JA-regulated glucosinolate biosynthesis (Chini et al., 2009). Interestingly, the subgroup 3E bHLH AtbHLH28 does not interact with the JAZ proteins, which has led to the suggestion that it may regulate GSL biosynthesis independently of JA signaling (Fernández-Calvo et al., 2011; Niu et al., 2011). In addition, the interaction strength between bHLH-JAZ protein pairs may be negatively correlated with that of bHLH-MYB pairs. For example, a strong interaction between AtMYB29 and AtMYC2 may produce a correspondingly weak interaction between AtMYC2 and JAZ proteins, allowing for AtMYB29 to function in JA-mediated control of GSL biosynthesis (Hirai et al., 2007; Gigolashvili et al., 2008).
While glucosinolates are lineage-specific secondary metabolites, the subgroup E bHLHs are present in diverse plant lineages. In plant species that do not synthesize glucosinolates, the subgroup E bHLHs may regulate other secondary metabolic pathways. For example, the pea subgroup E bHLH PsGBP1 has been shown to bind to the promoter of the CHS gene (Qian et al., 2007), possibly regulating flavonoid biosynthesis.
5. Developmental Pathway Regulation
5.1. Compartmentalization of glucosinolates and myrosinases
The defining feature of glucosinolate-producing plants is the cellular compartmentalization of glucosinolates and their hydrolyic enzymes, the myrosinases (also called thioglucoside glucohydrolase [TGG]) (Kissen et al., 2009; Ahuja et al., 2010). Normally in Arabidopsis, glucosinolates and myrosinases are synthesized and stored separately in adjacent cells termed S-cells and myrosin cells, respectively, at the leaf periphery and along veins (Andréasson et al., 2001; Husebye et al., 2002; Ueda et al., 2006; Shroff et al., 2008; Koroleva et al., 2010). Upon herbivore-induced release, the myrosinase, in the presence of water, cleaves the thioglucose bond in the glucosinolate, leading to the rapid generation of the unstable thiohydroximate-O-sulfate intermediate. Depending on the substrate, pH, availability of ferrous ions and presence of myrosinase-interacting proteins, a variety of biologically active breakdown products can be produced, including isothiocyanates, thiocyanates, nitriles and elemental sulfur, oxazolidine-2-thiones and/or epithioalkanes (Figure 4A). However, recent studies in Arabidopsis have shown that one particular glucosinolate species, 4-methoxyglucobrassicin (30) (Figure 4B), and the myrosinase PEN2 can be synthesized de novo in the same cells upon pathogen elicitation (Bednarek et al., 2009; Clay et al., 2009). The resulting breakdown products, as yet unknown, function as antifungals and as signaling molecules directing the deposition of callose polymers at the cell wall and thus restricting cellular entry by microbial pathogens (Bednarek et al., 2009; Clay et al., 2009). In addition, S-cells have been recently shown to synthesize both glucosinolates and myrosinases and then store them in separate subcellular compartments (Koroleva and Cramer, 2011).
5.2. bHLH heterodimers in myrosin cell development
There are two types of myrosinase-expressing myrosin cells: guard cells in stomata and idioblast myrosin cells. Idioblast myrosin cells are a recent developmental innovation that is restricted to the Brassicales order. By contrast, guard cells in stomata mediate gas exchange between the plant and the environment, and are present in nearly all extant land plants. In Arabidopsis, three “master regulator” subgroup 1A bHLHs, AtSPCH, AtMUTE and AtFAMA, regulate individual consecutive steps in stomatal development (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007). The final differentiation step in stomatal development is regulated by AtFAMA, which also confers guard cell fate (Ohashi-Ito and Bergmann 2006). Similarly, idioblast myrosin cell development and cell fate are also regulated by AtFAMA, which is transiently expressed in ground meristem cells before the cells differentiate into myrosinase-expressing idioblast myrosin cells (Li et al., 2014; Shirakawa et al., 2014). Differentiation of both myrosin cell types also requires two functionally redundant subgroup 3B bHLHs, AtICE1 (also called AtSCRM1) and AtSCRM2 (also called AtICE2), which are ubiquitously expressed in all cell types in the leaf and heterodimerize with AtFAMA (Kanaoka et al., 2008; Shirakawa et al., 2014). By contrast, guard cell differentiation additionally requires AtSPCH and AtMUTE, which also interact with AtICE1 and AtSCRM2 to regulate the first two differentiation steps in stomatal development (MacAlister et al., 2007; Pillitteri et al., 2007).
The subgroup 1A bHLHs are not functionally exchangeable in stomatal and idioblast myrosin cell differentiation (MacAlister et al., 2007; MacAlister and Bergmann, 2011; Shirakawa et al., 2014). In addition, the AtFAMA amino acid sequence is better conserved among land plants than those of AtSPCH or AtMUTE (Ran et al., 2013). In idioblast myrosin cell precursors, AtFAMA binds to its own promoter (Hachez et al., 2011) and positively regulates its own expression (Shirakawa et al., 2014), whereas in guard cell precursors, AtFAMA does not exhibit positive feedback regulation of its own expression (Ohashi-Ito and Bergmann, 2006). Stomatal development is also regulated by two R2R3-MYBs, AtFLP (also AtMYB124) and AtMYB88, that are coexpressed with AtFAMA, AtICE1 and its paralog AtSCRM2 in leaves, but AtFAMA does not interact with these MYBs (Ohashi-Ito and Bergmann 2006).
The recent data on myrosin cell differentiation indicate that despite differences between the two different developmental pathways that generate the different myrosin cell types, the underlying AtFAMA-dependent regulatory mechanisms appear to be shared. Because FAMA-expressing cells outside of the Brassicales do not express myrosinases (Liu et al., 2009), at some point during the evolution of the Brassicales, these cells were specified to accumulate myrosinases. Ongoing efforts to identify the downstream targets of AtFAMA may shed some light on whether this “myrosin specifying factor” is independent of AtFAMA expression.
5.3. bHLH heterodimers in laticifer cell development
S-cells are elongated phloem-associated cells that are rich in glucosinolates and thus in sulfur (Koroleva et al., 2000). S-cells are similar in structure and function to the non-articulated laticifer cells in latex plants. Laticifers have evolved repeatedly in nearly every major plant group (Evert et al., 2006; Hagel et al., 2008), and may share a common evolutionary origin with S-cells (Koroleva et al., 2010). Unlike myrosin cells, the molecular mechanisms regulating S-cell development remain elusive, largely due to a paucity of gene markers for S-cells. Recently, transcripts of the rubber tree (Hevea brasiliensis) subgroup 3E-like bHLH genes, HblMYC1 and HblMYC2, were found to increase or decrease, respectively, during mechanical wounding or JA treatment of laticifer cells, suggesting that these transcription factors may participate in JA-induced production of latex (Zhao et al., 2011). Parallels between laticifer and S-cell development suggest that S-cell development in Arabidopsis may also be regulated by the same subgroup 3E bHLHs that regulate glucosinolate biosynthesis. In addition to S-cells, both aliphatic and indole GSLs have been shown to accumulate in single-celled (non glandular) trichomes (Frerigmann et al., 2012), and indole GSLs in the cells of the outer leaf margin (Shroff et al., 2008; Koroleva et al., 2010).
6. Concluding Remarks
Plants are largely the basis for human nutrition and have widespread potential as a renewable source for fuel and chemical feedstocks. Technological advances in genomics and metabolomics combined with systems biology approaches have ushered plant metabolic research into a second golden age, at a time when there is increased awareness of diminishing food security and vanishing fossil fuels, the feedstocks of petrochemicals. A more fundamental understanding of plant secondary metabolism is needed to rationally redesign and predictably modify plant metabolic networks to address the many problems confronting humanity. This will entail not only the elucidation of the secondary metabolic networks, but also an increased understanding of the gene regulatory networks that are integrated with the metabolic networks and are responsible for generating chemical diversity in response to genetic and environmental perturbations. Presently, very little is known about the relationships between these regulatory proteins and their target sequences, as well as potential regulatory complexes that may exist for each transcription factor. A fuller understanding of the regulatory mechanisms that underlie secondary metabolism will enable rational engineering of these metabolic pathways through coordinated activation or repression of biosynthetic pathways by transcriptional regulators. In addition, it should be possible to engineer the regulation of heterologous genes and pathways to be activated alongside their precursors by modifying promoter sequences. Once more is known about the relationships between these regulators and their target promoter sequences, one could imagine the engineering of the regulators themselves to customize their gene targets. These tools could then be applied to improve biomass yields, increase stress tolerance, and enable large-scale production of compounds with industrial and pharmaceutical importance in order to meet the challenges of a growing population, climate change and bioenergy.
MYBs and bHLHs regulate secondary metabolism and epidermal cell differentiation
Multiple MYB subgroups regulate different pathways in phenylpropanoid metabolism
A single MYB subgroup regulates glucosinolate metabolism
bHLHs regulate specialized cell differentiation associated with secondary metabolism
Biographies

William R. Chezem received his B.S. in Biochemistry (2010) from Ball State University and is currently a Ph.D. candidate in the Department of Molecular, Cellular & Developmental Biology at Yale University. His research focuses on the transcriptional regulation of phenylpropanoid metabolism in plant immunity and the evolutionary conservation of pathway regulators. His research goal is to address needs in bioenergy, plant disease, and human health by engineering plant metabolism.
http://www.ncbi.nlm.nih.gov/myncbi/browse/collection/41467483/

Nicole K. Clay received her B.S. in Biology (1996) from Massachusetts Institute of Technology, and her Ph.D. in Biology (2005) from Yale University. She received her postdoctoral training in the field of plant-microbe interactions (2005–2010) at Massachusetts General Hospital, an affiliate of the Harvard Medical School. In 2011, she joined the faculty at Yale University as an Assistant Professor of Molecular, Cellular & Developmental Biology. Her research program exploits plant-pathogen interactions to discover new small molecules with antimicrobial and immune-signaling activities as well as their underlying biosynthetic “regulons” and regulatory networks. In addition, her program re-engineers plant immune receptors to discover novel protein modifications and quality-controls that regulate recombinant (and native) protein production and function.
Prof. Clay has served on the scientific advisory panel at the United States Department of Agriculture. She is the most recent recipient of the Elsevier/Phytochemistry Young Investigator Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, De Vos CHR, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O’Connell AP. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28:319–332. doi: 10.1046/j.1365-313x.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- Ahmad S, Veyrat N, Gordon-Weeks R, Zhang Y, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, Ton J. Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol. 2011;157:317–327. doi: 10.1104/pp.111.180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006;218:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Ahuja I, Rohloff J, Bones AM. Defense mechanisms of Brassicaceae: implications for plant-insect interactions and potential for integrated pest management. Agron. Sustain. Dev. 2010;30:311–348. [Google Scholar]
- Andersen JR, Zein I, Wenzel G, Darnhofer B, Eder J, Ouzunova M, Lübberstedt T. Characterization of phenylpropanoid pathway genes within European maize (Zea mays L.) inbreds. BMC Plant Biol. 2008;8:2. doi: 10.1186/1471-2229-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson E, Bolt Jørgensen L, Höglund AS, Rask L, Meijer J. Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus . Plant Physiol. 2001;127:1750–1763. doi: 10.1104/pp.010334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. LAF1, a MYB transcription factor for phytochrome A signaling. Genes Dev. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriére Y, Riboulet C, Méchin V, Maltese S, Pichon M, Cardinal A, Lapierre C, Lübberstedt T, Martinant JP. Genetics and genomics of lignification in grass cell walls based on maize as model species. Genes Genomes Genomics. 2007;1:133–156. [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze-Lefert P. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum anti-fungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- Beekwilder J, van Leeuwen W, van Dam NM, Bertossi M, Grandi V, Mizzi L, Soloviev M, Szabados L, Molthoff JW, Schipper B, Verbocht H, de Vos RC, Morandini P, Aarts MG, Bovy A. The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS One. 2008;3:e2068. doi: 10.1371/journal.pone.0002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards MA, Razem FA. The poly(phenolic) domain of potato suberin: a non-lignin cell wall bio-polymer. Phytochemistry. 2001;57:1115–1122. doi: 10.1016/s0031-9422(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 2003;130:6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- Bharti AK, Khurana JP. Mutants of Arabidopsis as tools to understand the regulation of phenylpropanoid pathway and UVB protection mechanisms. Photochem. Photobiol. 1997;65:765–776. doi: 10.1111/j.1751-1097.1997.tb01923.x. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouarab K, Melton R, Peart J, Baulcombe D, Osbourn A. A saponin-detoxifying enzyme mediates suppression of plant defences. Nature. 2002;418:889–892. doi: 10.1038/nature00950. [DOI] [PubMed] [Google Scholar]
- Broun P. Transcriptional control of flavonoid biosynthesis, a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis . Curr. Opin. Plant Biol. 2005;8:272–279. doi: 10.1016/j.pbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Byrne PF, Darrah LL, Snook ME, Musket TA, Theuri JM, Widstrom NW, Moellenbeck DJ, Barry BD. Maize silk-browning, maysin content, and antibiosis to the corn earworm, Helicoverpa zea (Boddie) Maydica. 1996a;41:13–18. [Google Scholar]
- Byrne PF, McMullen MD, Snook ME, Musket TA, Theuri JM, Widstrom NW, Wiseman BR, Coe EH., Jr Quantitative trait loci and metabolic pathways: Genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc. Natl. Acad. Sci. U.S.A. 1996b;93:8820–8825. doi: 10.1073/pnas.93.17.8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey CC, Strahle JT, Selinger DA, Chandler VL. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct penotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana . Plant Cell. 2004;16:450–464. doi: 10.1105/tpc.018796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A, Biggin MD. A comparison of in vivo and in vitro DNA-binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. EMBO J. 1999;18:1598–1608. doi: 10.1093/emboj/18.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB. The phenylpropanoid pathway is controlled at different braches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 2015;167:1448–1470. doi: 10.1104/pp.114.256172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D’Ascenzo MD, Fobert PR, Despres C, Martin GB. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell. 2003;15:3033–3050. doi: 10.1105/tpc.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Bishop KJ, Hall TC. Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J. 2003;33:853–866. doi: 10.1046/j.1365-313x.2003.01678.x. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R. The JAZ family of repressors is the missing link in jasmonate signaling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Clarke DB. Glucosinolates, structures and analysis in food. Anal. Methods. 2010;2:310–325. [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, Schuurink RC, Clark DG. PhMYB4 fine-tunes the floral volatile signature of Petunia x hybrida through PhC4H. J. Exp. Bot. 2011;62:1133–1143. doi: 10.1093/jxb/erq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuyisteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Davin LB, Jourdes M, Patten AM, Kim K, Vassão DG, Lewis NG. Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat. Prod. Rep. 2008;25:1015–1090. doi: 10.1039/b510386j. [DOI] [PubMed] [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012;17:349–359. doi: 10.1016/j.tplants.2012.03.001. [DOI] [PubMed] [Google Scholar]
- DellaPenna D, Last RL. Genome-enabled approaches shed new light on plant metabolism. Science. 2008;320:479–481. doi: 10.1126/science.1153715. [DOI] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006;140:499–511. doi: 10.1104/pp.105.067231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T, Fukuda H. Transcriptional regulation in wood formation. Trends Plant Sci. 2007;12:64–70. doi: 10.1016/j.tplants.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck-Reichhart D, DeLorenzo G, Ferrari S, Ausubel FM, Dewdney J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant. 2008;1:423–445. doi: 10.1093/mp/ssn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA. Natural products and plant disease resistance. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- Dooner HK, Robbins TP, Jorgensen RA. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 1991;25:173–199. doi: 10.1146/annurev.ge.25.120191.001133. [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana . Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis . Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Elliger CA, Chan BG, Waiss AC, Lundin RE, Haddon WF. C-Glycosyl flavones from Zea mays that inhibit insect development. Phytochemistry. 1980;19:293–297. [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis . Development. 2003;130:5885–5894. doi: 10.1242/dev.00812. [DOI] [PubMed] [Google Scholar]
- Evert RF, Esau K, Esau KPA. Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development. 3rd. Hoboken, NJ: Wiley-Interscience; 2006. Esau’s Plant Anatomy. [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, Pauwels L, Witters E, Puga MI, Paz-Ares J, Goossens A, Reymond P, De Jaeger G, Solano R. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornalé S, Sonbol FM, Maes T, Capellades M, Puigdoménech P, Rigau J, Caparrós-Ruiz D. Down-regulation of the maize and Arabidopsis thaliana Caffeic acid O-methyltransferase genes by two new maize R2R3-MYB transcription factors. Plant Mol. Biol. 2006;62:809–823. doi: 10.1007/s11103-006-9058-2. [DOI] [PubMed] [Google Scholar]
- Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, Fuguet E, Torres JL, Rovira P, Puigdoménech P, Rigau J, Grotewold E, Gray J, Caparrós-Ruiz D. ZmYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 2010;64:633–644. doi: 10.1111/j.1365-313X.2010.04363.x. [DOI] [PubMed] [Google Scholar]
- Fornalé S, Lopez E, Salazar-Henao JE, Fernández-Nohales P, Rigau J, Caparros-Ruiz D. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana . Plant Cell Physiol. 2014;55:507–516. doi: 10.1093/pcp/pct187. [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Sisó-Terraza P, Sudre D, Savirón M, Reyt G, Gamard F, Abadía A, Abadía J, Alvarez-Fernández A, Briat JF. Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 2013;201:155–167. doi: 10.1111/nph.12471. [DOI] [PubMed] [Google Scholar]
- Franken P, Schrell S, Peterson PA, Saedler H, Wienand U. Molecular analysis of protein domain function encoded by the myb-homologous maize genes C1, Zm1 and Zm38 . Plant J. 1994;6:21–30. doi: 10.1046/j.1365-313x.1994.6010021.x. [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Böttcher C, Baatout D, Gigolashvili T. Glucosinolates are produced in trichomes of Arabidopsis thaliana . Front. Plant Sci. 2012;3:242. doi: 10.3389/fpls.2012.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H, Berger B, Gigolashvili T. bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 2014a;166:349–369. doi: 10.1104/pp.114.240887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H, Gigolashvili T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana . Mol. Plant. 2014b;7:814–828. doi: 10.1093/mp/ssu004. [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG Gene Is Required to Specify Epidermal Cell Fate and Cell Patterning in the Arabidopsis Root. Dev. Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- Gerats T, Strommer J. Petunia: Evolutionary, Developmental and Physiological Genetics. 2nd. Berlin: Springer; 2009. [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Müller C, Weisshaar B, Flügge UI. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana . Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Müller C, Flügge UI. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana . Plant J. 2007;51:247–261. doi: 10.1111/j.1365-313X.2007.03133.x. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Müller C, Flügge UI. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana . New Phytol. 2008;177:627–642. doi: 10.1111/j.1469-8137.2007.02295.x. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Müller C, Flügge UI. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana . New Phytol. 2008;177:627–642. doi: 10.1111/j.1469-8137.2007.02295.x. [DOI] [PubMed] [Google Scholar]
- Goff SA, Klein TM, Roth BA, Fromm ME, Cone KC, Radicella JP, Chandler VL. Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 1990;9:2517–2522. doi: 10.1002/j.1460-2075.1990.tb07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. Functional analysis of the transcriptional activator encoded by the maize B gene, evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, Lapierre C, Pollet B, Verhaegen D, Chaubet-Gigot N, Grima-Pettenati J. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 2005;43:553–567. doi: 10.1111/j.1365-313X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Mendenhall J, Huo Y, Lloyd A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009;325:412–421. doi: 10.1016/j.ydbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Chamberlin M, Snook M, Siame B, Butler L, Swenson J, Maddock S, St Clair G, Bowen B. Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell. 1998;10:721–740. [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL. Identification of the residues in the MYB domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13579–13584. doi: 10.1073/pnas.250379897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb CD, Abel S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006;11:89–100. doi: 10.1016/j.tplants.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol. 2004;7:465–471. doi: 10.1016/j.pbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Hachez C, Ohashi-Ito K, Dong J, Bergmann DC. Differentiation of Arabidopsis guard cells: analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol. 2011;155:1458–1472. doi: 10.1104/pp.110.167718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel JM, Yeung EC, Facchini PJ. Got milk? The secret life of laticifers. Trends Plant Sci. 2008;13:631–639. doi: 10.1016/j.tplants.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt R. PHYTOALEXINS: What have we learned after 60 years? Annu. Rev. Phytopathol. 1999;37:285–306. doi: 10.1146/annurev.phyto.37.1.285. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Yamada K, Kosemura S, Yamamura S, Hasegawa K. Photropic stimulation induces the conversion of glucosinolate to phototropism-regulating substances of radish hypocotyls. Phytochemistry. 2000;54:275–279. doi: 10.1016/s0031-9422(00)00080-7. [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003;20:737–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- Heinze M, Brandt W, Marillonnet S, Roos W. “Self” and “non-self” in the control of phytoalexin biosynthesis: plant phospholipases A2 with alkaloid-specific molecular fingerprints. Plant Cell. 2015;27:448–462. doi: 10.1105/tpc.114.135343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, Goda H, Nishizawa OI, Shibata D, Saito K. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogge LR, Reed DW, Underhill EW, Haughn GW. HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thaliana and their identification using thermospray liquid chromatography/mass spectrometry. J. Chromatogr. Sci. 1988;26:551–556. [Google Scholar]
- Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, Rausch T, Lucchin M, Boss PK, Dry IB, Bogs J. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera . Plant Cell. 2013;25:4135–4149. doi: 10.1105/tpc.113.117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H, Chadchawan S, Winge P, Thangstad OP, Bones AM. Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 2002;128:1180–1188. doi: 10.1104/pp.010925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeandet P, Delaunois B, Conreux A, Donnez D, Nuzzo V, Cordelier S, Clément C, Courot E. Biosynthesis, metabolism, molecular engineering, and biological functions of stilbene phytoalexins in plants. Biofactors. 2010;36:331–341. doi: 10.1002/biof.108. [DOI] [PubMed] [Google Scholar]
- Jiang C, Gu J, Chopra S, Gu X, Peterson T. Ordered origin of the typical two and three repeat MYB genes. Gene. 2004;326:13–22. doi: 10.1016/j.gene.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens JJ, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai K, Mizutani M, Kawamura N, Yamamoto R, Tamai M, Yamaguichi H, Sakata K, Shimizu B. Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana . Plant J. 2008;55:989–999. doi: 10.1111/j.1365-313X.2008.03568.x. [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar P, Pajer P, Sedlak D, Tamaoki T, Dvorak M. c-Myb inhibits myogenic differentiation through repression of MyoD. Exp. Cell. Res. 2005;309:419–428. doi: 10.1016/j.yexcr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Kerwin RE, Jiménez-Gómez JM, Fulop D, Harmer SL, Maloof JN, Kliebenstein DJ. Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell. 2011;23:471–485. doi: 10.1105/tpc.110.082065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissen R, Rossiter JT, Bones AM. The “mustard oil bomb”: not so easy to assemble?! Localization, expression and distribution of the components of the myrosinase enzyme system. Phytochem. Rev. 2009;8:69–86. [Google Scholar]
- Kliebenstein D, Droymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 2001a;126:811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, Lambrix V, Reichelt M, Gershenzon J, Mitchell-Olds T. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001b;13:681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, D’Auria J, Behere A, Kim J, Gunderson K, Breen J, Lee G, Gershenzon J, Last R, Jander G. Characterization of seed-specific benzoyloxyglucosinolate mutations in Arabidopsis thaliana . Plant J. 2007;51:1062–1076. doi: 10.1111/j.1365-313X.2007.03205.x. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ. Making new molecules - evolution of structures for novel metabolites in plants. Curr. Opin. Plant Biol. 2013;16:112–117. doi: 10.1016/j.pbi.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Yamamoto NG, Hirochika H. Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin - color mutants. J. Japan Soc. Hort. Sci. 2005;74:196–203. [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Davies A, Deeken R, Thorpe MR, Tomos AD, Hedrich R. Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 2000;124:599–608. doi: 10.1104/pp.124.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Gibson TM, Cramer R, Stain C. Glucosinolate-accumulating S-cells in Arabidopsis leaves and flower stalks undergo programmed cell death at early stages of differentiation. Plant J. 2010;64:456–469. doi: 10.1111/j.1365-313X.2010.04339.x. [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Cramer R. Single-cell proteomic analysis of glucosinolate-rich S-cells in Arabidopsis thaliana . Methods. 2011;54:413–423. doi: 10.1016/j.ymeth.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Kosma DK, Murmu J, Razeq FM, Santos P, Bourgault R, Molina I, Rowland O. AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J. 2014;80:216–229. doi: 10.1111/tpj.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, Smeekens S, Tonelli C, Weisshaar B. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana . Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Lloyd AM, Paparozzi ET, Marks MD. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell. 1994;6:1065–1076. doi: 10.1105/tpc.6.8.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Walbot V. Translational genomics for bioenergy production from fuelstock grasses: Maize as the model species. Plant Cell. 2007;19:2091–2094. doi: 10.1105/tpc.107.053660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Lee MW, Qi M, Yang Y. A novel jasmonic acid-inducible rice MYB gene associates with fungal infection and host cell death. Mol. Plant-Microbe Interact. 2001;14:547–535. doi: 10.1094/MPMI.2001.14.4.527. [DOI] [PubMed] [Google Scholar]
- Lee S, Kaminaga Y, Cooper B, Pichersky E, Dudareva N, Chapple C. Benzoylation and sinapoylation of glucosinolate R-groups in Arabidopsis. Plant J. 2012;72:411–422. doi: 10.1111/j.1365-313X.2012.05096.x. [DOI] [PubMed] [Google Scholar]
- Legay S, Lacombe E, Giocoechea M, Briére C, Séguin A, Mackay J, Grima-Pettenati J. Molecular characterisation of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci. 2007;173:542–549. [Google Scholar]
- Li M, Sack FD. Myrosin idioblast cell fate and development are regulated by the Arabidopsis transcription factor FAMA, the auxin pathway, and vesicular trafficking. Plant Cell. 2014;26:4053–4066. doi: 10.1105/tpc.114.129726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development and trichome morphogenesis. Plant Cell. 2009;21:72–89. doi: 10.1105/tpc.108.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Ai Q, Yu D. MYB82 functions in regulation of trichome development in Arabidopsis . J. Exp. Bot. 2014;65:3215–3223. doi: 10.1093/jxb/eru179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana . Curr. Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Liu T, Ohashi-Ito K, Bergmann DC. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development. 2009;136:2265–2276. doi: 10.1242/dev.032938. [DOI] [PubMed] [Google Scholar]
- Luthy B, Matile P. The mustard oil bomb: rectified analysis of the subcellular organization of the myrosinase system. Biochem. Physiol. Pflanzen. 1984;179:5–12. [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- MacAlister CA, Bergmann DC. Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol. Dev. 2011;13:182–192. doi: 10.1111/j.1525-142X.2011.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malitsky S, Blum E, Less H, Venger I, Elbaz M, Morin S, Eshed Y, Aharoni A. The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 2008;148:2021–2049. doi: 10.1104/pp.108.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JW. Antimicrobial compounds and resistance: the role of phytoalexins and phytoanticipins. In: Slusarenko A, Fraser R, Loon van L, editors. Mechanisms of Resistance to Plant Diseases. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 325–370. [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Martínez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- Matile PH. ‘Die senfölbombe’: Sur kompartimentierung des myrosinase systems. Biochem. Physiol. Pflanzen. 1980;14:327–335. [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R. The BOTRYTIS SUSCEPTIBLE1 gene encodes and R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 2003;15:2551–2565. doi: 10.1105/tpc.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Morohashi K, Casas MI, Ferreyra MLF, Mejía-Guerra MK, Pourcel L, Yilmaz Al, Feller A, Carvalho B, Emiliani J, Rodriquez E, Pellegrinet S, McMullen M, Casati P, Grotewold E. A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell. 2012;24:2745–2764. doi: 10.1105/tpc.112.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LJ, Perazza DE, Juda L, Campbell MM. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 2004;37:239–250. doi: 10.1046/j.1365-313x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011;62:2143–2154. doi: 10.1093/jxb/erq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell. 2006;18:2493–2505. doi: 10.1105/tpc.106.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- Patra B, Schluttenhofer C, Wu Y, Pattanaik S, Yuan L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta. 2013;1829:1236–1247. doi: 10.1016/j.bbagrm.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, Campbell MM. Characterisation of a pine MYB that regulates lignification. Plant J. 2003a;36:743–754. doi: 10.1046/j.1365-313x.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, Bevan MW, Sederoff RR, Campbell MM. Characterisation of PtmYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 2003b;53:597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GLI and TTGI. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell. 2001;13:2777–2791. doi: 10.1105/tpc.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. AtMYB32 is required for normal pollen development in Arabidopsis thaliana . Plant J. 2004;40:979–995. doi: 10.1111/j.1365-313X.2004.02280.x. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BA, Goff SA, Klein TM, Fromm ME. C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites. Plant Cell. 1991;3:317–325. doi: 10.1105/tpc.3.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana . Plant Cell. 2011;23:1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Tan G, Liu H, He S, Gao Y, An C. Identification of a bHLH-type G-box binding factor and its regulation activity with G-box and Box I elements of the PsCHS1 promoter. Plant Cell Rep. 2007;26:85–93. doi: 10.1007/s00299-006-0202-x. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell. 2006;18:1274–1291. doi: 10.1105/tpc.105.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quidde T, Osbourn AE, Tudzynski P. Detoxification of alpha-tomatine by Botrytis cinerea . Physiol. Mol. Plant. Pathol. 1998;52:151–165. [Google Scholar]
- Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: Comparison of consensus versus nonconsensus binding sites. Genome Res. 2008;18:527–44. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, van der Peer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay N, Glover B. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Ran JH, She TT, Liu WJ, Wang XQ. Evolution of the bHLH genes involved in stomatal development: implications for the expansion of developmental complexity of stomata in land plants. PLoS One. 2013;8:e78997. doi: 10.1371/journal.pone.0078997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzka A, Vogel H, Kliebenstein DJ, Mitchell-Olds T, Kroymann J. Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11223–11228. doi: 10.1073/pnas.172112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt M, Brown P, Schneider B, Oldham N, Stauber E, Tokuhisa J, Kliebenstein D, Mitchell-Olds T, Gershenzon J. Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana . Phytochemistry. 2002;59:663–671. doi: 10.1016/s0031-9422(02)00014-6. [DOI] [PubMed] [Google Scholar]
- Reid LM, Mather DE, Arnason JT, Hamilton RI, Bolton AT. Changes in phenolic constituents of maize silk infected with Fusarium graminearum . Can. J. Bot. 1992;70:1697–1702. [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana . Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Meyer K, Cusumano JC, Chapple C. Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol. 1999;119:101–110. doi: 10.1104/pp.119.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S. Overexpression of the AtmybL2 gene represses trichome development in Arabidopsis. DNA Res. 2002;9:31–34. doi: 10.1093/dnares/9.2.31. [DOI] [PubMed] [Google Scholar]
- Schmid NB, Giehl RF, Doll S, Mock HP, Strehmel N, Scheel D, Kong X, Hider RC, von Wirén N. Feruloyl-CoA 6’-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 2014;164:160–172. doi: 10.1104/pp.113.228544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F, Fernández-Calvo P, Zander M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell. 2013;25:3117–3132. doi: 10.1105/tpc.113.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger DA, Chandler VL. A mutation in the pale aleurone color1 gene identifies a novel regulator of the maize anthocyanin pathway. Plant Cell. 1999;11:5–14. doi: 10.1105/tpc.11.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Joo J, Kim MJ, Kim YK, Nahm BH, Song SI, Cheong JJ, Lee JS, Kim JK, Choi YD. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011;65:907–921. doi: 10.1111/j.1365-313X.2010.04477.x. [DOI] [PubMed] [Google Scholar]
- Serna L, Martin C. Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 2006;11:1360–1385. doi: 10.1016/j.tplants.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Shelton D, Stranne M, Mikkelsen L, Pakserescht N, Welham T, Hiraka H, Tabata S, Sato S, Paquette S, Wang TL, Martin C, Bailey P. Transcription factors of Lotus: regulation of isoflavonoid biosynthesis requires coordinated changes in transcription factor activity. Plant Physiol. 2012;159:531–547. doi: 10.1104/pp.112.194753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa M, Ueda H, Nagano AJ, Shimada T, Kohchi T, Hara-Nishimura I. FAMA is an essential component for the differentiation of two distinct cell types, myrosin cells and guard cells, in Arabidopsis. Plant Cell. 2014;26:4039–4052. doi: 10.1105/tpc.114.129874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–671. doi: 10.1046/j.1365-313x.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- Shroff R, Vergara F, Muck A, Svatos A, Gershenzon J. Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6196–6201. doi: 10.1073/pnas.0711730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 2007;311:566–578. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Snyder BA, Nicholson RL. Synthesis of phytoalexins in sorghum as a site-specific response to fungal ingress. Science. 1990;248:1637–1639. doi: 10.1126/science.248.4963.1637. [DOI] [PubMed] [Google Scholar]
- Sonbol FM, Fornalé S, Capellades M, Encina A, Touriño S, Torres JL, Rovira P, Ruel K, Puigdoménech P, Rigau J, Caparrós-Ruiz D. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana . Plant Mol. Biol. 2009;70:283–296. doi: 10.1007/s11103-009-9473-2. [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ. A systems biology approach identified a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PloS One. 2007;2:e1322. doi: 10.1371/journal.pone.0001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 2010;15:283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. The Jasmonate-ZIM-domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis . Plant Cell. 2011;23:1000–1013. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Weber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana . Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors control flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles ED, Ceska O. Genetic control of 3-hydroxy- and 3-deoxy-flavonoids in Zea mays . Phytochemistry. 1975;14:413–415. [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R, Iwata M, Okada K, Wada T. Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell. 2007;19:2264–2277. doi: 10.1105/tpc.106.045732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerck JA, Fromm ME. Elements of the maize A1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell. 1994;6:1655–1663. doi: 10.1105/tpc.6.11.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Nishiyama C, Shimada T, Koumoto Y, Hayashi Y, Kondo M, Takahashi T, Ohtomo I, Nishimura M, Hara-Nishimura I. AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 2006;47:164–175. doi: 10.1093/pcp/pci232. [DOI] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidés C, Roby D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10179–10184. doi: 10.1073/pnas.152047199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia ER, Herrera MT, Gianzo C, Fidalgo J, Revilla G, Zarra I, Sampedro J. Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon . J. Exp. Bot. 2013;64:1333–1343. doi: 10.1093/jxb/ers394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. The jasmonate-indicible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 2001;25:43–53. doi: 10.1046/j.1365-313x.2001.00932.x. [DOI] [PubMed] [Google Scholar]
- van Meerder Meer IM, Stuitje AR, Mol JNM. Regulation of general phenylpropanoid and flavonoid gene expression. In: Verma DPS, editor. Control of Plant Gene Expression. Boca Raton, FL: CRC Press; 1993. pp. 125–155. [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010;153:895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, Welsh L, Haustraete J, McClellan C, Vanholme B, Ralph J, Simpson GG, Halpin C, Boerjan W. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science. 2013;341:1103–1106. doi: 10.1126/science.1241602. [DOI] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Waiss AC, Jr, Chan BG, Elliger CA, Wiseman BR, McMillian WW, Widstrom NW, Zuber MS, Keaster AJ. Maysin, a flavone glycoside from corn silks with antibiotic activity toward corn earworm. J. Econ. Entomol. 1979;72:256–258. [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1349. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007;49:772–785. doi: 10.1111/j.1365-313X.2006.02997.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, Chen JG. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development. 2007;134:3873–3882. doi: 10.1242/dev.009597. [DOI] [PubMed] [Google Scholar]
- Weston K. Myb proteins in life, death and differentiation. Curr. Opin. Genet. Dev. 1998;8:76–81. doi: 10.1016/s0959-437x(98)80065-8. [DOI] [PubMed] [Google Scholar]
- Williams CE, Grotewold E. Differences between plant and animal Myb domains are fundamental for DNA binding activity, and chimeric Myb domains have novel DNA binding specificities. J Biol. Chem. 1997;272:563–571. doi: 10.1074/jbc.272.1.563. [DOI] [PubMed] [Google Scholar]
- Wink M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry. 2003;64:3–19. doi: 10.1016/s0031-9422(03)00300-5. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoids in seeds and grains: Physiological function, agronomic importance and the genetics of biosynthesis. Seed Sci. Res. 1998;8:415–422. [Google Scholar]
- Wittstock U, Halkier BA. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002;7:263–270. doi: 10.1016/s1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- Xu Q, Yin XR, Zeng JK, Ge H, Song M, Xu CJ, Li X, Ferguson IB, Chen KS. Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Botany. 2014;65:4349–4359. doi: 10.1093/jxb/eru208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hasegawa T, Minami E, Shibuya N, Kosemura S, Yamamura S, Hasegawa KI. Induction of myrosinase gene expression and myrosinase activity in radish hypocotyls by phototropic stimulation. J. Plant Physiol. 2003;160:255–259. doi: 10.1078/0176-1617-00950. [DOI] [PubMed] [Google Scholar]
- Yan B, Stark RE. Biosynthesis, molecular structure, and domain architecture of potato suberin: a 13C NMR study using isotopically labeled precursors. J. Agric. Food Chem. 2000;48:3298–3304. doi: 10.1021/jf000155q. [DOI] [PubMed] [Google Scholar]
- Yang T, Perasso R, Baroin-Tourancheau A. MYB genes in ciliates: a common origin with the MYB proto-oncogene? Protist. 2003;154:229–238. doi: 10.1078/143446103322166527. [DOI] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Nishiyama MY, Garcia-Fuentes B, Souza GM, Janies D, Gray J, Grotewold E. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 2009;149:171–180. doi: 10.1104/pp.108.128579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Xu F, Zeng J, Zhan J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life. 2012;64:285–295. doi: 10.1002/iub.1005. [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Hanson J, Pieterse CMJ. β-glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 2014;204:368–379. doi: 10.1111/nph.12980. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis . Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y. SAD2, an importin β-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell. 2007;19:3805–3818. doi: 10.1105/tpc.106.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Gallego-Giraldo L, Wang H, Zeng Y, Ding SY, Chen F, Dixon RA. An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula . Plant J. 2010;63:100–114. doi: 10.1111/j.1365-313X.2010.04223.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhou LM, Chen YY, Yang SG, Tian WM. MYC genes with differential responses to tapping, mechanical wounding, ethrel and methyl jasmonate in laticifers of rubber tree (Hevea brasiliensis Muell. Arg.) J. Plant Physiol. 2011;168:1649–1658. doi: 10.1016/j.jplph.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 2007;10:564–572. doi: 10.1016/j.pbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007a;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. Transcriptional regulation of lignin biosynthesis. Plant Signal. Behav. 2009;4:1028–1034. doi: 10.4161/psb.4.11.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 2010a;152:1044–1055. doi: 10.1104/pp.109.148270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010b;15:625–631. doi: 10.1016/j.tplants.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye ZH. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 2011a;52:1856–1871. doi: 10.1093/pcp/pcr123. [DOI] [PubMed] [Google Scholar]
- Zhong R, McCarthy RL, Lee C, Ye ZH. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 2011b;157:1452–1468. doi: 10.1104/pp.111.181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 2012;53:368–380. doi: 10.1093/pcp/pcr185. [DOI] [PubMed] [Google Scholar]
- Zhong R, McCarthy RL, Haghighat M, Ye ZH. The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PloS One. 2013;8:e69219. doi: 10.1371/journal.pone.0069219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, Lee BH, Zhan X, Manabe Y, Sokolchik I, Zhu Y, Dong CH, Zhu JK, Hasegawa PM, Bressan RA. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9966–9971. doi: 10.1073/pnas.0503960102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like bHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]