Abstract
Cardiac induction of human embryonic stem cells (hESCs) is a process bearing increasing medical relevance, yet it is poorly understood from a developmental biology perspective. Anticipated technological progress in deriving stably expandable cardiac precursor cells or in advancing cardiac subtype specification protocols will likely require deeper insights into this fascinating system. Recent improvements in controlling hESC differentiation now enable a near-homogeneous induction of the cardiac lineage. This is based on an optimized initial stimulation of mesoderm-inducing signaling pathways such as Activin and/or FGF, BMP, and WNT, followed by WNT inhibition as a secondary requirement. Here, we describe a comprehensive data set based on varying hESC differentiation conditions in a systematic manner and recording high-resolution differentiation time-courses analyzed by genome-wide expression profiling (GEO accession number GSE67154). As a baseline, hESCs were differentiated into cardiomyocytes under optimal conditions. Moreover, in additional time-series, individual signaling factors were withdrawn from the initial stimulation cocktail to reveal their specific roles via comparison to the standard condition. Hence, this data set presents a rich resource for hypothesis generation in studying human cardiac induction, as we reveal numbers of known as well as uncharacterized genes prominently marking distinct intermediate stages in the process. These data will also be useful for identifying putative cardiac master regulators in the human system as well as for characterizing expandable cardiac stem cells.
1. Direct link to deposited data
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/hESC line HuES6/Differentiating human embryonic stem cells |
| Sex | Female |
| Sequencer or array type | HumanHT-12 v4 |
| Data format | Raw and processed |
| Experimental factors | Variation of cell signaling factor stimulation; Time-course expression profiling at daily intervals during hESC differentiation |
| Experimental features |
hESCs maintained in chemically defined culture medium were replated at high density and simultaneously treated with cardiac mesoderm-inducing factors for one day (20 ng/ml FGF2, 1 ng/ml BMP4, 1 μM CHIR99021). At 48 h, the differentiating cells were treated with WNT inhibitor IWP-2 for 2 days (2 μM). Overall differentiation was performed on Matrigel-coated plates in chemically defined medium devoid of serum, serum albumin, insulin, and complex additives. Furthermore, for investigating the significance of stimulating the three pathways at the beginning of the protocol, three additional time-series were recorded. In these, one signaling factor each was omitted from the initial stimulation cocktail, to reveal its significance for cardiac induction through comparison against the standard condition, respectively. |
| Consent | Not applicable |
| Sample source location | Münster, Germany |
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67154.
2. Experimental design, materials and methods
2.1. Generation and processing of samples
hESCs, cell line HuES6 [1], were maintained in FTDA medium [2], on Matrigel™-coated dishes. Cardiac differentiation was induced as described [3]. In brief, fully confluent hPSC cultures were harvested using Accutase™, and replated onto Matrigel-coated 24-well plates (500,000 cells per well in 2 ml of day 0 differentiation medium). An aliquot of cells was used for RNA isolation (“day 0” = undifferentiated hESCs). Day 0 differentiation medium contained Knockout™ DMEM, insulin/transferrin/selenium, 10 μM Y27632, penicillin/streptomycin/l-Glutamine, 20 ng/ml FGF2, 1 ng/ml BMP4, and 1 μM CHIR99021 (3 - standard condition). Alternatively, FGF2, BMP4, or CHIR were selectively omitted from the signaling stimulation cocktail (factor withdrawal time-courses). Day 1 medium contained Knockout™ DMEM, transferrin/selenium, penicillin/streptomycin/l-Glutamine, and 250 μM phospho-ascorbate (“TS medium”). On days 2 and 3, cells were fed with TS medium supplemented with 2 μM of WNT inhibitor IWP-2 (Fig. 1A). Hence after, cells were maintained in basal TS medium. Spontaneous beating was observed from day 6 onwards. CM differentiation efficiency in the standard time-course was approximately 90% as judged by FACS analysis for cardiac troponin T [4]. Samples were collected from replicate cultures at daily intervals. Total RNA was isolated using Qiagen RNeasy columns with on-column DNA digestion.
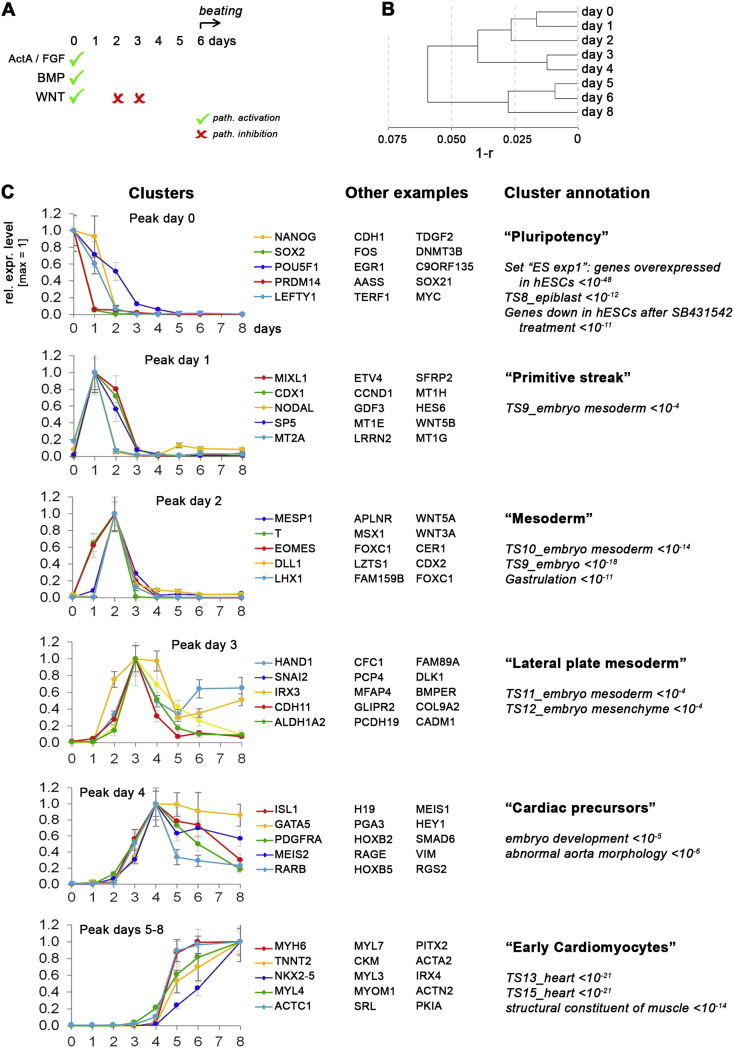
Fig. 1.
Basic analysis of standard cardiac induction time-course. (A) Illustration of the signaling factor treatment protocol used for promoting cardiac induction. (B) Global correlation-based dendrogram of the time-course samples; r = linear transcriptome correlation coefficient. (C) Clustering of gene sets according to temporal expression kinetics. Selected representatives of each cluster are plotted on the left. Error bars indicate bead standard deviation. Selected annotation terms revealed by GREAT are shown on the right along with corresponding P values.
500 ng of total RNA from each biological sample were used as input for the generation of biotin-labelled cRNA using an Illumina® TotalPrep™ RNA amplification kit (Life Technologies). Following the manufacturer's instructions, in-vitro transcription of double-stranded cDNA was performed for 14 h in a PCR cycler. Purified biotin-labelled cRNA was eluted in a volume of 100 μl and quality-checked on a 2100 Bioanalyzer device (Agilent Technologies). cRNA samples were adjusted to 150 ng/μl in water, and hybridized onto Illumina HumanHT-12 v4 bead arrays following the manufacturer's instructions throughout. Hybridization was carried out at 58 °C for 18 h. Staining with streptavidin-Cy3 (GE Healthcare #PA43001) was carried out as recommended, at a concentration 1 μg/ml in blocking buffer. Dried bead arrays were scanned on a HiScan SQ device (Illumina) using default settings.
2.2. Technical and biological data quality assessment
Scanned images were confirmed to show an overall clean fluorescence spot morphology with high signal-to-noise ratio, and array data were confirmed to display an average P95 intensity of > 700 (a.u.). Inspection of raw data in GenomeStudio suggested an overall high hybridization stringency, according to internal mismatch control probes, and no major hybridization artifacts. Following these routine checks, all separately hybridized samples were background-subtracted and normalized using the Cubic Spline algorithm in GenomeStudio. Processed data were filtered in MS Excel by setting experience-based thresholds for expression changes and minimal gene expression levels.
For instance, clusters in Fig. 1C were defined as follows: Peak expression level > 10-fold compared to at least one of the other time-points; Absolute expression level > 300 (a.u.). Similarly, the gene set underlying Fig. 2A was defined by a > 20-fold enrichment on either day 0, day 2, day 4, or day 8, while requiring an absolute signal intensity of > 500 (a.u.). Hierarchical clustering of genes was performed with standardized Euclidean metrics and the Ward linkage method [5]. For functional annotation of filtered gene sets, employing the Ensembl BioMart interface, array probe sequences were converted into GRCh37/hg19 genome coordinates which were then used as input for GREAT analysis [6]. Statistically significant hits were subjectively filtered for biological relevance. Expression levels of individual genes are presented as array intensity signals ± bead standard deviation.
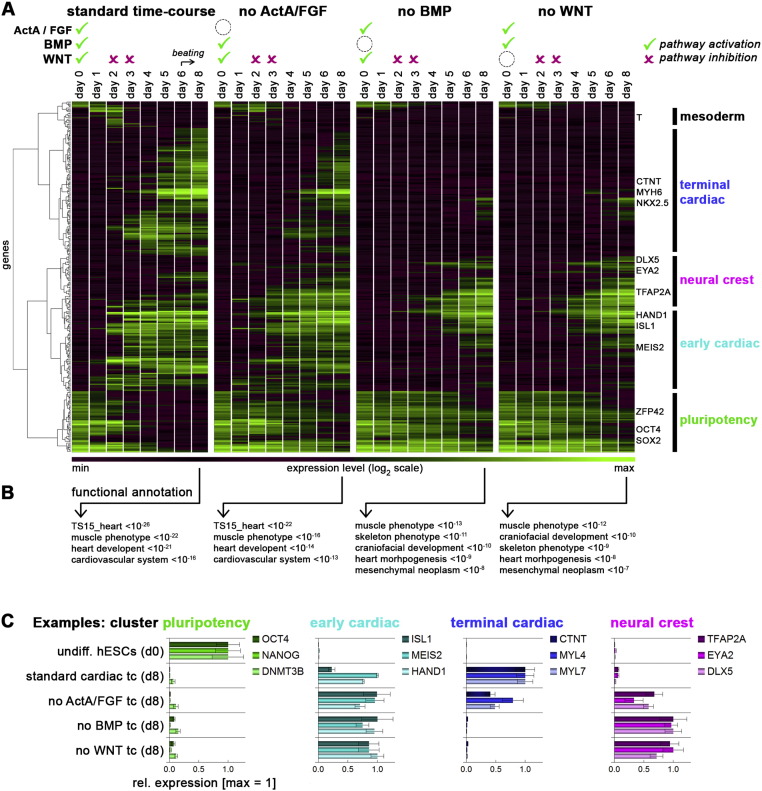
Fig. 2.
Signaling factor withdrawal time-courses. (A) Protocol illustrations (top) and hierarchical clustering of ~ 300 genes across all indicated samples. Designations of major clusters and key representative genes are given on the right. (B) Functional annotation based on enriched gene expression in all day 8 samples using GREAT. (C) Representative genes of color-coded clusters highlighted in part A. Note that the selected set of cardiac precursor genes becomes expressed in all time-series by day 8, whereas terminal differentiation fate differs as shown in the “terminal cardiac” and “neural crest” charts. Error bars indicate bead standard deviation.
As a biological quality control step, known markers were used to assess differential gene expression between samples. In line with the expectations, hPSC-specific genes like OCT4, NANOG and SOX2 were gradually downregulated under standard conditions, albeit with distinct kinetics [4]. Conversely, most known cardiac markers like MYH6, NKX2.5, and TNNT2 were only induced under standard cardiac and, to a lesser degree, under FGF2 withdrawal conditions. Accordingly, the latter samples only yielded a partially beating layer of cells towards the end of the time-course, whereas the BMP and WNT withdrawal cultures did not display any spontaneous beating. Furthermore, as already indicated, FACS analysis confirmed near-homogeneous differentiation into cardiomyocytes in the standard time-course. In addition, RT-qPCR analyses of independent experiments suggested an overall high degree of consistency and reproducibility with regards to kinetic profiles of stage-specific marker genes (data not shown).
2.3. Basic data analysis
Standard cardiac induction time-course. Fig. 1B shows a dendrogram based on global correlation analysis of the standard time-course samples. Of note, the day 5 to 8 samples formed a distinct cluster suggesting that as early as on day 5, the cells may already have reached a state similar to their final cardiomyocyte fate. In line with this idea, upon defining stage-specific clusters, we failed to identify a clear d5-specific gene expression signature. Hence, clusters were defined based on the day of peak expression - day 0, 1, 2, 3, or 4 - with the exception of the final cluster (“peak on days 5–8”). Fig. 1C shows the kinetic profiles of a number of “lead representative genes” and lists additional members, together with cluster-specific functional annotation. Not surprisingly, the first cluster was highly enriched for hESC-specific genes and their downregulation illustrates the loss of pluripotency in response to signaling changes (Fig. 1A). Interestingly, some genes such as PRDM14 displayed extremely rapid downregulation kinetics, perhaps suggesting a direct regulation by the BMP/WNT signaling pathways as in case of SOX2 [4].
Subsequently, based on unbiased functional annotation, the cells rapidly passed through physiological intermediate stages which we termed early “primitive streak-like”, “mesoderm”, and “lateral plate mesoderm-like”. Lead representatives of these clusters were prominent transcriptional regulators such as MIXL1/CDX1/SP5 (peaking on day 1), MESP1/T/EOMES (day 2), and HAND1/SNAI2/IRX3 (day 3), respectively (Fig. 1C). However, this figure also lists a number of uncharacterized genes that may warrant further investigation in the context of identifying cardiac master regulators, for instance. As early as by day 4, the differentiating hESCs then reached a cardiac precursor-like state marked by ISL1, PDGFRA, and MEIS2 which are well-known genes acting in multipotent cardiac progenitors [7], [8], [9]. Accordingly, from day 5 - or even from day 4 in some cases - various regulatory and structural cardiomyocyte-specific genes like NKX2.5, MYH6 (encoding myosin heavy chain 6), and TNNT2 (cardiac troponin T) became upregulated to substantial levels, to cause a spontaneous beating phenotype by day 6 (Fig. 1C, last cluster).
Factor withdrawal time-courses. Fig. 2A illustrates this exciting course of events at a more global scale, by highlighting expression profiles of approximately 300 marker genes. In addition, this figure also contains the signaling factor withdrawal time-courses, which allows to assess the importance and roles of ActA/FGF, BMP, and WNT stimulation at the very beginning of differentiation (see corresponding signaling treatments in the top panel of the figure). Hierarchical clustering of the filtered gene set revealed that BMP or WNT withdrawal essentially disabled all differentiation into cardiomyoctes, whereas omitting ActA/FGF from the day 0 stimulation cocktail only compromised cardiac induction without abolishing it entirely (see “terminal cardiac” cluster in Fig. 2A). Accordingly, the standard as well as the no-ActA/FGF time-courses yielded annotation terms closely linked to cardiomyocyte differentiation, whereas these results were statistically less significant in case of the ActA/FGF withdrawal time-series (Fig. 2B).
In comparison, the no-BMP and no-WNT time-courses displayed a delayed loss of pluripotency gene expression. Moreover, many - albeit interestingly not all - cardiac progenitor genes failed to become induced even until day 8 of differentiation. Instead, we noticed an induction of a small but prominent set of neural crest-associated genes in the BMP and WNT withdrawal samples, suggesting a redirection into this alternative differentiation fate (see “neural crest” cluster in Fig. 2A). This interpretation was also supported by functional annotation terms like “craniofacial development”, “muscle phenotype”, and “skeleton phenotype” (Fig. 2B). To substantiate these observations, Fig. 2C highlights the expression of key markers representing the aforementioned lineages: Strikingly, the d8 no-BMP and no-WNT samples seemed to coexpress neural crest genes TFAP2A/EYA2/DLX5 and cardiac precursor genes ISL1/MEIS2/HAND1, although this could also reflect heterogeneous differentiation in these cultures. In comparison, the standard cardiac time-course did not show any neural crest-like side-products, but the no ActA/FGF cultures displayed an intermediate phenotype characterized both by cardiomyocyte and neural crest-like cell identity (Fig. 2C).
3. Discussion
Our expression time-course analysis illustrates how defined perturbations of a few signaling pathways may induce a complex developmental program leading to the conversion of pluripotent human stem cells into beating cardiomyocyte-like cells within less than one week. This truly remarkable and medically relevant system is poorly studied at present, but the data set introduced here may serve as a useful resource for identifying important - and novel - players in this context. Indeed, in addition to prominent loci known from developmental studies in model organisms, the stage-specific intermediate clusters also contain a number of uncharacterized genes including, for instance, long non-coding RNAs.
Moreover, the signaling factor withdrawal time-courses reveal the importance of each of the extrinsic initiation factors of cardiac induction. Our analysis confirms that the stimulation of the BMP and WNT cascades is most crucial for promoting a cardiomyocyte identify, whereas the FGF and/or Activin/TGFβ pathways play a more accessory role [3], [4]. Somewhat surprisingly, though, both BMP and WNT withdrawal on day 0 still permitted an upregulation of several cardiac progenitor markers. However, some of these - like HAND1 for example - interestingly play dual roles both in cardiac mesoderm and in neural crest induction. Perhaps for this reason, neural crest appeared to be a major differentiation fate competing with cardiac induction in our experimental setting.
Acknowledgments
This work was supported by the Chemical Genomic Centre of the Max Planck Society.
References
- 1.Cowan C.A. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350(13):1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 2.Frank S., Zhang M., Scholer H.R., Greber B. Small molecule-assisted, line-independent maintenance of human pluripotent stem cells in defined conditions. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0041958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang M. Universal cardiac induction of human pluripotent stem cells in two and three-dimensional formats: implications for in vitro maturation. Stem Cells. 2015;33(5):1456–1469. doi: 10.1002/stem.1964. [DOI] [PubMed] [Google Scholar]
- 4.Rao J. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell. 2016;18(3):341–353. doi: 10.1016/j.stem.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Ward J.H. Hierarchical grouping to optimize an objective function. Journal of the American Statistical Association. 1963;58(301):236. [Google Scholar]
- 6.McLean C.Y. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paige S.L. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151(1):221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moretti A. Multipotent embryonic isl1 + progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127(6):1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 9.Kattman S.J. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]