SUMMARY
Cryptococcosis is a leading invasive fungal infection in immunocompromised patients. Considering the high prevalence and severity of these infections in immunocompromised patients attended at HC-FMRP-USP, the present research aimed to characterize the clinical isolates of Cryptococcus strains by biochemical and molecular methods and evaluate antifungal susceptibility of clinical isolates. Fifty isolates from 32 HIV-positive patients were obtained at HC-FMRP-USP. Most of the isolates (78.1%) were identified as C. neoformans, and 100% of C. neoformans and C. gattii strains were susceptible to amphotericin B, ketoconazole and fluconazole. All isolates were classified as serotype A (grubbii variety) by PCR and most of them were characterized in mating type MATa. PCR analysis of specific M13 microsatellite sequence revealed that VNI type was predominant among C. neoformans, while VGII was predominant among C. gattii. The strains did not show a significant resistance to the antifungals tested, and Canavanine-Glycine-Bromthymol Blue Agar (CGB) proved to be a reliable test presenting a good correlation with the molecular characterization. C. neoformans isolated from disseminated infections in the same patient showed molecular identity when different anatomical sites were compared; besides, the studied strains did not present a significant increase in resistance to antifungal agents. In addition, the homogeneity of the molecular types and detection of the mating types suggested a low possibility of crossing among the strains.
KEYWORDS: Cryptococcus neoformans, Serotyping, Genotyping, Fingerprinting, Antifungal agents, Mating type, HIV-patients
INTRODUCTION
Cryptococcus sp is a saprophyte encapsulated yeast that exhibits single or multiple asynchronous buds, and has five serotypes, A, B, C, D and AD. It is subdivided into two varieties known as C. grubii (serotype A) and C. neoformans (serotypes D and AD), while the serotypes B and C are grouped into C. gattii species 1 - 3 . However, there is a new nomenclature classifying C. neoformans as the strains considered var. grubii (serotype A) and C. deneoformans as the var. neoformans (serotype D). In addition, C. gattii will comprise five species: C.gattii, C. bacillisporus, C. deuterogattii, C. tetragattii and C. decagattii 4 , 5 .
Representatives of this genus grow at 37 °C, hydrolyze starch, produce urease and do not ferment lactose 6 . Cryptococcal infections occur worldwide in undefined endemic areas. However, the environmental serotype distribution shows some differences, presenting C. gattii as the prevalent species in tropical and subtropical areas 7 - 9 .
Routinely, C. neoformans and C. gatti cultures are distinguished in CGB-Agar, an enriched medium, which inhibits C. neoformans growth and favors C. gattii development. The color change from yellow-green to blue-cobalt indicates serotypes B and C 10 .
Another important identification method for this species is the sexual mating type analysis. The mating locus MAT of C. neoformans serotype A and D is unique. It is characterized by different regions with very similar structures and functions, which can be also found in other fungi. MAT locus has an average size of 100 Kb, comprising more than 20 genes, some being present in both alleles MATa and MATa, and other genes present in only one of these alleles 11 - 15 . Although diploid or aneuploidy strains are rare, they have already been isolated in nature 16 , 17 .
In C. gatii, it is only possible to determine the sexual type using oligonucleotides flanking the genes MFa1 and MFa2. On the other hand, the serotype B/C is identified by PCR using a primer pair specific for C. gattii superoxide dismutase gene 18 - 19 .
Serotypes A and D, mating type a, are the most common variety causing infections in humans. Although studies with serotypes A, D, AD, B and C have not demonstrated differences in the susceptibility to antifungal agents, the higher incidence of serotypes A, D and AD in HIV-infected patients have suggested a pattern behavior that is demonstrated by clinical and epidemiological studies 20 - 23 .
C. neoformans is the major cause of cryptococcosis affecting millions of people worldwide. In patients with the acquired immunodeficiency syndrome (AIDS) or immunosuppressive conditions, such as organ transplantation or submitted to chemotherapy treatment, cryptococcosis represents the most common fungal infection. During the disease, various strategies are employed to treat these infections, including amphotericin B alone or in combination with 5-flucytosine and azoles, as fluconazole, itraconazole or voriconazole, which are the standard reference drugs nowadays 24 , 25 . Considering the high frequency of cryptococcosis in HIV-infected patients attended at the University Hospital of Ribeirão Preto Medical School, University of Sao Paulo (HC-FMRP-USP), the present investigation aimed to characterize the clinical isolates of Cryptococcus strains by biochemical and molecular methods. The molecular typing, serotyping and mating type identification by molecular biology techniques were used to identify and distinguish the isolates. In addition, the strains were characterized regarding the sensitivity profile to the antifungal agents, amphotericin B, ketoconazole, itraconazole, fluconazole and 5-fluorocytosine.
MATERIAL AND METHODS
Cryptococcus isolates
Fifty isolates collected from 32 patients at HC-FMRP-USP and belonging to the Cryptococcus collection from the Mycology Laboratory - Department of Cell and Molecular Biology, and Pathogenic Bioagents (FMRP-USP) were selected for phenotypic and genotypic studies. The strains were maintained in Sabouraud Dextrose Agar medium (SDA) at room temperature. The strains were previously identified as belonging to the genus Cryptococcus using the classical identification tests such as capsule observation using Indian ink stain and biochemical tests, such as urea degradation and sugars assimilation analysis 26 .
The isolates were divided into three groups as follow: Group I - strains isolated from different patients; Group II - strains isolated from the same patient at the same period, in different anatomical sites; and Group III - strains isolated from the same patient at different periods.
Biochemical characterization of Cryptococcus
The isolates were subcultured in CGB-Agar, pH 5.6, in which the fungi culture presents the typical greenish yellow color according to Kwon-Chung et al. 10 . Isolates were incubated at 37 oC for 48 hours and the reading was performed according to the growth characteristic in this medium: the maintenance of the original medium color is an indication of C. neoformans (serotypes A, D, and AD), while the change to blue-cobalt shade characterizes C.gattii (serotypes B and C). Simultaneously, Cryptococcus reference strains were used as experimental controls: A (CDC 9759), B (ATCC 32269), C (ATCC 24066), and D (ATCC 28958).
DNA isolation
The genomic DNA extraction method was based on the methodology described by Bolano et al. 27 .
Determination of mating type and serotype
The amplification reactions for sexual type and serotype characterization were performed with Taq DNA Polymerase (Fermentas, Thermo Fisher Scientific, Waltham, Massachusetts, USA), according to the manufacturer's instructions in a PTC-200 thermal cycler (MJ Research, GMI Inc., Ramsey, Minnesota, USA), and it was stablish 32 cycles for the PCR. Table 1 presents the primer sequences, annealing temperatures and PCR product sizes according to each pair of pairs 28 , 17 . The amplification products were submitted to electrophoresis on 1% agarose gels in 1X TAE (Tris-Acetate-EDTA) at 75 volts for 2 hours. Gels were stained with ethidium bromide, visualized under UV light and the images were captured by the Alpha-Innotech Image System (Alpha-Innotech Corp., San Leandro, CA, USA) 29 .
Table 1. Identification of primers, primer sequences, annealing temperature, molecular weight of amplification products to determine mating type and serotype and references.
PCR-fingerprinting
The molecular typing of Cryptococcus isolates was performed according to the methodology described by Meyer 30 - 32 , which is based on random amplification of DNA fragments generated by primers recognizing specific minisatellite sequences, allowing a molecular classification defined as VNI, VNII, and VNIII VNIV to C. neoformans, and VGI, VGII, and VGIII to VGIV to C. gattii. The primers were M13 (5'-GAGGGTGGCGGTTCT-3') and (GACA)4 31 . All reactions were performed in a final volume of 50 µL containing 100 ng of genomic DNA, 10 pmol of each primer, 2 mM of MgCl2, 2.5 U of Taq DNA polymerase, 1x PCR Buffer and 10 mM of each dNTP. Amplification conditions were: initial denaturation step at 94 oC for 5 minutes, followed by 35 cycles: denaturation at 94 oC for 1 minute, annealing at 49 oC for 1 minute, extension at 72 oC for 1 minute, followed by a final extension at 72 oC for 10 minutes. The generated amplification products were submitted to electrophoresis on 1% agarose gels as previously described, and the interpretation of results was based on the number and size of the amplification products.
Antifungal susceptibility testing
The susceptibility of C. neoformans, C. gattii and the control strains to antifungal agents was determined by the microdilution plate method, with some modifications based on the protocol recommended by the National Committee for Clinical Laboratory Standards, document (CLSI M27-A2). The drugs used here were amphotericin B (Fungizon, Bristol-Myers-Squibb, Brazil), ketoconazole (Janssen Cilag, Brazil), itraconazole (Janssen Cilag, Brazil), fluconazole (Pfizer, Brazil) and 5-fluorocytosine (Roche, Brazil). The stock solution of each drug was prepared with sterilized water for amphotericin B, fluconazole and 5-fluorocytosine, and DMSO (dimethyl sulfoxide) for ketoconazole and itraconazole, divided in aliquots in sterilized microcentrifuge tubes and stored at -80 oC until used. Two serial dilutions of each antifungal agent were prepared with RPMI 1640 medium (Sigma Chemical Co., Saint Louis, Missouri, USA) with L-glutamine and without sodium bicarbonate, and buffered to pH 7.0 with 0.165 M MOPS (morpholinopropanesulfonic acid). The final concentrations ranged from 0.0312 to 16 mg/mL for amphotericin B; 0.0312 to 16 mg/mL for itraconazole; 0.0625 to 32 mg/mL ketoconazole; 0.25 to 128 mg/mL for 5-fluocytosine and 0.25 to 128 mg/mL fluconazole. The microdilution assay was performed in 96-well microdilution plates. Results were visually evaluated after 72 hours of incubation for C. neoformans isolates and 48 hours for Candida parapsilosis standard sample. In order to validate the test, the standard sample used in all reactions showed minimal inhibitory concentrations (MIC) for each antifungal, within known ranges. The MIC determined for ketoconazole, itraconazole, fluconazole and 5-fluorocytosine corresponded to the concentration that inhibited more than 50% of the fungal growth, compared with the positive control, and for amphotericin B the MIC was considered the concentration showing 100% of growth inhibition. Isolates were considered susceptible or resistant to the tested antifungal agents in accordance to breakpoint values defined by Clinical and Laboratory Standards Institute (CLSI), 2002 33 , 34 .
RESULTS
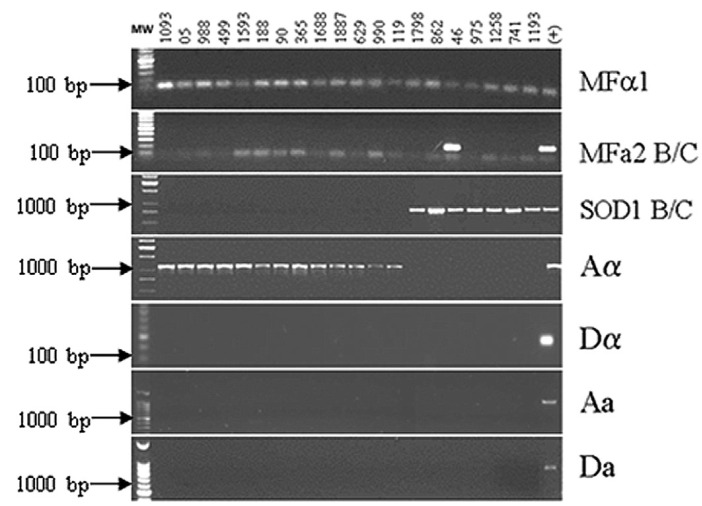
The strains collected from 32 HIV-positive patients comprised 50 isolates, from which 78.1% were phenotipically identified as C. neoformans and 21.9% corresponded to C. gattii species according to biochemical tests. Serotype and sexual type (mating type) characterizations were simultaneously performed by PCR, showing that every sample from group I was classified as MaTa type. From the seven isolates biochemically identified as C. gatti, only one sample (number 46) was positive for the MFa2 primer pair, which is specific for C. gattii sexual type "a". The amplifications for MFa1 and MFa2 showed that this sample is diploid a/a, serotype B/C. The remaining C. gattii isolates were compatible with serotype B/C which are sexual type a. The presence of bands for the gene SXI1Aa in the tested isolates, compared with bands for the genes STE20Da, STE20Aa and SXI2Da in the positive controls showed that all isolates in group I were classified as serotype A and mating type a, which is characteristic of C. neoformans variety grubbi (Fig. 1).
Fig. 1. Group I Cryptococcus isolates. Agarose gel of PCR reactions used for determination of sexual type and serotype. PCR products were obtained using specific primer pairs (MFa1, MFa2 B/C, SOD1 B/C, Aa, Da, Aa and Da). Molecular weight markers are shown on the left side; the lane on the right side indicates the (+) control of each reaction. The remaining lanes represent the studied isolates, identified at the top of the figure by numbers.
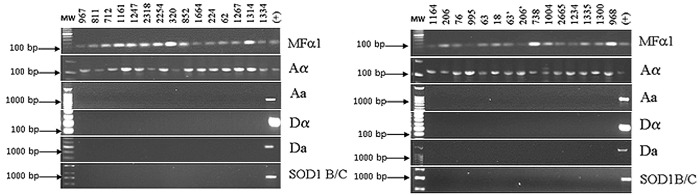
Using the same primers mentioned above, for the sexual typing and serotyping characterization, identical behavior was observed for the Groups II (strains isolated from the same patient in different anatomical sites) and III (strains isolated from the same patient at different periods of observation), all the isolates were characterized as MATa serotype A variety C. grubbi. Indeed, amplification with the SOD1/BC primers was not observed, corroborating the expectations of our previous characterization using the CGB method (Fig. 2).
Fig. 2. Agarose gels showing sexual types and serotypes of Cryptococcus isolates. 2A (on the left) corresponds to Group II samples and 2B (on the right) to Group III samples. PCR products were obtained using specific primers (MFa1, MFa2 B/C, SOD1 B/C, Aa, Da, Aa and Da). The first lane on the left side shows the molecular weight markers; the right lane on the right side shows the (+) control of each reaction. The remaining lanes represent the studied isolates, identified at the top of the figure by numbers.
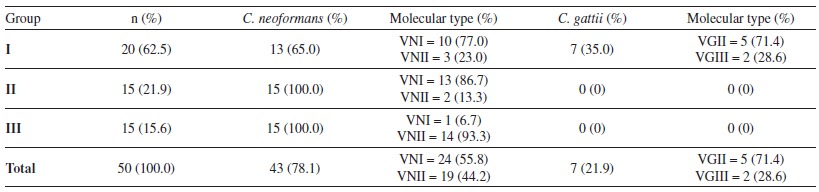
The technique with the specific microsatellite sequence M13, allowed the molecular typing of all the strains and revealed that the VNI molecular type predominated, occurring in 55% of the C. neoformans strains. Concerning C. gattii strains, the VGII molecular type was the most frequent, appearing in 71% of the strains (Table 2). Moreover, for almost all the patients that presented a new disease episode, even after one year without symptoms, the molecular typing results suggested an endogenous reactivation, due to the persistency of the original strain.
Table 2. Isolates of Cryptococcus species (C. neoformans or C. gattii) distributed according to the group (I, II, III), frequencies and molecular type.
The in vitro sensitivity tests performed by microdilution assays showed that from all the studied groups, 100% of C. neoformans and C. gattii strains were susceptible to amphotericin B, ketoconazole and fluconazole. C. neoformans strains presented a dose-dependent sensitivity profile to the antifungal 5-fluorocytosine. A high frequency of MIC values for itraconazole was also observed, corresponding to a susceptible dose-dependent and resistance pattern (Table 3 and 4).
Table 3. Frequency of Cryptococcus isolates identified as C. neoformans or C. gattii, distributed in groups I/II or III, with respect to their susceptibility to the three antifungal agents* (susceptible, intermediate and resistant).
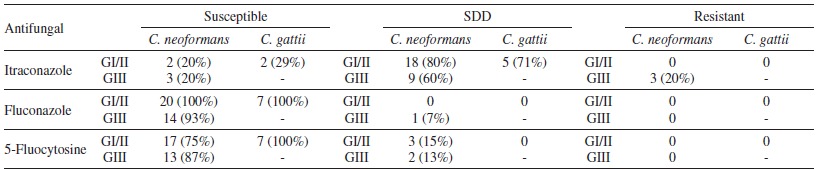
GI/II = group I and II; GIII = group III; SDD = susceptible-dose-dependent; - = group III does not present any C. gattii isolates. * All the isolates were susceptible to Amphotericin B and Fluconazole.
Table 4. Susceptibility test of the samples to five antifungals, and distribution according to the group (I/II and III), breakpoint concentrations, MIC range, MIC 50 and MIC 90.
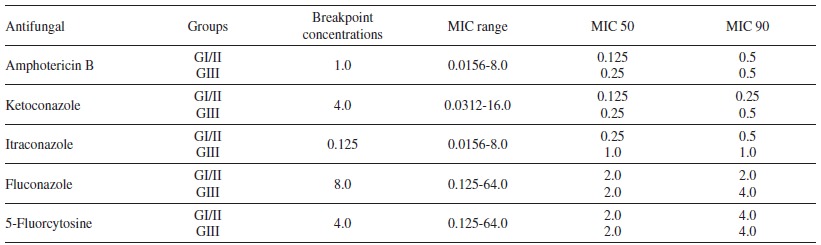
GI/II = groups I and II; GIII = group III. Values are presented in mg/mL. NCCLS MIC breakpoint concentrations.
DISCUSSION
Cryptococcus is a cosmopolitan fungus, isolated from soil, animals and bird excrement. The mycosis caused by this fungus, cryptococcosis, is one of the main diseases affecting especially immunocompromised individuals. In this study, the species characterization of isolates from HIV-infected patients living in the city of Ribeirão Preto, state of São Paulo, Brazil, was performed by analyses of fungal growth in CGB-Agar and molecular characterization. There was a 100% agreement between these two methods for the species identification, showing that the CGB biochemical test is reliable and offers advantages because it is not expensive, is little laborious and provides fast results 17 , 21 , 35 , 36 .
It was also observed that among the 32 patients, the majority of the isolates (78.1%) were identified as C. neoformans, and 21.9% were infected with C. gattii. In several studies in HIV patients, the isolation rate of C. neoformans is around 90%. Regarding C. gattii, overall rates of isolation from clinical material are around 11%, which is higher in immunocompetent patients than in patients with HIV. However, there is no definite explanation for this fact 37 - 44 . The high C. gatti rate found in this work can be explained by the proximity with its natural reservoirs or by the fact that HC-FMRP-USP is a reference center for the treatment of this pandemic infection, which may concentrate more exotic cases of cryptococcosis.
C. gattii was isolated only among patients from Group I, demonstrated by the positive amplification products of SOD1 sequence B/C. All of the groups (I, II and III) showed sexual type MATa prevalence (MFa1 gene recognition), which is consistent with reports of the literature for clinical isolates 35 , 45 . Meanwhile, in this study, only one C. gattii isolate (number 46) was identified and was also recognized by specific primers for the gene MFa1-B/C, resulting in a 213 bp amplification product, thus characterizing the sample as a diploid (MATa / a). Reports on this behavior are rare, especially for serotypes B and C, since the occurrence of diploidy is preferably checked in the case of serotypes A, D and AD isolates (AaDa or AaDa) 2 , 16 .
All the C. neoformans isolates (Groups I, II and III) were characterized as belonging to serotype A, MATa, a characteristic frequently observed in the literature 45 , 46 . For serotype A (C. neoformans var. grubii), Lengeler 16 and Keller 47 suggested that MATa is becoming extinct, because these isolates are usually non fertile or because they belong to an ecological niche that has not yet been found. In the present study, the serotype D was not identified in any of the studied groups, regarding MATa or MATa.
It is known that the mating type condition can influence the fungus virulence since there is a predominance of MATa cells in these patients. Furthermore, these cells are more pathogenic in the murine model, although the MATa type is also common in nature48. Because of the mating gene locus that is not the same among MATa, MATa alleles and serotypes, the molecular characterization is a safer identification method and requires the use of various combinations of primers to determine the serotype, as well as the corresponding species and sexual type. Therefore, this methodology is important since it can indicate differences in virulence and shows differences in the antifungal medical treatment responses. Besides, the knowledge of these variations is essential to understand the infection source, recurrence cases or reinfection and the host immune response 49 , 50 .
In this study, the molecular typing was performed using RAPD-PCR with the specific microsatellite sequences M13 and (GACA)4 according to the definition and molecular types standardization previously established by Meyer et al. 31 . Two C. neoformans molecular types were observed for the M13 specific primer, being the molecular type VNI the predominant, and for C. gattii two molecular types were detected, being the molecular type VGII the most frequent. It is important to emphasize that the C. neoformans serotype A (var. grubii) identification was confirmed, since the VNI and VNII molecular types correspond precisely to this strain, corroborating previous data 51 , 52 . It was possible to observe that with this analysis, four patients had recurrent episodes of cryptococcosis, and one patient was re-infected by a new C. neoformans 53 - 60 .
The MIC values observed for isolates from Groups I and II, regardless of the species, was consistent with the values reported in the literature, excepting for the values obtained for itraconazole 7 , 40 , 61 . No difference was observed between the MIC values for C. neoformans and C. gattii regarding the five antifungal agents tested. They were all in agreement with MIC values reported by Sorrel 39 , but in disagreement with those reported by Fernandes 40 that observed higher MIC values for C. gattii. For group III (patients with recurrent episodes of the disease) a discrepancy of MIC values was observed only for itraconazole, but for the other four drugs, MICs values indicated that 90% of the isolates had the MIC sensitivity profile defined for these drugs.
Amphotericin B alone or in combination with 5-fluorocytosine is the regimen of choice for the treatment of cryptococcal meningitis. This drug alone or the combination is used in the early stages of therapy and is associated with high toxicity. Fluconazole is primarily indicated for secondary prophylaxis of this disease, aiming to reduce the risk of infection recurrence in patients that still maintain the status of immunosuppression 41 , 42 . Ketoconazole and itraconazole are not first-line drugs for the treatment of this disease in Brazil, however, they were included in this research only to investigate the strains behavior regarding these antifungals and to evaluate possible cross-resistance among azoles.
Based on the results obtained in this study, it was possible to conclude that the strains isolated from HIV-patients living in the city of Ribeirão Preto, a region that did not present a significant increase in resistance to the antifungal agents used in the clinical practice, and also that the biochemical test CGB-Agar is reliable given the good correlation between this method and the molecular characterization. Furthermore, the homogeneity of the molecular and mating types detected in these strains indicate a low probability of crossing among strains.
ACKNOWLEDGEMENTS
The authors are thankful for the technical support of Marly de Castro, for the valuable help with the molecular characterization of strains provided by Mateus Terceti and for the financial support from the Coordination for the Improvement of Higher Education Personnel, Brazil - (CAPES - Institutional Quota).
REFERENCES
- 1.Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6:574–587. doi: 10.1111/j.1567-1364.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- 2.Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 3.Romeo O, Scordino F, Chillemi V, Criseo G. Cryptococcus neoformans/Cryptococcus gattii species complex in southern Italy: an overview on the environmental diffusion of serotypes, genotypes and mating-types. Mycopathologia. 2012;174:283–291. doi: 10.1007/s11046-012-9547-6. [DOI] [PubMed] [Google Scholar]
- 4.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Idnurm A, Lin X. Rising to the challenge of multiple Cryptococcus species and the diseases they cause. Fungal Genet Biol. 2015;78:1–6. doi: 10.1016/j.fgb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feder V, Kmetzsch L, Staats CC, Vidal-Figueiredo N, Ligabue-Braun R, Carlini CR, et al. Cryptococcus gattii urease as a virulence factor and the relevance of enzymatic activity in cryptococcosis pathogenesis. FEBS J. 2015;282:1406–1418. doi: 10.1111/febs.13229. [DOI] [PubMed] [Google Scholar]
- 7.Calvo BM, Colombo AL, Fischman O, Santiago A, Thompson L, Lazera M, et al. Antifungal susceptibilities varieties and electrophoretic karyotypes of clinical isolates of Cryptococcus neoformans from Brazil, Chile and Venezuela. J Clin Microbiol. 2001;39:2348–2350. doi: 10.1128/JCM.39.6.2348-2350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halliday CL, Bui T, Krockenberger M, Malik R, Ellis DH, Carter DA. Presence of a and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J Clin Microbiol. 1999;37:2920–2926. doi: 10.1128/jcm.37.9.2920-2926.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida F, Wolf JM, Casadevall A. Virulence-associated enzymes of Cryptococcus neoformans. Eukaryot Cell. 2015;14:1173–1185. doi: 10.1128/EC.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon-Chung KJ, Polacheck I, Benett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var gatti (serotypes B and C) J Clin Microbiol. 1982;15:535–537. doi: 10.1128/jcm.15.3.535-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickes BL. The role of mating type and morphology in Cryptococcus neoformans pathogenesis. Int J Med Microbiol. 2002;292:313–329. doi: 10.1078/1438-4221-00216. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Lin X. Mechanisms of unisexual mating in Cryptococcus neoformans. Fungal Genet Biol. 2011;48:651–660. doi: 10.1016/j.fgb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J. A MAP KINASE cascade composed of cell type specific and non-especific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2003;49:469–485. doi: 10.1046/j.1365-2958.2003.03563.x. [DOI] [PubMed] [Google Scholar]
- 14.Jones SK, Jr, Bennett RJ. Fungal mating pheromones: choreographing the dating. Fungal Genet Biol. 2011;48:668–676. doi: 10.1016/j.fgb.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun S, Hsueh YP, Heitman J. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 2012;8:69. doi: 10.1371/journal.pgen.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lengeler KB, Cox GM, Heitman J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun. 2001;69:115–122. doi: 10.1128/IAI.69.1.115-122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaturvedi S, Rodeghier B, Fan J, McClelland CM, Wickes BL, Chaturvedi V. Direct PCR of Cryptococcus neoformans MATa and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J Clin Microbiol. 2000;3:2007–2009. doi: 10.1128/jcm.38.5.2007-2009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans. var. gattii: implications for an outbreak on Vancouver island, Canada. Eucaryot Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escandón PQ, Quintero E, Granados D, Huérfano S, Castãneda E R. Isolation of Cryptococcus gattii serotype B from detritus of eucalyptus trees in Colombia. Biomedica. 2005;25:390–397. [PubMed] [Google Scholar]
- 20.D'Souza CA, Hagen F, Boekhout T, Cox GM, Heitman J. Investigation of the basis of virulence in serotype A strains of Cryptococcus neoformans from apparently immunocompetent individuals. Curr. Genet. 2004;46:92–102. doi: 10.1007/s00294-004-0511-y. [DOI] [PubMed] [Google Scholar]
- 21.Horta JA, Staats CC, Casali AK, Ribeiro ÂM, Schrank IS, Schrank A, Vainstein MH. Epidemiological aspects of clinical and enviromental Cryptococcus neoformans isolates in the Brazilian state Rio Grande do Sul. Med Mycol. 2002;40:565–571. doi: 10.1080/mmy.40.6.565.571. [DOI] [PubMed] [Google Scholar]
- 22.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, et al. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:1–15. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SH, Choi SC, Lee KW, Kim MN, Hwang SM. Genotypes of clinical and environmental isolates of Cryptococcus neoformans and Cryptococcus gattii in Korea. Mycobiology. 2015;43:360–365. doi: 10.5941/MYCO.2015.43.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loyse A, Dromer F, Day J, Lortholary O, Harrison TS. Flucytosine and cryptococcosis: time to urgently address the worldwide accessibility of a 50-year-old antifungal. J Antimicrob Chemother. 2013;68:2435–2444. doi: 10.1093/jac/dkt221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espinel-Ingroff A, Aller AI, Canton E, Castañón-Olivares LR, Chowdhary A, Cordoba S, et al. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother. 2012;56:5898–5906. doi: 10.1128/AAC.01115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lacaz CS, Porto E, Martins JEC, Heins-Vaccari EM, Takahashi de Melo N. Tratado de micologia médica. 9 ed. São Paulo: Savier; 2002. [Google Scholar]
- 27.Bolano A, Stinchi S, Preziosi R, Bistoni F, Allegrucci M, Baldelli F, Martini A, Cardinalli G. Rapid methods to extract DNA and RNA from Cryptococcus neoformans. FEMS Yeast Res. 2001;1:221–224. doi: 10.1111/j.1567-1364.2001.tb00037.x. [DOI] [PubMed] [Google Scholar]
- 28.Okabayashi K, Kano R, Watanabe T, Hasegawa A. Serotypes and mating types of clinical isolates from feline Cryptococcosis in Japan. J Vet Med Sci. 2006;68:91–94. doi: 10.1292/jvms.68.91. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 3nd ed. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 30.Meyer W, Mitchell TG, Freedman EZ, Vilgalys R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2274–2280. doi: 10.1128/jcm.31.9.2274-2280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methling K, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–1799. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1790::AID-ELPS1790>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E, IberoAmerican Cryptococcal Study Group Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen YC, Chang TY, Liu JW, Chen FJ, Chien CC, Lee CH, et al. Increasing trend of fluconazole-non-susceptible Cryptococcus neoformans in patients with invasive cryptococcosis: a 12-year longitudinal study. BMC Infect Dis. 2015;15:277–277. doi: 10.1186/s12879-015-1023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith KD, Achan B, Hullsiek KH, McDonald TR, Okagaki LH, Alhadab AA, et al. Increased antifungal drug resistance in clinical isolates of Cryptococcus neoformans in Uganda. Antimicrob Agents Chemother. 2015;59:7197–7204. doi: 10.1128/AAC.01299-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McClelland CM, Chang YC, Varma A, Kwon-Chung KJ. Uniqueness of the mating system in Cryptococcus neoformans. Trends in Microbiology. 2004;12:208–212. doi: 10.1016/j.tim.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Melo NT, Lacaz CS, Charbel CE, Pereira AD, Heins-Vaccari EM, Franca-Netto AS, et al. Quimiotipagem do Cryptococcus neoformans. Revisão da literatura. Novos dados epidemiológicos sobre a criptococose. Nossa experiência com o emprego do meio de C.G.B. no estudo daquela levedura. Rev Inst Med Trop Sao Paulo. 1993;35:469–478. [PubMed] [Google Scholar]
- 37.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS: 100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaller MA, Messer SA, Boyken L, Rice C, Tendolkar S, Hollis RJ, et al. Global trends in the antifungal susceptibility of Cryptococcus neoformans (1990 to 2004) J Clin Microbiol. 2005;43:2163–2167. doi: 10.1128/JCM.43.5.2163-2167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorrel TC. Cryptococcus neoformans variety gattii. Med Mycol. 2001;39:155–168. [PubMed] [Google Scholar]
- 40.Fernandes FLO, Passos XS, Souza LK, Miranda AT, Cerqueira CH, Silva MR. In vitro susceptibility characteristics of Cryptococcus neoformans varieties from AIDS patients in Goiania, Brazil. Mem Inst Oswaldo Cruz. 2003;98:839–841. doi: 10.1590/s0074-02762003000600022. [DOI] [PubMed] [Google Scholar]
- 41.Tay ST, Haryanty TT, NG KP, Rohani MY, Hamimah H. In vitro susceptibility of Malaysian clinical isolates of Cryptococcus neoformans var. grubii and Cryptococcus gattii to five antifungal drugs. Mycoses. 2006;49:324–330. doi: 10.1111/j.1439-0507.2006.01242.x. [DOI] [PubMed] [Google Scholar]
- 42.Tewari A, Behera B, Mathur P, Xess I. Comparative analysis of the Vitek 2 antifungal susceptibility system and E-test with the CLSI M27-A3 broth microdilution method for susceptibility testing of Indian clinical isolates of Cryptococcus neoformans. Mycopathologia. 2012;173(5-6):427–433. doi: 10.1007/s11046-012-9528-9. [DOI] [PubMed] [Google Scholar]
- 43.Bejar V, Tello M, García R, Guevara JM, Gonzales S, Vergaray G, et al. Molecular characterization and antifungal susceptibility of Cryptococcus neoformans strains collected from a single institution in Lima, Peru. Rev Iberoam Micol. 2015;32:88–92. doi: 10.1016/j.riam.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Mihara T, Izumikawa K, Kakeya H, Ngamskulrungroj P, Umeyama T, Takazono T, et al. Multilocus sequence typing of Cryptococcus neoformans in non-HIV associated cryptococcosis in Nagasaki, Japan. Med Mycol. 2013;51:252–260. doi: 10.3109/13693786.2012.708883. [DOI] [PubMed] [Google Scholar]
- 45.Barreto de Oliveira MT, Boekhout T, Theelen B, Hagen F, Baroni FA, Lazera MS, et al. Cryptococcus neoformans shows a remarkable genotypic diversity in Brazil. J Clin Microbiol. 2004;42:1356–1359. doi: 10.1128/JCM.42.3.1356-1359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Z, Li X, Xu J. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol. 2002;40:965–972. doi: 10.1128/JCM.40.3.965-972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller SM, Viviani MA, Esposto MC, Cogliati M, Wickes BL. Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolates. Microbiology. 2003;149:131–142. doi: 10.1099/mic.0.25921-0. [DOI] [PubMed] [Google Scholar]
- 48.Barchiesi F, Cogliati M, Esposto MC, Spreghini E, Schimizzi AM, Wickes BL, et al. Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. J Infect. 2005;51:10–16. doi: 10.1016/j.jinf.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Mlinaric-Missoni E, Hagen F, Chew WH, Vazic-Babic V, Boekhout T, Begovac J. In vitro antifungal susceptibilities and molecular typing of sequentially isolated clinical Cryptococcus neoformans strains from Croatia. J Med Microbiol. 2011;60:1487–1495. doi: 10.1099/jmm.0.031344-0. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan D, Haynes K, Moran G, Shanley D, Coleman D. Persistence, replacement, and microevolution of Cryptococcus neoformans strains in recurrent meningitis in AIDS patients. J Clin Microbiol. 1996;34:1739–1744. doi: 10.1128/jcm.34.7.1739-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liaw SJ, Wu HC, Hsueh PR. Microbiological characteristics of clinical isolates of Cryptococcus neoformans in Taiwan: serotypes, mating types, molecular types, virulence factors, and antifungal susceptibility. Clin Microbiol Infect. 2010;16:696–703. doi: 10.1111/j.1469-0691.2009.02930.x. [DOI] [PubMed] [Google Scholar]
- 52.Bertout S, Drakulovski P, Kouanfack C, Krasteva D, Ngouana T, Dunyach-Rémy C, et al. Genotyping and antifungal susceptibility testing of Cryptococcus neoformans isolates from Cameroonian HIV-positive adult patients. Clin Microbiol Infect. 2013;19:763–769. doi: 10.1111/1469-0691.12019. [DOI] [PubMed] [Google Scholar]
- 53.Wu SY, Lei Y, Kang M, Xiao YL, Chen ZX. Molecular characterisation of clinical Cryptococcus neoformans and Cryptococcus gattii isolates from Sichuan province, China. Mycoses. 2015;58:280–287. doi: 10.1111/myc.12312. [DOI] [PubMed] [Google Scholar]
- 54.Kangogo M, Bader O, Boga H, Wanyoike W, Folba C, Worasilchai N, et al. Molecular types of Cryptococcus gattii/Cryptococcus neoformans species complex from clinical and environmental sources in Nairobi, Kenya. Mycoses. 2015;58:665–670. doi: 10.1111/myc.12411. [DOI] [PubMed] [Google Scholar]
- 55.Brito-Santos F, Barbosa GG, Trilles L, Nishikawa MM, Wanke B, Meyer W, et al. Environmental isolation of Cryptococcus gattii VGII from indoor dust from typical wooden houses in the deep Amazonas of the Rio Negro basin. PLoS One. 2015;10:69. doi: 10.1371/journal.pone.0115866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Illnait-Zaragozi MT, Martínez-Machín GF, Fernández-Andreu CM, Perurena-Lancha MR, Hagen F, Meis JF. Cryptococcus and cryptococcosis in Cuba. A minireview. Mycoses. 2014;57:707–717. doi: 10.1111/myc.12275. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y, Litvintseva AP, Frazzitta AE, Haverkamp MR, Wang L, Fang C, et al. Comparative analyses of clinical and environmental populations of Cryptococcus neoformans in Botswana. Mol Ecol. 2015;24:3559–3571. doi: 10.1111/mec.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Badali H, Alian S, Fakhim H, Falahatinejad M, Moradi A, Mohammad Davoudi M, et al. Cryptococcal meningitis due to Cryptococcus neoformans genotype AFLP1/VNI in Iran: a review of the literature. Mycoses. 2015;58:689–693. doi: 10.1111/myc.12415. [DOI] [PubMed] [Google Scholar]
- 59.González GM, Casillas-Veja N, Garza-González E, Hernández-Bello R, Rivera G, Rodríguez JA, et al. Molecular typing of clinical isolates of Cryptococcus neoformans/Cryptococcus gattii species complex from Northeast Mexico. Folia Microbiol (Praha) 2016;61:51–56. doi: 10.1007/s12223-015-0409-8. [DOI] [PubMed] [Google Scholar]
- 60.Dou HT, Xu YC, Wang HZ, Li TS. Molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii in China between 2007 and 2013 using multilocus sequence typing and the DiversiLab system. Eur J Clin Microbiol Infect Dis. 2015;34:753–762. doi: 10.1007/s10096-014-2289-2. [DOI] [PubMed] [Google Scholar]
- 61.Souza LK Fernandes Ode F, Kobayashi CC Passos, XS Costa CR, Lemos JA, et al. Antifungal susceptibilities of clinical and environmental isolates of Cryptococcus neoformans in Goiania city, Brazil. Rev Inst Med Trop. Sao Paulo. 2005;47:253–256. doi: 10.1590/s0036-46652005000500003. [DOI] [PubMed] [Google Scholar]


