Abstract
Inactivation of ptmB1, ptmB2, ptmT2, or ptmC in Streptomyces platensis SB12029, a platensimycin (PTM) and platen-cin (PTN) overproducer, revealed that PTM and PTN biosynthesis features two distinct moieties that are individually constructed and convergently coupled to afford PTM and PTN. A focused library of PTM and PTN analogues was generated by mutasynthesis in the ΔptmB1 mutant S. platensis SB12032. Of the 34 aryl variants tested, 18 were incorporated with high titers.
Graphical Abstract
Platensimycin (PTM, 1m) and platencin (PTN, 1n) are selective and potent bacterial and mammalian fatty acid synthase inhibitors. They are effective against a broad range of bacterial pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococci (VRE), and Myco-bacterium tuberculosis.1 PTM and PTN have also demonstrated efficacy in animal models as a new class of antibacterial and antidiabetic drug leads.2 However, it is imperative to generate PTM and PTN analogues to improve their poor pharmacokinetics, as they had to be continuously infused into mice in order to achieve antibiotic efficacy in vivo.1b
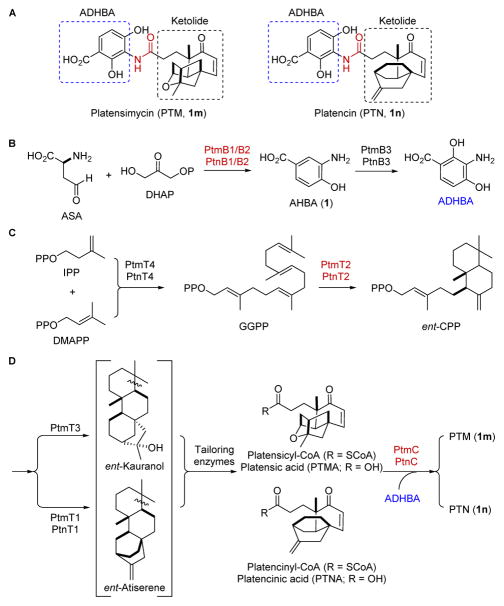
PTM and PTN are comprised of two distinct motifs, a highly modified diterpene-derived aliphatic cage (ketolide) and a 3-amino-2,4-dihydroxybenzoic acid (ADHBA), linked together by an amide bond (Figure 1A).3 We previously cloned and sequenced the biosynthetic gene clusters encoding PTM and PTN production from the PTM-PTN dual producer Streptomyces platensis MA7327 and the PTN producer Streptomyces platensis MA7339, respectively (Figure S1).4 The high similarities between the ptm and ptn gene clusters suggested that a conserved set of enzymes was able to process two different ketolide scaffolds into PTM and PTN. We initially proposed a convergent biosynthetic pathway where (i) one set of enzymes biosynthesizes the ADHBA moiety, (ii) another set of enzymes biosynthesizes the ent-kaurene and ent-atiserene scaffolds and processes them into the penultimate intermediates platensicyl- and platencinyl-CoA, and (iii) a final coupling reaction links ADHBA with platensicyl- or platencinyl-CoA to form PTM and PTN, respectively (Figure 1B–D).4
Figure 1.
A convergent biosynthesis for PTM and PTN. (A) Structures of PTM (1m) and PTN (1n) highlighting the ADHBA and diterpene-derived ketolides moieties linked by an amide bond. (B) Biosynthesis of ADHBA, (C) ent-copalyl diphosphate (ent-CPP), and (D) the ketolides of PTM and PTN, which are coupled to ADHBA in the final step. PtmB1/PtnB1, PtmB2/PtnB2, PtmT2/PtnT2, and PtmC/PtnC are highlighted in red.
We recently discovered six additional dual PTM-PTN producing Streptomyces platensis strains.5 By inactivating the negative transcriptional regulator ptmR1 in S. platensis CB00739, we established a model strain, SB12029, for the study of PTM and PTN.6 The dramatic overproduction of PTM and PTN (up to 300 mg L−1), along with the ability to genetically manipulate SB12029, gave us outstanding opportunities to carry out functional characterization of the PTM and PTN biosynthetic pathway and to explore the promiscuity of the tailoring enzymes found within the biosynthetic machinery to generate PTM and PTN structural diversity.
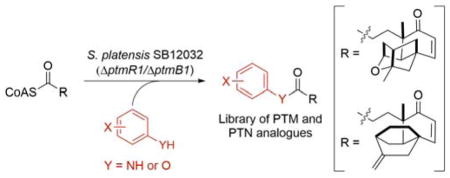
Herein, we first report three production phenotypes that support the convergent nature of the PTM and PTN biosynthetic pathways. Deletion of key genes involved in the biosynthesis of the ADHBA (ptmB1 or ptmB2), ketolide moieties (ptmT2), or the linkage between these two moieties (ptmC), within the PTM-PTN overproducing strain SB12029, resulted in mutant strains that all abolished PTM and PTN production but accumulated ADHBA only (ΔptmT2), ketolides only (ΔptmB1 or ΔptmB2), or both (ΔptmC), respectively, in high titers. We then utilized SB12032, which was unable to biosynthesize 3-amino-4-hydroxybenzoic acid (AHBA, 1), the immediate precursor of ADHBA, to test the capability of the PTM and PTN biosynthetic machinery to couple unnatural benzoate analogues, in vivo, to the ketolide moieties for the preparation of a focused PTM and PTN library.
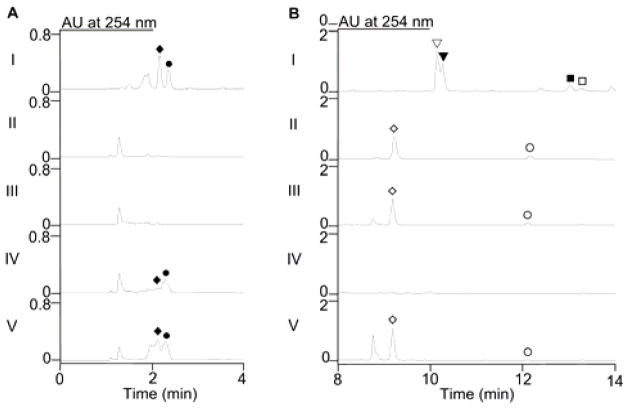
The structures of PTM and PTN both possess an ADHBA moiety, which is biosynthetically constructed by PtmB1/PtnB1, PtmB2/PtnB2, and PtmB3/PtnB3 (Figure 1B). PtmB1/PtnB1 and PtmB2/PtnB2 are proposed to catalyze the formation of 1 from aspartate semialdehyde (ASA) and dihydroxyacetone phosphate (DHAP).4,7 PtmB3/PtnB3 then regiospecifically hydroxylates the C-2 position of 1 to afford ADHBA for both PTM and PTN biosynthesis.8 In an effort to construct an AHBA–blocked mutant in the engineered PTM and PTN overproducer SB12029 (i.e., ΔptmR1), we used λRED-mediated PCR-targeting mutagenesis9 [see Supplementary Information (SI) for experimental details and Tables S1–S3] to inactivate ptmB1 or ptmB2 in SB12029 to afford the double mutants SB12032 (i.e., ΔptmR1/ΔptmB1) and SB12033 (i.e., ΔptmR1/ΔptmB2), respectively (Figures S2 and S3). When cultured under previously described conditions for PTM and PTN production, as well as the sulfur-containing analogues PTM S1 and PTN S1, in SB12029 (Figure 2B, panel I),10 analysis of the crude extract of SB12032 and SB12033 showed that the PTM and PTN production was completely abolished; only platensic (PTMA) and platencinic (PTNA) acids, the hydrolyzed products of platensicyl- and platencinyl-CoA, respectively (Figure 1D), were detected (Figure 2B, panels II and III). Neither 1 nor ADHBA was detected in the aqueous layers of SB12032 or SB12033 (Figure 2A, panels II and III) after extraction (SI). Although the PTNA titers were low in both SB12032 (4.7 ± 0.8 mg L−1) and SB12033 (3.2 ± 0.5 mg L−1), the PTMA titers were high, with 147 ± 17 mg L−1 in SB12032 and 145 ± 9 mg L−1 in SB12033, respectively. Both strains were chemically complemented upon addition of 1 to the fermentation medium, restoring PTM and PTN production and yielding small amounts of 3′-dehydroxy–PTM and –PTN (Figure S6).
Figure 2.
LC-MS analysis of S. platensis recombinant strains. Aqueous layers (A) or crude extracts (B) of each strain were analyzed using UV detection at λ254. (I) SB12029 (CB00739 ΔptmR1); (II) SB12032 (SB12029 ΔptmB1); (III) SB12033 (SB12029 ΔptmB2); (IV) SB12034 (SB12029 ΔptmT2); (V) SB12035 (SB12029 ΔptmC).●, AHBA; ◆, ADHBA; ◇, PTMA; ○, PTNA; ▽, PTM S1; ▼, PTM; □, PTN; ■, PTN S1.
Biosynthesis of the diterpenoid moieties begins with geranyl-geranyl diphosphate (GGPP) synthase (PtmT4/PtnT4) followed by ent-CPP synthase (PtmT2/PtnT2) to form ent-CPP (Figure 1C).4 Two dedicated diterpene synthases, PtmT3 and PtmT1/PtnT1, then partition PTM and PTN biosynthesis by catalyzing the cyclization of ent-CPP into ent-kauranol and ent-atiserene, respectively (Figure 1D).4,11 A group of tailoring enzymes, which must inherently process relaxed substrate specificity, processes both diterpene scaffolds to furnish the final ketolide moieties PTMA and PTNA (Figure 1D). Inactivation of ptmT2 therefore should abolish the production of all ent-CPP-derived diterpenes (i.e., PTMA and PTNA); production of 1 and ADHBA, however, should remain unaffected. Inactivation of ptmT2 in SB12029 afforded SB12034 (i.e., ΔptmR1/ΔptmT2, Figure S4), fermentation of which indeed yielded neither PTM, PTN, PTMA, PTNA, nor any intermediates or congeners previously isolated from mutants or wild-type strains (Figure 2B, IV); however, both 1 and ADHBA were produced (Figure 2A, panel IV). When SB12034 was chemically complemented with (11S,16S)-ent-kauran-11,16-epoxy-19-oic acid,12 a PTMA intermediate isolated from SB12030,6 PTM production was restored (Figure S6).
PtmC/PtnC belong to the arylamine N-acetyltransferases superfamily13 and was proposed to catalyze the final coupling step in PTM and PTN biosynthesis.4 We inactivated ptmC in SB12029 to afford SB12035 (i.e., ΔptmR1/ΔptmC, Figure S5). As expected, fermentation of SB12035 revealed the production of 1 and ADHBA (Figure 2A, panel V), as well as PTMA and PTNA (Figure 2B, panel V). As the ketolide and ADHBA moieties were both present, but the coupled final products PTM and PTN were not detected, PtmC is supported as the amide synthase responsible for the final coupling step in the PTM and PTN biosynthesis.
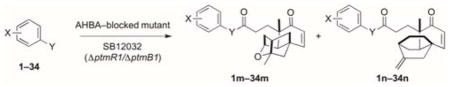
Inspired by the findings that PTM and PTN are biosynthesized in a convergent manner, we set out to produce a focused PTM and PTN library, with varying ADHBA moieties, by a mutasynthesis strategy (Table 1). Thirty-four commercially available aryl variants (Figure S7) were selected to test the capability of the PTM and PTN biosynthetic machinery to couple them to the ketolide moieties in vivo. We supplemented SB12032 with 0.2 mg mL−1 of each aryl variant and analyzed the crude extracts of each of the fermentations by LC-MS to determine whether the coupling was successful. For clarity, each aryl variant is numbered 1–34 and coupled products are numbered 1–34 with an “m” or “n” denotation according to which ketolide scaffold, PTMA or PTNA, was coupled (e.g., 1m and 1n for PTM and PTN, respectively) (Table 1).
Table 1.
Mutasynthesis with aryl variants in SB12032
| |||
|---|---|---|---|
| nr.a | compound | productb | yieldc |
| 3-, 4-, or 2-aminobenzoic acids | |||
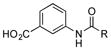
| 1 |
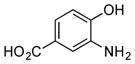
|
|
+++ |
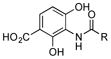
|
++ | ||
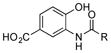
| 2 |
|
|
+++ |
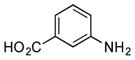
| 3 |
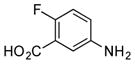
|
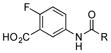
|
+++ (115) |
| 6 |
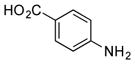
|
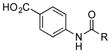
|
+++ (67) |
| 7 |
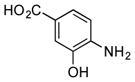
|
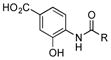
|
+++ (168) |
| 8 |
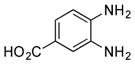
|
|
+++ (22) |
| 13 |
|
|
++ (33) |
| amino/hydroxymethylbenzoic acids | |||
| 17 |
|
|
+ (3.5) |
| 18 |
|
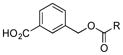
|
+ |
| 19 |
|
|
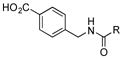
+ (4.2) |
| 20 |
|
|
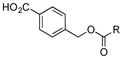
+ |
| heteroarylamines | |||
| 24 |
|
|
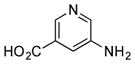
++ (14) |
| arylamines | |||
| 27 |
|
|
++ (14) |
|
+ (3.0) | ||
| 29 |
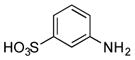
|
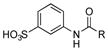
|
+ |
| 31 |
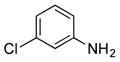
|
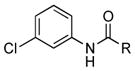
|
+ |
| 32 |
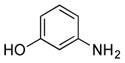
|

|
++ (23) |
| 33 |
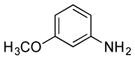
|
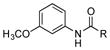
|
+ |
| 34 |
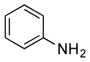
|
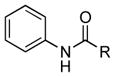
|
++ (21) |
See Figure S7 for full list of tested compounds.
R = PTM or PTN ketolides; see Figure S9 for full structures; see Tables S4–S9 and Figures S11–S34 for NMR data.
Qualitative titers based on LC-MS analysis (+, ++, or +++); titers (mg L−1) are given in parentheses for each isolated PTM product.
Of the 34 aryl variants tested, 18 were successfully coupled to the ketolides (Table 1, Figure S8). The aryl variants tested could be divided into six categories: 3-aminobenzoic acids (meta), 4-amino-benzoic acids (para), 2-aminobenzoic acids (ortho), aminomethyl- or hydroxymethylbenzoic acids (both meta and para), heteroarylamines, and arylamines. Surprisingly, members (1–3, 6–8, 13, 17–20, 24, 27, 29, and 31–34) of each of the six categories were successfully incorporated, indicating that the biosynthetic machinery of PTM and PTN, and specifically PtmC/PtnC, is extremely flexible in regard to its aryl variant substrate. The varying production titers of the coupled products, together with the inability to incorporate the other 16 aryl variants tested, revealed insights into the substrate preference of the coupling reaction.
Considering ADHBA contains a meta amino group, it was unsurprising that 3-aminobenzoic acids (1–3) were readily coupled to the ketolides (Table 1). As an example, when 5-amino-2-fluorobenzoic acid (3) was added to a fermentation of SB12032, two new peaks were detected in the LC-MS chromatogram (Figure S8), the m/z values of which were consistent with the PTM- and PTN-coupled products of 3m and 3n, respectively. The high ketolide titers of SB12032 and excellent incorporation of 3 facilitated the isolation of 3m (115 mg L−1), the structure of which was subsequently confirmed by 1D and 2D NMR spectroscopy (SI). Not all 3-aminobenzoic acids, however, were incorporated into PTM and PTN derivatives. When the 4-OH in AHBA was substituted by 4-OCH3 (4) or 4-Cl (5), no coupled products were detected.
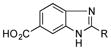
4-Aminobenzoic acids 6–8 were also efficiently incorporated into PTM and PTN analogues. Compounds 6m (67 mg L−1) and 7m (168 mg L−1) were isolated and their structures were confirmed by 1D and 2D NMR spectroscopy (SI). Since both 3-amino and 4-aminobenzoic acids were successfully incorporated with high efficiencies, we tested the preference of meta and para amino substitutions by feeding 3,4-diaminobenzoic acid (8) to SB12032. LC-MS analysis showed high levels of 8m-a and 8n-a (Figure S8). Two minor products (8m-b and 8n-b) were additionally detected, the m/z of which were 18 Da less than to those of 8m-a and 8n-a, respectively, suggesting they may be dehydrated analogues. Unfortunately, 8m-a was extremely unstable during purification and spontaneously transformed into 8m-b. The structure of 8m-b, possessing a unique benzimidazole moiety, was determined by a combination of HRMS and 1D and 2D NMR spectroscopy and confirmed that 8m-b is likely the non-enzymatic dehydration product of 8m-a (SI). Due to the interconversion of the benzimidazole group, we were unable to establish whether the 3- or 4-amide was the dominant coupled product (Figure S10).
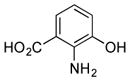
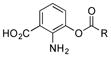
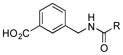
Of the five 2-aminobenzoic acids (9–13) tested, only 2-amino-3-hydroxybenzoic acid (13) was incorporated (Figure S8). Analogue 13m (33 mg L−1) was subsequently isolated and its structure was fully characterized (SI). Consistent with the inability to incorporate 2-aminobenzoic acids, 13m did not contain an amide linkage. Instead, it contained an ester linkage at 3-OH and is the first PTM or PTN analogue with an ester linkage. Encouraged by the ability of the biosynthetic machinery to form ester linkages, three additional 3-hydroxybenzoic acids (14–16) were tested. LC-MS analysis revealed, however, that none of these analogues was converted into PTM or PTN analogues by SB12032. However, two hydroxymethylbenzoic acids (18, 20) and two aminomethylbenzoic acids (17, 19), with either meta and para substitutions, were each successfully incorporated with comparable titers (SI).
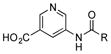
Six heteroarylamines, including a thiazole (21), four pyridines (22–25), and a pyrimidine (26), were added to a culture of SB12032. Only 5-aminonicotinic acid (24) was successfully incorporated with moderate efficiency and led to the isolation of 24m (14 mg L−1, SI), the first pyridine-derived PTM analogue.
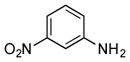
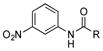
Finally, we investigated whether the carboxylic acid group is compulsory for the coupling reaction. When SB12032 was supplemented with arylamines containing nitro (27), sulfonic acid (29), chloro (31), hydroxyl (32), or methoxyl (33) groups in place of the carboxylic acid, coupled products were detected (Figure S8). Again, nitro- and sulfonic acid-containing analogues with 4–OCH3 groups, 28 and 30, failed to be incorporated. As it appeared that the carboxylic acid was not essential for coupling, we hypothesized that aniline (34) might also be incorporated. This was indeed confirmed by the isolation of 34m (21 mg L−1, SI) after feeding 34 to a fermentation culture of SB12032.
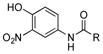
We initially envisioned that if we only inactivated ptmB1 or ptmB2, PtmB3 may hydroxylate 1, or 2–34 should it be promiscuous. However, only two compounds, 1 and 27, appeared to be hydroxylated, leading to additional PTM and PTN analogues. Not surprisingly, 1, the native substrate of PtmB3, was hydroxylated to yield ADHBA. Addition of 3-nitroaniline (27) yielded the expected analogue containing the 3-nitroaniline moiety (27m-a, 14 mg L−1), as well as an analogue containing an additional hydroxyl group on the aniline ring at C-6 (27m-b, 3 mg L−1).
Our efforts to construct a focused library of PTM and PTN analogues also revealed key insights into the permissibility and limitations of the coupling reaction. Overall, this in vivo study supports PtmC as an amide synthase with extreme flexibility for its aryl variant substrate as 18 of the 34 tested aryl variants were incorporated in the biosynthetic pathway and led to the generation of more than 30 PTM and PTN analogues. Three mechanistic implications regarding the coupling reaction were discovered: (i) the benzoic acid is not compulsory; (ii) amide and ester linkages can be formed from meta and para positions, but not ortho; and (iii) large functional groups ortho to the amine prevent coupling. Further biochemical evidence is needed, however, to fully understand the substrate and chemical specificity of the coupling reaction.
Each of the PTM analogues isolated (3m, 6m, 7m, 8m-b, 13m, 17m, 19m, 24m, 27m-a, 27m-b, 32m, and 34m) was tested for antibacterial activity against Micrococcus luteus ATCC 9431 and Staphylococcus aureus ATCC 25923 (SI). Although none of the isolated PTM analogues showed antibacterial activity up to 80 μg per disk, with PTM, at 5 μg per disk, as a positive control, these findings provided additional experimental data supporting the current structure-activity relationship (SAR) for PTM.10,14
In summary, we reported three distinct phenotypes in the PTM and PTN overproducer S. platensis SB12029 by the inactivation of key genes involved in the biosynthesis of the ADHBA (ΔptmB1 or ΔptmB2), ketolide moieties (ΔptmT2), or the linkage (ΔptmC) between these two moieties. The convergent nature and flexibility of the PTM and PTN biosynthetic pathways facilitated construction of a focused library of analogues via mutasynthesis. Although none of the PTM analogues isolated in this study retained the antibacterial activity of PTM, they provide important experimental evidence supporting the current SAR for PTM. Importantly, this study sets the stage to explore additional aryl variants and investigate the feasibility of preparing designer PTM and PTN analogues by a mutasynthetic strategy. Finally, after successfully generating PTM and PTN analogues with modified aminobenzoic acids, one can easily envisage generating structural diversity in the diterpene-derived ketolide moiety. The high and facile production of PTM and PTN analogues by mutasynthesis encourages the continued development of novel unnatural products that will focus on understanding the SAR of PTM and PTN and addressing the poor pharmacokinetics of these natural product drug leads. A focused library of PTM and PTN analogues will also provide opportunities to explore the privileged scaffolds of PTM and PTN for other targets of biological significance.
Supplementary Material
Acknowledgments
We thank the John Innes Center, Norwich, UK, for providing the REDIRECT Technology kit. This work is supported in part by the National Institutes of Health Grants GM114353 (BS). JDR is supported by an Arnold O. Beckman Postdoctoral Fellowship.
Footnotes
The authors declare no competing financial interest.
Complete experimental procedures, Tables S1–S9, Figures S1–S34. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Brown AK, Taylor RC, Bhatt A, Fütterer K, Besra GS. PLoS One. 2009;4:e6306. doi: 10.1371/journal.pone.0006306. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, et al. Proc Natl Acad Sci U S A. 2007;104:7612–7616. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, et al. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 2.Wu M, Singh SB, Wang J, Chung CC, Salituro G, Karanam BV, Lee SH, Powles M, Ellsworth KP, Lassman ME, et al. Proc Natl Acad Sci U S A. 2011;108:5378–5383. doi: 10.1073/pnas.1002588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Jayasuriya H, Herath KB, Zhang C, Zink DL, Basilio A, Genilloud O, Diez MT, Vicente F, Gonzalez I, Salazar O, et al. Angew Chem, Int Ed. 2007;46:4684–4688. doi: 10.1002/anie.200701058. [DOI] [PubMed] [Google Scholar]; (b) Singh SB, Jayasuriya H, Ondeyka JG, Herath KB, Zhang C, Zink DL, Tsou NN, Ball RG, Basilio A, Genilloud O, et al. J Am Chem Soc. 2006;128:11916–11920. doi: 10.1021/ja062232p. [DOI] [PubMed] [Google Scholar]
- 4.(a) Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR, Shen B. Proc Natl Acad Sci U S A. 2011;108:13498–13503. doi: 10.1073/pnas.1106919108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rudolf JD, Dong LB, Cao H, Hatzos-Skintges C, Osipiuk J, Endres M, Chang CY, Ma M, Babnigg G, Joachimiak A, et al. J Am Chem Soc. 2016;138:10905–10915. doi: 10.1021/jacs.6b04317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hindra, Huang T, Yang D, Rudolf JD, Xie P, Xie G, Teng Q, Lohman JR, Zhu X, Huang Y, et al. J Nat Prod. 2014;77:2296–2303. doi: 10.1021/np5006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudolf JD, Dong L-B, Huang T, Shen B. Mol BioSyst. 2015;11:2717–2726. doi: 10.1039/c5mb00303b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki H, Ohnishi Y, Furusho Y, Sakuda S, Horinouchi S. J Biol Chem. 2006;281:36944–36951. doi: 10.1074/jbc.M608103200. [DOI] [PubMed] [Google Scholar]
- 8.(a) Waldman AJ, Pechersky Y, Wang P, Wang JX, Balskus EP. ChemBioChem. 2015;16:2172–2175. doi: 10.1002/cbic.201500407. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sugai Y, Katsuyama Y, Ohnishi Y. Nat Chem Biol. 2016;12:73–75. doi: 10.1038/nchembio.1991. [DOI] [PubMed] [Google Scholar]
- 9.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. Proc Natl Acad Sci U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong L-B, Rudolf JD, Shen B. Bioorg Med Chem. 2016 doi: 10.1016/j.bmc.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smanski MJ, Peterson RM, Huang SX, Shen B. Curr Opin Chem Biol. 2012;16:132–141. doi: 10.1016/j.cbpa.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami T, Iida H, Tanaka N, Saiki Y, Chen CM, Iitaka Y. Chem Pharm Bull. 1981;29:657–662. [Google Scholar]
- 13.Sim E, Sandy J, Evangelopoulos D, Fullam E, Bhakta S, Westwood I, Krylova A, Lack N, Noble M. Curr Drug Metabol. 2008;9:510–519. doi: 10.2174/138920008784892100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicolaou K, Stepan AF, Lister T, Li A, Montero A, Tria GS, Turner CI, Tang Y, Wang J, Denton RM, et al. J Am Chem Soc. 2008;130:13110–13119. doi: 10.1021/ja8044376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.