Abstract
In homeostasis, whether blood cells are derived from committed progenitor or mutipotent stem cell activity remains controversial. In this issue of Immunity, Sawai et al. (2016) describe murine HSCs as the major contributor to the maintenance of multilineage hematopoiesis, both in the steady state and during cytokine response.
Hematopoietic stem cell (HSC) transplantation is an important therapeutic strategy in the treatment of immune and hemato-logical disorders, including leukemia and aplastic anemia, and a key driver for generating new insights on stem cell behavior. Accumulating evidence from in vivo analyses of the cellular properties of various bone marrow fractions has uncovered a complex hematopoietic hierarchy, through which all hematopoietic cells, including progenitors and mature cells, are perpetually regenerated from rare and mostly quiescent HSCs in the adult bone marrow (Seita and Weissman, 2010).
By definition, functional HSCs have the capacity to engraft the recipient bone marrow after transplantation. However, whether HSCs contribute to produce blood cells under steady-state conditions is unclear. In fact, recent gene-marking studies have suggested that committed progenitors could produce all circulating blood cells during homeostasis. Transposon-based cellular tracking was engineered to show the uncoupling of cellular barcodes between HSCs and granulocytes, accompanied by clonal diversity, in steady-state hematopoiesis (Sun et al., 2014). A subsequent lineage tracing study using inducible Cre recombinase also implied that a large number of long-lived progenitors or short-term HSCs (ST-HSCs), rather than classically defined long-term HSCs (LT-HSCs), are the main drivers of steady-state hematopoiesis during most of adulthood (Busch et al., 2015). Technical considerations, however, might influence the conclusions derived from these experiments. For example, clonal labeling by inducible transposon could be affected by leaky transposase expression after initial induction or the low labeling efficiency (0.5%–1% of the total HSC fraction) of the CreER inducible system, might have reduced the probability of observable HSC differentiation (Sun et al., 2014; Busch et al., 2015).
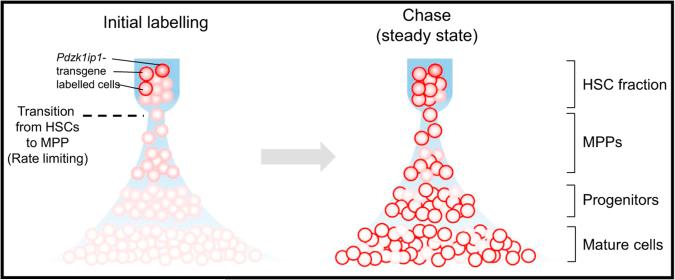
In this issue of Immunity, taking advantage of the enrichment of Pdzk1ip1 (also known as Map17) expression in HSCs, Sawai et al. have developed genetic systems to label self-renewing HSCs and trace the progeny over several months (Sawai et al., 2016). During serial transplantation, HSCs expressing GFP driven by a BAC Pdzk1ip1 transgene exhibited multilineage reconstitution capacity, along with the ability to give rise to GFP-negative subsets (but not vice versa). To label and trace the Pdzk1ip1 transgene-expressing HSCs, they generated a Pdzk1ip1-CreER reporter mice crossed with a Rosa26tdTomato strain, in which the specific labeling of HSCs was confirmed within the most immature subset. While a constant frequency of labeled cells should be observed after random HSC sampling by transgene (Busch et al., 2015), the initial fraction of labeled HSCs (~30%) increased ~2-fold within several weeks, and finally reached ~85% in 6 months to 1 year. These results thus suggest that the Pdzk1ip1 transgene labels the higher (or top) level HSC subset and that a majority of this subset is labeled with high efficiency (Figure 1).
Figure 1. HSCs Provide Major Contribution of Steady-State Hematopoiesis.
HSCs to MPP (dashed line) might be the rate-limiting transition. This transition is followed by rapid and massive amplification at progenitor stages for continuous production of large numbers of mature cells (left). Pdzk1ip1-CreER transgene efficiently marks self-renewing HSC cells, and its labeling is rapidly accumulated in most of committed progenitors and mature cells during unperturbed hematopoiesis. MPP, multi-potent progenitor.
The differentiation spectrum was then further assessed. Labeled HSCs rapidly contributed to committed progenitor populations in all lineages (erythroid, megakaryocytic, myeloid, common lymphoid, and early thymic T cell progenitors) and mature myeloid cells and lymphocytes (except for B-1a cells and tissue macrophages). These results suggest a major contribution of labeled HSCs to all lineages, although the contribution of HSCs to immune cell types derived from the developmental stage might be limited and might vary among tissues. Serial bone marrow biopsy showed that the labeling kinetics of multipotent progenitors (MPPs) and myeloid progenitors (MyPs) were identical and that differentiated granulocytes increased from < 3% at the outset to ~70% by 1 year. Thus, the rate-limiting factor in the hierarchical model might be the transition from HSC to MPP, which is followed by rapid and linear differentiation, and this subsequent massive amplification of progenitor cells enables continuous production of large numbers of mature cells by the few cycling HSCs (Figure 1). Further, labeled HSCs were found to give rise to ~70% of all myeloid cells and platelets in adult mice, and this contribution with its associated labeling kinetics, were accelerated by the induction of an interferon response. In this model, the results suggest that classically defined HSCs might serve as the ultimate source of multilineage hematopoiesis and adult immune cell development, both at the steady state and during a systemic cytokine response.
While these findings represent an important step forward in our understanding of the fundamental properties of HSCs in unperturbed animals, new questions emerge. A remaining issue is the mechanism by which Pdzk1ip1-expressing HSCs are able to exhibit superior self-renewal capacity. In that regard, Pdzk1ip1 itself is unlikely to play a functional role in HSCs similar to that of other genetic HSC markers (e.g., CD150 and α-catulin). Although some mechanistic assays are provided, more detailed transcriptomic assessments (e.g., single cell expression data) will be needed to determine the molecular differences distinguishing Pdzk1ip1-labeled “top-level” HSCs from cells at the next level.
Another interesting issue will be to map the location of Pdzk1ip1-labeled HSCs in the bone marrow. The microenvironment that maintains HSCs is currently the subject of intense study, where several imaging techniques have defined the spatial distribution of HSCs in the bone marrow (Kunisaki et al., 2013; Acar et al., 2015). Although Pdzk1ip1-expressing HSCs seem to lodge near sinusoidal blood vessels, most hematopoietic cells would show the same distributions since sinusoids are broadly and regularly interspersed, sandwiched by only few (i.e., 4–6) layers of cells. More extensive imaging assays for spatial distributions (e.g., arteriole versus sinusoid, perivascular versus endosteal) of Pdzk1ip1-expressing or not-expressing HSCs, both in steady-state and cytokine-stimulated hematopoiesis, will enable the identification of the different niche types for “top-level” HSCs, and will also lead to a deeper understanding of how HSCs respond to the needs of the organism.
While HSCs have been identified retrospectively by clonal assays after single-cell transplantation, such assays have also demonstrated the heterogeneity of current HSC-enriched fractions (Dykstra et al., 2007). This heterogeneity has made it difficult to assess the behavior of individual HSCs, and researchers have long sought the validation of a reliable marker of individual long-term repopulating HSCs. In most of the assays the Reizis team performed, multiple Pdzk1ip1-labeled cells (e.g., 20 cells per transplantation) were used. Although molecular heterogeneity was not yet evaluated, single-cell transplantation assays have revealed that 5 out of 43 (11.6%) single Pdzk1ip1-expressing HSCs displayed multilineage hematopoietic reconstitution capacity, with 3 (7.0%) exhibiting β-type (balanced) pattern and 10 (23.3%) and 1 (2.3%) producing α (myeloid dominant) and γ/δ (lymphoid dominant) patterns, respectively (Dykstra et al., 2007). Pdzk1ip1-labeled fractions are thus heterogeneous, and although this method exclusively and efficiently labels superior self-renewing HSCs with minimal “leak” to mature cells, a majority of Pdzk1ip1-expressing cells could still be the “next-level down” cells. Further, Pdzk1ip1-GFPneg HSCs can also exhibit a multi-lineage repopulation capacity once transplanted, although these cells are not serially transplantable, and might correspond to intermediate-term (IT) or short-term (ST) HSCs (Yamamoto et al., 2013). The establishment of more specific labeling for top- or next-level HSCs will be needed to provide a more definite picture of unperturbed hematopoiesis.
Sawai et al. performed a detailed analysis of steady-state hematopoiesis in adult animals, but can the Pdzk1ip1 transgene also label HSCs during the developmental stage? The contribution kinetics of HSCs to adult hematopoiesis reflect the speed of labeling turnover (myeloid cells are quickly labeled, whereas T cell or immature B cells in bone marrow have slow labeling). The HSC contribution to fetal-derived immune cell types (e.g., B-1a cells) was found to be limited in the study by Sawai et al., consistent with the notion that plasticity toward the B-1a lineage is lost and that initially B-1a potent HSC clones become B-2 (adaptive)-restricted lymphocytes over time. B-1a cells were shown to be relatively well reconstituted in HSC fractions sorted from neonatal bone marrow at 2.5 weeks of age (Barber et al., 2011). It will therefore be interesting to explore how HSC contributions differ when labeling is induced at neonatal (and/or fetal) stages.
Because Pdzk1ip1 is not expressed in human HSCs, it will also be important to dissect through other methods whether the findings from these murine models are applicable to human steady-state hematopoiesis. Recent studies using integration-site-mediated marking to track the human hematopoietic system in vivo and assess clonal dynamics during post-transplant phases have revealed that multipotent “hematopoietic stem and progenitor cells” provided a major sustained contribution for years (Biasco et al., 2016). Additional markers enhancing the purity of human HSCs will no doubt allow researchers to determine the roles of the top hierarchical human HSCs in native and post-transplant hematopoiesis. Although great advances over the past decade have led to the isolation highly HSC-enriched fractions, evidence of heterogeneity persists even in the most purified and rare isolates. It could be that, like us as individuals, each HSC is unique. Finding precisely the family that makes steady-state blood will be an important future challenge.
REFERENCES
- Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, Jaiyeola C, Zhao Z, Luby-Phelps K, Morrison SJ. Nature. 2015;526:126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber CL, Montecino-Rodriguez E, Dorsh-kind K. Proc. Natl. Acad. Sci. USA. 2011;108:13700–13704. doi: 10.1073/pnas.1107172108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasco L, Pellin D, Scala S, Dionisio F, Basso-Ricci L, Leonardelli L, Scaramuzza S, Baricordi C, Ferrua F, Cicalese MP, et al. Cell Stem Cell. 2016;19:107–119. doi: 10.1016/j.stem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, Rodewald HR. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee SJ, Brinkman R, Eaves C. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, et al. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai CM, Babovic S, Upadhaya S, Knapp DJHF, Lavin Y, Lau CM, Goloborodko A, Feng J, Fujisaki J, Ding L, et al. Immunity 45. 2016:597–609. doi: 10.1016/j.immuni.2016.08.007. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J, Weissman IL. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, Camargo FD. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, Ema H, Nakauchi H. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]