Highlight
The Albino Leaf 2 (AL2) gene is involved in the splicing of chloroplast group I and II introns and is responsible for chloroplast development in rice.
Key words: Albino leaf, chloroplast development, CRS1, group I intron, group II intron, rice, RNA splicing.
Abstract
Chloroplasts play an essential role in plant growth and development through manipulating photosynthesis and the production of hormones and metabolites. Although many genes or regulators involved in chloroplast biogenesis and development have been isolated and characterized, identification of novel components is still lacking. We isolated a rice (Oryza sativa) mutant, termed albino leaf 2 (al2), using genetic screening. Phenotypic analysis revealed that the al2 mutation caused obvious albino leaves at the early developmental stage, eventually leading to al2 seedling death. Electron microscopy investigations indicated that the chloroplast structure was disrupted in the al2 mutants at an early developmental stage and subsequently resulted in the breakdown of the entire chloroplast. Molecular cloning illustrated that AL2 encodes a chloroplast group IIA intron splicing facilitator (CRS1) in rice, which was confirmed by a genetic complementation experiment. Moreover, our results demonstrated that AL2 was constitutively expressed in various tissues, including green and non-green tissues. Interestingly, we found that the expression levels of a subset of chloroplast genes that contain group IIA and IIB introns were significantly reduced in the al2 mutant compared to that in the wild type, suggesting that AL2 is a functional CRS1 in rice. Differing from the orthologous CRS1 in maize and Arabidopsis that only regulates splicing of the chloroplast group II intron, our results demonstrated that the AL2 gene is also likely to be involved in the splicing of the chloroplast group I intron. They also showed that disruption of AL2 results in the altered expression of chloroplast-associated genes, including chlorophyll biosynthetic genes, plastid-encoded polymerases and nuclear-encoded chloroplast genes. Taken together, these findings shed new light on the function of nuclear-encoded chloroplast group I and II intron splicing factors in rice.
Introduction
Chloroplasts are essential for plant development and growth, through manipulating the fixation of CO2 and biosynthesis of carbon skeletons and other physiological processes (Sakamoto et al., 2008; Jarvis and Lopez-Juez, 2013). Accumulating evidence has shown the importance of chloroplast biogenesis and development during germination for plant vitality, seed set and growth (Lopez-Juez and Pyke, 2005; Pogson and Albrecht, 2011). Chloroplast biogenesis is initiated from proplastids through endosymbiosis from an ancestor of extant cyanobacteria and is dependent on the coordinated expression of genes encoded in both nuclear and plastid genomes (Lopez-Juez and Pyke, 2005; Sakamoto et al., 2008; Kessler and Schnell, 2009). The development of chloroplasts differs between organs and species depending on the specialization of tissues and stage of development. For example, distinct phenotypes between cotyledons and true leaves were observed in variegated (var) and snowy cotyledon (sco) mutants in Arabidopsis, respectively (Albrecht et al., 2006, 2008, 2010; Liu et al., 2010), i.e. chlorotic true leaves but green cotyledons in the var and chlorotic or bleached cotyledons but green true leaves in the sco mutants. It is evident that nucleus-encoded polymerases (NEPs) and plastid-encoded polymerases (PEPs) involved in gene transcription, RNA maturation, protein translation and modification have great effect on the biogenesis of chloroplasts (Hedtke et al., 1997; Pogson and Albrecht, 2011; Yu et al., 2014). Recent studies indicated that a large group of nuclear-encoded pentatricopeptide repeats proteins (PPRs) involved in RNA processing, splicing, editing, stability, maturation and translation are critical for chloroplast development (Pogson and Albrecht, 2011). Besides PPRs, a group of chloroplast RNA splicing and ribosome maturation (CRM) domain proteins were also shown to be required for chloroplast development, which regulate the splicing of certain introns in chloroplasts and/or mitochondria (de Longevialle et al., 2010). In general, primary RNA transcripts in chloroplasts are spliced by a group of ribozymes via the same chemical steps as spliceosome-mediated splicing in the nucleus (de Longevialle et al., 2010; Borner et al., 2015). Based on the conserved structures and different splicing mechanisms, the introns of certain chloroplast and mitochondrial genes are classified into two main families in plants, group I and group II. Twenty group II introns and only one group I intron have been identified in the chloroplast genome of Arabidopsis, whereas 17 group II introns and one group I intron have been described in maize and rice (de Longevialle et al., 2010; Bonen and Vogel, 2001).
Splicing of group I introns is mediated through a two-step phosphoryl transfer reaction (Kruger et al., 1982). So far, only the intron of trnL has been characterized as a group I intron in chloroplasts and its splicing requires the function of matK, ZmRNC1, ZmWTF1 and AtCFM2 (Asakura and Barkan, 2007; Watkins et al., 2007; Asakura et al., 2008; Kroeger et al., 2009; Zoschke et al., 2010). Group II introns are divided into four subgroups: IIA, IIB, IIC and IID (Michel et al., 1989; Toor et al., 2001). Many group II intron splicing factors have been identified and characterized, including AtnMat2 (Keren et al., 2009), ORGANELLAR TRANSCRIPT processing 43 (OTP43) (de Longevialle et al., 2007; Keren et al., 2012), mCSF1 (Zmudjak et al., 2013), RUG3 (Kuhn et al., 2011) and ABA overly-sensitive 5 (ABO5) (Liu et al., 2010) in mitochondria, and chloroplast RNA splicing 1 (CRS1) (Ostersetzer et al., 2005; Till et al., 2001), ZmRNC1 (Watkins et al., 2007), AtCFM2 (Asakura and Barkan, 2007) and Zm-mTERF4 (Hammani and Barkan, 2014) in chloroplasts. Among these proteins, CRS1 is the first defined CRM protein from maize, which contains three CRM domains (Jenkins et al., 1997; Till et al., 2001). Sixteen and fourteen proteins containing one or more CRM domains are identified in Arabidopsis and rice, respectively. Besides CRS1, four more CRM proteins have been characterized as chloroplast splicing factors: CRM Family Member 2 (CFM2), CFM3, CRS2-associated factors 1 (CAF1) and CAF2 (Barkan et al., 2007; de Longevialle et al., 2010). CRS1 only functions in the splicing of the atpF intron (group IIA), whereas the function of maize CFM2 is associated with one group I intron (trnL intron) and two group II introns (ndhA intron, ycf3 intron 1), and Arabidopsis CFM2 also promotes the splicing of the additional group II intron (clpP intron) (Asakura and Brakan, 2007; Asakura et al., 2008). Differing from other splicing factors in the CRM family, CFM3 is dual-targeted to chloroplasts and mitochondria. In chloroplasts, the function of CFM3 is associated with a subset of group II introns (ndhB, rpl16, rps16, trnG, petB and petD introns) and regulates their splicing in rice (de Longevialle et al., 2010). CSR2 binds to CAF1 and CAF2 to form the complexes CRS2-CAF1 and CRS2-CAF2, respectively, which are required for the splicing of a subset of group IIB introns, including the introns of ndhA, ndhB, petB, petD, rpl16, rps16, trnG and ycf3 in maize and rpoC1and ClpP in Arabidopsis (Ostheimer et al., 2003; Asakura and Barkan, 2006; de Longevialle et al., 2010). To the best of our knowledge, most of the group II intron splicing factors were identified from Arabidopsis or maize, whereas these factors, particularly the CRM proteins in rice, are either not identified or their functions are not fully characterized.
We describe the function of a nuclear-encoded splicing factor termed Albino Leaf 2 (AL2) that encodes a chloroplast group IIA intron splicing factor CRS1, which is required for chloroplast development in rice. Our results suggest that in contrast to the function of the orthologous CRS1 in maize and Arabidopsis, AL2 probably participates in the splicing of both chloroplast group I and II introns in rice. In addition, AL2 appears to coordinate the expression of a subset of chloroplast-associated genes to regulate chloroplast development in rice.
Materials and methods
Plant materials and growth conditions
The rice japonica cultivar Zhonghua 11 was used as the wild type. The T-DNA insertion library with Zhonghua 11 background was obtained from the Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences. All rice seeds in this study were propagated in a paddy field in Guangzhou, China. For laboratory work, rice plants were grown in the greenhouse under a 16-h light/8-h dark cycle at 30 ºC with the given light intensity (1000 mmol m−2 s−1). No significant differences were observed when albino leaf (al2) mutants were grown in the greenhouse versus the paddy field.
Electron microscopy
The samples prepared for electron microscopy were fixed overnight at 4 °C with 2.5% glutaraldehyde in 0.1M phosphate buffer (pH 7.4), and then the samples were dehydrated in a graded ethanol series followed by substitution with a graded isoamyl acetate series. The samples were critical point dried, sputter coated with gold and observed using a HITACHI S-3 000N scanning electron microscope at 10kV. For transmission electron microscopy analysis, samples were fixed overnight at 4 °C with 2.5% glutaraldehyde in 0.1M phosphate buffer (pH 7.4). After washing with phosphate buffer three times, samples were fixed overnight in 2% (v/v) OsO4 in phosphate buffer. After staining and dehydration in a grade ethanol series, the samples were submerged with LR White resin and polymerized for 2 d. Ultrathin cross-sections were prepared with a Leica EM UC6 ultra-microtome and observed under a Tecnai Spirit (120kV) transmission electron microscope.
Analysis of the T-DNA insertion locus in al2 mutant
Inverse polymerase chain reaction (IPCR) was used to isolate the flanking sequence of T-DNA (Ochman et al., 1988). Nested primers of the T-DNA right border primers were C1 and C2, and those of the left border primers were H1 and H2. The genomic DNA was digested by HindIII. Primers for testing of the T-DNA inserting locus were AL2-11310 and 5TF1 for the left site and AL2-11655 and 5TR1 for the right. Primers sequences are listed in Supplementary Table S1 at JXB online.
Plasmid construct and transformation
To produce the AL2 complementation transgenic plants, a 6.2kb DNA fragment of AL2 was amplified from wild type using their corresponding primer pairs listed in Supplementary Table S1. The amplified DNA fragment was confirmed by DNA sequencing and cloned into the binary vector pCAMBIA1301 (Promega, Madison, WI, USA) and then transformed into al2 heterozygotes. To generate the interfering AL2 construct, a 549-bp fragment from the specific coding region of AL2 was amplified from wild-type complementary DNA (cDNA) templates using their corresponding primer pairs listed in Supplementary Table S1. The DNA fragment was confirmed by sequencing and cloned into the binary vector pCAMBIA1301-35S. The final constructs were electroporated into Agrobacterium tumefaciens strain EHA105 for rice transformations that were conducted as previously described (Toki et al., 2006).
Histological β-glucuronidase (GUS) assay
GUS activity analysis was performed following a standard protocol (Jefferson et al., 1987). Transgenic plant tissues were incubated in X-gluc buffer [0.1mol l−1 K2HPO4 (pH 7.0), 0.1mol l−1 KH2PO4 (pH 7.0), 5 mmol l−1 K3Fe (CN)6, 5 mmol l−1 K4Fe(CN)6•3H2O, 0.1% Triton X-100, 20% methanol, 1mg ml−1 X-Gluc] at 37 °C for 2h and then cleared by ethanol. Stained samples were photographed using a Cannon digital camera and stereoscope (OLYMPUS SZX12) and further sliced using resin sections (Leica HistoResin) and analysed under a light microscope (OLYMPUS BX51).
RNA extraction and quantitative real-time (qRT)-PCR assay
Total RNA was extracted using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer’s instructions. The first strand of cDNA was synthesized using TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech) and qRT-PCR was performed as previously described (Chen et al., 2015). The relative expression level of the target gene was normalized to that of rice gene actin1. All primers used in qRT-PCR are listed in Supplementary Table S1.
Chlorophyll detection
Total chlorophyll was determined in triplicate according to the method described previously (Hartmut, 1983). Extracts were obtained from the sixth leaves or seedlings at different growth stages. Approximately 0.2g of fresh tissue was homogenized in 5ml of 80% acetone for 12h in darkness. Spectrophotometric quantification was carried out in a Gene Quant spectrophotometer (GE Healthcare) and followed with calculations: Chl a=12.21×A663−2.81×A646, and Chl b=20.13×A646–5.03×A663 (μg ml−1).
Results
Phenotype of al2 mutants
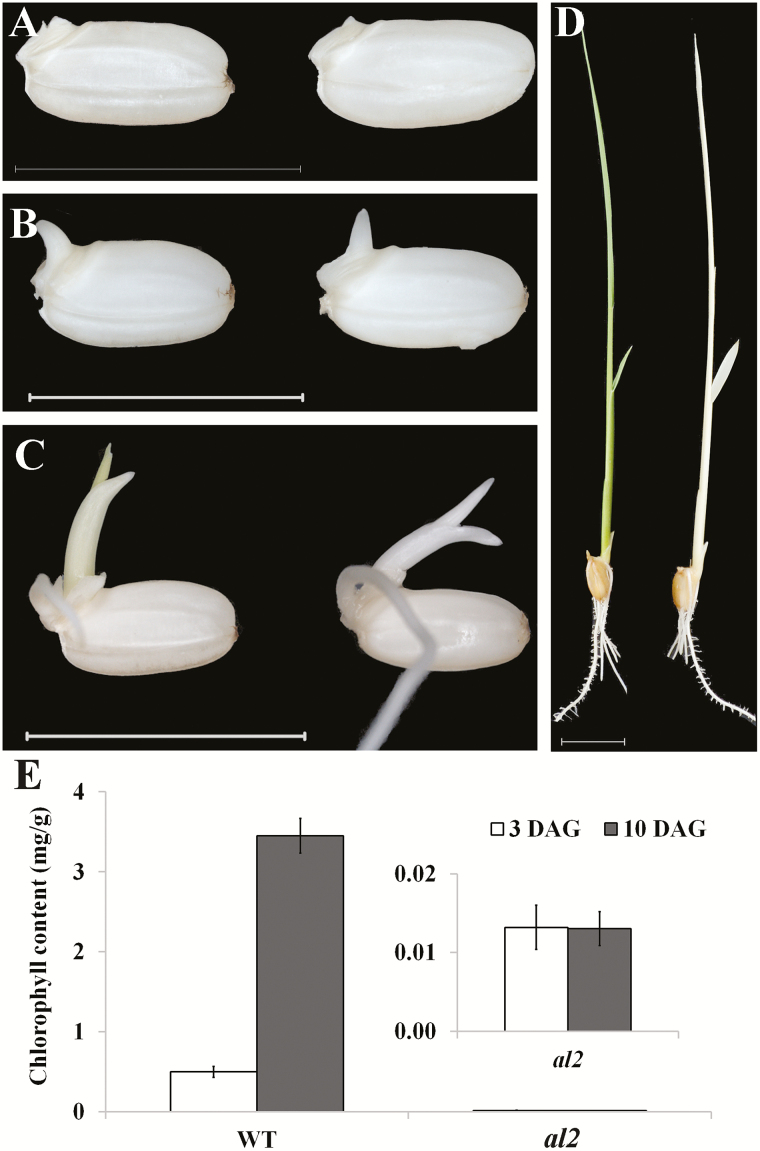
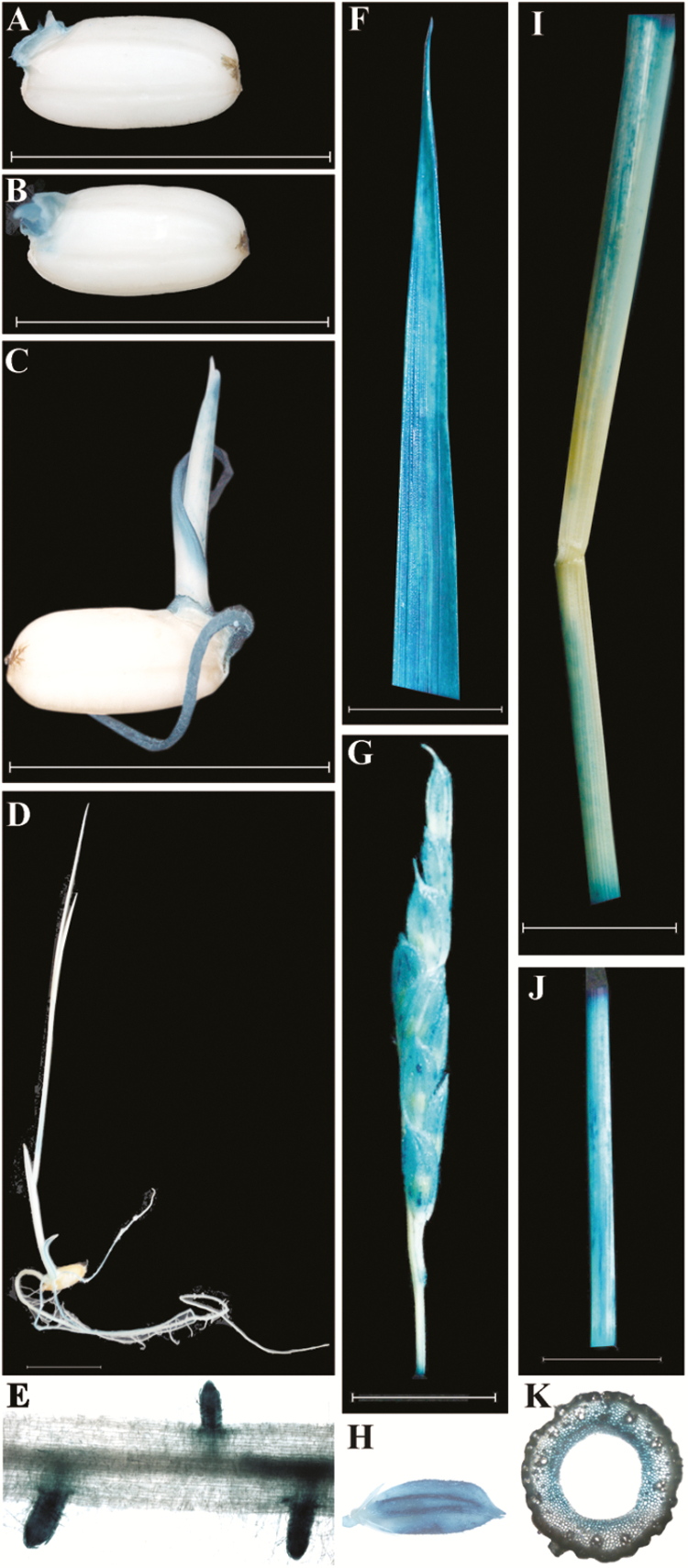
To identify novel genes or regulators involved in chloroplast development, a rice T-DNA insertion population (Zhonghua 11 background) was screened for mutants that exhibited defects in leaf colour, termed albino leaf mutants (als). Dozens of als were obtained through phenotypic investigation. Among these als, the al2 mutant has an interesting and distinct phenotype compared to the others. The al2 mutant showed no significant difference compared to the wild type at the germination stage (Fig. 1A, B), but it had an apparent albino phenotype in the young buds (Fig. 1C). During seedling growth, the albino phenotype became more obvious and spread around the entire leaf at the third-leaf stage in the al2 mutant (Fig. 1D), which eventually led to the seedling lethal al2 mutant. To quantify the changes in this albino phenotype, chlorophyll contents were measured in al2 mutant and wild type. Consistent with their phenotypes, the chlorophyll contents were significantly reduced in al2 compared with that of the wild type (Fig. 1E). These data suggested that the albino phenotype of al2 mutants might be caused by defects in chlorophyll metabolism or the breakdown of entire chloroplasts.
Fig. 1.
Characterization of al2. Phenotype of al1 during germination at (A) 1 d after germination (DAG), (B) 2 DAG, (C) 4 DAG and (D) 10 DAG. (E) Chlorophyll content of al2 at 4 and 10 DAG. In each panel, from left to right is wild type and al2, respectively. Bar: 1cm (A–C); 2cm (D).
Chloroplasts are disrupted in al2 mutants
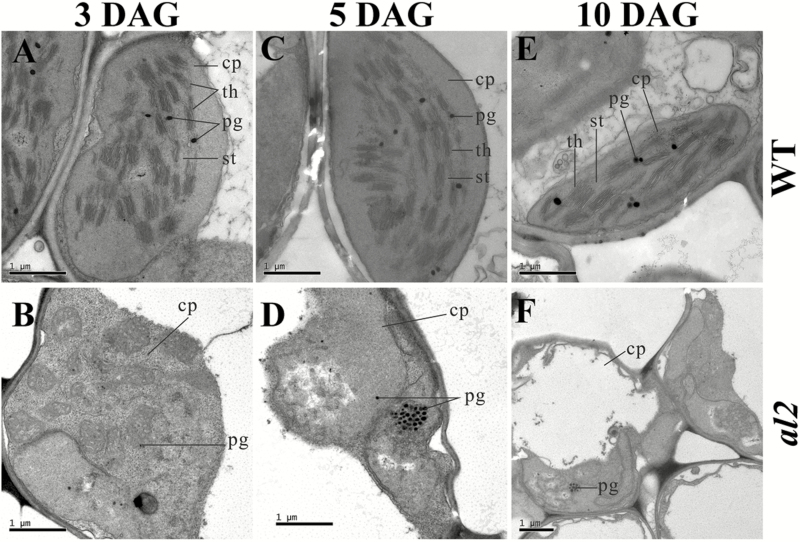
To further investigate the albino leaf phenotype of al2, we observed the chloroplast structure of al2 mutants and wild type in leaves by electron microscopy. Because the albino phenotype of the al2 mutant was developed from the germination to the third-leaf stage, leaves of al2 and wild type from 3 d after germination (DAG), 5 DAG and 10 DAG were selected for microscopy analysis. Our results showed that the development of chloroplasts was significantly affected in al2 leaves at 3 DAG (Fig. 2A, D), and chloroplasts were severely disrupted in al2 at 5 DAG, particularly in the formation of thylakoids (Fig. 2B, E). The most apparent defective phenotype of chloroplasts in al2 was observed at 10 DAG compared to that in wild type (Fig. 2C, F). Taken together, our results indicated that the albino leaf phenotype of the al2 mutant resulted from the abnormal development of chloroplasts, suggesting that the AL2 gene is required for chloroplast development in rice.
Fig. 2.
Electron microscopy observation of al2 leaf. Chloroplasts in wild type mesophyll cells at the (A) 3 DAG, (C) 5 DAG and (E)10 DAG, respectively. Chloroplast in al2 mutant mesophyll cells at (B) 3 DAG, (D) 5 DAG and (F) 10 DAG, respectively. Chloroplasts of the wild type have abundant and well-ordered thylakoid, whereas those of the al2 mutant have no similar structures. cp, chloroplast; DAG, day after germination; pg, plastoglobuli; st, stroma; th, thylakoid. Bar, 1 µm.
Molecular cloning of the AL2 gene
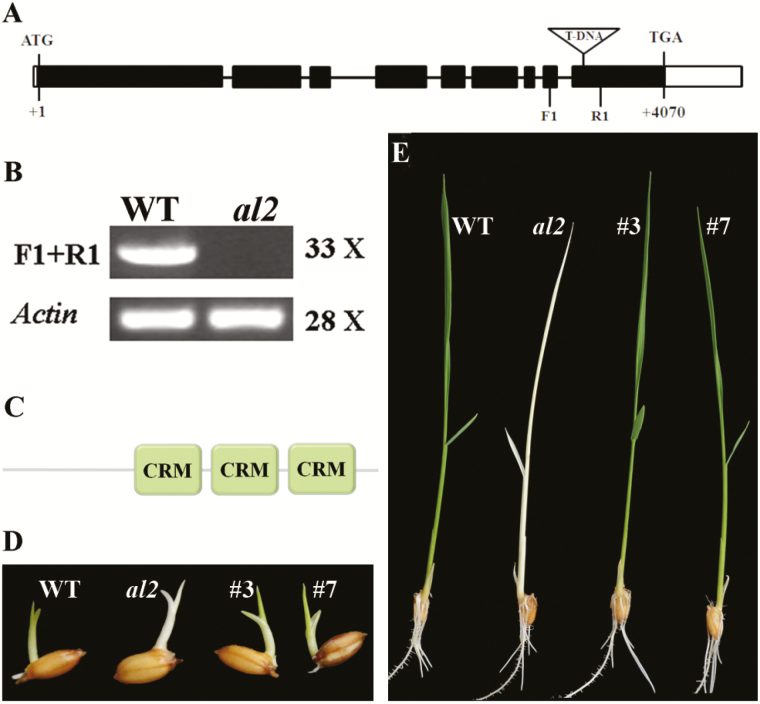
We backcrossed the al2 mutant with wild type and did a segregation analysis in the F2 population based on the al2 mutant phenotype. The segregation ratio between al2 mutant and wild type was ~1:3 (χ2=0.120), indicating that al2 was controlled by a single recessive gene. We then performed inverse PCR and isolated the genomic DNA fragment flanking the T-DNA insertion region in the al2 mutant. Analysis of the resulting sequences via the BLAST function in the NCBI database revealed that T-DNA was inserted into the ninth exon of the Os09g19850 gene, which is comprised of nine exons and eight introns (Fig. 3A). Semi-quantitative PCR analysis indicated that the transcript level of Os09g19850 was completely eliminated in the al2 mutants (Fig. 3B). These data all suggested that Os09g19850 is a strong candidate for the AL2 gene. Gene annotation showed that Os09g19850 encodes a chloroplast group IIA intron splicing facilitator CRS1 in rice (http://rice.plantbiology.msu.edu) containing three CRS1-YhbY domains (also called the CRM domain) (Fig. 3C). Protein alignment analysis indicated that AL2/CRS1 proteins were highly conserved among various organisms. Moreover, phylogenetic analysis demonstrated that the AL2-like proteins in the plant kingdom were grouped into two clusters: monocot plants and dicot plants (Supplementary Fig. S1).
Fig. 3.
Molecular cloning of AL2. (A) Schematic representation of the AL2 gene. Black boxes represent the exon, the lines represent the intron and the white boxes represent the untranslated region (UTR). The putative star codon (ATG) and stop codon (TGA) are located at the +1 and +4070 positions, respectively. The T-DNA is inserted into the ninth exon of Os09g19850. (B) Transcript level of AL2 in wild type and al2 as determined by semi-quantitative PCR with the specific primers indicated in panel A. (C) Distribution of three CRM domains in AL2 protein. (D) Germination phenotype of complementation lines. (E) Seedling phenotype of complementation lines. Panels D, E: #3 and #7 are the al2 complementation lines 3 and 7, respectively. X, PCR cycle.
To verify the identity of AL2, a genetic complementation assay was performed. A 6225-bp wild-type genomic DNA fragment of Os09g19850, containing the putative promoter (2318bp), the coding sequences (including the intron sequences) and a part of the 3ʹ-untranslated region (246bp), was cloned into a binary vector and transformed into al2 heterozygous plants due to the lethality of the al2 homozygous plants. Five independent transgenic lines within the al2 homozygote background were obtained, and two of them were used as representatives in the following studies. In these two transgenic lines, the albino leaf phenotype of al2 was fully rescued by the AL2 transgene (Fig. 3D, E), and the chlorophyll changes (Supplementary Fig. S2). Therefore, we concluded that Os09g19850 is the AL2 gene, and the disruption of its function results in the albino leaf phenotype.
Disruption of AL2 leads to an al2-like phenotype
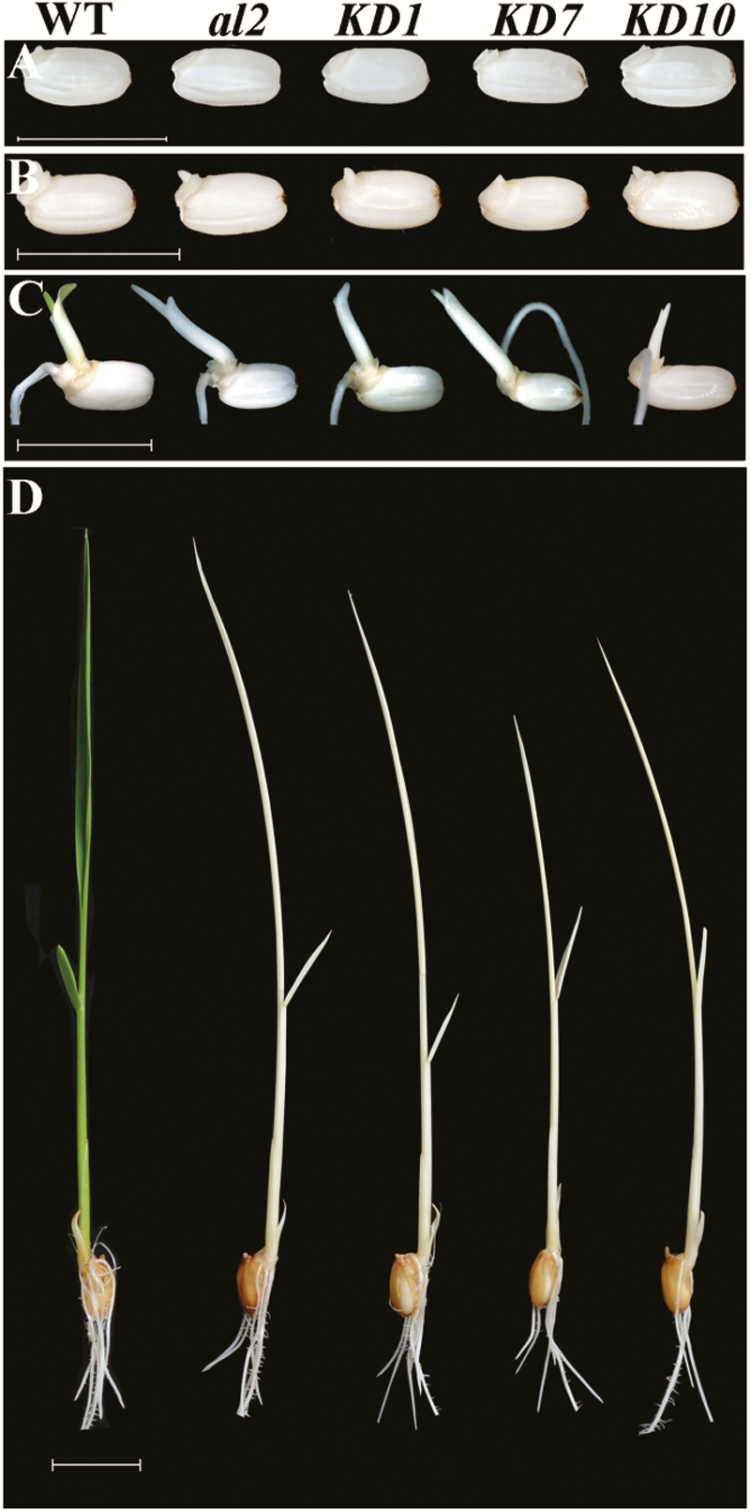
The al2 mutants were seedling lethal, which caused difficulty in the further study of the AL2 functions. To address this issue, we produced weak al2 alleles by specific knockdown of AL2 gene expression. Finally, six independent knockdown lines (KD) were obtained with different expression levels of the AL2 gene (Supplementary Fig. S3). In accordance with their relevant expression levels of AL2, the KD plants exhibited different degrees of the colourless phenotype. Of these 11 KD lines, KD1, KD7 and KD10, with the most significant down-regulation of AL2, showed a highly similar phenotype to that of the al2 mutants (Fig. 4A–D). These results further confirmed that AL2 is required for chloroplast development.
Fig. 4.
Knockdown of AL2 leads to al2-like phenotype. Phenotypic analysis of the AL2 knockdown lines (KD) at (A) 1 d after germination (DAG), (B) 2 DAG, (C) 4 DAG and (D) the third-leaf stage. From left to right is wild type, al2, KD1, KD7 and KD10, respectively. Bar, 2cm.
Expression pattern of AL2
To explore the expression pattern of the AL2 gene, its promoter was fused with the GUS reporter and transformed into wild-type plants. Consistent with the appearance of the al2 phenotype, GUS staining revealed that the AL2 gene was expressed during germination (Fig. 5A–D). Because plastids are also present in the root and root hairs, GUS staining was also observed in the root and root hairs in addition to the chloroplasts (Fig. 5E). Subsequently, the AL2 gene was highly expressed in the mature leaf (Fig. 5F) and also detected in the panicle, spikelet, culm and leaf sheath (Fig. 5G–J). Histological analysis indicated that AL2 was specifically expressed in the endodermis of the culm (Fig. 5K). Additionally, a qRT-PCR analysis of the AL2 gene expression in various tissues revealed the similar expression pattern observed in the promoter analyses (Supplementary Fig. S4). Taken together, our results demonstrate that AL2 is constitutively expressed in various tissues and mainly functions in the green tissues.
Fig. 5.
Expression pattern of AL2. GUS staining indicates that AL2 is expressed at (A) 1 d after germination (DAG), (B) 2 DAG, (C) 4 DAG and (D) 10 DAG. Expression pattern of AL2 in the (E) root, (F) leaf blade, (G) panicle, (H) spikelet, (I) leaf sheath, (J) culm and (K) endodermis of culm.
AL2 is likely to be involved in the splicing of chloroplast group I and II introns
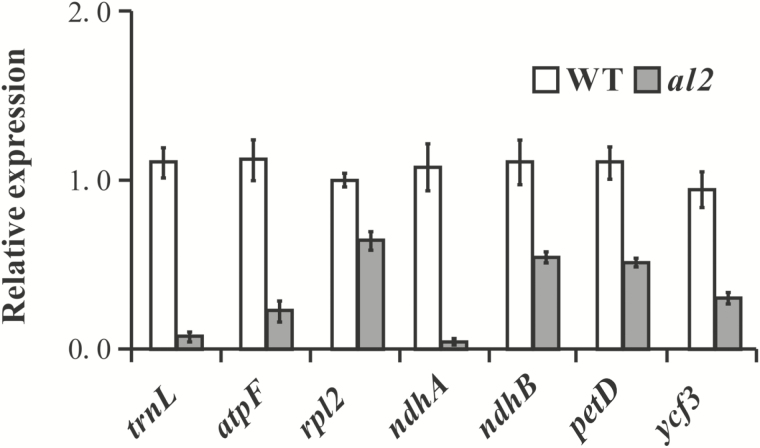
According to the gene annotation (http://rice.plantbiology.msu.edu), AL2 encodes a putative chloroplast group IIA intron splicing facilitator CRS1 in rice. It has been implicated that CRS1 is required for the splicing of the group IIA atpF intron via direct interaction (Till et al., 2001). To validate the function of AL2, we detected the expression level of chloroplast group IIA atpF and rpl2. Our results indicated that the expression level of atpF and rpl2 was significantly reduced in al2 compared with that in wild type, suggesting that AL2 is a functional CRS1. We also analysed the expression levels of chloroplast-associated genes containing group IIB introns, including ndhA, ndhB, petD and ycf3, and found that the expression of these genes was also significantly reduced in al2 (Fig. 6), suggesting that AL2 may also be involved in the splicing of chloroplast group IIB introns. We further tested the expression level of trnL containing the chloroplast group I introns. Interestingly, trnL expression was also predominately eliminated in the al2 mutant (Fig. 6). Taking these results together, we propose that AL2 is likely to be involved in the splicing of both chloroplast group I and II introns.
Fig. 6.
Relative expression of chloroplast genes that contain group I and II introns in al2. Relative expression of trnL (containing group I introns), and atpF and rpl2 (containing group IIA introns), and ndhA, ndhB, petD and ycf3 (containing group IIB introns). The values are the mean of three biological repeats with SD.
Altered expression of chloroplast-associated genes in the al2 mutants
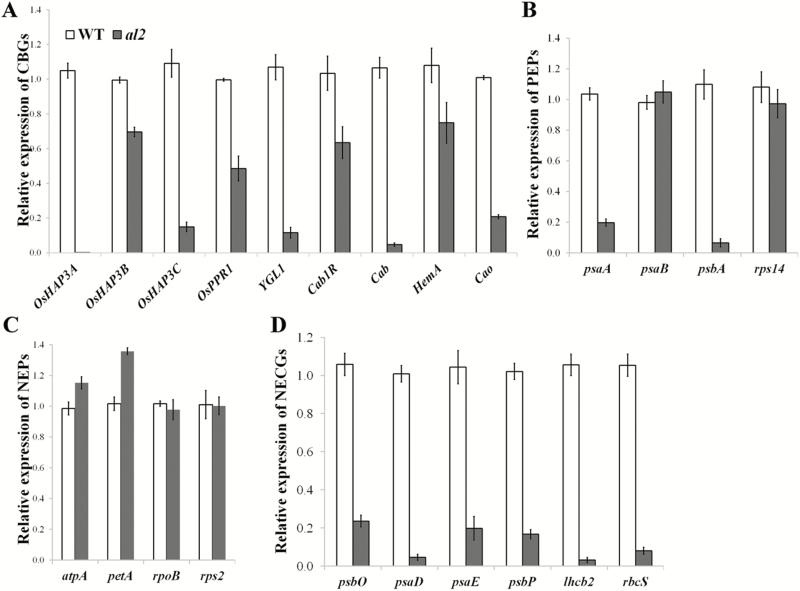
To further determine the role of AL2 in the biological process of chloroplast development, multiple chloroplast-associated genes were investigated. Our results indicated that the expression levels of chlorophyll biosynthetic genes (CBGs) were attenuated in the al2 mutant (Fig. 7A). Two types of genes, PEP and NEP, play a pivotal role in chloroplast biogenesis and development, (Hedtke et al., 1997; Yu et al., 2014). In the al2 mutant, the expression levels of two PEP genes were down-regulated whereas that of the other two were not significantly changed (Fig. 7B). In contrast, all four of the detected NEP genes expressed normally in al2 compared to that in wild type (Fig. 7C), implying that AL2-mediated chloroplast development is independent of NEPs. Nevertheless, the transcript levels of the nuclear-encoded chloroplast genes were significantly impaired in the al2 mutants (Fig. 7D). Taking these results together, we proposed that AL2 coordinates the expression of a subset of chloroplast-associated genes to regulate chloroplast development in rice.
Fig. 7.
Relative expression of chloroplast-associated genes in al2. Relative expression of the chloroplast-associated genes in wild type and al1, including (A) chlorophyll biosynthetic genes (CBGs), (B) plastid-encoded polymerases (PEPs), (C) nucleus-encoded polymerases (NEPs) and (D) nuclear-encoded chloroplast genes (NECGs). Values are the mean of three biological repeats with SD.
Discussion
RNA splicing is a modification process of the nascent precursor messenger RNA transcript in which the introns are removed and exons are joined. In plants, certain chloroplast and mitochondrial genes, encoding either tRNAs or proteins, are interrupted by introns. Based on their conservation of structure and different splicing mechanisms, these introns are divided into group I and group II introns (de Longevialle et al., 2010). We characterized the function of the AL2 gene encoding CRS1 in rice. Our results suggested that AL2 is likely to be involved in the splicing of both chloroplast group I and II introns, and it coordinates a subset of chloroplast-associated genes to regulate the development of chloroplast in rice.
The maize CRS1 contains three CRM domains and is required for the splicing of group IIA atpF intron via direct interaction (Ochman et al., 1988; Till et al., 2001; Ostheimer et al., 2003). AL2 also contains three typical CRM domains. Moreover, multiple alignments of AL2-like proteins among various species indicated that AL2 protein is highly conserved in evolution. The albino leaf phenotype is found not only in the rice al2 mutant but also in the Arabidopsis crs1 mutant (Asakura and Barkan, 2006), suggesting that AL2 may be a functional CRS1 in rice. Validation of the expression of atpF suggested that AL2 is involved in regulating the splicing of chloroplast group IIA introns. We thus propose that AL2 is a functional CRS1 in rice. Surprisingly, our results also showed that the expression of ndhA, ndhB, petD, ycf3 and trnL, was significantly reduced in al2, suggesting that AL2 probably participates in the splicing of group IIB and I introns. Therefore, we also propose that AL2 not only has an overlapping role as the Arabidopsis CRS1 but also functions distinctly. However, further studies are still required to understand the function of AL2 in the splicing of group I and IIB introns. In addition, two additional orthologous CRS1 are present in rice, including Os08g27150 and Os05g47850 (de Longevialle et al., 2010). Characterization of these two genes would extend our knowledge about the function of CRS1 in chloroplast development in rice.
Phylogenetic analysis revealed that AL2-like proteins in the plant kingdom were grouped into two clusters – monocots and dicots – implying that AL2 may also have distinct roles across these plant groups. However, studies of CRS1 in the two representative monocot and dicot plants, maize and Arabidopsis, have implicated both maize and Arabidopsis CRS1 in the specific function of splicing of the chloroplast group II intron (Ostheimer et al., 2003; Ostersetzer et al., 2005; Asakura and Barkan, 2006). Therefore, we propose that the role of AL2 in intron splicing in rice may be different from the above plants. This assumption would be an interesting issue to explore in the future. So far, numerous genes have been shown to be crucial for chloroplast biogenesis and development, and mutations of these genes cause chlorotic or bleached leaf phenotypes. Similar to al2, snow-white leaf1 (swl1) and albino lethal 1 (al1) also exhibit albino leaf and seedling lethality phenotype in rice (Hayashi-Tsugane et al., 2014; Zhao et al., 2016), whereas other mutants, such as sco and var, generate a leaf phenotype restricted to one leaf organ in Arabidopsis. Therefore, characterization of many more monocot mutants will facilitate the discovery of genes functioning in chloroplast biogenesis and/or development that have not yet been clarified in dicot research.
The most abundant transcript of AL2 is found in green tissues, such as the leaf and culm. However, AL2 is also expressed in non-green tissues, such as the root. This suggests a possible involvement of AL2 in the regulation of RNA processing in different kinds of plastids in addition to the chloroplast. However, this hypothesis requires further determination as it remains unclear whether chloroplast intron splicing factors participate in the regulation of other chloroplast-associated genes. In this study, we showed that AL2 is also involved in the regulation of PEPs and nuclear-encoded chloroplast genes. By contrast, our results indicate that AL2 is not involved in the regulation of the NEPs, suggesting that AL2-mediated chloroplast development is independent of NEPs. Additionally, we found that a group of CBGs was significantly down-regulated in the al2 mutants, which may be due to the absence of apoproteins for chlorophyll rather than to the direct involvement of AL2 in regulating these CBGs.
Overall, our results showed that AL2 is likely to be involved in the splicing of both group I and II introns in rice, which is distinct from the function of orthologous CRS1 in maize and Arabidopsis. Moreover, our results also indicated that AL2 coordinates a subset of chloroplast-associated genes to regulate the development of chloroplast in rice.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phylogenetic tree of AL2-like proteins among multiple organisms.
Figure S2. Chlorophyll contents of complementation lines.
Figure S3. Expression pattern of the AL2 gene in the AL2 knock-down lines.
Figure S4. Expression pattern of the AL2 gene in various tissues.
Table S1. Primers used in this study.
Acknowledgements
We thank Prof. Jingliu Zhang and Dr Jiang Wang for providing the T-DNA insertion lines, are grateful to Prof. Guiquan Zhang for his discussions on the data analysis, and thank Dr Zhilong Bao (Cornell University) for his useful comments on the manuscript. This work was supported by grants from the National Natural Science Foundation of China (30571145 and 31071389) and the Natural Science Foundation of Guangdong Province (China) (10151064201000024).
References
- Albrecht V, Ingenfeld A, Apel K. 2006. Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Molecular Biology 60, 507–518. [DOI] [PubMed] [Google Scholar]
- Albrecht V, Ingenfeld A, Apel K. 2008. Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Molecular Biology 66, 599–608. [DOI] [PubMed] [Google Scholar]
- Albrecht V, Simkova K, Carrie C, Delannoy E, Giraud E, Whelan J, Small ID, Apel K, Badger MR, Pogson BJ. 2010. The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. The Plant Cell 22, 3423–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. 2006. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiology 142, 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. 2007. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. The Plant Cell 19, 3864–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Bayraktar OA, Barkan A. 2008. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA 14, 2319–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins KP. 2007. The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA 13, 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L, Vogel J. 2001. The ins and outs of group II introns. Trends in Genetics 17, 322–331. [DOI] [PubMed] [Google Scholar]
- Borner T, Aleynikova AY, Zubo YO, Kusnetsov VV. 2015. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochimica et Biophysica Acta 1847, 761–769. [DOI] [PubMed] [Google Scholar]
- Chen Q, Xie Q, Gao J, et al. 2015. Characterization of Rolled and Erect Leaf 1 in regulating leave morphology in rice. Journal of Experimental Botany 66, 6047–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andres C, Taylor NL, Lurin C, Millar AH, Small ID. 2007. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana . The Plant Cell 19, 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Small ID, Lurin C. 2010. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Molecular Plant 3, 691–705. [DOI] [PubMed] [Google Scholar]
- Hammani K, Barkan A. 2014. An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Research 42, 5033–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmut K. 1983. Determinations of total carotenoids and chlorophylls b of leaf extracts in different solvents. Analysis 4, 142–196. [Google Scholar]
- Hayashi-Tsugane M, Takahara H, Ahmed N, Himi E, Takagi K, Iida S, Tsugane K, Maekawa M. 2014. A mutable albino allele in rice reveals that formation of thylakoid membranes requires the SNOW-WHITE LEAF1 gene. Plant & Cell Physiology 55, 3–15. [DOI] [PubMed] [Google Scholar]
- Hedtke B, Borner T, Weihe A. 1997. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277, 809–811. [DOI] [PubMed] [Google Scholar]
- Jarvis P, Lopez-Juez E. 2013. Biogenesis and homeostasis of chloroplasts and other plastids. Nature Reviews Molecular Cell Biology 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BD, Kulhanek DJ, Barkan A. 1997. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. The Plant Cell 9, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Bezawork-Geleta A, Kolton M, Maayan I, Belausov E, Levy M, Mett A, Gidoni D, Shaya F, Ostersetzer-Biran O. 2009. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15, 2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araujo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O. 2012. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant Journal 71, 413–426. [DOI] [PubMed] [Google Scholar]
- Kessler F, Schnell D. 2009. Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Current Opinion in Cell Biology 21, 494–500. [DOI] [PubMed] [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A. 2009. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proceedings of the National Academy of Sciences, USA 106, 4537–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. 1982. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157. [DOI] [PubMed] [Google Scholar]
- Kuhn K, Carrie C, Giraud E, Wang Y, Meyer EH, Narsai R, des Francs-Small CC, Zhang B, Murcha MW, Whelan J. 2011. The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant Journal 67, 1067–1080. [DOI] [PubMed] [Google Scholar]
- Liu X, Yu F, Rodermel S. 2010. Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. Journal of Integrative Plant Biology 52, 750–761. [DOI] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z. 2010. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant Journal 63, 749–765. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA. 2005. Plastids unleashed: their development and their integration in plant development. The International Journal of Developmental Biology 49, 557–577. [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. 1989. Comparative and functional anatomy of group II catalytic introns – a review. Gene 82, 5–30. [DOI] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O, Cooke AM, Watkins KP, Barkan A. 2005. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. The Plant Cell 17, 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A. 2003. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. The EMBO Journal 22, 3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V. 2011. Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiology 155, 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Miyagishima SY, Jarvis P. 2008. Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. The Arabidopsis Book 6, e0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A. 2001. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. 2006. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant Journal 47, 969–976. [DOI] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S. 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins KP, Kroeger TS, Cooke AM, Williams-Carrier RE, Friso G, Belcher SE, van Wijk KJ, Barkan A. 2007. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. The Plant Cell 19, 2606–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Huang C, Yang ZN. 2014. Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Frontiers in Plant Science 5, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DS, Zhang CQ, Li QF, Yang QQ, Gu MH, Liu QQ. 2016. A residue substitution in the plastid ribosomal protein L12/AL1 produces defective plastid ribosome and causes early seedling lethality in rice. Plant Molecular Biology 91, 161–177. [DOI] [PubMed] [Google Scholar]
- Zmudjak M, Colas des Francs-Small C, Keren I, Shaya F, Belausov E, Small I, Ostersetzer-Biran O. 2013. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytologist 199, 379–394. [DOI] [PubMed] [Google Scholar]
- Zoschke R, Nakamura M, Liere K, Sugiura M, Borner T, Schmitz-Linneweber C. 2010. An organellar maturase associates with multiple group II introns. Proceedings of the National Academy of Sciences, USA 107, 3245–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.