Highlight
The brown fibre cotton Lc1 locus is linked to a 1.4Mb genomic inversion that activates GhTT2_A07. This mutation upregulates flavonoid biosynthesis and confers natural flame retardancy.
Key words: Lc1 locus, brown fibre, cotton, fame retardant, flavonoid, proanthocyanidin, textiles, transparent testa.
Abstract
Some naturally coloured brown cotton fibres from accessions of Gossypium hirsutum L. can be used to make textiles with enhanced flame retardancy (FR). Several independent brown fibre loci have been identified and mapped to chromosomes, but the underlying genes have not yet been identified, and the mechanism of lint fibre FR is not yet fully understood. In this study, we show that both the brown colour and enhanced FR of the Lc1 lint colour locus are linked to a 1.4Mb inversion on chromosome A07 that is immediately upstream of a gene with similarity to Arabidopsis TRANSPARENT TESTA 2 (TT2). As a result of the alternative upstream sequence, the transcription factor GhTT2_A07 is highly up-regulated in developing fibres. In turn, genes in the phenylpropanoid metabolic pathway are activated, leading to biosynthesis of proanthocyanidins and accumulation of inorganic elements. We show that enhanced FR and anthocyanin precursors appear in developing brown fibres well before the brown colour is detectible, demonstrating for the first time that the polymerized proanthocyanidins that constitute the brown colour are not the source of enhanced FR. Identifying the particular colourless metabolite that provides Lc1 cotton with enhanced FR could help minimize the use of synthetic chemical flame retardant additives in textiles.
Introduction
Naturally coloured cotton fibres exist in various hues including light to dark brown, red, rust, and green, and are found among new world tetraploid Gossypium species including G. hirsutum L., G. barbadense L., G. darwinii G. Watt, and G. mustelinum Miers ex G. Watt., as well as their diploid progenitors G. arboreum L. and G. herbaceum L. Studies on the inheritance of brown lint colour in Upland cottons date back to the early 20th century when it was determined that plants producing fibre in various shades of brown and green were allelomorphic to plants producing white fibres, with F1 plants producing an intermediate fibre colour phenotype (Ware, 1932). In a cross with a brown fibre phenotype and white fibre phenotype the fibre colour segregation in the F2 generation was 1:2:1 as brown, intermediate brown, and white (Ware, 1932). Currently, researchers have characterized at least six genetic loci that are linked to brown fibre cotton (Lc1–Lc6) and result in various shades from light to dark brown. These genetic loci were demonstrated to be incompletely dominant to white cotton (Endrizzi and Kohel, 1966; Kohel, 1985). More recently, The Lc1 locus of G. hirsutum was mapped to within 3.8 cM on the long arm of chromosome A07, and the Lc2 locus was mapped to within 4.4 cM on the short arm of chromosome A06 using simple sequence repeat (SSR) and expressed sequence tag (EST)-SSR markers (Wang et al., 2014).
Naturally coloured cottons are typically grown as a source of fibre for niche textile markets that promote the use of natural colours in textiles as an alternative to scouring, bleaching, and dyeing cotton fibres. This use has great merit considering that global textile processing generates a large toxic chemical waste stream with negative environmental impacts (Khatri et al., 2015). Despite the appeal of naturally coloured cottons as an environmentally friendly source of staple fibre for the textile industry, they still occupy only a small market due to inferior agronomic traits. The fibres of brown cotton in particular are typically lower yielding, shorter, and weaker compared with conventional white fibres, which causes difficulty during modern high speed textile processing into yarns, threads, and woven fabrics due to breakage and fibre loss (Dutt et al., 2004; Yuan et al., 2013).
Another limitation to the commercialization of naturally coloured cottons is the lack of colour uniformity and diversity. To that end, and to address negative correlations with fibre quality traits, a large number of studies have been conducted to elucidate the biosynthetic pathways leading to colour formation in brown fibre cottons and determine the source of brown colour. The overall results have demonstrated upregulation of structural genes in the phenylpropanoid and flavonoid biosynthetic pathways and proanthocyanidins (PAs), or condensed tannins, as the source of the brown colour (Xiao et al., 2007; Li et al., 2012; Feng et al., 2013; Gong et al., 2014; Xiao et al., 2014). This was initially demonstrated through dimethylaminocinnamaldehyde (DMACA) staining of brown fibres compared with white fibres from different cotton lines (Xiao et al., 2007). It was also demonstrated through DMACA and toluidine blue O (TBO) staining that PA precursors begin to accumulate as early as 3 days post-anthesis (DPA) in developing brown fibres (Li et al., 2012). Concurrent with staining techniques the presence of flavonoids during brown fibre developmental time points, structural genes in the early and late committed PA biosynthetic pathways were shown by semi-quantitative PCR to be upregulated in developing brown fibres compared with white fibres with peak expression levels during the elongation stage of fibre development (Xiao et al., 2007; Li et al., 2012). Upregulation of the flavanoid pathway in brown cotton fibres was further confirmed by detection of elevated levels of specific flavanoids by high performance liquid chromotagraphy (HPLC) in developing brown cotton fibres (Feng et al., 2013). Further studies have utilized transcriptome analysis by RNA-seq to demonstrate flavonoid structural gene upregulation in developing brown cotton fibres (Gong et al., 2014; Xiao et al., 2014). It was postulated based on liquid chromatography–mass spectrometry (LC-MS) analysis of PA extracts from 20 DPA developing brown and white fibres that leucoanthocyanidin reductase (LAR) represents the primary flow of the PA pathway in brown fibres with catechin and gallocatechin providing the primary precursors to condensed tannins (Xiao et al., 2014). Conversely, it was indicated by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) analysis of brown and white cotton fibres that anthocyanidins reductase (ANR) was a key structural gene in the brown fibre PA pathway based on detection of epigallocatechin and epicatechin precursors in 20 DPA developing brown fibres (Feng et al., 2014). Taken together, most of these results have demonstrated the source of the pigmentation in brown cotton fibre to be PAs in fibres of the brown cotton lines. However, it has also been proposed that quinones are the source of colour development in brown cotton fibres and accumulate due to oxidation of PAs (Feng et al., 2014). Regardless of the source of colour in brown fibres, the unifying finding was upregulation of structural genes in the metabolic pathways leading to PA biosynthesis. Interestingly, no analyses have been conducted on the regulatory genes that are known to control structural gene expression in the committed PA pathway and which have been extensively studied in Arabidopsis and well reviewed in Xu et al. (2014a ).
The interest of our laboratory in brown cotton fibres is presently focused on the enhanced flame retardancy (FR) of brown cotton fibres and fabrics (Fox, 1996; VanZandt et al., 1997; Kimmel et al., 2000; Parmar and Chakraborty, 2001; Corradini et al., 2009; Zhang et al., 2009; Hinchliffe et al., 2015; Nam et al., 2016). In its current state, this level of FR has specific end-use applications including automobile interiors where the burn rates of materials are limited according to standardized testing procedures (ASTM D5132-11; ASTM, 2011). Woven and needlepunched nonwoven fabrics produced from commercially available brown fibres were recently demonstrated to pass standardized testing for automotive interiors (Fox, 1996; VanZandt et al., 1997; Hinchliffe et al., 2015). The brown cotton FR also has the potential to reduce the amount of FR chemical additives required for textiles to pass more stringent tests such as those required by the Federal Aviation Administration for aircraft interior components (FAA, 1986).
The source of brown fibre FR is not yet understood and is not absolutely correlated to the intensity of colour in different cultivars (Hinchliffe et al., 2015). Recently, our laboratory demonstrated that the greater FR observed in brown naturally coloured cottons is likely the result of inorganic salts such as sodium being sequestered through ionic interactions with the partial negative charges generated by adjacent hydroxyl units on the B-ring of flavonoid units in PAs and the possible formation of flavonoid–metal complexes (Nam et al., 2016). The previous studies conducted by Hinchliffe et al. (2015) and Nam et al. (2016) indicate a synergistic process of enhanced FR in brown cotton fibres. Based on these observations, elucidating the regulatory mechanisms of the cotton brown fibre PA and upstream precursor biosynthetic pathways, and identification of the causative brown fibre mutation will provide greater insight to the molecular basis of enhanced FR in brown fibres.
Here we report the results of (i) characterization of the combustion properties of developing and mature fibres and nonwoven fabrics derived from two white fibre and two Lc1 brown fibre cotton lines; (ii) fibre transcriptome analyses of developing white and brown cotton fibres by mRNA-seq and RT-qPCR, which revealed upregulation of many of the structural genes and regulatory genes in the PA and upstream precursor biosynthetic pathways; (iii) identification of the causative mutation by next-generation genomic sequencing and genetic mapping as a 1.4Mb sequence inversion directly upstream of the GhTT2_A07 flavonoid transcription factor gene.
Materials and methods
Cotton lines and cultivation
A brown fibre cotton (G. hirsutum) line of unknown background was observed to have a reversion in fibre colour from brown to white on one branch. White and brown fibre seed cotton was collected from this single plant and homozygous lines, respectively designated MC-WL and MC-BL, were advanced and maintained in Starkville, MS. Both lines continued to breed true for their respective fibre colours. The lines were further advanced by forced self-pollination single seed descent (SSD) for three generations in New Orleans, LA. The cotton lines PD-3 (Culp et al., 1988) and PD 93002 (May et al., 1994) were planted and advanced by SSD. The cotton line PD-3 is a conventional cotton variety with white fibres. The cotton line PD 93002 is an improved fibre quality brown cotton derived from a cross of an unknown brown fibre cotton with PD-3 followed by single plant selections in the F2 generation (May et al., 1994). Pure lines of MC-WL, MC-BL, PD-3, and PD 93002 were planted in an experimental plot in New Orleans, LA in 2012 and 2013 and grown using standard agricultural practices for cotton cultivation. The soil type in the field was Aquents dredged over alluvian in an elevated location to provide for proper drainage. The parent lines MC-BL and MC-WL were crossed, and the parent lines PD-3 and PD 93002 were crossed to produce two F2 populations of 496 and 99 individuals, respectively, that were planted in the same experimental plot in New Orleans, LA in 2014 and 2015 along with the parent lines. Plants that were grown under greenhouse conditions were cultivated in 18.9L pots with Metro-Mix 366 potting soil (Sun Gro Horticulture Canada Ltd, Agawam, MA, USA).
Nonwoven cotton fabric production
Cotton fibres were collected from plants grown in the 2012 growing season and the fibres were converted into hydroentangled nonwoven fabrics on a 1 m wide Fleissner pilot-scale hydroentanglement system (Trützschler Nonwovens GmbH, Dülmen, Germany) running at a constant production speed of 5 m min−1. The hydroentanglement system utilized three pressure heads: one low pressure for fabric wet-out maintained at a constant pressure of 3MPa during fabric production; and two high-pressure heads both maintained at 10MPa during fabric production. Each strip on the pressure heads consisted of 16 orifices per centimetre with an orifice pore size of 120µm. The water used for the hydroentangled fabric production was ambient temperature, which was approximately 25 °C. After hydroentanglement, the fabrics were fed directly through a gas-fired fabric drying oven (Trützschler Nonwovens GmbH) at ~170 °C and wound into rolls.
Flammability testing
The flammability characteristics of the hydroentangled fabrics manufactured from brown and white cotton fibres were determined using a 45 ° flammability tester (Model TC 45, Govmark Ltd) according to test method ASTM D1230-10 (ASTM, 2010) with the following modifications to the protocol. The exposure time of the flame source to the test specimen was increased from the 1s exposure time specified in the test method to 10s. This was necessary to achieve ignition of the white greige fibre hydroentangled fabrics and establish a baseline flame exposure time. Five replications were performed for fabrics manufactured from each cotton line.
Microscale combustion calorimetry and fibre DMACA staining
Fibres from the four cotton lines were tested for enhanced flame retardancy using microscale combustion calorimetry (MCC), which is known to correlate with flammability of textiles and is used as a standardized test by the Federal Aviation Administration for aircraft interior components (ASTM D7309-13; FAA, 1986; Lyon et al., 2007; ASTM, 2013). Approximately 4mg of cotton fibre was placed in an MCC ceramic specimen cup, and the sample mass measured prior to and after pyrolysis on a Sartorius CP2P-F micro balance (Satorius, Bohemia, NY, USA) stationed on a Scienceware Vibrasorb vibration damping mount (Bel-Art Products, Wayne, NJ, USA) mounted on a marble slab table. The heat release capacity (HRC: Jg−1 K−1), peak heat release rate (pHRR: W g−1), total heat release (tHR: kJg−1), temperature at pHRR (°C), and percentage mass of sample not consumed (% char yield) were measured on an MCC model MCC-2 (Deatak, McHenry, IL, USA) and analysed using the MCC Curve Fit v.2 software (Deatak). The specimen heating rate was held constant at 1.2 °C s−1 and data were recorded from 90 to 550 °C. For analysis of immature fibres, developing bolls were harvested in the greenhouse at 20, 28, 36, and 44 DPA and the carpels removed leaving the locules in place. To detect the presence of flavonoids, 50 µl of DMACA (Becton, Dickinson and Co., Sparks, MD, USA) was carefully distributed onto the surface of one to two locules and incubated for 5min at room temperature before image acquisition. Unstained locules were removed from the boll and fibres hand ginned from the ovules. The fibres were dried at room temperature and used for MCC analyses as described.
Colourspace measurement of cotton fibres
The coloration of naturally coloured cotton and white cotton fibres were measured in CIE (International Commission on Illumination, Vienna, Austria) L*a*b* colour space (Ibraheem, 2012), which includes all colours perceivable to the human eye. In CIE L*a*b* colour space, L* indicates whiteness (0=black; 100=white); a* indicates colours from greenish (negative values) to reddish (positive values); and b* indicates colours from bluish (negative values) to yellowish (positive values).
Sample collection, RNA, and DNA isolations
Mature seed cotton in all years from the parent cotton lines and F2 population was hand-harvested and ginned using a laboratory roller gin. During the 2012 growing season, fibre samples representing developmental time points were collected from the four cotton lines at 6, 8, 10, 12, 14, 16, 18, and 20 DPA as previously described (Hinchliffe et al., 2010). Total RNA was extracted and an on-column DNase I digestion performed using the Sigma Spectrum Plant Total RNA Kit (Sigma-Aldrich, St Louis, MO, USA) as per the manufacturer’s protocol and RNA quantity and integrity evaluated as previously described (Hinchliffe et al., 2011). Genomic DNA from the cotton parents and mapping populations was isolated from young leaf tissues as previously described (Fang et al., 2010).
DNA sequencing and linkage mapping
Genomic DNA from the parents used in this study, MC-WL, MC-BL, PD-3, and PD-93002, was isolated from ~100mg of fresh root radical tissue as previously described (Fang et al., 2010) and used for Illumina HiSeq X Ten Sequencing (Novogene Corp., Chula Vista, CA, USA) using 150bp paired-end sequencing runs. Equal amounts of the four indexed libraries were run together on three lanes of an Illumina flow cell. Sequence reads were aligned to the draft G. hirsutum TM-1 reference genome as well as a pseudo-tetraploid genome composed of the reference sequences from two diploids, Gossypium raimondii Ulbr., and G. arboreum (Paterson et al., 2012; Li et al., 2014; Zhang et al., 2015) with GSNAP software (Wu and Nacu, 2010). Polymorphisms were identified with bcftools software and by manual inspection of alignment files (Li et al., 2009; Li, 2011). Select single nucleotide polymorphisms (SNPs) were converted to subgenome specific primers and scored on the F2 populations as before (Thyssen et al., 2014), which also allowed detection of the structural variation using primers flanking the boundaries of the 1.4Mb inversion (Supplementary Tables S1 and S2 at JXB online).
Gene expression analysis
Total RNA from 8 and 20 DPA MC-BL and MC-WL cotton fibres harvested in 2012 were utilized for library preparation and high-throughput sequencing by mRNA-seq (LC Sciences, Houston, TX, USA). Two biological replications for each cotton line and developmental time point were used for a total of eight samples. Sample preparation and library constructions was performed using TruSeq Stranded mRNA Library Prep Kit (Illumina Inc., San Diego, CA, USA) as per the manufacturer’s protocols. Samples were sequenced using a HiSeq 2000 (Illumina Inc.) with 100bp paired-end reads. Equal amounts of the eight indexed libraries were run together on two lanes of an Illumina flow cell. Raw sequence reads were filtered for quality and trimmed by SICKLE (Joshi and Fass, 2011) and aligned to the draft G. hirsutum TM-1 reference genome (Zhang et al., 2015) with GSNAP software (Wu and Nacu, 2010). Reads mapping to annotated genes were counted using BEDTools software (Quinlan and Hall, 2010). Differential expression was determined by data normalization and ANOVA as previously described (Naoumkina et al., 2014). Selected genes from the flavonoid pathway were studied with RT-qPCR using specific primers (Supplementary Table S3) according to standard procedures (Bustin et al., 2009). The reference genes used in the RT-qPCR reactions were the 18S rRNA (GenBank accession U42827), ubiquitin-conjugating protein E2 (GhUCP E2; GenBank AI730710), and α-tubulin 4 (GhTubA4; GenBank AF106570) (Whittaker and Triplett, 1999).
Elemental analysis using inductively coupled plasma mass spectrometry (ICP-MS)
Mature fibre samples from the cotton lines MC-BL, MC-WL, PD 93002, and PD-3 were collected from the field during the 2012 and 2013 growing seasons. Triplicate samples for each line from each year were digested in 8M TraceMetal Grade nitric acid (Thermo Fisher Scientific Inc., Waltham, MA, USA) and analysed by ICP-MS as previously described (Hinchliffe et al., 2015). The ICP-MS analysis was performed by the Nanomedicines Characterization Core Facility (University of North Carolina, Chapel Hill, NC, USA) using a Nexion 300-D ICP-MS (Perkin Elmer, Waltham, MA, USA).
Results
Inherent flame retardancy of naturally coloured brown cottons
The MCC analysis revealed significantly lower heat release capacity, peak heat release rate, and total heat release from fibres of both brown cotton lines compared with the white lines and indicated enhanced FR properties of brown cotton fibres that were consistent with previous studies by this laboratory using commercially available brown and white cotton fibres and nonwoven fabrics (Hinchliffe et al., 2015; Table 1). The percentage char yield, which is a measurement of inorganic compounds remaining after pyrolysis, was also significantly higher from fibres of both brown lines (MC-BL and PD 93002) compared with the white lines (MC-WL and PD 93002). This was previously demonstrated to correlate with higher levels of inorganic salts, which accumulated synergistically with condensed tannins and resulted in greater FR in cotton brown fibres compared with white fibres (Nam et al., 2016). The fibre colour phenotypes were distinguished in CIE L*a*b* colour space (Table 1). These data confirmed our selection of the cotton lines for further studies on the genetics of fibre colour and linkage with enhanced FR in brown fibres.
Table 1.
Microscale combustion calorimetry and CIE colour space values (means and standard deviations) obtained from fibres of two white and two brown cottons
| Cotton line | Colour | Microscale combustion calorimetry | CIE colour space | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Char yield (%) | HRC (J g−1 K−1) | pHRR (W g−1) | tHR (kJ g−1) | Temp (°C) at pHRR | L* | a* | b* | ||
| MC-BL | Brown | 24.6±0.2 | 112±2 | 144.0±1.4 | 6.1±0.1 | 359.1±1.4 | 49.39±0.48 | 12.50±0.30 | 29.96±0.47 |
| MC-WL | White | 16.6±0.8 | 150±7 | 195.7±9.2 | 8.2±0.3 | 362.0±1.1 | 89.40±0.20 | -0.62±0.12 | 4.44±0.23 |
| PD-3 | White | 18.1±0.5 | 134±4 | 175.9±6.4 | 7.8±0.1 | 356.1±1.5 | 91.93±0.90 | -0.60±0.04 | 7.19±0.53 |
| PD 93002 | Brown | 23.2±0.4 | 108±2 | 140.5±2.5 | 6.6±0.1 | 359.9±1.7 | 52.50±0.50 | 12.39±0.05 | 29.64±0.15 |
HRC: heat release capacity; pHRR: peak heat release rate; tHR: total heat release. L* indicates whiteness (0=black; 100=white); a* indicates colours from greenish (negative values) to reddish (positive values); and b* indicates colours from bluish (negative values) to yellowish (positive values).
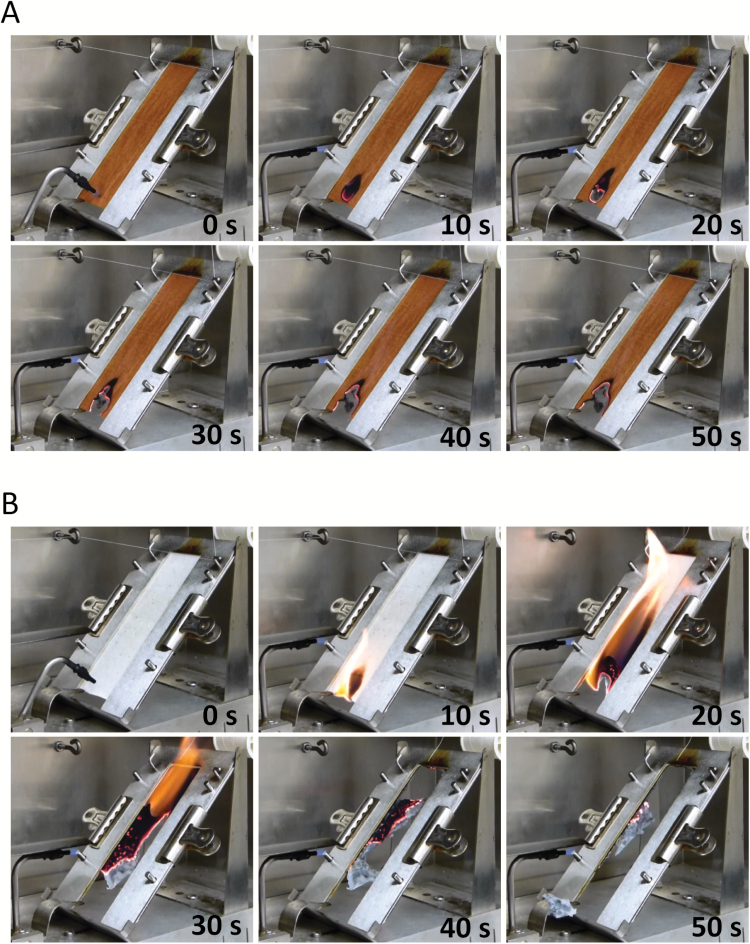
Nonwoven hydroentangled fabrics manufactured from fibres of white MC-WL and brown MC-BL were subjected to ASTM D1230-10 (ASTM, 2010) standardized testing for measuring the flammability of apparel textiles (Fig. 1). Fabrics manufactured from fibres of MC-BL self-extinguished immediately following removal of the flame source in all five replications (Fig. 1A). Afterglow, which is the continuation of glowing parts of a specimen after flame exposure has ceased, continued for 577.4±52.7s until the MC-BL fabrics were completely consumed and only char remained. Nonwoven hydroentangled fabrics manufactured from fibres of MC-WL ignited and the fabric was completely consumed with the time to break the thread recorded at 20.1±2.1s (Fig. 1B).
Fig. 1.
Time lapse depiction in 10s increments of the ASTM D1230-10 (45-degree) flammability test with nonwoven fabrics produced by hydroentanglement of cotton fibres harvested from Lc1 and white cotton lines. (A) Following a 10s exposure to the ignition source, nonwoven fabrics produced from MC-BL fibres immediately self-extinguished with afterglow visible in the remaining panels. (B) Nonwoven fabrics produced from MC-WL fibres ignited and stopped the test timer at ~20s followed by complete consumption of the material by ~50s.
Flame retardancy (FR) is independent of brown colour
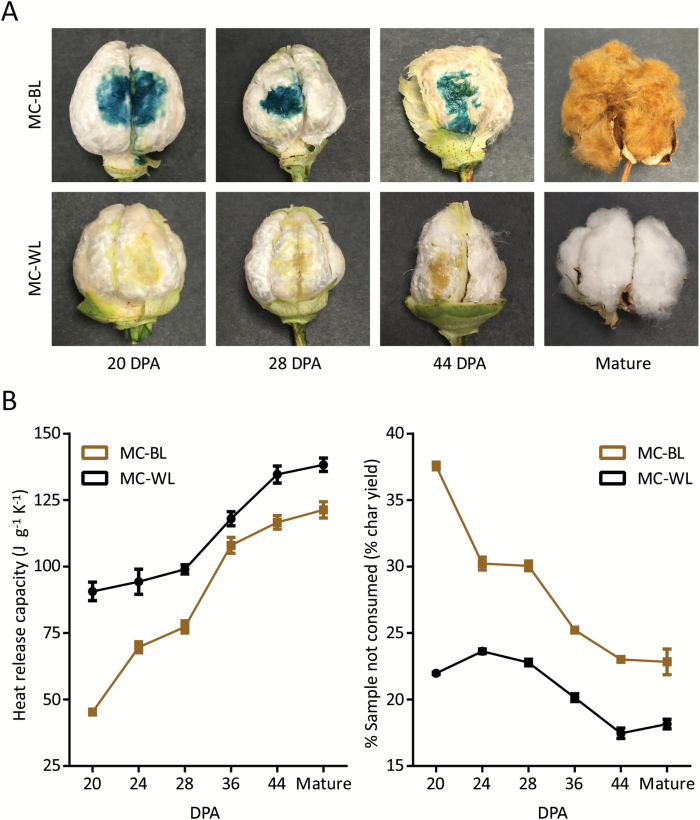
Developing Lc1 fibres from the cotton lines MC-BL and PD 93002 revealed no brown colour development until approximately 36–44 DPA (Fig. 2). Immature cotton locules from a developmental time course were stained using DMACA, which revealed the presence of flavonoids in developing, and still colourless, Lc1 fibres (Fig. 2A; Supplementary Fig. S1A). Unstained fibres harvested from the same bolls were analysed using MCC, which revealed the enhanced FR of Lc1 fibres was already present while the fibres were colourless (Fig. 2B and Supplementary Fig. S1B). During development, the fibres of MC-BL and PD 93002 maintained consistently greater FR compared with their white counterparts (Fig. 2B and Supplementary Fig. S1B). Gravimetric data obtained from the MCC analysis also revealed a consistently higher percentage of unconsumed sample following pyrolysis (% char yield) (Fig. 2B and Supplementary Fig. S1B).
Fig. 2.
Accumulation of flavonoid PA precursor molecules demonstrated by DMACA staining is correlated with enhanced flame retardancy (lower heat release capacity) and elevated levels of inorganic material after pyrolysis (% char yield). (A) Bolls from the cotton lines MC-BL and MC-WL with the carpels removed and fibres stained with DMACA and mature fibres. (B) Comparative analysis of fibre flammability (heat release capacity) and inorganic material remaining after pyrolysis (% char yield) from developing and mature fibre of the cotton lines MC-BL and MC-WL. Developmental stages are indicated by days post-anthesis (DPA) on the x-axis. Error bars represent standard deviation of three biological replicates.
Flame retardancy (FR) and brown colour are linked to 1.4Mb inversion
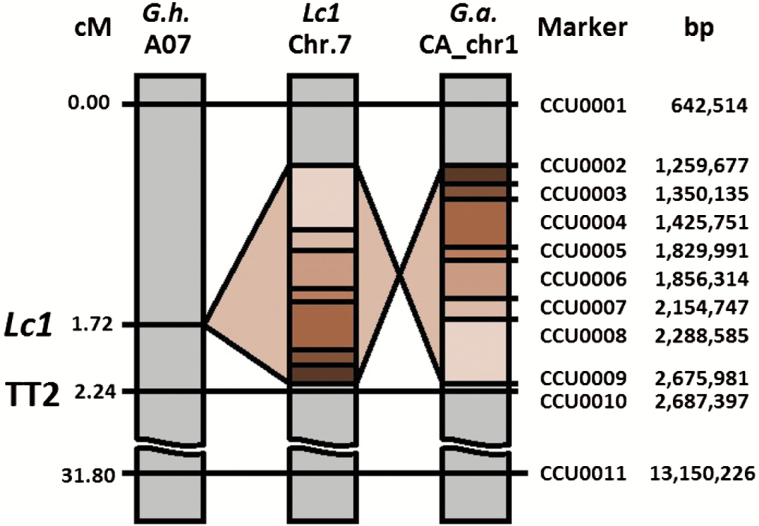
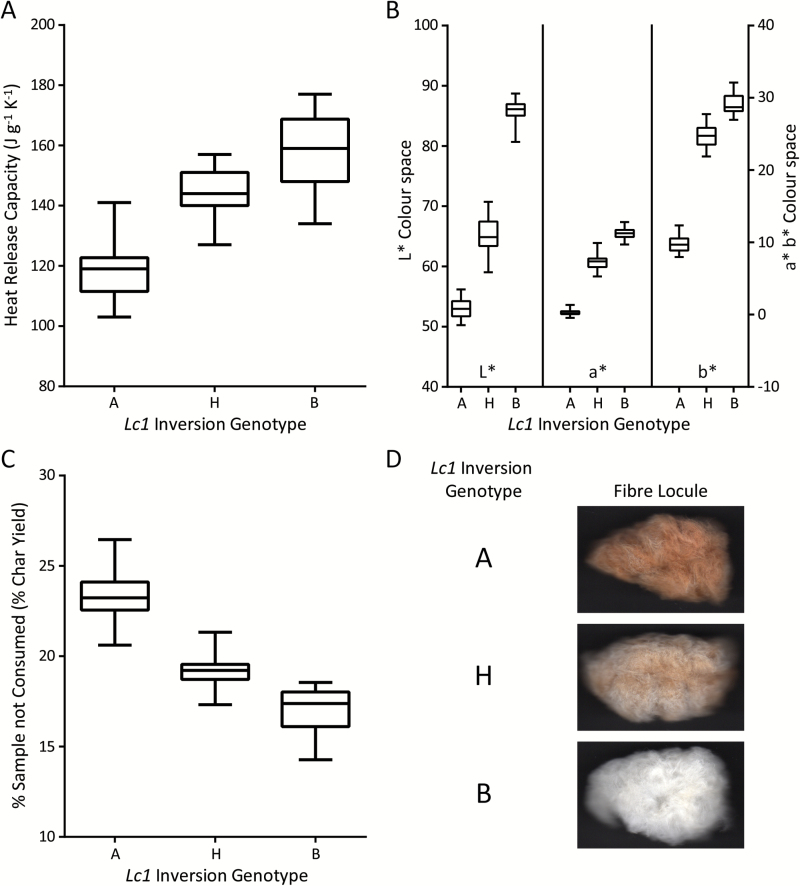
Whole-genome sequencing identified SNPs between the brown and white parental lines. The genomic sequencing data were submitted to the National Center for Biotechnology Information–Sequence Read Archive (NCBI-SRA) under the BioProject accession PRJNA326737. We designed SNP markers that we screened for polymorphism on a subset of the F2 populations. One marker, in GhTT2_A07 (CCU0011), showed incomplete linkage with the brown phenotype. Manual inspection of the alignment files revealed the presence of a large 1.4Mb inversion, just upstream of GhTT2_A07. The inversion was present in the A-subgenomes of both Lc1 cotton lines MC-BL and PD 93002, but was present in the wild-type orientation in the genomes of the white fibre cotton lines MC-WL and PD-3 (Supplementary Fig. S2). We designed and tested markers for the inversion junctions (CCU0002 and CCU0009) along with additional SNPs. We found that the inversion and the SNPs within the inversion exhibited complete genetic linkage to the brown phenotype in all 595 F2 plants (Fig. 3). The brown parental allele was homozygous in all 166 dark brown plants, heterozygous in all 273 light brown plants, and absent in all 157 white F2 progeny (Fig. 4B). Heterozygocity for the inversion also resulted in an intermediate flame retardancy as measured by MCC, as well as an intermediate level of inorganic material as measured by percentage char yield in the segregating progeny (Fig. 4).
Fig. 3.
Linkage and physical maps of the Lc1 genetic locus in Gossypium hirsutum. Genetic distances on G. hirsutum chromosome A07 are shown in centimorgans (cM) and physical distance along the orthologous G. arboreum reference sequence is shown in base pairs (bp). See also Supplementary Fig. S2 for sequencing-based evidence for the 1.4Mb genomic inversion.
Fig. 4.
Correlations between CIE L*a*b* colour space and fibre flammability (heat release capacity) in a subset of the F2 segregating population. A lower HRC indicated a more flame retardant material. Whisker bars represent the maximum and minimum of three measurements. (A) Heat release capacity increased in order from homozygous Lc1 fibres (A genotype) to heterozygous Lc1 fibres (H phenotype) to homozygous wild-type (B genotype). (B) The CIE L*a*b* colour space coordinated with increased HRC starting with dark brown fibres (A genotype) to light brown fibres (H genotype) to white wild-type fibres (B genotype). (C) Percentage of sample not consumed (% char yield), which indicates the level of inorganic material remaining after pyrolysis, decreased in order from homozygous Lc1 fibres (A genotype) to heterozygous Lc1 fibres (H phenotype) to homozygous wild-type (B genotype). (D) Scanned images of single locules representative of the indicated Lc1 inversion genotype. In CIE L*a*b* colour space, L* indicates whiteness (0=black; 100=white); a* indicates colours from greenish (negative values) to reddish (positive values); and b* indicates colours from bluish (negative values) to yellowish (positive values).
Activation of GhTT2_A07 results in activation of flavonoid metabolic pathway
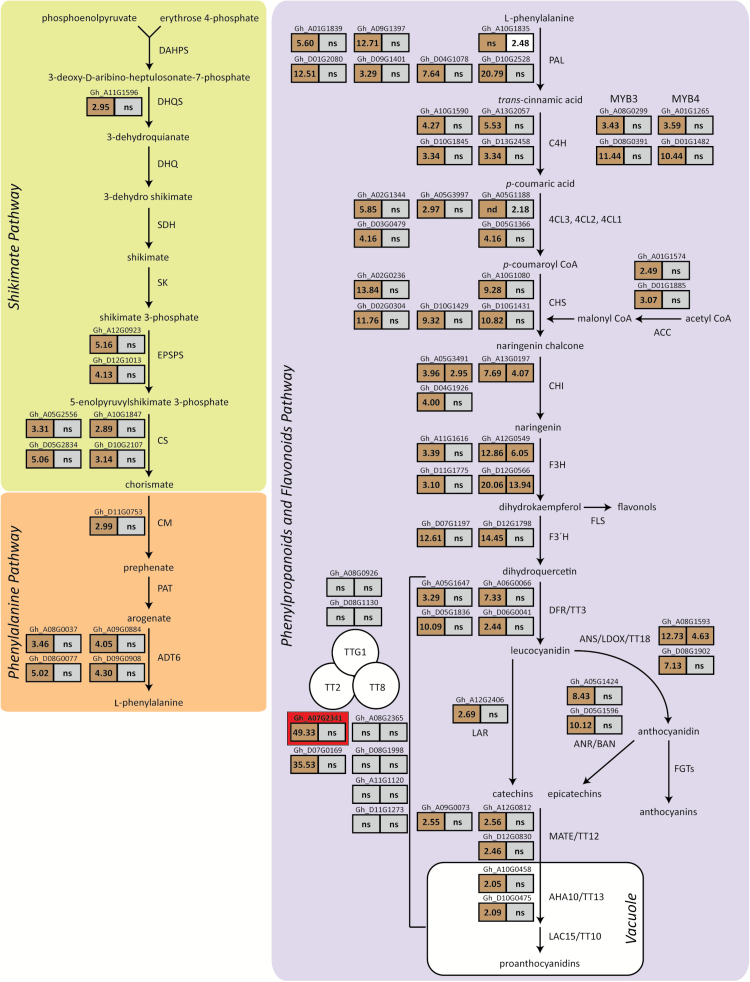
Transcriptome analysis by mRNA-seq revealed many of the genes in the phenylpropanoids pathway, beginning with the first committed step of phenylalanine ammonia-lyase (PAL), were significantly upregulated in developing fibres of MC-BL compared with MC-WL (Fig. 5). Genes in the shikimate and phenylalanine metabolic pathways upstream of the phenylpropanoids pathway were similarly upregulated at 8 DPA in developing fibres of MC-BL compared with MC-WL (Fig. 5). Twelve regulatory and structural genes in the flavonoids biosynthetic pathway were selected for verification of the mRNA-seq data and analysed by RT-qPCR that included additional developmental time points from all four cotton lines used in the study. We observed coordinated upregulation of many of the analysed genes in Lc1 fibres, which peaked during fibre elongation and declined after 14 DPA (Supplementary Figs S3 and S4). An annotated list of all genes that exhibited statistically significant and differential expression (≥2-fold) at 8 and/or 20 DPA in fibres of the cotton lines MC-BL and MC-WL based on the mRNA-seq analysis are shown in Supplementary Table S4. The mRNA-seq data was submitted to the NCBI-SRA under the BioProject accession PRJNA326737.
Fig. 5.
Differential expression of structural and regulatory genes in the committed phenylpropanoid and flavonoid pathways, and in the upstream phenylalanine and shikimate pathways. Genes that were statistically significant and upregulated more than 2-fold based on the mRNA-seq analysis are indicated along with the accession numbers based on the Gossypium hirsutum cv. TM-1 draft genome. The fold differences in expression between MC-BL and MC-WL are indicated in the boxes below the accession numbers, with 8 days post anthesis (DPA) in the left box and 20 DPA in the right. The boxes are colour-coded brown for upregulation in MC-BL and white for upregulation in MC-WL. No statistically significant difference in gene expression is indicated by ns. The genes known to be directly regulated by the TT2-TT8-TTG1 regulon in Arabidopsis are indicated within the bracket next to the regulatory complex in the illustration. The GhTT2_A07 gene (Gh_A07G2341) activated by the upstream genomic inversion is indicated within the red box.
Altered metabolome in Lc1 fibres
Gravimetric data obtained from the MCC analysis revealed a higher percentage of unconsumed sample following pyrolysis (% char yield). Increased levels of inorganic elements were previously observed in mature brown cotton fibres compared with white fibres and were observed to correlate with enhanced FR presumably through the formation of flavonoid–metal complexes (Hinchliffe et al., 2015; Nam et al., 2016). In the present study, iron, potassium, magnesium, and sodium were significantly higher in fibres of both Lc1 cotton lines compared with the fibres of the white lines, with potassium being the most concentrated in the fibres among the elements analysed (Table 2).
Table 2.
Element concentrations (means and standard deviations) in parts per million (ppm) in mature fibre of Lc1 cotton lines MC-BL and PD 93002 compared with wild-type lines MC-WL and PD-3
| Element | MC-BL | MC-WL | PD 93002 | PD-3 |
|---|---|---|---|---|
| B | 14.8±5.1 | 6.8±2 | 18.7±7.2 | 11.4±6.3 |
| Ca | 1503.3±192.5 | 1233.6±228 | 2993.8±1207.8 | 1211.1±361.6 |
| Cu | 2.1±1.7 | 0.7±0.1 | 1±0.1 | 0.5±0.2 |
| Fe* | 6.2±0.9 | 4.2±1.1 | 5.5±0.7 | 4.3±1.6 |
| K* | 14249.9±3319.4 | 8859.7±1815.4 | 16738.9±3470.7 | 7342±1595 |
| Mg* | 1358.8±196 | 1022.6±155.1 | 2031.5±580.6 | 1137.5±202 |
| Mn | 4.1±0.4 | 1.8±0.2 | 3.3±0.4 | 2.6±0.7 |
| Na* | 51.4±4.6 | 39±5.1 | 59.4±18.1 | 32.1±3.9 |
| P | 211.5±23.1 | 175.2±31 | 201.7±28.9 | 164.4±31.5 |
| Si | 16.3±11.6 | 10.3±3.4 | 12.3±2.8 | 7.8±0.8 |
| Zn | 2.6±0.6 | 1.6±0.2 | 2.3±0.8 | 1.6±0.4 |
*Significantly higher in Lc1 lines compared with wild-type at the 0.05 probability level as determined by 2-tailed, paired t-test.
Data are representative of fibres collected in triplicate from two growing seasons. B: boron; Ca: calcium; Cu: copper; Fe: iron; K: potassium; Mg: magnesium; Mn: manganese; Na: sodium; P: phosphorous; Si: silicon; Zn: zinc.
Discussion
The primary objective of this study was to identify the causative mutation resulting in brown colour and enhanced FR in the fibres and fabrics produced from Lc1 cotton plants. The more long term objectives of this project are to try and develop a white fibre cotton plant with enhanced FR properties for use in specific textile applications that require a certain level of FR as dictated by regulatory authorities. This would include, but is not limited to, apparel, bedding, and automotive and aircraft interiors. This scope of application is dictated by standardized testing procedures and could include direct usage or minimization of additive FR compounds to a textile substrate. Identification of the gene responsible for the Lc1 phenotypes was the logical first step in this process in order to better understand the regulation of the PA pathway and access the feasibility of separating brown colour from the FR phenotype of Lc1 cotton fibres.
A colourless flavonoid is the likely cause of natural flame retardancy in Lc1 brown cotton
The accumulation of both flavonoid precursors and polymerized flavonoids that form PAs may contribute to enhanced FR in Lc1 brown cotton fibres by coordination of metals with the B-ring of the flavonoid structure and the formation of flavonoid–metal complexes (Nam et al., 2016). In this study the enhanced FR of fibres and nonwovens textiles of two white fibre and two brown fibre Lc1 cotton lines was examined by MCC and confirmed by standardized textile flammability testing (Table 1 and Fig. 1). The enhanced FR of Lc1 fibres was further demonstrated to exhibit consistently enhanced FR during development and prior to polymerization of flavonoid precursors into PAs (Fig. 2 and Supplementary Fig. S1). This novel observation of an immature white cotton fibre exhibiting enhanced FR independent of the brown fibre colour implicates a colourless compound in the mechanism of enhanced FR. While flavonoid–metal complexes that include brown coloured PAs may contribute to the enhanced FR in mature brown fibres, the existence of enhanced FR in the colourless immature fibres suggests that one or more colourless flavonoids or PA precursors can impart natural FR to developing cotton fibres.
The causative Lc1 mutation was mapped to a non-coding genomic inversion
Here we report the complete linkage of a 1.4Mb genomic inversion with the brown colour and the enhanced FR phenotypes in Lc1 fibres. This is the first report that identified the causative mutation of a naturally coloured cotton line, and also provided the first insights into the regulatory mechanism of the PA biosynthetic pathway in cotton fibres. Upregulation of the GhTT2_A07 transcription factor gene just downstream of the inversion suggests this non-coding region of the genome may contain regulatory elements that either suppress GhTT2_A07 expression in the wild-type orientation, or activate its expression in the mutant Lc1 orientation.
Upregulation of metabolic pathways by activation of GhTT2_A07 in Lc1 fibres
In the current study, as a result of the activation of GhTT2_A07, additional upstream effects were observed in the PA pathway that included upregulation of all genes in the phenylpropanoid and flavonoid pathways, as well as genes in the shikimate pathways and phenylalanine pathways that provide the initial precursor compounds (Fig. 5). A discussion of the regulatory mechanisms of these pathways is beyond the scope of this research and extensively reviewed in Maeda and Dudareva (2012). The TT2 transcription factor is part of the TT2–TT8–TTG1 regulon that has been extensively studied in Arabidopsis and directly regulates late biosynthetic pathway genes in the PA pathway (Fig. 5 and Supplementary Table S5) (Xu et al., 2014a ,b). Orthologues of TT8 and TTG1 were expressed at much lower levels than GhTT2_A07 and their transcript abundances were not significantly different in Lc1 fibres compared with white fibres in the mRNA-seq and RT-qPCR analyses (Fig. 5 and Supplementary Figs S3 and S4). The absence of coordinated expression of the genes comprising this regulatory complex suggests that GhTT2_A07 may act independently or in conjugation with unknown transcription factors in upregulating the PA pathway in Lc1 fibres or that the accumulation of downstream metabolites initiates positive feedback regulation. The activation of GhTT2_A07 by the genomic inversion in Lc1 cotton results in the expression of many genes that may play critical roles in the development of natural flame retardance.
Future work will target downstream genes to restore colourless fibres but maintain FR
Currently, we are targeting late biosynthetic genes in the committed PA pathway using a virus-induced gene silencing (VIGS) system that was previously demonstrated to be usable as a functional assay in cotton fibres (Wan et al., 2016). Based on our results that demonstrated enhanced FR in developing colourless Lc1 fibres, inhibition of specific late biosynthetic genes involved in PA precursor biosynthesis, transport, and/or polymerization to PAs may allow accumulation of flavonoid precursors and flavonoid–metal complexes and result in the enhanced FR phenotype independent of brown colour.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Enhanced flame retardancy develops prior to brown colour in Lc1 fibres
Figure S2. Alignment of DNA reads to pseudo-tetraploid reference genome reveals the 1.4Mb genomic inversion upstream of GhTT2_A07 in Lc1 cotton lines.
Figure S3. A RT-qPCR comparative analysis of the transcript abundances of selected genes in the phenylpropanoid and flavonoid pathways from developing fibres of the cotton lines MC-BL and MC-WL.
Figure S4. A RT-qPCR comparative analysis of the transcript abundances of selected genes in the phenylpropanoid and flavonoid pathways from developing fibres of the cotton lines PD 93002 and PD-3.
Table S1. Inversion-specific marker scheme and amplification results.
Table S2. SNP genetic markers used to map the 1.4Mb genomic inversion in Lc1 cotton lines.
Table S3. Nucleotide sequences of primer pairs utilized for RT-qPCR, gene annotations, and database accession numbers.
Table S4. Significantly and differentially expressed genes from mRNA-seq analysis.
Table S5. List of enzyme abbreviations used in Fig. 5 and Supplementary Figs S3 and S4.
Acknowledgements
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
References
- ASTM 2010. ASTM D1230-10. Standard test method for flammability of apparel textiles. West Conshohocken, PA, USA: ASTM International, http://www.astm.org. [Google Scholar]
- ASTM 2011. ASTM D5132-11. Standard test method for horizontal burning rate of polymeric materials used in occupant compartments of motor vehicles. West Conshohocken, PA, USA: ASTM International, http://www.astm.org. [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative realtime PCR experiments. Clinical Chemistry 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Corradini E, Teixeira EM, Paladin PD, Agnelli JA, Silva ORRF, Mattoso LHC. 2009. Thermal stability and degradation kinetic study of white and colored cotton fibers by thermogravimetric analysis. Journal of Thermal Analysis and Calorimetry 97, 415–419. [Google Scholar]
- Culp TW, Moore RF, Harvey LH, Pitner JB. 1988. Registration of ‘PD-3’ cotton. Crop Science 28, 190. [Google Scholar]
- Dutt Y, Wang XD, Zhu YG, Li YY. 2004. Breeding for high yield and fibre quality in coloured cotton. Plant Breeding 123, 145–151. [Google Scholar]
- Endrizzi JE, Kohel RJ. 1966. Use of telosomes in mapping three chromosomes in cotton. Genetics 54, 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAA 1986. Federal Aviation Regulations Part 25–Airworthiness standards: transportation category airplanes, section 25.853, appendix F. Washington, DC, USA: Federal Aviation Administration. [Google Scholar]
- Fang DD, Xiao J, Canci PC, Cantrell RG. 2010. A new SNP haplotype associated with blue disease resistance gene in cotton (Gossypium hirsutum L.). Theoretical and Applied Genetics 120, 943–953. [DOI] [PubMed] [Google Scholar]
- Feng H, Tian X, Liu Y, Li Y, Zhang X, Jones BJ, Sun Y, Sun J. 2013. Analysis of flavonoids and the flavonoid structural genes in brown fiber of upland cotton. PLoS ONE 8, e58820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Li Y, Wang S, Zhang L, Liu Y, Xue F, Sun Y, Wang Y, Sun J. 2014. Molecular analysis of proanthocyanidins related to pigmentation in brown cotton fibre (Gossypium hirsutum L.). Journal of Experimental Botany 65, 5759–5769. [DOI] [PubMed] [Google Scholar]
- Fox SV. 1996. Naturally flame resistant cotton. Patent US 5496623 A. [Google Scholar]
- Gong W, He S, Tian J, Sun J, Pan Z, Jia Y, Sun G, Du X. 2014. Comparison of the transcriptome between two cotton lines of different fiber color and quality. PLoS ONE 9, e112966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe D, Condon B, Delhom DD, Chang S, Montalvo J, Madison C, Reynolds M, VonHoven T, Cintrón MS. 2015. Physical and combustion properties of nonwoven fabrics produced from conventional and naturally colored cottons. Textile Research Journal 85, 1666–1680. [Google Scholar]
- Hinchliffe DJ, Meredith WR, Yeater KM, Kim HJ, Woodward AW, Chen ZJ, Triplett BA. 2010. Near-isogenic cotton germplasm lines that differ in fiber-bundle strength have temporal differences in fiber gene expression patterns as revealed by comparative high-throughput profiling. Theoretical and Applied Genetics 120, 1347–1366. [DOI] [PubMed] [Google Scholar]
- Hinchliffe DJ, Turley RB, Naoumkina M, Kim HJ, Tang Y, Yeater KM, Li P, Fang DD. 2011. A combined functional and structural genomics approach identified an EST-SSR marker with complete linkage to the Ligon lintless-2 genetic locus in cotton (Gossypium hirsutum L.). BMC Genomics 12, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheem NA, Hasan MM, Khan RZ, Mishra PK. 2012. Understanding color models: a review. ARPN Journal of Science and Technology 2, 265–275. [Google Scholar]
- Joshi NA, Fass JN. 2011. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software], https://github.com/najoshi/sickle.
- Khatri A, Peerzada MH, Mohsin M, White M. 2015. A review on developments in dyeing cotton fabrics with reactive dyes for reducing effluent pollution. Journal of Cleaner Production 87, 50–57. [Google Scholar]
- Kimmel L, Tao W, Yachmenev V, Calamari T., Jr 2000. Naturally colored brown cotton for needlepunched nonwoven fabrics. In: Proceedings of the 2000 Beltwide Cotton Conference. Memphis, TN, USA: National Cotton Council of America, 846–851. [Google Scholar]
- Kohel RJ. 1985. Genetic analysis of fiber color variants in cotton. Crop Science 25, 793–797. [Google Scholar]
- Li F, Fan G, Wang K, et al. 2014. Genome sequence of the cultivated cotton Gossypium arboreum . Nature Genetics 46, 567–572. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Fan H, Li Z, Wei J, Lin Y, Cai Y. 2012. The accumulation of pigment in fiber related to proanthocyanidins synthesis for brown cotton. Acta Physiologiae Plantarum 34, 813–818. [Google Scholar]
- Lyon RE, Walters RN, Stoliarov SI. 2007. Thermal analysis of flammability. Journal of Thermal Analysis and Calorimetry 89, 441–448. [Google Scholar]
- Maeda H, Dudareva N. 2012. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annual Review of Plant Biology 63, 73–105. [DOI] [PubMed] [Google Scholar]
- May OL, Green CC, Roach SH, Kittrell BU. 1994. Registration of PD 93001, PD 93002, PD 93003, and PD 93004 germplasm lines of upland cotton with brown lint and high fiber quality. Crop Science 34, 542. [Google Scholar]
- Nam S, Kim HJ, Condon BD, Hinchliffe DJ, Chang S, McCarty JC, Madison CA. 2016. High resistance to thermal decomposition in brown cotton is linked to tannins and sodium content. Cellulose 23, 1137–1152. [Google Scholar]
- Naoumkina MA, Thyssen GN, Fang DD, Hinchliffe DJ, Florane CB, Yeater KM, Page JJ, Udall JA. 2014. The Li2 mutation results in reduced subgenome expression bias in elongating fibers of allotetraploid cotton (Gossypium hirsutum L.). PLoS ONE 9, e90830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar MS, Chakraborty M. 2001. Thermal and burning behavior of naturally colored cotton. Textile Research Journal 71, 1099–1102. [Google Scholar]
- Paterson AH, Wendel JF, Gundlach H, et al. 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibers. Nature 492, 423–427. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyssen GN, Fang DD, Turley RB, Florane CB, Li P, Naoumkina MA. 2014. Next generation genetic mapping of the Ligon-lintless-2 (Li2) locus in upland cotton (Gossypium hirsutum L.). Theoretical and Applied Genetics 127, 2183–2192. [DOI] [PubMed] [Google Scholar]
- VanZandt MJ, Horridge P, Dever JK. 1997. Flame resistance and physical characteristics of upholstery-weight naturally colored cotton. Textile Research Journal 15, 246–251. [Google Scholar]
- Wan G, Guan X, Yang et al. 2016. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytologist 210, 1298–1310. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu H, Li X, Xiao X, Ai X, Luo C, Zhu L, Li X. 2014. Genetic mapping of fiber color genes on two brown cotton cultivars in Xinjiang. SpringerPlus 3, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware JO. 1932. Inheritance of lint colours in upland cotton. Journal of the American Society of Agronomy 24, 550–562. [Google Scholar]
- Whittaker DJ, Triplett BA. 1999. Gene-specific changes in alpha-tubulin transcript accumulation in developing cotton fibers. Plant Physiology 121, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y-H, Yan Q, Ding H, et al. 2014. Transcriptome and biochemical analyses revealed a detailed proanthocyanidin biosynthesis pathway in brown cotton fiber. PLoS ONE 9, e86344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y-H, Zhang Z-S, Yin M-H, Luo M, Li X-B, Hou L, Pei Y. 2007. Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochemical and Biophysical Research Communications 358, 73–78. [DOI] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. 2014. a Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends in Plant Science 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Xu W, Grain D, Bobet S, et al. 2014. b Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytologist 202, 132–144. [DOI] [PubMed] [Google Scholar]
- Yuan SN, Malik W, Bibi N, Wen GJ, Ni M, Wang XD. 2013. Modulation of morphological and biochemical traits using heterosis breeding in coloured cotton. Journal of Agricultural Science 151, 57–71. [Google Scholar]
- Zhang L, He J, Wang S-Y. 2009. Structure and thermal properties of natural colored cottons and bombax cotton. Journal of Thermal Analysis and Calorimetry 95, 653–659. [Google Scholar]
- Zhang T, Hu Y, Jiang W, et al. 2015. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnology 33, 531–537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.