Abstract
The carboxyl‐terminal sequence of tau composes the framework for its intracellular inclusions that appear in diverse neurodegenerative disorders known as tauopathies. However, microtubule‐associated protein 2 (MAP2), which contains a homologous carboxyl‐terminal sequence of tau, is undetectable in the mature tau inclusions. The mechanisms underlying this phenomenon have remained largely unknown. Here, we show that tau and MAP2 have different aggregation properties: tau aggregates to form filaments but MAP2 remains to be granules. Exchanging 221 YKPV 224 of tau (0N3R) near the PHF6 motif for 340 TKKI 343 of MAP2c profoundly changed aggregation properties, suggesting that the YKPV motif is important for filament formation, whereas the TKKI motif is for granule formation. Thus, these minimal sequences may determine the different fates of tau and MAP2 in the formation of inclusions in tauopathies.
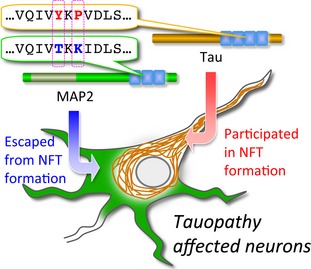
Tau and microtubule‐associated protein 2 (MAP2) are homologous microtubule‐associated proteins in neurons. So far, it is largely unknown why tau but not MAP2 is selectively involved in the filamentous inclusions (neurofibrillary tangles, NFT) formation in tauopathies, including Alzheimer's disease. In this study, we found that the difference of only two amino acids in tau and MAP2 sequences may determine their different fates in tauopathies. These results may lead to the elucidation of tau deregulation in pathological conditions.
Keywords: aggregation, inclusion, MAP2, microtubule‐associated protein 2, tau, tauopathies
Abbreviations used
- AD
Alzheimer's disease
- MAP2
microtubule‐associated protein 2
- NFTs
neurofibrillary tangles
- ThT
thioflavin T
Abnormal protein aggregation is observed in a variety of diseases such as Huntington's disease, Creutzfeldt–Jakob disease, Parkinson's disease, and tauopathies. The aggregations of these distinct proteins appear to represent the major pathological process (Ross and Poirier 2004). The intracellular neurofibrillary tangles (NFTs) comprising aggregates of tau in the central nervous system are the major pathological hallmarks of tauopathies including Alzheimer's disease (AD), frontotemporal dementia, and Parkinsonism linked to chromosome 17 (FTDP‐17) (Lee et al. 2001; Mandelkow and Mandelkow 2012).
The microtubule‐associated protein tau has the microtubule‐binding domains (MTBDs) consisting of three or four imperfect repeats, which are called 3 repeat (3R)‐tau or 4 repeat (4R)‐tau, respectively, and are located in the carboxyl‐terminal region. Neurons contain a tau homolog, named microtubule‐associated protein 2 (MAP2). MAP2 also has MTBDs, which are typically of three imperfect repeats. Thus, both tau and MAP2 have homologous carboxyl‐terminal regions and are considered to evolve from the same ancient protein (Dehmelt and Halpain 2004). Previous studies have indicated that the core of NFTs is made up of the carboxyl‐terminal sequence of tau (Kondo et al. 1988; Wischik et al. 1988). This raises the possibility that MAP2 is also involved in NFT formation. In recent studies, we examined the relationship between the carboxyl‐terminal sequences of tau and MAP2 and their roles in the inclusion formation in AD brains. Our results showed that the carboxyl‐terminal sequence of tau is involved in the mature inclusions, whereas the homologous carboxyl‐terminal sequence of MAP2 is not detected (Xie et al. 2014). These findings led us to investigate what causes the different fates of tau and MAP2 in tauopathies.
Here, we compared the aggregation properties of full‐length tau and MAP2 containing wild‐type or mutant sequences. We found that the aggregation properties of tau and MAP2 are determined solely by their homologous carboxyl‐terminal sequences. Tau aggregated to form filaments, whereas MAP2 aggregated to granules. According to the previous study, the motif PHF6 comprising six‐amino acid sequence VQIVYK is required for tau fibril formation (von Bergen et al. 2001). Therefore, we compared the corresponding amino acid sequences of tau and MAP2 around the PHF6 motif, and found a similar sequence of 11 amino acids (VQIVXKXXDLS) in tau and MAP2. To our surprise, the different aggregation properties of tau and MAP2 could be changed by exchanging a few amino acids in the 11 amino acids. This finding indicated that these amino acids are the key to determine the aggregation properties of the two microtubule‐associated proteins.
Experimental procedures
Chemicals and proteins
Heparin sodium was obtained from ACROS Organics (Geel, Belgium). Thioflavin T (ThT) was purchased from Waldeck GmbH & Co. KG (Münster, Germany). Human tau cDNA cloned in the pRK172 vector was a gift from Dr M. Goedert. Human MAP2c cDNA was amplified and cloned into NdeI‐ and EcoRI‐digested sites of the pRK172 vector. Site‐directed mutagenesis of constructs was performed by PCR and confirmed by DNA sequencing. Proteins were purified as described in a previous study (Xie et al. 2014). Briefly, the recombinant proteins expressed in Escherichia coli strain BL21(DE3) were applied to a phosphocellulose column and eluted with a gradient of 0.1–0.3 M NaCl. Protein‐containing fractions were purified further by ammonium sulfate precipitation. Finally, the heat‐stable supernatant was fractioned by reversed‐phase HPLC (Cosmosyl Protein‐R; Nacalai Tesque, Inc., Kyoto, Japan). The purified proteins were quantified by sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by Coomassie brilliant blue staining.
Aggregation assay
The aggregation of recombinant proteins was induced with heparin as described previously (Xie et al. 2014). Briefly, 10 μM recombinant proteins were mixed with 60 μg/mL heparin sodium, 100 mM NaCl, 10 μM ThT, and 10 mM HEPES (pH 7.4). ThT fluorescence of the mixture was measured at the indicated time points at 465 ± 35 nm (excitation) and 535 ± 25 nm (emission).
Atomic force microscopy
Morphology of the aggregated proteins was viewed using a modification of a previously described protocol (Maeda et al. 2007). After the aggregation assay, samples that included the aggregates were loaded onto mica and incubated at room temperature (22–26°) for 30 min in a moist box. The mica was washed with Milli‐Q water, and the aggregates were imaged by atomic force microscope (SPM9700; Shimadzu Corp., Kyoto, Japan). Two or three images (2 × 2 μm) were obtained from the surface of the mica coated with the aggregates.
Molecular dynamic analysis
Molecular dynamics (MD) of peptides were performed by CHARMM (Brooks et al. 1983) in DS modeling program package (Accelrys Software Inc., San Diego, CA, USA). The simulation was performed in vacuum using the CHARMM force field. Two methods, Steepest Descents and Conjugate Gradients, were used in the energy minimization. The total calculation number was 2000 with 1 fs for one calculation. The equilibration dynamics of the experimental temperature were calculated 10 000 times with 1 fs for one calculation. Finally, the dynamics of 100 productions were saved for each system.
Results
The carboxyl‐terminal sequences of tau and MAP2 determine their aggregation properties
Because MAP2 typically contains 3R MTBDs and the shortest isoform MAP2c has a similar molecular weight as tau, we compared MAP2c and 3R‐tau in this study. Our previous studies suggested that tau and MAP2 have different aggregation properties (Xie et al. 2014). However, it was unknown which regions of tau and MAP2 determine the different aggregation properties. Thus, we asked whether the homologous carboxyl‐terminal sequence of tau and MAP2 is important for their aggregation properties. To address this issue, we expressed and purified recombinant full‐length 3R‐tau and MAP2c from E. coli. The homologous 160‐amino acid carboxyl‐terminal sequences of tau and MAP2 were exchanged with each other to obtain two chimeric proteins, which were named tauN–MAP2C and MAP2N–tauC (Fig. 1a). Aggregations of the following proteins, tau, MAP2, tauN–MAP2C, and MAP2N–tauC were induced with heparin and monitored by the ThT fluorescence, which reflects the amount of β‐pleated sheet structure (Fig. 1b). The ThT fluorescence value of tau increased constantly to a plateau in 24 h, whereas that of MAP2 stayed at a lower level. This suggested that tau and MAP2 have different aggregation properties, which is also consistent with the finding in our previous studies (Xie et al. 2014).
Figure 1.
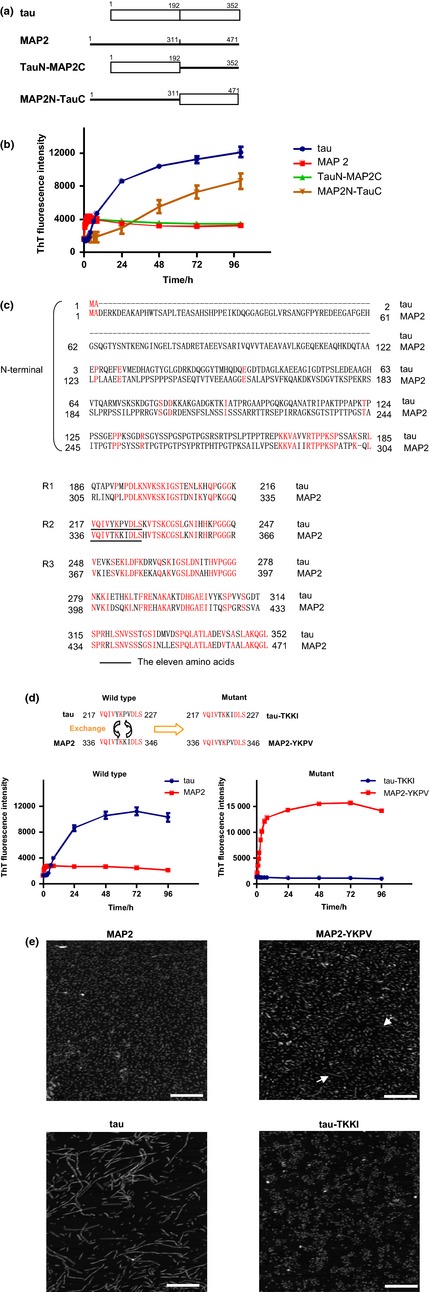
The aggregation properties of tau and MAP2 are determined by the homologous carboxyl‐terminal sequences. (a) Bar diagrams of tau (0N3R), MAP2c, and their chimeric proteins. The numbers shown above indicate the position of amino acids. (b) Thioflavin T (ThT) fluorescence of tau, MAP2, and chimeric protein aggregations was measured at the indicated time (h). Error bars represent SD; n = 3. (c) Comparison of the amino acid sequences of tau (0N3R) and MAP2c. The identical amino acids are shown in red. R, repeat; (d) ThT fluorescence of tau, MAP2, tau–TKKI, and MAP2–YKPV aggregations was measured at the indicated time points. Error bars represent SD; n = 4. (e) (AFM) imaging of tau, MAP2, tau–TKKI, and MAP2–YKPV aggregations. Arrows indicate the filaments that are formed from MAP2–YKPV. Bars = 400 nm. Note that exchanging the corresponding amino acids between tau and MAP2 (T and Y, K and P, and I and V) reversed their aggregation properties and that MAP2–YKPV aggregated to filaments but tau–TKKI remained granules.
To identify which region, the amino‐terminal or the carboxyl‐terminal sequence, is the key that determines the aggregation properties, the two chimeric proteins were incubated with heparin. The aggregation properties of MAP2N–tauC were similar to that of tau, whereas the aggregation properties of tauN–MAP2C were similar to that of MAP2. This suggested that the aggregation properties of tau and MAP2 depend on the homologous carboxyl‐terminal sequence (Fig. 1b).
Exchanging three amino acids between tau and MAP2 remarkably alters their aggregation properties
The carboxyl‐terminal sequences of tau and MAP2 are very similar and about 70% of the amino acids match completely (Fig. 1c). We next asked which amino acids in the carboxyl terminus are important for the different aggregation properties. Because the PHF6 motif (VQIVYK) is required for tau fibril formation (von Bergen et al. 2001), we compared the corresponding amino acid sequences of tau and MAP2 around the PHF6 motif and found a particular sequence of 11 amino acids (VQIVXKXXDLS) in tau and MAP2, as shown in Fig. 1(c). In the 11‐amino acid sequence, only three amino acids differed between tau and MAP2; that is, the corresponding sequence of 221YKPV224 in tau (0N3R) was replaced by 340TKKI343 in MAP2c.
To examine whether the small changes regulate the different aggregation properties of tau and MAP2, we constructed, expressed, and purified site‐directed mutant proteins in which YKPV of tau and TKKI of MAP2 were exchanged to obtain two mutant proteins, tau–TKKI and MAP2–YKPV (Fig. 1d, upper). To our surprise, the aggregation properties of tau and MAP2 monitored by ThT fluorescence were greatly altered by exchanging just the three amino acids (Fig. 1d, lower). The aggregation products after 7‐day incubation were imaged by atomic force microscopy. As shown in Fig. 1(e), for the wild‐type proteins, tau clearly aggregated to form filaments, whereas MAP2 formed only granules. For mutants, tau–TKKI, which contained three amino acids of MAP2, showed MAP2‐like aggregation properties in that only granules were formed. By contrast, MAP2–YKPV, which contained three amino acids of tau, led to filament formation, although the filaments were shorter than those formed by tau (Fig. 1e). These results clarified that the small difference between YKPV and TKKI determined the different aggregation properties of tau and MAP2 whether or not forming filaments.
Tyrosine and proline together are sufficient for the full filament formation
We next examined which amino acid is important for filament formation. Tyrosine 221 (Y221) has been proven to be essential for fibril formation (Nishiura et al. 2010; Sogawa et al. 2012). Mutating Y221 to another amino acid such as alanine abolished the aggregation of tau. However, it remained unclear whether the difference between Y221 of tau and threonine 340 (T340) of MAP2 is important to the regulation of the aggregation properties. We exchanged Y221 and T340 to obtain two site‐directed mutant proteins, tau–TKPV and MAP2–YKKI, and examined their aggregation properties by monitoring ThT fluorescence, as described in the method section. The ThT fluorescence value of tau–TKPV stayed at the baseline level, suggesting that tau–TKPV lost the ability to form filaments. Similarly, MAP2–YKKI did not aggregate to filaments either, which suggests that Y is essential but not sufficient for filament formation (Fig. 2a). Therefore, the other two amino acids, proline 223 (P223) and/or valine 224 (V224) of tau may be necessary for filament formation. We constructed and purified the other two mutant proteins MAP2–YKPI and MAP2–YKKV in which Y, and P or V were received from the tau sequence. The aggregation properties of MAP2–YKPI and MAP2–YKKV were examined. The fluorescence value of MAP2–YKPI increased constantly to a plateau comparable to that of tau, indicating the filament formation. By contrast, this increase in the fluorescence was not observed in MAP2–YKKV (Fig. 2b). Moreover, the MAP2‐TKPI mutant with only P substitution showed weaker filament formation ability, suggesting that only P is not sufficient for the full filament formation like MAP2‐YKPI. Taken together, these results indicated that Y and P together are sufficient for the full filament formation.
Figure 2.
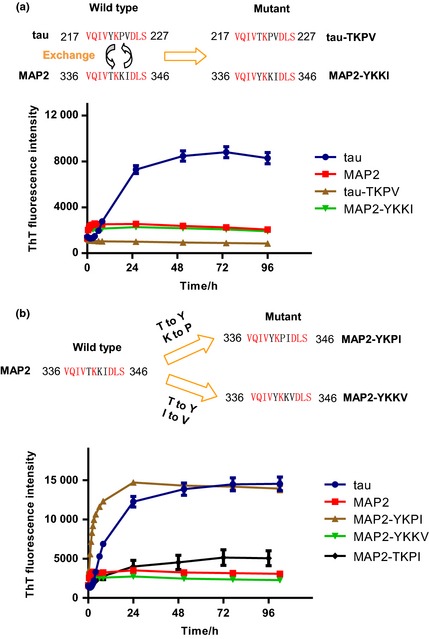
Key amino acids are responsible for the filament formation of tau and MAP2. Thioflavin T (ThT) fluorescence of tau, MAP2, tau‐TKPV, MAP2‐YKKI, MAP2‐YKPI, MAP2‐YKKV and MAP2‐TKPI were measured at the indicated time points. Error bars represent SD; n = 3. (a) Tyrosine 221 (Y) is essential but not sufficient for the filament formation. (b) Tyrosine 221 (Y) and proline 223 (P) together were sufficient for the full filament formation.
Molecular characteristics of the sequences
Because the two motifs YKPV and TKKI determined the aggregation properties, we speculate that the sequences containing YKPV and TKKI motifs may have completely different molecular conformations. To test this speculation, molecular dynamics simulation was performed to calculate the structural changes of two peptides, 217VQIVYKPVDLS227 of tau (0N3R) and 336VQIVTKKIDLS346 of MAP2c. The distance between the alpha carbon atoms was calculated at 37°C (Fig. 3a and b). In Fig. 3(a) for the peptide 1VQIVYKPVDLS11, all conformers showed a shorter distance between alpha carbon atoms of V1 and S11 than the distance between V1 and K6 or P7 suggested that the peptide has a relatively non‐extended conformation. On the other hand, in Fig. 3(b) for the peptide 1VQIVTKKIDLS11, all conformers showed a longer distance between alpha carbon atoms of V1 and S11 than the distance between V1 and K6 or K7, suggesting that the peptide has an extended conformation.
Figure 3.
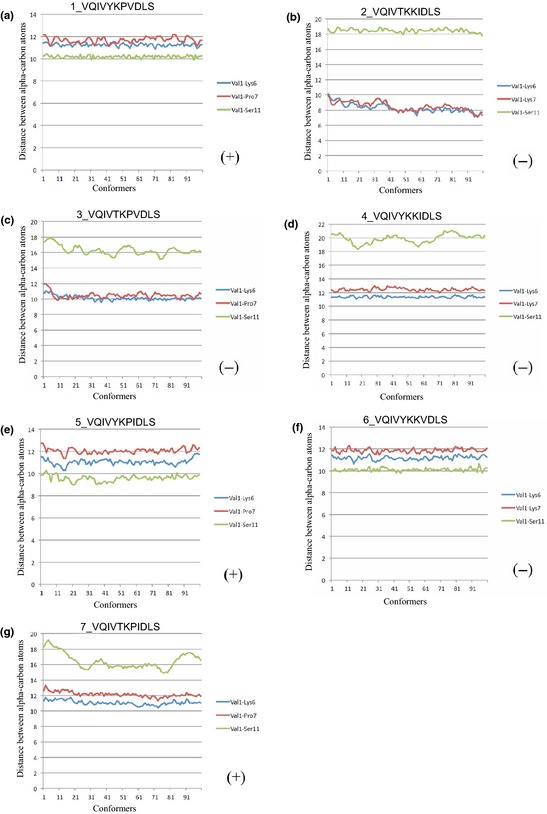
Molecular characteristics analysis of the sequences. The horizontal axes indicate conformers and the vertical axes indicate the distance between alpha carbon atoms (in Å) at 37°C. (+) indicates that the peptide have filament formation ability and (−) indicates no. The peptides (a, e, and f) showed non‐extended conformation and the peptides (b, c, d, and g) showed extended conformation.
To examine the relationship between molecular conformation and aggregation properties, we further analyzed the other sequences used in this study by molecular dynamics simulation. The peptides A, E, and G have filament formation ability, tended to show non‐extended conformation except for the peptide G, which showed an extended conformation. And the peptides that have no filament formation ability, including B, C, D, and F tended to show extended conformation except for the peptide F, which showed a non‐extended conformation (Fig. 3). It seems that there are no rules in the relationship between molecular conformation and aggregation properties here; however, our data give an insight into the conformations of these peptides and suggest that the filaments formation may be easier when the non‐extended molecular conformation exists and may be prevented relatively by the extended molecular conformation.
Discussion
We focused on MAP2 because it has a homologous carboxyl‐terminal sequence to that of tau. The carboxyl‐terminal region of tau is characterized by the localization of most mutations causing the hereditary tauopathy FTDP‐17 and forming the core of NFTs, suggesting a close relationship between the sequences and tauopathies. However, little is known about the relationship between MAP2 and tauopathies. Our previous study showed that tau and MAP2 have different fates in the formation of inclusions and prompted us to investigate the cause of this phenomenon (Xie et al. 2014). Tau and MAP2 are the two major microtubule‐associated proteins and, therefore, the different fates of both in NFT formation may be considered as one pathological hallmark of tauopathies. Moreover, the homologous sequence between tau and MAP2 suggests that MAP2 may be considered as a mutation of tau that is not involved in the inclusion‐forming process. Therefore, comparing the properties of MAP2 and tau may lead us to a better understanding of tauopathies.
The most important properties of tau inclusions found in AD brains are the β‐pleated sheet‐rich filaments. In Fig. 1, despite the MAP2‐YKPV and wild‐type tau forming different filaments and showing distinct aggregation kinetics, both formed β‐pleated sheet‐rich filaments, as judged by atomic force microscopy and ThT fluorescence. This suggests that the major properties of tau inclusions solely depend on the short YKPV motif.
Thus far, increasing evidence suggests that the mechanisms responsible for neuronal degeneration may not be limited to the formation of inclusions, which is the final step of the pathology (Gomez‐Isla et al. 1997; Miyasaka et al. 2005; Santacruz et al. 2005). Many studies showed that soluble species of tau such as oligomers were toxic to neurons (Cowan and Mudher 2013). Although MAP2 is incapable of forming filaments, our previous studies have reported that soluble MAP2 is neurotoxic, which raises the possibility that MAP2 may also be involved in the mechanism of neurodegeneration (Xie et al. 2014).
Author Contribution
C.X. and T.M. designed the study; C.X., T.M., and Y. Shinzaki performed the research; Y. Soeda performed the atomic force microscopic observation; Y. In and K.T. performed the molecular dynamics simulation; C.X., T.M., and Y. Ihara wrote the manuscript.
Acknowledgments and conflict of interest disclosure
This work was supported in part by Grant‐in‐Aid for Scientific Research (KAKENHI; Grant‐in‐Aid for Young Scientists (B); 20700324; T.M. and 24700368; C.X. and Grant‐in‐Aid for Challenging Exploratory Research; 22650074; T.M.), Core Research for Evolutional Science and Technology (CREST; T.M., C.X., and Y. Ihara), the Japan Science and Technology Agency (JST), the Japan Society for the Promotion of Science (JSPS) and Grant‐in‐Aid for Scientific Research on Innovative Areas (Brain Protein Aging and Dementia Control; T.M. 26117004).
All experiments were conducted in compliance with the ARRIVE guidelines. The authors have no conflict of interest to declare.
References
- von Bergen M., Barghorn S., Li L., Marx A., Biernat J., Mandelkow E. M. and Mandelkow E. (2001) Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta‐structure. J. Biol. Chem. 276, 48165–48174. [DOI] [PubMed] [Google Scholar]
- Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S. and Karplus M. (1983) A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217. [Google Scholar]
- Cowan C. M. and Mudher A. (2013) Are tau aggregates toxic or protective in tauopathies? Front. Neurol. 4, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmelt L. and Halpain S. (2004) The MAP2/Tau family of microtubule‐associated proteins. Genome Biol. 6, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Isla T., Hollister R., West H., Mui S., Growdon J. H., Petersen R. C., Parisi J. E. and Hyman B. T. (1997) Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer's disease. Ann. Neurol. 41, 17–24. [DOI] [PubMed] [Google Scholar]
- Kondo J., Honda T., Mori H., Hamada Y., Miura R., Ogawara M. and Ihara Y. (1988) The carboxyl third of tau is tightly bound to paired helical filaments. Neuron 1, 827–834. [DOI] [PubMed] [Google Scholar]
- Lee V. M., Goedert M. and Trojanowski J. Q. (2001) Neurodegenerative tauopathies. Annu. Rev. Neurosci. 24, 1121–1159. [DOI] [PubMed] [Google Scholar]
- Maeda S., Sahara N., Saito Y., Murayama M., Yoshiike Y., Kim H., Miyasaka T., Murayama S., Ikai A., Takashima A. (2007) Granular tau oligomers as intermediates of tau filaments. Biochemistry 46, 3856–3861. [DOI] [PubMed] [Google Scholar]
- Mandelkow E. M. and Mandelkow E. (2012) Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb. Perspect Med. 2, a006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka T., Ding Z., Gengyo‐Ando K., Oue M., Yamaguchi H., Mitani S. and Ihara Y. (2005) Progressive neurodegeneration in C. elegans model of tauopathy. Neurobiol. Dis. 20, 372–383. [DOI] [PubMed] [Google Scholar]
- Nishiura C., Takeuchi K., Minoura K., Sumida M., Taniguchi T., Tomoo K. and Ishida T. (2010) Importance of Tyr310 residue in the third repeat of microtubule binding domain for filament formation of tau protein. J. Biochem. 147, 405–414. [DOI] [PubMed] [Google Scholar]
- Ross C. A. and Poirier M. A. (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10(Suppl), S10–S17. [DOI] [PubMed] [Google Scholar]
- Santacruz K., Lewis J., Spires T. et al (2005) Tau suppression in a neurodegenerative mouse model improves memory function. Science 309, 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogawa K., Okuda R., In Y., Ishida T., Taniguchi T., Minoura K. and Tomoo K. (2012) C‐H. π interplay between Ile308 and Tyr310 residues in the third repeat of microtubule binding domain is indispensable for self‐assembly of three‐ and four‐repeat tau. J. Biochem. 152, 221–229. [DOI] [PubMed] [Google Scholar]
- Wischik C. M., Novak M., Thøgersen H. C., Edwards P. C., Runswick M. J., Jakes R., Walker J. E., Milstein C., Roth M. and Klug A. (1988) Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc. Natl Acad. Sci. USA 85, 4506–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Miyasaka T., Yoshimura S., Hatsuta H., Yoshina S., Kage‐Nakadai E., Mitani S., Murayama S. and Ihara Y. (2014) The homologous carboxyl‐terminal domains of microtubule‐associated protein 2 and TAU induce neuronal dysfunction and have differential fates in the evolution of neurofibrillary tangles. PLoS ONE 9, e89796. [DOI] [PMC free article] [PubMed] [Google Scholar]


