Abstract
The oxyntic proliferative isthmus zone contains the main stem/progenitor cells that provide for physiological renewal of the distinct mature cell lineages in the oxyntic epithelium of the stomach. These cells are also proposed to be the potential cells‐of‐origin of gastric cancer, although little is known about their molecular characteristics and specific biological markers are lacking. In this study, we developed a method for serial section‐navigated laser microdissection to isolate cells from the proliferative isthmus zone of rat gastric oxyntic mucosa for genome‐wide microarray gene expression analysis. Enrichment analysis showed a distinct gene expression profile for the isthmus zone, with genes regulating intracellular processes such as the cell cycle and ribosomal activity. The profile was also related to stem cell transcriptional networks and stomach neoplasia. Genes expressed uniquely in the isthmus zone were associated with E2F transcription factor 1 (E2F1), which participates in the self‐renewal of stem cells and in gastric carcinogenesis. One of the unique genes was Aspm [Asp (abnormal spindle) homologue, microcephaly‐associated (Drosophila)]. Here we show ASPM in single scattered epithelial cells located in the proliferative isthmus zone of rat, mouse and human oxyntic mucosa, which do not seem to be actively dividing. The ASPM‐expressing cells are mainly mature cell marker‐deficient, except for a limited overlap with cells with neuroendocrine and tuft cell features. Further, both ASPM and E2F1 were expressed in human gastric cancer cell lines and increased and correlated in human gastric adenocarcinomas compared to non‐tumour mucosa, as shown by expression profile analyses and immunohistochemistry. The association between ASPM and the transcription factor E2F1 in gastric tissue is relevant, due to their common involvement in crucial cell fate‐regulatory mechanisms. Our results thus introduce ASPM as a novel possible oxyntic stem/progenitor cell marker that may be involved in both normal gastric physiology and gastric carcinogenesis. © 2015 Authors. Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: microarray, stomach, neoplasia, immunocytochemistry, cell culture, in situ hybridization, immunofluorescence, navigated laser microdissection, cancer stem cells, mucosa
Introduction
The acid‐producing oxyntic mucosa of the stomach consists of glands that are divided into pit, isthmus, neck and base zones, based on the presence of characteristic cell types. To supply the constant turnover of the distinct mature cell lineages, the glands have multipotent stem cells that self‐renew and give rise to progenitor cells that differentiate further 1, 2, 3. These oxyntic stem/progenitor cells have been described morphologically and are believed to reside in the isthmus zone, which has been shown to be the predominant location of immature cells and cellular proliferation 2, 4, 5. Many putative markers for gastric stem cells have been proposed, although none have proved to completely fulfil the requirements of adult stem cells in the mature oxyntic mucosa 6, 7, 8, 9. LGR5 is a promising stem cell marker in all gastric mucosal areas during embryonic development and in adult antrum, but apparently not in adult corpus 10, 11. However, a recent report shows Lgr5 expression in a small subset of fully differentiated chief cells located at the oxyntic gland base. These cells also express the new stem cell marker Troy and can serve as quiescent ‘reserve’ stem cells activated by tissue damage. Troy is not expressed in the proliferating oxyntic isthmus zone and does not seem to mark the main physiological renewing stem cells, which are yet to be identified specifically 9. This indicates more plasticity in the oxyntic mucosa than was previously recognized. In general, much less is known about the gastric oxyntic stem/progenitor cells, which seem to differ from the more studied intestinal and antral stem cells 6, 7, 8.
A predominant concept is that resident adult stem/progenitor cells can accumulate mutations, undergo transformation and thereby become cancer‐initiating and cancer stem cells. These are defined as having the ability to self‐renew and to provide all the heterogeneous cells that comprise a tumour, thus being responsible for its maintenance and growth 12, 13. Despite the progress in unravelling the molecular carcinogenesis of gastric adenocarcinomas (GAs), clinical outcome is little improved and the disease still remains a worldwide health problem. The role of cancer stem cells is not fully understood 7, 8. Increased knowledge about the gastric stem cell niche is important to improve understanding of normal cellular homeostasis and to elucidate the underpinning mechanisms involved in common diseases of the stomach. Due to the lack of specific biomarkers and methods to isolate stem/progenitor cells, eg for gene expression analysis, these cells have been difficult to study. Laser microdissection (LM) enables rapid and precise sampling of defined morphological areas from heterogeneous tissue sections. This is a valuable approach for specific high‐throughput analysis and identification of candidate molecular markers, which have an important place in understanding cell position, type and relations in the dynamic renewal of gastric glands. Such knowledge is also required to recognize markers that would be suitable for further studies, eg by clonal lineage tracing.
Thus, we have developed a method using serial section‐navigated LM to isolate cells from the proliferative isthmus zone of rat gastric oxyntic mucosa for genome‐wide gene expression analysis. We introduce ASPM [Asp (abnormal spindle) homologue, microcephaly‐associated (Drosophila)] as a novel gastric cell marker, possibly for the oxyntic stem/progenitor cells, and are the first to investigate ASPM in association with the stem cell‐related E2F transcription factor 1 (E2F1) in normal and malignant gastric tissues.
Materials and methods
Gastric cell lines, animal and human tissues
The study was approved by the Norwegian National Animal Research Authority and the animals were handled according to guidelines described previously 14. Oxyntic mucosal biopsies were taken from the greater curvature and either immediately frozen in liquid nitrogen for the preparation of frozen sections or fixed in formalin for immunohistochemistry (IHC). The human material used was extracted RNA and formalin fixed, paraffin‐embedded (FFPE) biopsies from GAs, adjacent non‐tumour mucosa and matched normal mucosa. The patients gave written informed consent and the study was approved by the Regional Medical Research Ethics Committee of Central Norway (Approval No. 018–02). The human gastric adenocarcinoma cell lines AGS [American Type Culture Collection (ATCC), Rockville, MD, USA], MKN‐45 (gift from Queens Medical Centre, University Hospital, Nottingham, UK) and KATO‐III (ATCC) were grown under standard conditions (see supplementary material, Supplementary materials and methods). Whole‐cell extracts were prepared and protein concentrations measured, as described previously 15.
Laser microdissection and RNA isolation
Biopsies from rat oxyntic mucosa (n = 6) were mounted in Tissue‐Tek OCT (Sakura, Alpena an den Rijn, The Netherlands) and from each biopsy a 5 µm guiding section was cut, followed by three 16 µm sections which were mounted on membrane slides [Molecular Machines and Industries (MMI), Zurich, Switzerland]. The guiding section was stained for the proliferation marker Ki67 to identify the isthmus zone and placed in the MMI CellCut Plus instrument (with Olympus IX71 inverted microscope) together with the sequential unstained sections from which the tissue was harvested. The isthmus zone was located using the software's serial section function (CellTools, v. 4.0.9) that allowed the operator to transfer information about the cutting area from one stained guiding section to unstained serial sections. RNA was extracted from the microdissected tissues using RNAqueous Micro Kit (Life Technologies, Grand Island, NY, USA); the first incubation was performed with the tubes upside down, to ensure lysis of the harvested tissue adherent to the MMI cap. RNA integrity was measured as a RIN score using the Bioanalyser system (Agilent Technologies, Santa Clara, CA, USA) (see supplementary material, Supplementary materials and methods).
Genome‐wide gene expression analysis
The cRNA was prepared with Ambion's Illumina® TotalPrep™‐96 RNA Amplification Kit (Life Technologies), using all extracted total RNA from the laser‐microdissected samples as input material. For each sample, the biotin‐labelled cRNA concentrations were checked (NanoDrop, Thermo Fisher Scientific, Wilmington, DE, USA) and adjusted to 150 ng/µl before hybridization to RatRef‐12 Expression BeadChip (Illumina, San Diego, CA, USA). Only probes abiding to the Illumina detection p value of 0.01 in at least one sample were included in further analyses, using the limma (v. 3.12.1) Bioconductor package 16 for paired t‐tests. A FDR adjusted p value < 0.05 was taken as significant. Differentially expressed genes, with expression above the Illumina detection limit in ≥ 5 samples from the isthmus zone and expression above background in ≤ 1 sample from the remaining oxyntic mucosa, were defined as genes unique for the isthmus zone. The microarray data are available from the ArrayExpress Repository 17 (Accession No. E‐MTAB‐2542). Canonical pathways, gene‐disease association and transcriptional networks associations were determined by MetaCoreTM 18 and enrichment analysis, with a p < 0.05 threshold for the data inputs (see supplementary material, Supplementary materials and methods). Expression profile analyses of ASPM and E2F1 mRNA in multiple human gastric cancer datasets were performed using the Oncomine database (www.oncomine.org) and an in‐house gene expression dataset, where RNA isolation and microarray analysis followed standard protocols using 300 ng total RNA/sample on Human HT‐12 Expression Bead Chips (Illumina; see Accession No. E‐MTAB‐1338).
Immunohistochemistry and in situ hybridization
The frozen guiding sections (5 µm) were fixed in precooled ethanol and endogenous peroxidases quenched prior to incubation with a Ki67 antibody for 2 h at room temperature (RT). Standard pretreatments such as permeabilization, quenching of endogenous peroxidases and antigen retrieval of FFPE sections (4 µm) were followed by incubation with the primary antibodies for 1–2 h at RT, or overnight at 4 °C. All single‐stained sections were prepared using the rabbit/mouse EnVision‐HRP/DAB+ kit (Dako, Glostrup, Denmark) and counterstaining with haematoxylin. For double‐stained sections, primary antibodies were incubated simultaneously and visualized following 1 h of incubation at RT with fluorescent secondary antibodies or Lectin GS‐II (Life Technologies) and counterstaining with DAPI. Tyramide Signal Amplification (TSA kits, Life Technologies) was used for ASPM and E2F1 in combination with other antibodies. In situ hybridization (ISH) for ASPM mRNA was performed using an RNAscope 2.0 HD Reagent Kit (Brown) for FFPE tissue (310035; Advanced Cell Diagnostics (ACD), Hayward, CA, USA) and probeset (427881, ACD) according to the manufacturer's instructions, except for increased boiling time (Pretreat 2; see supplementary material, Supplementary materials and methods).
Western blot and immunocytochemistry
Proteins (60 µg/well) were separated on 3–8% NuPage Novex Tris/acetate gels (Invitrogen) and electroblotted onto Immobilon PVDF membranes (Millipore). The membranes were blocked and binding of the primary antibodies (ASPM, E2F1 and β‐actin) and HRP‐conjugated or fluorescent secondary antibodies were visualized using the Odyssey Fc imaging system (LI‐COR Biosciences, Lincoln, NE, USA). For immunocytochemistry, cells were grown in eight‐well Lab‐Tek™ Chambered Coverglass (NUNC, Thermo Scientific, Rockford, IL, USA), fixed with paraformaldehyde, permeabilized (Triton‐X) and blocked with bovine serum albumin (BSA). The cells were incubated overnight at 4 °C with the primary antibodies, reblocked and incubated for 1 h with fluorescent secondary antibodies. DNA was stained with DAPI (see supplementary material, Supplementary materials and methods).
Histomorphometry, statistical analysis and imaging
Immunoreactivity in GAs and normal human mucosa was evaluated by mean index value, by multiplying staining intensity (weak = 1, moderate = 2, strong = 3) and percentage of immunoreactive cells (<10% = 1, 10–50% = 2, > 50% = 3) from five randomly selected areas (500 × 500 µm) in each sample. Immunofluorescent staining was evaluated using an Olympus IX71 inverted microscope with a digital monochrome XM10 camera and CellP software (Olympus Co., Tokyo, Japan), or a Leica SP8 inverted microscope (Leica Microsystems, Mannheim, Germany) equipped with an HC plan‐apochromat 63×/1.4 CS2 oil‐immersion objective, with light laser and hybrid (HyD) detectors. Statistically significant differences (ANOVA with Bonferroni's multiple comparison test and two‐sided Fisher's exact test) and correlation (Pearson) were analysed using Prism 5 (GraphPad Software, San Diego, CA, USA).
Results
Genome‐wide analysis shows a distinct gene expression profile of the oxyntic proliferative isthmus zone compared to the remaining mucosa
Serial section‐navigated LM provided enough RNA of sufficient quality (mean RIN 8.0; see supplementary material, Supplementary results) to compare the gene expression profiles in the isthmus zone to the remaining mucosa (Figure 1Ai–iii). Initial interpretation of the data was done using principal component analysis (PCA); this showed that the principal variation was within each animal, with the gene expression profiles of the isthmus zone and the remaining mucosa constituting two distinctly different tissue zones (Figure 1B). Of the 1131 differentially expressed transcripts, 599 were more and 532 were less abundant in the isthmus zone compared to the remaining mucosa (see supplementary material, Table S1).
Figure 1.
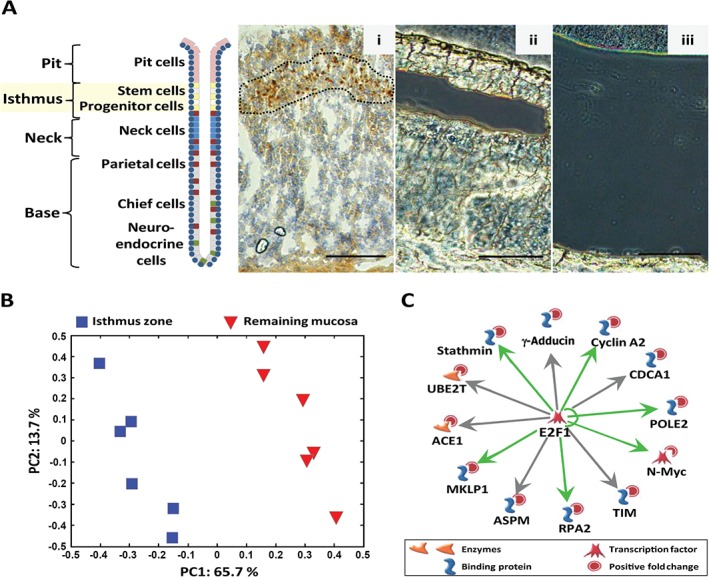
Gene expression analysis of the rat oxyntic proliferative isthmus zone sampled by laser microdissection. (A) Schematic illustration of the architectural organization of the gastric oxyntic gland, showing positions of major cell types in the different regions – pit, isthmus, neck and base. Stem and progenitor cells in the isthmus region are highlighted. Frozen sections of rat oxyntic mucosa showing: (i) immunostaining for the proliferation marker Ki67 (brown), used as a guide for the isthmus zone during laser microdissection, and representative examples of unstained frozen sections after laser microdissection of (ii) the isthmus zone and (iii) the remaining mucosa used for RNA extraction and genome‐wide gene expression analysis. (B) Plot from principal component analysis (PCA) visualizing variation between the expression profiles of the 12 different samples; each point represents one sample from the microdissected isthmus zone (Aii) (blue  ) and the remaining mucosa (Aiii) (red
) and the remaining mucosa (Aiii) (red  ); the axis indicates the percentage of total variance explained in each component. (C) 'MetaCore transcriptional regulation' algorithm with default settings was used to generate the top‐scored E2F1‐centred transcriptional network for the genes (n = 51) uniquely expressed in the microdissected isthmus zone (Aii); The transcriptional regulation is indicated as known (green arrows) or possible (grey arrows); other symbols are as specified in the figure. Scale bars = 100 µm
); the axis indicates the percentage of total variance explained in each component. (C) 'MetaCore transcriptional regulation' algorithm with default settings was used to generate the top‐scored E2F1‐centred transcriptional network for the genes (n = 51) uniquely expressed in the microdissected isthmus zone (Aii); The transcriptional regulation is indicated as known (green arrows) or possible (grey arrows); other symbols are as specified in the figure. Scale bars = 100 µm
To search for molecular processes that might characterize the two tissue zones, differentially expressed genes were subjected to enrichment analysis. For the genes (513 identified IDs) over‐expressed in the isthmus zone compared to the remaining mucosa, we found that the top‐scoring canonical pathways and networks were related to cell cycle phases and protein translation (Table 1). In contrast, the genes (447 identified IDs) that were under‐expressed in the isthmus zone were significantly enriched in pathways and networks related to growth‐factor regulation (eg the stomach hormone gastrin), differentiation and unfolded protein response (UPR)/ER‐stress (Table 1). Many of these under‐expressed genes (see supplementary material, Table S1) are known gastrin‐responsive genes and are expressed by mature cells located at the base of oxyntic glands 14, 19, 20.
Table 1.
Canonical pathways and networks most significantly associated with genes expressed at higher or lower levels in the oxyntic proliferative isthmus zone compared to the remaining mucosa
| Enrichment by MetaCore™ | p | Network objects from active data a , b |
|---|---|---|
| Genes over‐expressed in the isthmus zone | ||
| Pathway maps | ||
| Cell cycle_Sister chromatid cohesion | 1.22E‐08 | PCNA, DNA polymerase sigma, Rad21, DCC1, RFC3, Separase, Histone H1, CDK1 (p34) |
| Cell cycle_Role of APC in cell cycle regulation | 3.33E‐07 | BUB1, Geminin, Cyclin A, PLK1, Aurora‐B, Kid, CDC20, CDK1 (p34) |
| Cell cycle_Spindle assembly and chromosome separation | 4.316E‐07 | TPX2, CSE1L, KNSL1, Aurora‐B, Kid, CDC20, Separase, CDK1 (p34) |
| Process networks | ||
| Cell cycle_S‐phase | 2.12E‐12 | BUB1, β‐Tubulin, SPBC25, TPX2, CSE1L, Cyclin A, PARP‐2, PLK1, Actin, KNSL1, Aurora‐B, HP1γ, ASPM, Kid, CDCA1, HP1, CDC20, Rad21, Separase, MKLP1, Histone H1, CDK1 (p34) |
| Cell cycle_Mitosis | 4.05E‐07 | BUB1, β‐Tubulin, SPBC25, TPX2, CSE1L, Cyclin A, PARP‐2, PLK1, Actin, KNSL1, Aurora‐B, HP1γ, ASPM, Kid, CDCA1, HP1, CDC20, Rad21, Separase, MKLP1, Histone H1, CDK1 (p34) |
| Translation_Translation initiation | 3.70E‐07 | rpL23a, RPL39, RPL19, RPL26, RPS5, RPL3, RPL29, eIF5A, RPS8, PABPC1, RPS15, RPS21, RPL23, RPL27, eIF3S5, RPL35, RPS2, eIF3S3, RPS7, RPLP0, RPS25 |
| Genes under‐expressed in the isthmus zone | ||
| Pathway maps | ||
| Cell cycle_Sister chromatid cohesion protein folding and maturation_Angiotensin system maturation\rodent version | 1.84E‐07 | Angiotensin II, Angiotensin IV, Angiotensin I, Angiotensin III, Angiotensin (2–10), CD13*, Angiotensin (1–7), Angiotensinogen**, Angiotensin (1–9) |
| Development_Gastrin in differentiation of the gastric mucosa | 5.09E‐06 | VMAT2, HDC, PKC‐β, ICRF, IP3 receptor, PKC, cPKC (conventional) |
| Translation_Regulation of EIF4F activity | 4.92E‐05 | p38 MAPK, 4E‐BP1, PDK (PDPK1), p70 S6 kinase2, eIF4E, PAK1, EGF |
| Process networks | ||
| Protein folding_Response to unfolded proteins | 1.06E‐08 | HSP90, ATF‐4, Endoplasmin, CHIP, GRP78, HSP70, UFD2, Calnexin, Glutaredoxin, PDI, HERP, XBP1, SELS |
| Cell cycle_G1–S growth factor regulation | 6.33E‐06 | STAT3, PDGF‐A, IGF‐2, G‐protein α‐i family, Inhibin‐α, NDPK‐A, IKKβ, TGFβ3, LTBP3, Tob1, PKCβ, FGFR1, PKC, EGF, TGFβ, ActRIIB, cPKC (conventional) |
| Development_Regulation of angiogenesis | 9.85E‐06 | IRAK1/2, STAT3, Angiotensin II, PDGF‐A, G‐protein α‐i family, Angiotensin III, PDK (PDPK1), Calnexin, IRAK1, BTG1, PLCδ, CD13, IP3 receptor, PKC, PAK1, PLCδ, Clusterin, ITGA7 |
The objects are generated by MetaCore tools 18 based on 513 uploaded gene IDs.
The objects are generated by MetaCore tools based on 447 uploaded gene IDs (see supplementary material, Table S1).
CD13 alias, Anpep (ILMN_1350090).
Agt (ILMN_1361016).
In accordance with earlier reports 4, 6, 7, 8, 10, 21, the mRNA levels of eg Prom1, Tff2, Dclk1, Pcna, Muc6 and Adam17 were higher in the oxyntic isthmus zone, while others were not differentially expressed (Sox2) or not detected (Lgr5, Msh‐1, Sox9 or CD44) (see supplementary material, Table S1). Further analysis of ADAM17 by IHC showed strong expression in possibly preparietal cells in the isthmus zone, which agrees with our gene expression data but not with earlier reported localization 22, 23 (see supplementary material, Supplementary results, Figure S1). Overall, we find that our gene expression data corroborate previous results (see supplementary material, Table S2). This validates our experimental set‐up using serial section‐navigated LM and shows a distinctly different expression profile of the proliferative isthmus zone compared to the remaining oxyntic mucosa.
Genes expressed in the oxyntic proliferative isthmus zone are associated with stem cell transcriptional networks and stomach neoplasia
To further characterize the gene expression signature of cells in the oxyntic proliferative isthmus zone, the genes over‐expressed in this zone compared to the remaining mucosa were analysed for transcriptional networks. We identified sub‐networks with a centre of transcription factors including c‐MYC, OCT‐3/4, E2F1, NANOG and KLF4. These are important in maintaining pluripotency 24, 25 and self‐renewal of pluripotent stem cells 26, 27 and were predicted to be regulators of several (n = 165, 84, 83, 71 and 57, respectively) of the 513 over‐expressed genes (data not shown). In addition, when the genes uniquely expressed (n = 51) in the isthmus zone (ie below detection limit in the remaining mucosa) (see supplementary material, Table S3) were analysed separately, the top scored transcriptional network was centred on the transcription factor E2F1 (Table 2, Figure 1C). Some of the 12 unique genes in this network (N‐Myc, Rpa2, Mklp1, Stathmin, Pole2 and Cyclin A) are known to be transcriptionally regulated by E2F1 28, 29, 30 (Figure 1C, green arrows), while others (Ace1, Ube2t, γ‐Adductin, Cdca1, Tim and Aspm) have been identified as possible E2F1 target genes by ChIP–chip analysis 27, 31, 32, 33, 34 (Figure 1C, grey arrows). A gene–disease association analysis indicated that the genes over‐expressed in the isthmus zone compared to the remaining mucosa were significantly associated with neoplasm in the gastrointestinal system. Interestingly, the transcription factor E2F1 was predicted to regulate many of the genes associated with stomach neoplasm, including all 12 unique genes (Figure 1C) (see supplementary material, Supplementary materials and methods, Table S4). Thus, from our genome‐wide data we can deduce that the genes expressed in the proliferative isthmus zone of oxyntic mucosa are associated with stem cell transcriptional networks and stomach neoplasm.
Table 2.
Top scored transcriptional networks for genes unique for the oxyntic proliferative isthmus zonea
| Key network objects | p | Network objects from active data b |
|---|---|---|
| E2F1 | 2.07e‐39 | γ‐Adductin, Cyclin A2, CDCA1, POLE2, n‐Myc, TIM, RPA2, ASPM, MKLP1, ACE1, UBE2T, Stathmin |
| CREB1 | 4.77e‐36 | Cyclin A2, Stathmin, RPA2, RHAMM, MARCKS, C19orf6, CDA1, STAT5B, LTBP1, PFTAIRE‐1, KIAA1712 |
| c‐Myc | 2.21e‐29 | BUB1, ACE1, Stathmin, MKLP1, PFTAIRE‐1, Cyclin A, C6orf108, N‐Myc, RHAMM |
| E2F4 | 4.46e‐26 | UBE2T, ASPM, Stathmin, N‐Myc, CDCA1, MKLP1, RHAMM, Cyclin A2 |
| c‐Jun | 1.57e‐19 | LTBP1, Cyclin A2, STAT5B, MARCKS, RHAMM, THAS |
| ZNF143 | 1.57e‐19 | FANCB, C19orf6, Stathmin, BUB1, MKLP1, Cyclin A2 |
| P53 | 1.57e‐19 | THAS, Cyclin A2, Stathmin, DCC1, RHAMM, N‐Myc |
The 51 genes were below detection threshold in the remaining mucosa, as described in Materials and methods; see also supplementary material, Table S3.
Analysis in MetaCore 18 using the 'Transcriptional regulation algorithm' with default settings resulted in sub‐networks with a centre of various transcription factors. The seven top scored networks (by the number of pathways > 5) are listed.
ASPM is expressed in scattered epithelial cells in the oxyntic proliferative isthmus zone
Among the possible E2F1 target genes (Figure 1C), Aspm seemed particularly interesting to investigate further, as it is associated with stem cells in other tissues 35, 36, 37, 38, 39, 40, 41 and is a suggested target gene for c‐MYC 27, 42 and KLF4 27. In addition, E2F1 and ASPM share a functional association with cell cycle regulation 26, 35, 40, 43, 44, 45, 46. IHC showed strong ASPM expression mostly in small, scattered cells with relatively limited cytoplasm, located primarily in the proliferative isthmus zone of rat and human oxyntic mucosa (Figure 2A, B). A similar expression pattern was also found in mouse oxyntic mucosa (Figure 2C) and confirmed by ISH in human mucosa (Figure 2D). The cells were of epithelial origin, as shown by consistent co‐localization with E‐cadherin, and most ASPM‐expressing cells did not seem to be actively dividing, as we could not find compelling co‐localization with Ki67 (or PCNA) (n = 250) (Figure 2E, F). Still, morphologically we observed a very few ASPM‐expressing cells (∼0.5%) in division (Figure 2A, E).
Figure 2.
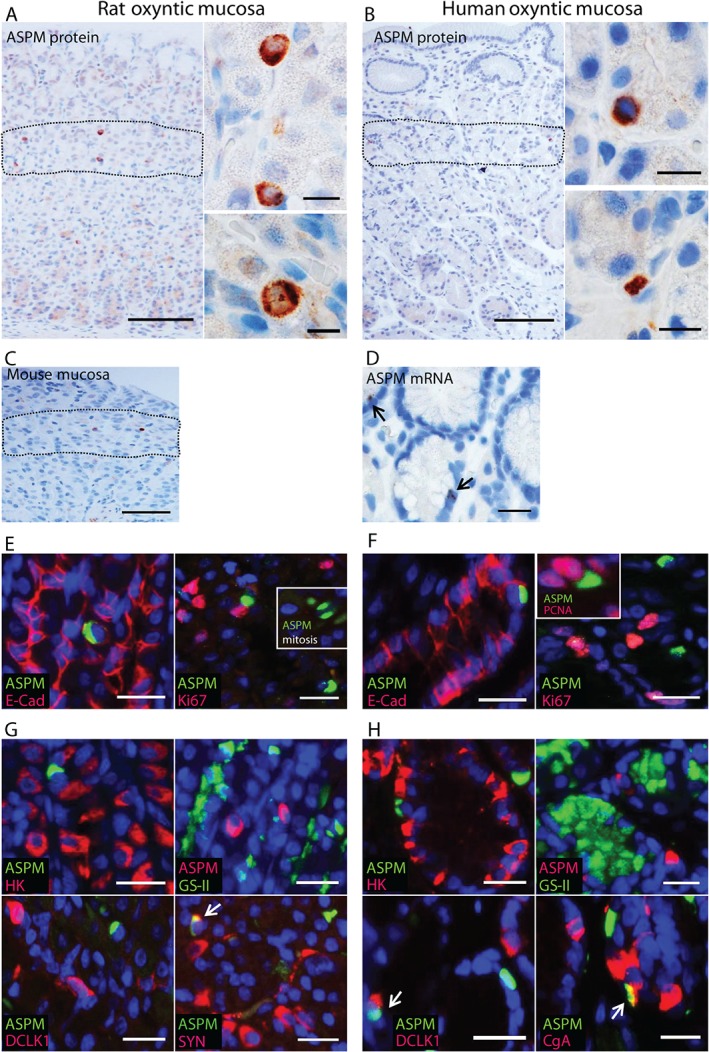
ASPM is expressed in scattered epithelial cells in the oxyntic proliferative isthmus zone. (A–C) Immunostaining showing ASPM expression (brown) in single cells in the isthmus zone (dashed line) of (A) rat oxyntic mucosa (n = 14), (B) human oxyntic mucosa (n = 14) and (C) mouse oxyntic mucosa (n = 4); Lower right inset in (A) shows a rat ASPM‐expressing cell in mitosis. (D) In situ hybridization showing a similar pattern of ASPM mRNA expression in single cells in human oxyntic mucosa (n = 3). (E, F) Double immunofluorescence staining showing co‐expression of ASPM (green) and the epithelial marker E‐cadherin (E‐Cad, red), and no obvious co‐localization of ASPM (green) and the proliferation marker Ki67 (red) in (E) rat oxyntic mucosa (n = 3–10) and (F) human oxyntic mucosa (n = 3–8); insets show an ASPM‐expressing cell (green) in mitosis (rat) and possible co‐localization with PCNA (red) in human. (G, H) Double immunofluorescence staining showing no co‐localization, or limited overlap, as indicated by the arrows, of ASPM (green) and the parietal cell marker H+/K+‐ATPase (HK, red), ASPM (red) and the mucous neck cell marker GS‐II (green), ASPM (green) and the tuft cell marker DCLK1(red), and ASPM (green) and the neuroendocrine markers synaptophysin (SYN, red) or chromogranin A (CgA, red) in (G) rat oxyntic mucosa (n = 3) and (H) human oxyntic mucosa (n = 3–10). Nuclei were counterstained with DAPI (blue); scale bars = (A, B) 100 µm; (insets) 20 µm; (C–H) 50 µm
Established markers were used to determine whether ASPM was expressed by any of the distinct cell types present in the oxyntic isthmus zone. We did not detect co‐localization (each n = 100) with H+/K+‐ATPase (parietal cells) or GS‐II (mucous neck cells) in rat or human, or with DCLK1 (tuft cells 47) in rat (Figure 2G, H). Conversely, a few ASPM‐expressing cells seemed co‐stained for DCLK1 (tuft cells) or chromogranin A (neuroendocrine cells) (both n = 2–3/100) in human stomach (Figure 2H) and somewhat more for synaptophysin (neuroendocrine cells) (n = 29/100) in rat (Figure 2G). Even though there could be doubts about the explicit localization of synaptophysin, this suggests that some of the ASPM‐expressing cells could have neuroendocrine or tuft cell features. Taken together, our results verify the microarray results at the protein level and show expression of ASPM in single, mainly mature cell marker‐deficient epithelial cells that apparently are not actively dividing and located in the isthmus zone of oxyntic mucosa.
ASPM and E2F1 are over‐expressed in human gastric cancer
Given our findings, we investigated further the relevance of ASPM and the possible association with the E2F1 transcription factor in gastric cancer. First, we used western blotting to show that ASPM and E2F1 are indeed expressed in the human gastric adenocarcinoma cell lines AGS, MKN‐45 and KATO‐III; ASPM at high levels in all three cell lines and E2F1 most distinctly in KATO‐III (Figure 3A). Correspondingly, in all cell lines, immunocytochemistry showed ASPM localized to both nuclei and cytoplasm during interphase (Figure 3B) and condensed to the spindle poles in mitotic cells (Figure 3C, D), as previously described 40, 41, 43, 46, 48. E2F1 was mainly observed in the nuclei of non‐mitotic cells, with varying intensity and degree of co‐localization with ASPM (Figure 3B, C). Thus, ASPM and E2F1 are both expressed in gastric adenocarcinoma cells.
Figure 3.
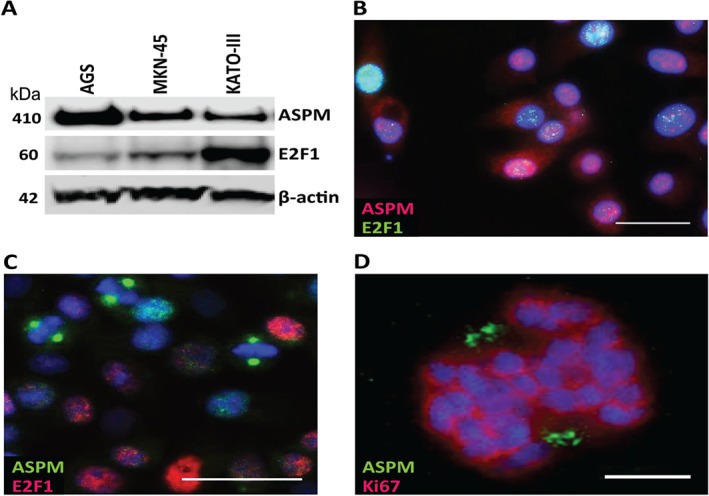
ASPM and E2F1 expression in human gastric cancer cell lines. (A) Representative western blot (n = 4) of ASPM (full‐length) and E2F1 in the gastric cancer cell lines AGS, MKN‐45 and KATO‐III; β‐actin was used as loading control. (B) Immunocytochemical staining of MKN‐45 cells (n = 3), showing cytoplasmic and nuclear localization of ASPM (red; Millipore) and nuclear localization of the transcription factor E2F1 (green) in cells in interphase. (C) Immunocytochemical staining of MKN‐45 cells (n = 3), showing ASPM (green; Bethyl) condensed to the spindle poles in mitotic cells that do not seem to express E2F1 (red). (D) Confocal imaging showing ASPM (green) localized to the spindle poles during metaphase, as indicated by the position of Ki67 (red); nuclear DNA is stained with DAPI (blue). For the confocal image, DAPI, Alexa488 and Alexa647 were excited by 405, 500 and 640 nm laser lines and the fluorescence detected in the ranges 421–475, 510–564 and 653–711 nm, respectively. Sequential imaging was performed to minimize crosstalk. Deconvolution was performed using Huygens Professional software (v 143.06.1p3; Scientific Volume Imaging). Scale bars = (B, C) 25 µm; (D) 5 µm
Next, we analysed the mRNA expression profiles of human gastric cancers. Four relevant datasets (346 samples) from Oncomine demonstrated an over‐expression of both ASPM and E2F1 mRNA in gastric cancers compared with non‐tumour gastric tissues, also when distinguishing between different histological subtypes of GAs (149 samples) in datasets from Cho et al 49 and D'Errico et al 50. This was further supported by our in‐house dataset (Figure 4A, B), which also revealed a significant correlation (r = 0.5707, p < 0.0001) between the ASPM and E2F1 expression levels in both non‐tumour tissues and GAs (Figure 4C). To examine this at the protein level, we immunostained a selection of GAs (n = 36) and compared the mean index value with that of normal mucosa (n = 14) using Fisher's exact test. A significant over‐expression (index value > 3) of ASPM (p = 0.0221) and E2F1 (p = 0.0044) in tumour cells was found in 11/36 samples (30.6%; Figures 2B, 5A) and 15/36 samples (41.7%; Figure 5A, B), respectively. In normal mucosa, few E2F1‐expressing cells were detected, also mainly in the isthmus zone (Figure 5B). ISH demonstrated that ASPM mRNA was present in tumour cells but apparently not in stromal cells, in contrast to the ASPM immunostaining (Figure 5A, C). In our samples we found no correlation between tumours over‐expressing ASPM and E2F1, which might reflect different post‐transcriptional regulation of the transcription factor and the target gene. Still, we observed co‐localization of ASPM and E2F1 in tumour cells (Figure 5D). ASPM and Ki67 on adjacent sections co‐localized to the same areas; however, most of the proliferating cells were not ASPM‐immunoreactive (Figure 5E). Considering the proportion of GAs arising from antral mucosa, the single cell‐expression of ASPM in the isthmus zone was also confirmed in normal antrum (see supplementary material, Figure S2) and the mRNA over‐expression was independent of tumour localization (see supplementary material, Supplementary materials and methods; data not shown). Taken together, our findings show over‐expression of both ASPM and E2F1 in human GAs compared to non‐tumour tissue. A possible association between ASPM and the transcription factor E2F1 was further supported by correlated mRNA levels.
Figure 4.
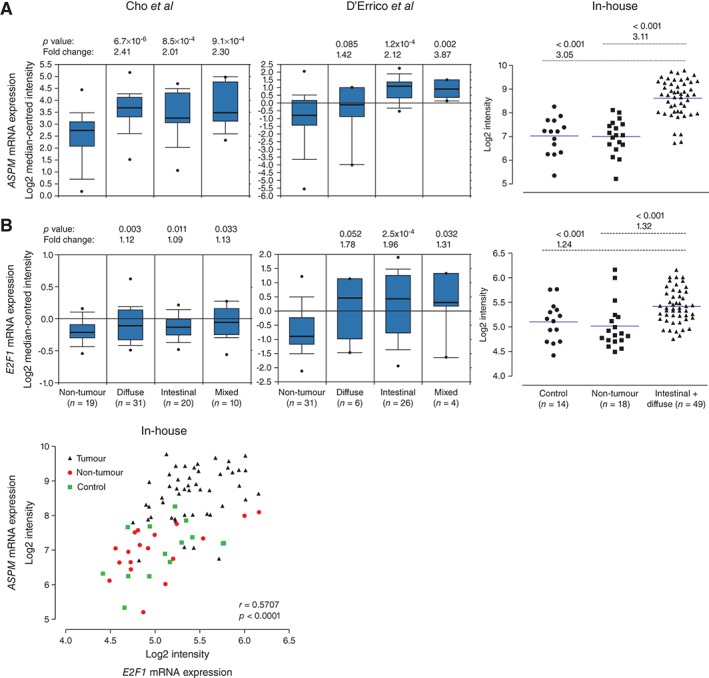
Expression profile meta‐analysis of ASPM and E2F1 mRNA in human gastric adenocarcinomas, using Oncomine and in‐house datasets. (A) Cho et al
49 (ILMN_1815184 Illumina HumanWG‐6 v3.0), D'Errico et al
50 (reporter 232238, Human Genome U133 Plus 2.0 Array) and our in‐house gene expression datasets (ILMN_1815184 Illumina HumanHT‐12) showing increased expression of ASPM mRNA levels in human gastric adenocarcinomas of diffuse, intestinal and mixed type compared with non‐tumour gastric tissues. (B) Cho et al
49 (reporter; ILMN_12051469 Illumina Human WG‐6 v. 3.0), D'Errico et al
50 (reporter 204947, Human Genome U133 Plus 2.0 Array) and our in‐house gene expression datasets (ILMN_2051469 Illumina Human HT‐12), showing increased E2F1 mRNA levels in human gastric adenocarcinomas of diffuse, intestinal and mixed type compared to non‐tumour gastric tissues. (C) The mRNA expression levels of ASPM and E2F1 were significantly correlated in the in‐house gene expression dataset; fold changes, symbols, correlation coefficient (r) and p values are as specified in the figure. The Oncomine data show log2 median‐centred intensities, the box plots indicate median and 90% central range, and  represents maximum and minimum values. The symbols in the in‐house dataset represent individual mRNA expression levels (log2 intensities) in the designated groups, with the blue line indicating median
represents maximum and minimum values. The symbols in the in‐house dataset represent individual mRNA expression levels (log2 intensities) in the designated groups, with the blue line indicating median
Figure 5.
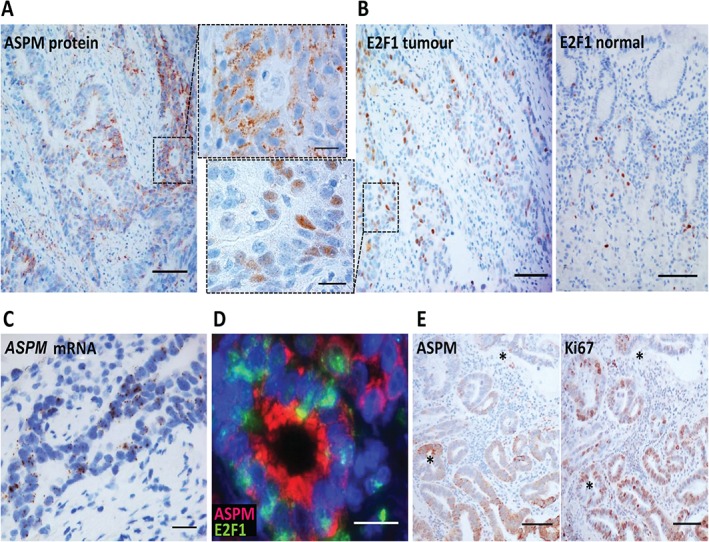
Immunohistochemical analysis of ASPM and E2F1 protein expression in human gastric adenocarcinomas. (A) Immunostaining showing significantly higher ASPM expression (brown; calculated index value > 3) compared to normal expression (Figure 2B) in patches of tumour cells with a mainly granular‐like and cytoplasmic pattern (insets with higher magnification). (B) Immunostaining showing significantly higher E2F1 expression (brown; calculated index value > 3) in nuclei of tumour cells compared to normal expression (calculated index value ≤ 3). (C) In situ hybridization (n = 3) showing a similar pattern of ASPM mRNA expression in tumour cells. (D) Double immunofluorescence staining of adenocarcinomas (n = 2), showing example of co‐expression of ASPM (red) and E2F1 (green) in tumour cells. (E) Immunostaining of adenocarcinomas (n = 3) showing partial co‐localization of ASPM expression (brown) and the proliferation marker Ki67 (brown) in serial sections ( identical positions); scale bars = (A, B, E) 100 µm; (C, D) 50 µm; (insets) 20 µm
identical positions); scale bars = (A, B, E) 100 µm; (C, D) 50 µm; (insets) 20 µm
Discussion
LM is a valuable approach for supporting high‐throughput assays of sub‐samples from complex tissues. In this study, we developed a method for serial section‐navigated LM of the oxyntic proliferative isthmus zone that enabled us to do genome‐wide gene expression analysis without confounding additional amplification. LM has been used previously for genome‐wide analysis of gastric progenitor cells from transgenic parietal cell‐ablated mice (Atpb4–tox176) with expansion of the isthmus progenitor cell populations 4, 51. Those reports showed enrichment of GO terms corresponding to those found by us, such as nucleus function, DNA binding/replication, cell cycle/proliferation and translation, but also additional genes involved in growth factor response pathways, mRNA processing and cytoplasmic localization. This could be explained by principal experimental differences, such as the use of transgenic and embryonic mice, three‐way comparisons, whole‐stomach samples and different microarray technology. Our set‐up enabled a more direct analysis of the isthmus, which is generally believed to represent the main stem cell zone for the physiological renewal of gastric oxyntic glands 1, 2, 3, 5, 9. Also, higher‐order analysis showed that different adult tissue stem/progenitor cells share functional properties and patterns of gene expression for internal functions that distinguish them from differentiated cells 52. Our enrichment analysis revealed that the isthmus zone was indeed enriched for genes regulating intracellular processes such as cell cycle and ribosomal activity. Thus, we can deduce that the microdissected isthmus zone contains stem/progenitor cells and that our genome‐wide data can be analysed for features related to stemness.
We showed that genes expressed both at a higher level and uniquely in the isthmus zone compared to the remaining mucosa, are associated with stem cell‐related transcriptional networks and gastric neoplasia. Of particular interest are the unique isthmus zone target genes in the top‐scored E2F1 transcription factor network, which was also top‐scored for the genes associated with stomach neoplasia. The relation of the E2F1 network to both stem cells 26, 27 and gastric cancer 53, 54, 55, 56 is relatively novel, and the network plays dichotomous roles in controlling counteracting functions, such as cell‐cycle progression versus arrest, apoptosis versus survival and stemness versus differentiation, depending on context 26, 44, 45, 53. One of the genes in the E2F1 network, Aspm, was particularly intriguing, being a mitotic spindle pole component and already associated with controlling self‐renewal and symmetrical cell division in neural stem/progenitor cells 35, 36, 37, 38, 40. The expression of ASPM is higher in fetal tissues (including the stomach) 38, 43, 58 and it declines markedly during differentiation 35, 36, 38, 41. In the mouse intestine it was found in all epithelial cells during embryonic development but was confined to the stem cell crypts in the adult intestine 39, and it is part of the Lgr5 intestinal stem cell molecular signature 59. ASPM also regulates Wnt signalling 41, 60, which is probably important in gastric homeostasis 6, 7, 9, 51, 61. The association with E2F1 is relevant, since both seem to be involved in balancing whether stem/progenitor cells proliferate or differentiate 26, 35, 40, 43, 44, 45.
The present study is the first to investigate ASPM in gastric tissues. We identify ASPM as a new interesting cell marker strongly expressed in scattered epithelial cells in the proliferative isthmus zone of oxyntic mucosa and a candidate marker for stem/progenitor cells. The cells are mainly mature cell marker‐deficient, except for a limited overlap with cells with neuroendocrine or tuft cell features. The exact stepwise maturation of these lineages is least established in the oxyntic mucosa 6, 7, 62 and ASPM‐expressing cells could represent immature sub‐populations. As ASPM is associated with cell division 35, 36, 40, 41, 43, 48, it was somewhat unexpected not to find clear co‐localization with Ki67, although morphologically the proliferation rate was not zero. Thus, apparently there are either few proliferating ASPM‐expressing cells in oxyntic mucosa or the localization during division makes it difficult to detect. One might speculate that ASPM‐expressing cells could represent quiescent adult stem cells or slow‐cycling progenitor cells for lineages with a long life span, which is a characteristic feature of the oxyntic mucosa 5, 6, 7. Other putative oxyntic stem/progenitor cell markers have also been found intermingled with, but not fully co‐localized to, proliferating cells 5, 63, 64, 65.
ASPM is over‐expressed and correlated with increased malignancy in many human cancers 38, 41, 43, 48, 57, 58, 66. In pancreatic ductal adenocarcinomas, ASPM was recently found negatively associated with glandular differentiation and tubulogenesis, whereas strong expression of ASPM promoted stem cell features and tumour aggressiveness 41. Here, we identify expression of ASPM as relevant for gastric carcinogenesis, showing expression in gastric cancer cell lines and higher levels in GAs. The staining pattern of ASPM in GAs was patchy, similar to the antral stem cell marker LGR5 11, 23. The partial overlap with proliferating cells could reflect distinct characteristics, fitting with the dynamic and heterogenic nature of the cancer stem cells pool within and between tumours 12, 13. Additionally, aberrant expression of E2F1 is also relevant for gastric cancer, and we are the first to show increased E2F1 mRNA in GAs by genome‐wide analysis, together with increased E2F1 protein by IHC, as previously reported 53, 54, 55, 56. Our results could imply an association between ASPM and the transcription factor E2F1 in gastric tissue, which is particularly relevant due to their common involvement in crucial cell fate‐regulatory mechanisms. This suggests diverse possible roles for these in tumours, and more functional experiments are needed to further elucidate this association and possible biological relevance.
In summary, we introduce ASPM as a new oxyntic gland cell marker, possibly of stem/progenitor cells, that may be involved in both normal gastric physiology and gastric carcinogenesis. ASPM expression in certain cells could somehow participate in the cellular turnover in the oxyntic mucosa, perhaps by regulating symmetrical versus asymmetrical divisions; thus, ASPM could be an intriguing candidate molecular marker for clonal lineage tracing. Our findings represent a significant contribution to the understanding of the still unidentified oxyntic stem/progenitor cells and their role in tissue homeostasis and cancer.
Author contributions
PV, TB, VB, SEE, AF, BD, AKS and IB contributed to the conception and design of different parts of the study; PV, TB, VB, SEE, BD and IB performed the experiments; PV, TB, VB, SEE, AF, BD, AKS and IB contributed to the assembly, analysis and interpretation of data; PV, TB and IB wrote the manuscript, and PV, TB, VB, SEE, AF, BD, AKS and IB were involved in revision and final approval of the submitted and published versions.
SUPPLEMENTARY MATERIAL ON THE INTERNET.
The following supplementary material may be found in the online version of this article:
Supplementary materials and methods
Supplementary results
Figure S1. IHC of ADAM17 in rat and human oxyntic mucosa
Figure S2. IHC of ASPM in rat and human antral mucosa
Table S1. Gene expression analysis, showing genes higher and lower expressed in the microdissected oxyntic proliferative isthmus zone compared to the remaining mucosa
Table S2. Verification of differentially expressed genes in rat oxyntic proliferative isthmus zone compared to the remaining mucosa
Table S3. Genes uniquely expressed in the microdissected oxyntic proliferative isthmus zone
Table S4. Genes higher expressed in the oxyntic proliferative isthmus zone are associated with diseases in the gastrointestinal system
Supporting information
Supplementary materials and methods
Figure S1.ADAM17 is mainly expressed in cells of the parietal cell lineage located in the proliferative isthmus zone, confirming the gene expression analysis. Immunostaining for ADAM17 in rat (A) and human (B) oxyntic mucosa shows expression (brown) in parietal cells located throughout the gastric oxyntic glands, with a stronger expression in possible preparietal cells located in the isthmus zone, as indicated by the dashed line. (C) Double immunofluorescence analysis in rat oxyntic mucosa shows an almost complete co‐localization between ADAM17‐expressing cells (green) and cells immunoreactive for the parietal cell marker H+/K+‐ATPase (red); DAPI‐stained nuclei in blue. Scale bars = 100 µm
Figure S2.ASPM is expressed in scattered single cells in the isthmus zone of rat and human gastric antral mucosa. Immunostaining for ASPM in rat (A) and human (B) antral mucosa show expression (brown) in scattered single cells located in the isthmus zone (dashed line). Scale bars = 100 µm
Table S1.Gene expression analysis, showing genes higher and lower expressed in the microdissected oxyntic proliferative isthmus zone compared to the remaining mucosa
Table S2.Verification of differentially expressed genes in rat oxyntic proliferative isthmus zone compared to the remaining mucosa
Table S3.Genes uniquely expressed in the microdissected oxyntic proliferative isthmus zone
Table S4.Genes higher expressed in the oxyntic proliferative isthmus zone are associated with diseases in the gastrointestinal system
Acknowledgements
We thank Wahida Afroz, Anne Kristensen and Bjørn Munkvold for skilful technical assistance and Professor Helge L Waldum for providing access to cancer material from the in‐house GA Biobank. The microarray and bioinformatic analysis were provided in collaboration with the Genomics Core Facility (GCF), NTNU. The imaging analyses were performed in collaboration with Bjørnar Sporsheim at the Cellular and Molecular Imaging Core Facility (CMIC), NTNU. We thank the Comparative Medicine Core Facility (CoMed), NTNU for assistance with the animal experiments. All core facilities, GCF, CMIC and CoMed are funded by the Faculty of Medicine at NTNU and the Central Norway Regional Health Authority. This work was supported by the Liaison Committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology (NTNU), the Faculty of Medicine at NTNU, the Cancer Fund at St. Olav's University Hospital, Trondheim, Norway, and 'Familien Blix fond til fremme av medisinsk forskning'.
No conflicts of interest were declared.
References
- 1. McDonald SA, Greaves LC, Gutierrez‐Gonzalez L, et al. Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells. Gastroenterology 2008; 134: 500–510. [DOI] [PubMed] [Google Scholar]
- 2. Karam SM, Straiton T, Hassan WM, et al. Defining epithelial cell progenitors in the human oxyntic mucosa. Stem Cells 2003; 21: 322–336. [DOI] [PubMed] [Google Scholar]
- 3. Bjerknes M, Cheng H. Multipotential stem cells in adult mouse gastric epithelium. Am J Physiol Gastrointest Liver Physiol 2002; 283: G767–777. [DOI] [PubMed] [Google Scholar]
- 4. Mills JC, Andersson N, Hong CV, et al. Molecular characterization of mouse gastric epithelial progenitor cells. Proc Natl Acad Sci USA 2002; 99: 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pan Q, Nicholson AM, Barr H, et al. Identification of lineage‐uncommitted, long‐lived, label‐retaining cells in healthy human esophagus and stomach, and in metaplastic esophagus. Gastroenterology 2013; 144: 761–770. [DOI] [PubMed] [Google Scholar]
- 6. Mills JC, Shivdasani RA. Gastric epithelial stem cells. Gastroenterology 2011; 140: 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qiao XT, Gumucio DL. Current molecular markers for gastric progenitor cells and gastric cancer stem cells. J Gastroenterol 2011; 46: 855–865. [DOI] [PubMed] [Google Scholar]
- 8. Singh SR. Gastric cancer stem cells: a novel therapeutic target. Cancer Lett 2013; 338: 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stange DE, Koo BK, Huch M, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 2013; 155: 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barker N, Huch M, Kujala P, et al. Lgr5+ stem cells drive self‐renewal in the stomach and build long‐lived gastric units in vitro . Cell Stem Cell 2010; 6: 25–36. [DOI] [PubMed] [Google Scholar]
- 11. Jang BG, Lee BL, Kim WH. Distribution of LGR5+ cells and associated implications during the early stage of gastric tumorigenesis. PLoS One 2013; 8: e82390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer 2013; 13: 727–738. [DOI] [PubMed] [Google Scholar]
- 13. Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10: 717–728. [DOI] [PubMed] [Google Scholar]
- 14. Fjeldbo CS, Bakke I, Erlandsen SE, et al. Gastrin upregulates the prosurvival factor secretory clusterin in adenocarcinoma cells and in oxyntic mucosa of hypergastrinemic rats. Am J Physiol Gastrointest Liver Physiol 2012; 302: G21–33. [DOI] [PubMed] [Google Scholar]
- 15. Akbari M, Otterlei M, Pena‐Diaz J, et al. Repair of U/G and U/A in DNA by UNG2‐associated repair complexes takes place predominantly by short‐patch repair both in proliferating and growth‐arrested cells. Nucleic Acids Res 2004; 32: 5486–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smyth GK. Limma: linear models for microarray data In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Gentleman R, Carey V, Dudoit S, et al. (eds). Springer: New York, 2005; 397–420. [Google Scholar]
- 17. Parkinson H, Sarkans U, Shojatalab M, et al. ArrayExpress – a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 2005; 33: D553–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MetaCore . Thomson Reuters Systems Biology Solutions: http://lsresearch.thomsonreuters.com/pages/solutions/1/metacore (accessed 15 December 2014).
- 19. Selvik LK, Fjeldbo CS, Flatberg A, et al. The duration of gastrin treatment affects global gene expression and molecular responses involved in ER stress and anti‐apoptosis. BMC Genom 2013; 14: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Capoccia BJ, Huh WJ, Mills JC. How form follows functional genomics: gene expression profiling gastric epithelial cells with a particular discourse on the parietal cell. Physiol Genomics 2009; 37: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kouznetsova I, Kalinski T, Meyer F, et al. Self‐renewal of the human gastric epithelium: new insights from expression profiling using laser microdissection. Mol Biosyst 2011; 7: 1105–1112. [DOI] [PubMed] [Google Scholar]
- 22. Schmuck R, Warneke V, Behrens HM, et al. Genotypic and phenotypic characterization of side population of gastric cancer cell lines. Am J Pathol 2011; 178: 1792–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simon E, Petke D, Boger C, et al. The spatial distribution of LGR5+ cells correlates with gastric cancer progression. PLoS One 2012; 7: e35486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aoi T, Yae K, Nakagawa M, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 2008; 321: 699–702. [DOI] [PubMed] [Google Scholar]
- 25. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676. [DOI] [PubMed] [Google Scholar]
- 26. Yeo HC, Beh TT, Quek JJ, et al. Integrated transcriptome and binding sites analysis implicates E2F in the regulation of self‐renewal in human pluripotent stem cells. PLoS One 2011; 6: e27231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- 28. Bracken AP, Ciro M, Cocito A, et al. E2F target genes: unraveling the biology. Trends Biochem Sci 2004; 29: 409–417. [DOI] [PubMed] [Google Scholar]
- 29. Chen YL, Uen YH, Li CF, et al. The E2F transcription factor 1 transactivates stathmin 1 in hepatocellular carcinoma. Ann Surg Oncol 2013; 20: 4041–4054. [DOI] [PubMed] [Google Scholar]
- 30. Vella P, Barozzi I, Cuomo A, et al. Yin Yang 1 extends the Myc‐related transcription factors network in embryonic stem cells. Nucleic Acids Res 2012; 40: 3403–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu X, Bieda M, Jin VX, et al. A comprehensive ChIP‐chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res 2007; 17: 1550–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin VX, Rabinovich A, Squazzo SL, et al. A computational genomics approach to identify cis‐regulatory modules from chromatin immunoprecipitation microarray data – a case study using E2F1. Genome Res 2006; 16: 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith AK, Maloney EM, Falkenberg VR, et al. An angiotensin‐1 converting enzyme polymorphism is associated with allostatic load mediated by C‐reactive protein, interleukin‐6 and cortisol. Psychoneuroendocrinology 2009; 34: 597–606. [DOI] [PubMed] [Google Scholar]
- 34. Bieda M, Xu X, Singer MA, et al. Unbiased location analysis of E2F1‐binding sites suggests a widespread role for E2F1 in the human genome. Genome Res 2006; 16: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fish JL, Kosodo Y, Enard W, et al. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA 2006; 103: 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marinaro C, Butti E, Bergamaschi A, et al. In vivo fate analysis reveals the multipotent and self‐renewal features of embryonic AspM expressing cells. PLoS One 2011; 6: e19419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gurok U, Loebbert RW, Meyer AH, et al. Laser capture microdissection and microarray analysis of dividing neural progenitor cells from the adult rat hippocampus. Eur J Neurosci 2007; 26: 1079–1090. [DOI] [PubMed] [Google Scholar]
- 38. Horvath S, Zhang B, Carlson M, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA 2006; 103: 17402–17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Luers GH, Michels M, Schwaab U, et al. Murine calmodulin binding protein 1 (Calmbp1): tissue‐specific expression during development and in adult tissues. Mech Dev 2002; 118: 229–232. [DOI] [PubMed] [Google Scholar]
- 40. Higgins J, Midgley C, Bergh AM, et al. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol 2010; 11: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang WY, Hsu CC, Wang TY, et al. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology 2013; 145: 1110–1120. [DOI] [PubMed] [Google Scholar]
- 42. Agrawal P, Yu K, Salomon AR, et al. Proteomic profiling of Myc‐associated proteins. Cell Cycle 2010; 9: 4908–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kouprina N, Pavlicek A, Collins NK, et al. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum Mol Genet 2005; 14: 2155–2165. [DOI] [PubMed] [Google Scholar]
- 44. Chong JL, Wenzel PL, Saenz‐Robles MT, et al. E2f1–3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 2009; 462: 930–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Magri L, Swiss VA, Jablonska B, et al. E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. J Neurosci 2014; 34: 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhong X, Liu L, Zhao A, et al. The abnormal spindle‐like, microcephaly‐associated (ASPM) gene encodes a centrosomal protein. Cell Cycle 2005; 4: 1227–1229. [DOI] [PubMed] [Google Scholar]
- 47. Saqui‐Salces M, Keeley TM, Grosse AS, et al. Gastric tuft cells express DCLK1 and are expanded in hyperplasia. Histochem Cell Biol 2011; 136: 191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bikeye SN, Colin C, Marie Y, et al. ASPM‐associated stem cell proliferation is involved in malignant progression of gliomas and constitutes an attractive therapeutic target. Cancer Cell Int 2010; 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cho JY, Lim JY, Cheong JH, et al. Gene expression signature‐based prognostic risk score in gastric cancer. Clin Cancer Res 2011; 17: 1850–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. D'Errico M, de Rinaldis E, Blasi MF, et al. Genome‐wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer 2009; 45: 461–469. [DOI] [PubMed] [Google Scholar]
- 51. Giannakis M, Stappenbeck TS, Mills JC, et al. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem 2006; 281: 11292–11300. [DOI] [PubMed] [Google Scholar]
- 52. Doherty JM, Geske MJ, Stappenbeck TS, Mills JC. Diverse adult stem cells share specific higher‐order patterns of gene expression. Stem Cells 2008; 26: 2124–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xanthoulis A, Tiniakos DG. E2F transcription factors and digestive system malignancies: how much do we know? World J Gastroenterol 2013; 19: 3189–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu JG, Yu JW, Wu HB, et al. Expressions and clinical significances of c‐MET, p‐MET and E2f‐1 in human gastric carcinoma. BMC Res Notes 2014; 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yan LH, Wang XT, Yang J, et al. Reversal of multidrug resistance in gastric cancer cells by E2F‐1 downregulation in vitro and in vivo . J Cell Biochem 2014; 115: 34–41. [DOI] [PubMed] [Google Scholar]
- 56. Yan LH, Wei WY, Cao WL, et al. Overexpression of E2F1 in human gastric carcinoma is involved in anti‐cancer drug resistance. BMC Cancer 2014; 14: 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Komatsu M, Yoshimaru T, Matsuo T, et al. Molecular features of triple negative breast cancer cells by genome‐wide gene expression profiling analysis. Int J Oncol 2013; 42: 478–506. [DOI] [PubMed] [Google Scholar]
- 58. Lin SY, Pan HW, Liu SH, et al. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2008; 14: 4814–4820. [DOI] [PubMed] [Google Scholar]
- 59. Munoz J, Stange DE, Schepers AG, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+ 4’ cell markers. EMBO J 2012; 31: 3079–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Buchman JJ, Durak O, Tsai LH. ASPM regulates Wnt signaling pathway activity in the developing brain. Genes Dev 2011; 25: 1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bartfeld S, Bayram T, van de Wetering M, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 2015; 148: 126–136 e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li HJ, Johnston B, Aiello D, et al. Distinct cellular origins for serotonin‐expressing and enterochromaffin‐like cells in the gastric corpus. Gastroenterology 2014; 146: 754–764, e753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arnold K, Sarkar A, Yram MA, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 2011; 9: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fukui T, Kishimoto M, Nakajima A, et al. The specific linker phosphorylation of Smad2/3 indicates epithelial stem cells in stomach; particularly increasing in mucosae of Helicobacter‐associated gastritis. J Gastroenterol 2011; 46: 456–468. [DOI] [PubMed] [Google Scholar]
- 65. Akasaka Y, Saikawa Y, Fujita K, et al. Expression of a candidate marker for progenitor cells, Musashi‐1, in the proliferative regions of human antrum and its decreased expression in intestinal metaplasia. Histopathology 2005; 47: 348–356. [DOI] [PubMed] [Google Scholar]
- 66. Kabbarah O, Nogueira C, Feng B, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One 2010; 5: e10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Figure S1.ADAM17 is mainly expressed in cells of the parietal cell lineage located in the proliferative isthmus zone, confirming the gene expression analysis. Immunostaining for ADAM17 in rat (A) and human (B) oxyntic mucosa shows expression (brown) in parietal cells located throughout the gastric oxyntic glands, with a stronger expression in possible preparietal cells located in the isthmus zone, as indicated by the dashed line. (C) Double immunofluorescence analysis in rat oxyntic mucosa shows an almost complete co‐localization between ADAM17‐expressing cells (green) and cells immunoreactive for the parietal cell marker H+/K+‐ATPase (red); DAPI‐stained nuclei in blue. Scale bars = 100 µm
Figure S2.ASPM is expressed in scattered single cells in the isthmus zone of rat and human gastric antral mucosa. Immunostaining for ASPM in rat (A) and human (B) antral mucosa show expression (brown) in scattered single cells located in the isthmus zone (dashed line). Scale bars = 100 µm
Table S1.Gene expression analysis, showing genes higher and lower expressed in the microdissected oxyntic proliferative isthmus zone compared to the remaining mucosa
Table S2.Verification of differentially expressed genes in rat oxyntic proliferative isthmus zone compared to the remaining mucosa
Table S3.Genes uniquely expressed in the microdissected oxyntic proliferative isthmus zone
Table S4.Genes higher expressed in the oxyntic proliferative isthmus zone are associated with diseases in the gastrointestinal system


