Abstract
The habenula, located on the dorsal thalamic surface, is an emotional and reward processing center. As in the mammalian brain, the zebrafish habenula is divided into dorsal (dHb) and ventral (vHb) subdivisions that project to the interpeduncular nucleus and median raphe (MR) respectively. Previously, we have shown that kisspeptin 1 (Kiss1) expressing in the vHb, regulates the serotonin (5‐HT) system in the MR. However, the connectivity between the Kiss1 neurons and the 5‐HT system remains unknown. To resolve this issue, we generated a specific antibody against zebrafish Kiss1 receptor (Kiss‐R1); using this primary antibody we found intense immunohistochemical labeling in the ventro‐anterior corner of the MR (vaMR) but not in 5‐HT neurons, suggesting the potential involvement of interneurons in 5‐HT modulation by Kiss1. Double‐fluorescence labeling showed that the majority of habenular Kiss1 neurons are glutamatergic. In the MR region, Kiss1 fibers were mainly seen in close association with glutamatergic neurons and only scarcely within GABAergic and 5‐HT neurons. Our findings indicate that the habenular Kiss1 neurons potentially modulate the 5‐HT system primarily through glutamatergic neurotransmission via as yet uncharacterized interneurons.
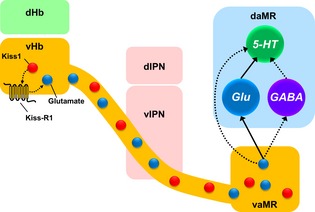
The neuropeptide kisspeptin (Kiss1) play a key role in vertebrate reproduction. We have previously shown modulatory role of habenular Kiss1 in the raphe serotonin (5‐HT) systems. This study proposed that the habenular Kiss1 neurons modulate the 5‐HT system primarily through glutamatergic neurotransmission, which provides an important insight for understanding of the modulation of 5‐HT system by the habenula‐raphe pathway.
Keywords: GPR54, kisspeptin 1, serotonin, ventro‐anterior MR, zebrafish
Abbreviations used
- 5‐HT
serotonin
- GABA
γ‐aminobutyric acid
- GFP
green fluorescent protein
- Kiss1
kisspeptin1
- Kiss‐R1
Kiss1 receptor
- MR
median raphe
- vHb
ventral habenula
The habenula constitutes the dorsal diencephalic conduction system, relaying information from forebrain regions to the boundary dividing the midbrain from hindbrain regions, further connecting to monoaminergic neurons including serotonergic and dopaminergic neurons (Sutherland 1982). The basic connectivity and structure of the habenula is well conserved throughout the mammalian lineage with subdivisions of the medial habenula (MHb) projecting to the interpeduncular nucleus (IPN) and the lateral habenula (LHb) to midbrain regions and the raphe nuclei, implicated in serotonin (5‐hydroxytryptophan, 5‐HT) release (Wang and Aghajanian 1977; Hikosaka 2010). Similarly, in non‐mammalian vertebrates, the habenula is divided into the dorsal (dHb) and ventral (vHb) subnuclei (Braitenberg and Kemali 1970; Kemali and Guglielmotti 1977). The dHb is further subdivided into the medial (dHbM) and lateral (dHbL) subnuclei projecting to the ventral (vIPN) and dorsal IPN (dIPN) respectively, while the vHb projects to the median raphe (MR), accounting for the homologous nature of the dHb to the MHb and the vHb to the LHb (Amo et al. 2010). It is well known that significant homologies exist between mammals and fish in terms of neuroanatomy, physiology and behaviours (Chowdhary et al. 1998; Lieschke and Currie 2007). In most teleost, the 5‐HT‐containing raphe region is divided into the superior (SR) and inferior (IR) raphe nuclei (Lillesaar 2011) and the SR is further subdivided into the MR (caudal of the IPN) and dorsal raphe (DR, superior to the dIPN) based on cytoarchitectural structuring (Lillesaar et al. 2007; Amo et al. 2010, 2014; Okamoto et al. 2012). In the zebrafish (Danio rerio), the dHb expresses transmitters such as tachykinin, glutamate (DeCarvalho et al. 2014) and acetylcholine (Hong et al. 2013) along with their receptors while the vHb neurons has been shown to project mainly to the vIPN potentially including the MR region (Servili et al. 2011; Ogawa et al. 2012), suggesting its potential role in 5‐HT modulation (Ogawa et al. 2012).
Kisspeptin and its receptor (Kiss‐R) homologues have been identified in several mammalian and non‐mammalian vertebrates (Felip et al. 2009; Lee et al. 2009; Um et al. 2010; Gopurappilly et al. 2013). Zebrafish possess two genes encoding kisspeptin in the brain, kiss1, predominantly expressed in the habenula (Ogawa et al. 2012) and kiss2, in the hypothalamus (Kitahashi et al. 2009), with preferential affinity for their respective receptors, Kiss‐R1 and ‐R2 (Lee et al. 2009). While Kiss2 in teleosts is a major regulator of reproduction (Kitahashi et al. 2009; Akazome et al. 2010; Tena‐Sempere et al. 2012; Gopurappilly et al. 2013), our recent findings showed that kisspeptin 1 (Kiss1)/Kiss‐R1 system modulates alarm substance‐evoked fear potentially via the 5‐HT system (Ogawa et al. 2014), involving the interaction of 5‐HT1A and 5‐HT2 receptors (Nathan et al. 2015). However, the neural pathway underlying this modulatory effect is poorly understood, as morphological analysis of Kiss1 in the zebrafish failed to identify dense connections between the vHb and 5‐HT neurons in the SR regions, although a sparse plexus of Kiss1‐positive fibers was seen (Rink and Wullimann 2001; Amo et al. 2010; Servili et al. 2011; Ogawa et al. 2012; Song et al. 2015). Based on recent evidences on the projection of the vHb to the MR (Amo et al. 2010, 2014; Ogawa et al. 2012; Broms et al. 2015), we hypothesize that Kiss1 neurons indirectly act on 5‐HT neurons via non‐serotonergic interneurons in the MR, similar to pathways of LHb neurons to the 5‐HT neurons located within the DR via the MR in rats (Wang and Aghajanian 1977; Vertes et al. 1999).
In this study, to map out the potential neuronal pathway underlying the neuroanatomical association of Kiss1 terminals in the MR and the raphe 5‐HT neuronal population in the zebrafish brain, we generated a specific antibody for zebrafish Kiss‐R1 which we used to reveal the cellular location of its expression. The specificity of Kiss‐R1 immunoreactivity was confirmed by comparison with expression patterns of kissr1 gene. Furthermore, as the epitope of the Kiss‐R1 antibody is also found in kissr1b‐derived protein 2 (KRBDP2), an alternative splice variant of the kissr1 gene (Onuma and Duan 2012), we performed in situ hybridization of KRBDP2 mRNA transcripts. Using double‐label immunofluorescence in transgenic (Tg) zebrafish lines, we determined the associations between Kiss1 neurons and the MR neuronal populations. To further identify the potential mechanism of Kiss1 signal transmission, we co‐localized habenular Kiss1 and Kiss‐R1 in neurons also expressing markers for glutamatergic, γ‐ aminobutyric acid (GABA)‐ergic and acetylcholinergic neurons.
Materials and methods
Animals
Adult (6 month) wild‐type male short‐finned zebrafish were obtained from local suppliers. Transgenic zebrafish, [Tg (brn3a‐hsp70:GFP)rw0110b; expressing green fluorescent protein (GFP) in dHbM‐vIPN pathway] (Aizawa et al. 2005; Sato et al. 2007), [Tg (–3.2pet1:eGFP)ne0214; expresses enhanced GFP in 5‐HT neurons in the SR] (Lillesaar et al. 2009) and [Tg (gad1b:GFP); expressing GFP in GABAergic neurons] (Satou et al. 2013) were provided by Prof. Hitoshi Okamoto (RIKEN, Japan), Dr Christina Lillesaar (Institute of Developmental Genetics, Germany), and Dr Shin‐ichi Higashijima (National Institute of Natural Science, Japan) respectively (Table 1). Fish were housed in 20 L tanks with a controlled temperature of 28°C ± 0.5°C and a 14/10 h light/dark phase. Fish were killed by immersion in 0.01% MS222 (tricaine methanesulfonate, Sigma, St Louis, MO, USA) solution before dissecting out the brain regions. All fish were maintained and research was carried out after obtaining ethical approval from Monash University Animal Ethics Committee (MARP/2012/093).
Table 1.
List of transgenic zebrafish lines used in this study
| Abbreviation of transgenic fish used in the manuscript | Name of transgenic line | Characteristic | References | Source |
|---|---|---|---|---|
| Tg brn3a | (brn3a‐hsp70:GFP)rw0110b | Express GFP in the dHbM‐vIPN pathway under enhancer element control of brn3a, expressed in the habenula as a POU domain transcription‐factor‐encoding gene, upstream of the basal promoter at ambient temperature, hsp70 | Aizawa et al. (2005), Sato et al. (2007) | Dr Hitoshi Okamoto (RIKEN, Japan) |
| Tg pet1 | (3.2pet1:eGFP)ne0214 | Express enhanced GFP (eGFP) at 3.2‐kb fragment upstream of the pet1 gene in 5‐HT neurons in the SR | Lillesaar et al. (2007) | Dr Christina Lillesaar (Institute of Developmental Genetics, Germany) |
| Tg gad1b | gad1b:GFP | Express GFP in GABA‐producing neurons | Satou et al. (2013) | Dr Shin‐ichi Higashijima (National Institute of Natural Science, Japan) |
Generation of antibody for zebrafish Kiss‐R1
Polyclonal antiserum for zebrafish Kiss‐R1 (Antibody ID, #PA4217) was generated in rabbits against a synthetic peptide, QRSTEPLATYNREMNFLSS (the carboxy‐terminus of zebrafish Kiss‐R1: GenBank accession number: EU047918). The antigen was conjugated with keyhole limpet haemocyanine which was used to immunize rabbits according to standard commercial procedures by Open Biosystems, USA. After an immune response had been verified, blood samples were centrifuged, and the antibody fraction was purified by the affinity purification column that was constructed by coupling the antigen to a gel. The purified antibody fraction was stored at −80°C. Specificity of the Kiss‐R1 immunoreactivity was confirmed by comparison with expression patterns of kissr1 and KRBDP2 mRNA transcripts (see below).
Immunohistochemistry of zebrafish Kiss1 and Kiss‐R1
Immunohistochemical localization of zebrafish Kiss1 and Kiss‐R1 were performed as previously described (Soga et al. 2005). Anti‐zebrafish Kiss1 (Antibody ID, #PAS 15133/15134) previously generated in our lab, specifically recognizes the Kiss1 cells in the vHb and axons in the MR (Ogawa et al., 2014). For the controls for immunohistochemistry, alternate coronal brain sections, 15 μm (n = 1) were incubated with either anti‐zebrafish Kiss1 or anti‐zebrafish Kiss‐R1 antisera or those pre‐absorbed with 10 μg/mL of respective antigens at the working dilution for a 24 h period. For Kiss‐R1 detection, signals were enhanced with tyramide signal amplification (TSA) Plus Biotin Kit (PerkinElmer/NEN Life Science Products, Wellesley, MA, USA). Sections were scanned and images captured using a Zeiss MIRAX Slide scanning system (Zeiss, GmBH, Göttingen, Germany) with the Mirax Viewer Image Software (3DTech, Budapest, Hungary) at a resolution of 230 nm with 20× and 40× objectives. Nomenclature of brain regions were adopted from Aizawa et al., 2011; Amo et al. 2010, 2014; Yamamoto and Ito 2008; Wullimann et al., 1996 and Wullimann and Rink 2002.
DIG‐in situ hybridization
Expression of kissr1 and KRBDP2 mRNA transcripts and marker genes for glutamatergic (slc17a6b), GABAergic (gad1b and gad2), and cholinergic (chat) neurons were examined by DIG‐in situ hybridization. DIG‐labeled riboprobes for kissr1 were prepared as described previously (Ogawa et al. 2012). As for marker genes of neurotransmitters, RNA probes were synthesized via in vitro transcription from pGEM‐T Easy vector (Promega, Madison, WI, USA) containing fragments from zebrafish cDNA of slc17a6b, gad1b, gad2 and chat, with DIG or biotin RNA Labeling Mix (Roche Diagnostics, Mannheim, Germany). Gene abbreviations, the primer sequences used for probe synthesis, probe size and GenBank accession numbers are listed in Table S1. DIG‐in situ hybridization was performed as previously described (Ogawa et al. 2012).
Immunofluorescence
Kiss1 and Kiss‐R1 immunoreactivities were analyzed in the brain of Tg brn3a zebrafish that express GFP in the dHbM‐vIPN pathway (Aizawa et al. 2005; Sato et al. 2007), Tg pet1 zebrafish that express GFP in 5‐HT neurons in the SR (Lillesaar et al. 2009) Sagittal [Tg brn3a] and coronal [Tg pet1] brain sections (15 μm) were prepared as described above and immunoreacted with anti‐zebrafish Kiss1 (1 : 500; #PAS 15133/15134) and anti‐zebrafish Kiss‐R1 antiserum (1 : 500). The zebrafish Kiss1 and Kiss‐R1 immunoreactivity was visualized after development with Alexa Fluor 594‐labeled anti‐rabbit IgG (Cat# A11012; 1 : 500 dilution; Invitrogen, Eugene, OR, USA). To visualize 5‐HT neurons in Tg pet1 zebrafish, GFP signals were enhanced using a mouse anti‐GFP monoclonal antibody (Cat# A11120; Invitrogen) at 1 : 250 dilution followed by development with Alexa‐Fluor 488‐labeled anti‐mouse IgG (1 : 500 dilution; Invitrogen) and counterstained with 300 nM 4′,6‐diamino‐2‐phenylindole (Cat# D3571; DAPI; Invitrogen).
Co‐localization of marker genes for glutamatergic and GABAergic neurons in habenular Kiss1/Kiss‐R1 neurons and 5‐HT neurons in the MR were examined by double‐labeling combining fluorescence in situ hybridization and immunofluorescence. Tg gad1b zebrafish (Satou et al. 2013) was also used to examine association between Kiss1/Kiss‐R1 with GABAergic neurons in the MR as above. DIG‐ and biotin‐labeled slc17a6b (glutamate) and gad2 (GABA) riboprobes were applied to the brain sections of wild type fish (for Kiss1, n = 3) and Tg pet1 zebrafish (for 5‐HT, n = 3), respectively. slc17a6b and gad2 riboprobes were detected with TSA Plus kit (PerkinElmer/NEN Life Science Products) according to the manufacturer's instructions. For double‐labeling with Kiss1 and Kiss‐R1, TSA‐labeled sections were rinsed in 0.01 M phosphate‐buffered saline and then incubated with either zebrafish Kiss1/Kiss‐R1 antiserum (1 : 500) or Alexa Fluor 594‐labeled anti‐rabbit IgG (1 : 500; Invitrogen). For double‐labeling with GFP‐labeled 5‐HT neurons, signals for the slc17a6b and gad2 probes were detected using TSA Plus Cyanin3 (Cy3) Kit (PerkinElmer/NEN Life Science Products), while 5‐HT neurons were detected as GFP signals.
Image and data analysis
The sections were coverslipped with Vectashield (Vector Laboratories Inc., Burlingame, CA, USA), viewed and images were captured under a fluorescent microscope (90i, Nikon, Tokyo, Japan) attached to a digital camera (DXM 1200c, Nikon) with commercial software (NIS Elements, D v4.0, Nikon). For this examination we used the appropriate excitation and emission wavelengths for GFP, Alexa Fluor 488, Alexa Fluor 594 and DAPI. Higher magnification (60× water immersion plus optical zooming 1.5×) images were obtained with confocal microscopy (C1si, Nikon) attached to an inverted microscope (ECLIPSE TE2000‐E, Nikon). Neurons of interest were aligned in X, Y and Z planes and optical sections through the z‐axis were obtained at 0.15 μm intervals. For high‐resolution imaging, 60× water immersion objective (N.A. = s1.4) with an additional 1.5 × optical zoom to give a final magnification of 79.8 × was utilized at a single‐plane with z‐axis optical sections at 0.15 μm z‐step size involving 7 steps each. Co‐localization was determined as described by Corson and Erisir 2013. Z‐stacks containing the neuron of interest were aligned in X, Y and Z planes. An apposition was identified by the presence of a two‐ or three‐voxel wide overlap between two fluorophore‐filled objects. If the labeled voxels seemed apposed but did not overlap, the presence of an actual space of at least 0.15 μm between the cell and the dendrite indicated this (Corson and Erisir 2013). This yielded a voxel size of 0.15 μm3. Percentage of co‐localization of Kiss1/Kiss‐R1 cells with neurotransmitter makers was determined by the formula, (co‐localizing cells/total number of cells in area) × 100% for a total number of 4 (habenula) and 3 (raphe) sections for each examination.
Color modification (adjusting gamma parameter and input intensity range) of multi‐channel images was set with the look‐up tables, and color rendering and color merging was performed using commercial software (NIS Elements, AR v4.1, Nikon). The brightness, contrast, color balance, and final size of the images were adjusted using Adobe Photoshop CS5.1 (Adobe Systems, San Jose, CA) and figures were prepared with Adobe Illustrator CS5.1 (Adobe Systems).
Results
Localization of zebrafish Kiss1‐ and Kiss‐R1‐immunoreactivity in the brain
Zebrafish Kiss1‐immunoreactive (‐ir) cells were observed in the vHb, giving rise to axonal projections bypassing the vIPN and terminating at the ventro‐anterior corner of the MR (vaMR) through the fasciculus retroflexus (FR) (Fig. 1a–c). Double‐immunofluorescence in the brain of Tg brn3a zebrafish confirmed the presence of axon terminals of Kiss1‐ir fibers in the vaMR but not as a part of the vIPN (Fig. 1d–j). No co‐localization was noted between Kiss1 immunoreactivities and GFP in the dHb or vIPN (Fig. 1h and i). Specificity of the antiserum against zebrafish Kiss‐R1 was further confirmed via expression the of DIG‐labeled kissr1 mRNA‐expressing cells in the vHb of the anti‐sense riboprobe (Fig. 2a) in comparison to the sense riboprobe (Fig. 2b). Similar to the expression patterns of Kiss1‐ir, zebrafish Kiss‐R1‐ir cells were predominantly observed in the vHb with axons projecting through the FR and terminating in the vaMR (Fig. 2c–f). In addition, some Kiss1 or Kiss‐R1‐ir fibers and varicosities were also observed in the dorsal MR (dMR) region (Fig. 2f). Preabsorption of the primary antiserum with antigen for zebrafish Kiss‐R1 peptide successfully abolished all immunoreactivity in the zebrafish brain (Fig. 2c inset).
Figure 1.
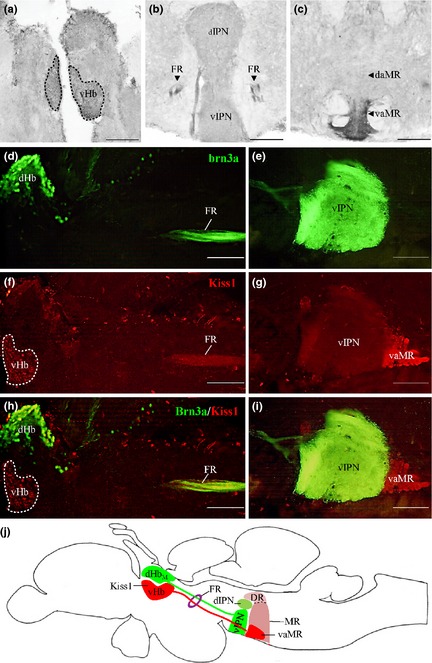
Kisspeptin 1 (Kiss1) projection in the zebrafish brain. (a–c) Kiss1‐immunoreactive cells observed in the vHb (a) project through the fasciculus retroflexus (FR) (b) down to the vaMR (c). (d and e), Photomicrograph of sagittal section of Tg brn3a zebrafish expressing green fluorescent protein (GFP) in dHbM‐vIPN pathway. (f–i) Kiss1‐immunoreactive cells noted in vHb (red) and not in the dHb (green) with axonal projections coursing through the FR and terminating at the vaMR, a structure following the GFP‐expressing vIPN. (j) Illustration depicts projections of dHbM and vHb through the FR to the vIPN and vaMR (a subregion of the MR) respectively as a representation of the sagittal images. Scale bars, 100 μm.
Figure 2.
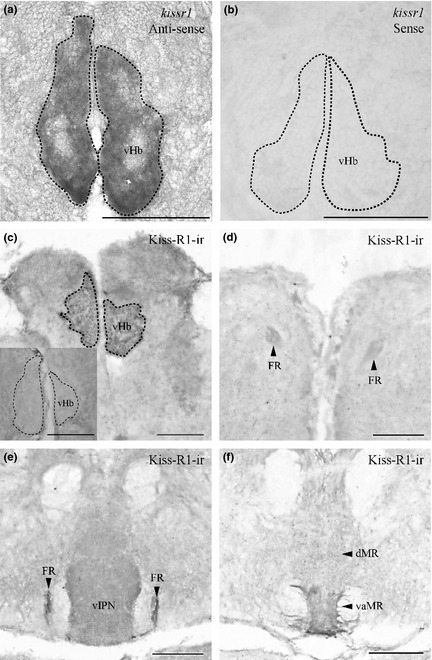
Expression of kissr1 mRNA and Kiss‐R1 projection in the zebrafish brain. Coronal sections where kissr1 mRNA is noted in the ventral subnuclei of the habenula (a) and no cells were observed in the sense strand (b). Kiss‐R1‐immunoreactive (‐ir) cells observed in the vHb (c) send projections through the fasciculus retroflexus (FR) (d and e) down to the vaMR (f). Preabsorption with antigen showed no Kiss‐R1–ir fibers or cells (C inset). Scale bars, 100 μm.
Some Kiss‐R1‐ir cells were observed in the telencephalon, diencephalon and spinal cord regions with staining at varying intensities (Fig. 3a1–d1 and Figure S1a1–c1), as described below. The distribution patterns of Kiss‐R1‐ir cells are as illustrated in Fig. 4. Confirming the expression of Kiss‐R1 neurons in these regions, we also saw the expression of KRBDP2 mRNA at varying intensities in these neurons (Fig. 3a2–d2 and Figure S1a2–d2). The specificity of the riboprobe was confirmed by the absence of expression in the corresponding regions by the sense riboprobe (Fig. 3a3–d3 and S1a3–d3). In the telencephalic region, Kiss‐R1–ir and KRBDP2 mRNA‐expressing cells were observed in the medial, central and posterior zones of the dorsal telencephalon as well as in the medial olfactory tract (Fig. 3a1, a2, b1, b2, Figure S1a1–c1 and 4). In the diencephalon, some Kiss‐R1 immunoreactivity was observed in the central posterior thalamic nucleus (Fig. 3c1 and c2). Some cells were noted in regions surrounding the FR and in the posterior commissure (Fig. 3c1). A few Kiss‐R1‐ir cells were noted in the dorsal region of the trigerminal motor nucleus and the lateral longitudinal fascicle (Fig. 3d1 and d2, and 4). Both Kiss‐R1–ir and KRBDP2 cell somata were present in the inner arcuate fibers and the intermediate reticular formation, although the intensity was relatively low (Figure S1d1 and d2, and 4).
Figure 3.
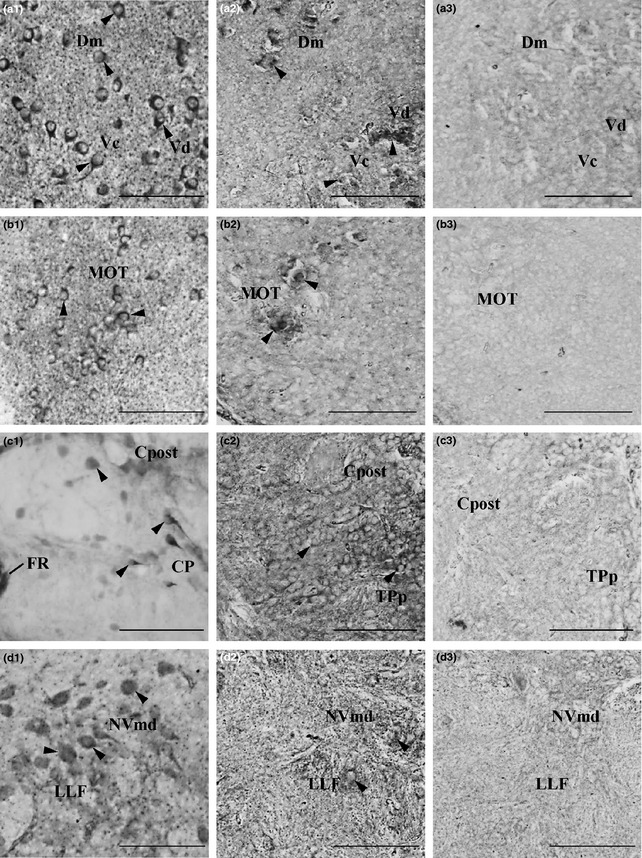
Kiss‐R1‐ir localization and kissr1b‐derived protein 2 (KRBDP2) expression in other regions of the zebrafish brain. a1–d1, Kiss‐R1‐immunoreactive (‐ir) cell somata observed outside of the habenula such as the forebrain (a and b), midbrain (c) and hindbrain (d) regions. a2–d2, KRBDP2 mRNA expression was also observed in same regions as Kiss‐R1‐ir cells. a3–d3, The sense riboprobe showed no expression of KRBDP2 mRNA. For abbreviations, see (Table 2). Scale bars, 100 μm.
Figure 4.
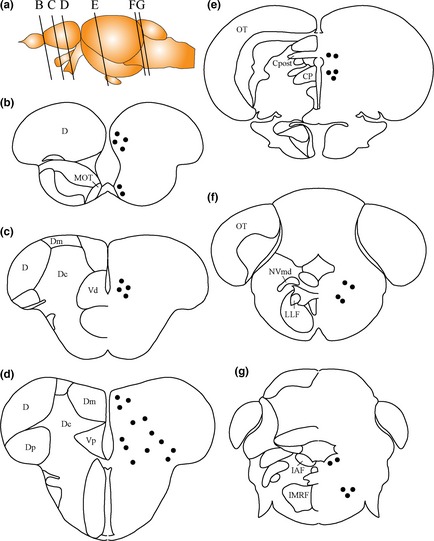
Schematic representation of Kiss‐R1‐ir and kissr1b‐derived protein 2 (KRBDP2) cells in the zebrafish brain. Black dots indicate Kiss‐R1‐immunoreactive cells and the same sites of expression of KRBDP2. For abbreviations, see (Table 2). Schematic diagram of brain and naming of regions adapted from Yamamoto and Ito (2008), Wullimann and Rink (2002) and Wullimann et al. (1996).
Table 2.
List of abbreviations
| 5‐HT | 5‐hydroxytryptamine; serotonin |
| CP | Central posterior thalamic nucleus |
| Cpost | Posterior commissure |
| D | Dorsal telencephalic area |
| daMR | Dorso‐anterior corner of the MR |
| Dc | Central zone of dorsal telencephalic area |
| dHb | Dorsal habenula |
| Dm | Medial zone of dorsal telencephalic area |
| Dp | Posterior zone of dorsal telencephalic area |
| DR | Dorsal raphe |
| FR | Fasciculus retroflexus |
| Gad | Glutamic acid decarboxylase |
| IAF | Inner arcuate nucleus |
| IMRF | Intermediate reticular formation |
| LLF | Lateral longitudinal fascicle |
| MOT | medial olfactory tract |
| MR | Median raphe |
| NVmd | Dorsal region of trigeminal motor nucleus |
| OT | Optic tectum |
| Slc17a6b | Solute carrier family 17 sodium‐dependent inorganic phosphate cotransporter 6 transcript variant 2 |
| SR | Superior raphe |
| TPp | periventricular nucleus of posterior tuberculum |
| vaMR | Ventro‐anterior corner of the MR |
| Vc | Caudal nucleus of ventral telencephalic area |
| Vd | Dorsal nucleus of ventral telencephalic area |
| vHb | Ventral habenula |
| Vp | Postcommissural nucleus of ventral telencephalic area |
Kiss1/Kiss‐R1‐ir in relation to 5‐HT neurons in the MR
Triple‐label immunofluorescence in the brain of Tg pet1 zebrafish showed neither direct associations with Kiss1‐ir fibers (Fig. 5a–c) nor co‐expression of Kiss‐R1‐ir in GFP‐labeled 5‐HT neurons (Fig. 5f–h) in the MR. However, at a higher magnification (79.8×), in the MR, we found a few 5‐HT neurons in close association (no actual space observed more than 0.15 μm between the fibers and cells as explained above) with Kiss1 and Kiss‐R1‐ir fibers (Fig. 5d and i). Confocal imaging (60× plus 1.5× optical zoom; N.A. = 1.4; z‐step = 0.15 μm) showed associations between both Kiss1 and Kiss‐R1‐ir fibers in DAPI‐positive (non‐serotonergic) cells in the vaMR region (Fig. 5e and j). In addition, confocal imaging showed close associations between 5‐HT fibers with Kiss1‐ir fibers within the vaMR and dMR regions (Fig. 5k and l). There were some 5‐HT neurons in the ventral region of the dMR region, but not in the vaMR region (Fig. 5m).
Figure 5.
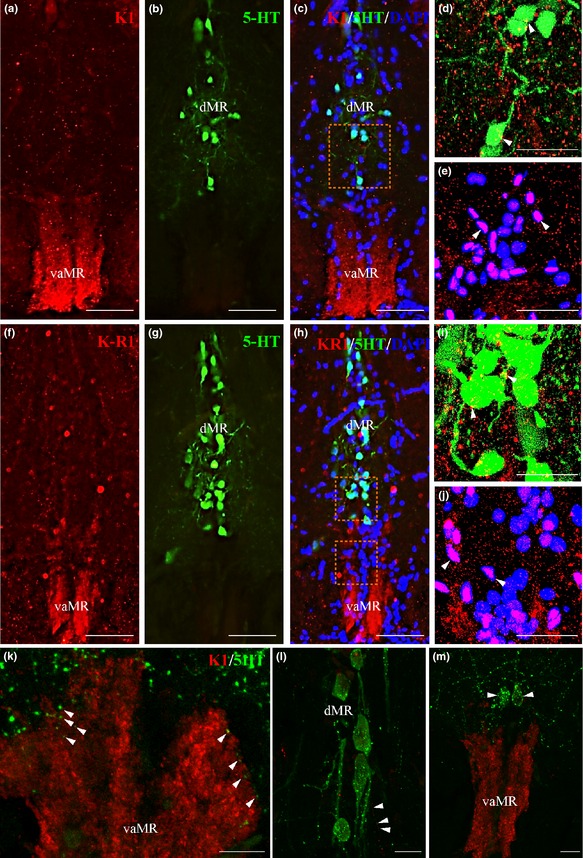
Neural association of kisspeptin 1 (Kiss1) and Kiss‐R1 fibers (red) with serotonergic (green) and non‐serotonergic (blue) raphe neurons in pet1 Tg zebrafish. (a–c) and F‐H: Kiss1 (a–c) and Kiss‐R1 (f–h) fiber terminal was seen in the vaMR, while pet1‐green fluorescent protein (GFP) labeled 5‐HT neurons were noted in the MR. (d, e, i and j) Confocal image of double‐labeling showed close association of Kiss1 (d and e) and Kiss‐R1 (i and j) fibers with 5‐HT neurons in the dMR (d and i) and non‐5‐HT cells (DAPI) in the vaMR (e and j). There was no co‐expression of Kiss‐R1 in 5‐HT cells. (60× plus 1.5× optical zoom; N.A. = 1.4; z‐step = 0.15 μm). (k–m) 5‐HT fibers were seen in close association with Kiss1 fibers in the vaMR (k) and in the dMR (l). There were few 5‐HT cells seen near the vaMR region, but not inside the vMR (m). Scale bars, (a–c) and (f–g) 100 μm; (d, e, i and j) 20 μm; (k–m) 10 μm.
Expression of markers for neurotransmitters in habenular Kiss1 neurons and raphe nucleus
In the habenular region, slc17a6b (glutamatergic neuronal marker) mRNA expression was observed in the dHb and vHb (Fig. S2a). As for GABAergic neurons, while gad1b signals were not detected by our probe (data not shown), gad2 mRNA expression was seen only in the most ventral region of the vHb and in the lateral dorsal nucleus of the thalamus (Fig. S2b). A low expression of chat (acetylcholinergic) mRNA was observed in the dHb but not in the vHb (Fig. S2c). In the raphe region, slc17a6b mRNA‐containing cells were observed in the MR, and sparsely in the DR (Fig. S2d). In the Tg gad1b, cells were observed only in the dMR region (Fig. 7e), while gad2 was only observed in the dMR (Fig. S2e). There was weak expression of chat mRNA in the SR (MR and DR) (Fig. S2f).
Dual‐fluorescence in situ hybridization coupled with immunofluorescence showed 80% co‐expression (approximately 40/50 cells per section of area examined) of slc17a6b mRNA with Kiss1‐ir cells in the vHb (Fig. 6a–c), while no co‐expression of gad2 mRNA in Kiss1‐ir cells (Fig. 6d–f) was observed. In the raphe nuclei, double‐immunofluorescence showed Kiss1‐ir fibers in close association with slc17a6b mRNA‐containing neurons in the dMR (Fig. 7a–c). However, no close association was observed between Kiss‐ir fibers with GFP‐labeled Gad1b neurons (Fig. 7e–g), but there rarely are close associations with gad2 mRNA (Fig. 7h–j) expressing GABAergic neurons in the dMR. Similar expression patterns were noted for Kiss‐R1 in relation to these neurotransmitter markers in the raphe nucleus (Figure S3). In addition, double‐labeling in the brain of Tg pet1 zebrafish showed 25% (approximately 5/20 cells) of slc17a6b (Figure S4a–c) and 25.8% (approximately 6/20 cells) of gad2 (Figure S4d–f) mRNA co‐expressed in some GFP‐labeled 5‐HT neurons.
Figure 6.
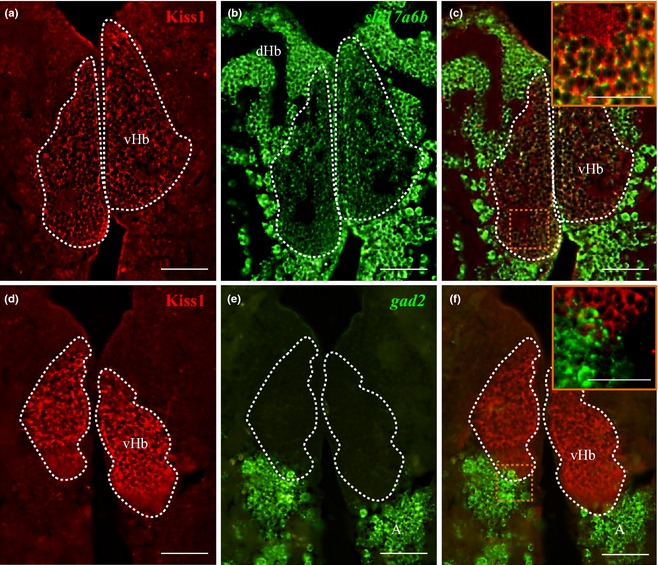
Dual‐fluorescence labeling of kisspeptin 1 (Kiss1) and Kiss‐R1 (red) in glutamatergic and GABAergic neurons (green) in the habenula. (a–c) Photomicrographs showing some populations of Kiss1‐immunoreactive (‐ir) cells in the vHb that co‐localized with cells expressing slc17a6b mRNA. Some individual Kiss1‐ir fibers originating from the vHb co‐localized with slc17a6b mRNA (as seen in the magnified confocal inset representing the region enclosed by the orange‐dotted box; 60× plus 1.5× optical zoom; N.A. = 1.4; z‐step = 0.15 μm). No co‐localization was observed between Kiss1‐ir cells and gad2 mRNA‐expressing cells. A: anterior thalamic nucleus. Scale bars, (a–f) 100 μm and inset c and f: 50 μm.
Figure 7.
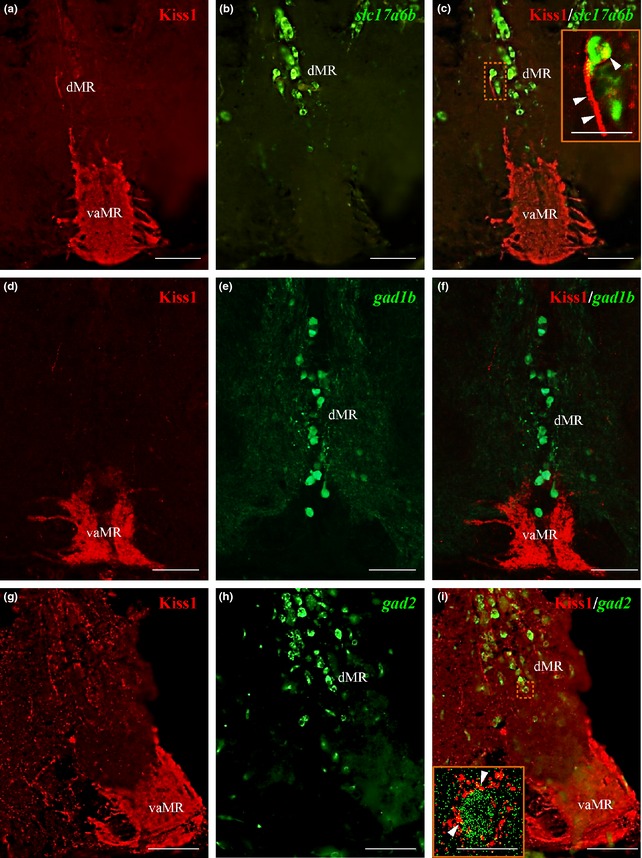
Dual‐fluorescence labeling of kisspeptin 1 (Kiss1) (red) in glutamatergic and GABAergic neurons (green) in raphe nuclei. (a–c) In the raphe nuclei, Kiss1‐immunoreactive (‐ir) fibers were seen in close association with slc17a6b mRNA expressing neurons in the dMR as denoted by the confocal image (inset C). (d–f) green fluorescent protein (GFP)‐labeled Gad1b neurons were observed only in the dMR with no close associations observed with Kiss1‐ir fibers. G‐I: Kiss1‐ir fibers were seen in close association with gad2 mRNA expressing neurons in the dMR, denoted by the confocal image (inset I; 60× plus 1.5× optical zoom; N.A. = 1.4; z‐step = 0.15 μm). Presence of actual space of at least 0.15 μm noted between fibers and cells. Scale bars, (a–i): 100 μm; inset C and I: 50 μm.
Discussion
We report the connectivity of neurons in the vHb that co‐expresses Kiss1 and Kiss‐R1 in the zebrafish and describe the localization of some of the important neurotransmitters involved in Kiss1 modulation of the 5‐HT system in the MR alongside with the localization of Kiss‐R1 immunoreactivity in the zebrafish.
In teleosts, the localization of multiple Kiss‐R types in the brain have been demonstrated by RT‐PCR (Biran et al. 2008; Filby et al. 2008; Shahjahan et al. 2010) and in situ hybridization (Ogawa and Parhar 2012; Ogawa et al. 2012; Escobar et al. 2013; Kanda et al. 2013) but there is limited information available on their neuroanatomical distribution. In addition, subcellular localization of G‐protein coupled receptor (GPCR) cannot be defined by mRNA localization because GPCR protein products translocate within subcellular regions due to the post‐translational GPCR trafficking (Ulloa‐Aguirre et al. 2006). The present study is the first report demonstrating the detailed neuroanatomical as well as subcellular localization of Kiss‐R1 in any species, either mammalian or non‐mammalian. We also observed additional Kiss‐R1‐ir cells in the telencephalon, synencephalon and spinal cord regions where no kissr1 mRNA was detected by in situ hybridization. However, in our previous report employing laser‐capture microdissection coupled with real‐time PCR, we have shown low amounts of kissr1 mRNA expressions in the preoptic area, midbrain, hypothalamus, vIPN, cerebellum, optic tectum and spinal cord (Ogawa et al. 2012), much of which could potentially correspond to the very same Kiss‐R1‐ir cells detected by our antibody. Recently, zebrafish kissr1 (also referred as kiss1rb) gene has been shown to produce 4 additional alternative splice variants encoding different protein lengths (kiss1rb‐derived protein; KRBDP 1–4) (Onuma and Duan 2012). All the truncated forms can be translated, but none are functionally capable of mediating kisspeptin‐derived cellular responses (Namba et al. 1993). KRBDP2 consists of 4 exons including the C‐terminal region of the Kiss‐R1 protein, the epitope of our Kiss‐R1 antibody. Hence, our antibody may recognize the full‐length of Kiss‐R1 and KRBDP2 in the zebrafish brain, explaining the presence of Kiss‐R1 immunoreactive cells outside the habenula‐MR region, which only express KRBDP2 mRNA, justified by our in situ hybridization findings.
Fluorescence labeling in Tg brn3a zebrafish confirmed the presence of Kiss1 in the vaMR. Our results indicate Kiss1 neurons in the vHb mainly project to the MR, particularly to the vaMR. These findings implicate the vHb‐vaMR as the main pathway of habenular Kiss1 neurons; this corresponds to the projection of vHb neurons in the zebrafish (Amo et al. 2010), a finding that was recently confirmed to involve specifically the vaMR (Amo et al. 2014) as well as Kiss1 projection seen in kiss1:mCherry transgenic zebrafish (Song et al. 2015). Projections from these neurons with soma in the habenula converged at the vaMR with some fibers observed projecting towards the dMR (Bianco et al. 2008). The co‐localization of Kiss‐R1 and Kiss1 in the vHb‐vaMR corresponds with previous findings (Ogawa et al. 2010, 2012; Servili et al. 2011). Although these studies report the projection to the vIPN, we emphasize on the basis of the present results in the Tg brn3a that the MR is not part of the vIPN, but instead a division of the SR (Lillesaar et al. 2007; Amo et al. 2010), and though the projections bypass the vIPN, the fibers terminate at the MR (Amo et al. 2010), specifically the ventro‐anterior corner, a subregion which has also been identified and recently re‐affirmed (Amo et al. 2014). Our previous conclusion (Ogawa et al. 2012) was based on previous findings by Amo et al. (2010) and Servili et al. (2011), which however failed to clearly describe the terminating region due to lack of information on the MR but instead more on the IPN. The Tg brn3a zebrafish showed clear distinction between both these structures with the vHb‐expressing Kiss‐ir fibers terminating at the MR and not the vIPN. The current findings corroborates with our previous findings (Ogawa et al. 2012) where we mentioned the potential inclusion of the MR. The presence of Kiss1 and Kiss‐R1 in the same cells and axon terminals at the MR denote their possible presynaptic role, although it remains unknown whether Kiss1 peptides are released from Kiss1 terminals and act presynaptically at Kiss‐R1 (i.e. autoreceptor or retrograde transmission). Co‐localization of a neuropeptide and its cognate receptor in the same neurons has been also observed for kisspeptin (Arai 2009; Ogawa et al. 2012), oxytocin (Freund‐Mercier et al. 1994) and vasopressin (Hurbin et al. 2002).
The SR 5‐HT system comprises of the median (MR) and dorsal (DR) divisions (Lillesaar et al. 2007; Amo et al. 2010) where MR neurons occupy the midline region and DR neurons are located more dorsally, and lateral to the midline region (Lillesaar et al. 2007). Although 5‐HT neurons are the main population occupying these regions, a recent study in the zebrafish shows that the vaMR region, where the vHb terminates, is absent of 5‐HT neuronal cell bodies (Amo et al. 2014). Functional studies have implicated the teleost raphe serotonergic system in tolerance, resistance, adaptation or appropriate ‘coping’ strategies to acute or chronic stress (Deakin 1996). These findings indicate that during basal and stressful conditions the habenular Kiss1 system that modulates the raphe 5‐HT system (Ogawa et al. 2014) may play a role in the hypothalamic‐pituitary‐adrenal axis, which is extensively and selectively modulated by serotonergic and non‐serotonergic neuronal projections.
Our findings in the Tg pet1 zebrafish show that very few Kiss1 fibers are in close association with 5‐HT cell bodies in the MR, whereas there was no expression of Kiss‐R1 in 5‐HT neurons. This suggests that Kiss1 is likely to act indirectly on 5‐HT neurons. However, a few individual fibers potentially originating from a subset of Kiss1 cells in the vHb were in direct contact with 5‐HT neuronal cell bodies. We observed the presence of non‐serotonergic cells surrounded by Kiss1 and Kiss‐R1–ir axonal varicosities in the vaMR. However, based on the recent findings by Amo et al. (2014) that showed non‐serotonergic cells in the MR do not respond to the vHb axonal stimulation in comparison to some of the 5‐HT neurons that responded (Amo et al. 2014), these cells may not be involved in the regulation of the 5‐HT system. Based on our immunohistochemical findings, Kiss1 occupies the entire vHb with some of the axons terminating on 5‐HT neurons while a sparse population terminate on non‐5HT neurons which could further act on the 5‐HT neurons in the dMR. Additionally, we also observed close associations between Kiss1‐fibers with 5‐HT fibers in the vaMR, suggesting the possibility of their axon‐axon interactions, which remains to be examined.
Our in situ hybridization study revealed that the majority of habenular Kiss1 cells are glutamatergic and not GABAergic. LHb projections utilize glutamate as a main excitatory neurotransmitter (Kalen et al. 1986; Behzadi et al. 1990; Ferraro et al. 1996) and there are cases in which the autocrine regulation of Kiss1 has been found to activate glutamate release (Arai 2009), further supporting the idea that the majority of Kiss1 neurons in the zebrafish vHb are glutamatergic. Recent observations in zebrafish have shown that glutamatergic neuronal projections from the vHb exclusively target the vaMR (Amo et al. 2014), where we observe the majority of Kiss1/Kiss‐R1 axons. However, we also found some additional Kiss1/Kiss‐R1 fibers and varicosities in the dorso‐anterior corner of the MR, which are making contact with glutamatergic cells. This discrepancy may indicate that the majority of glutamatergic habenular Kiss1 neurons are terminating in the vaMR, but additional fibers derived from another subset of Kiss1 neurons further extend their projections to the dorso‐anterior corner of the MR. Additionally, Amo et al. (2014) show activation of MR 5‐HT neurons through excitatory glutamatergic projections from the vHb, where we also observed glutamatergic expression. Co‐occurrence as well as co‐release of neuropeptides and neurotransmitters in a single neural cell have been reported in various neurons (Reiner and Anderson 1990). The co‐existence is necessary and seen in higher brain regions where a neuropeptide enhances or depresses the release of amino acid transmitters in order to produce a subset of responses to a similar modulatory outcome (van den Pol et al. 1998). Co‐existing transmitters may work together to evoke physiological responses such that they may activate pre‐ and post‐synaptic receptors, while the second transmitter may work in an inhibitory manner. Based on similar expression patterns observed for gad1b and gad2 (Martin et al. 1998), double‐labeling with gad1b and Kiss1/Kiss‐R1 was only carried out in the MR to further clarify the extent of GABAergic neurotransmission involvement in 5‐HT modulation. Due to the relatively weak staining of chat (marker for cholinergic neurons) observed in comparison to that of glutamate in the habenula, and based on recent findings in the zebrafish vHb (Amo et al. 2014), we did not progress with double‐labeling of Kiss1/Kiss‐R1 and chat. Hence, we speculate that Kiss1 could presynaptically activate Kiss‐R1 that could alter glutamatergic levels in co‐localizing cells in the vHb. This could further regulate 5‐HT levels in the MR directly or indirectly, via GABAergic or glutamatergic MR neurons (Valentino et al. 2003; Ebner and Singewald 2006).
Our in situ hybridization showed some 5‐HT neurons co‐express with markers of glutamatergic and GABAergic neurons in the MR indicating that these neurons could transmit information to fear‐regulating regions via the co‐release of GABA and glutamate (El Mestikawy et al. 2011). Subsets of 5‐HT neurons co‐expressing the serotonin transporter project to various regions involved in depressive and frightful behaviours, such as the periaqueductal gray, amygdala and hypothalamus in rats (Shutoh et al. 2008; El Mestikawy et al. 2011). Co‐expression of glutamic acid decarboxylase (GAD) and 5‐HT has also been reported in various studies (Gaspar and Lillesaar 2012; Gocho et al. 2013), although a very small population is noted in the raphe nuclei, explaining the lesser extent of co‐expression that we noted of gad2 and 5‐HT neurons in the MR.
In conclusion, Kiss‐R1 expressing cells in the vHb project down through the FR where they terminate specifically at the vaMR. This projection pattern, along with that of Kiss1 and the lack of Kiss‐R1 expressing cell soma in the MR suggests a presynaptic action of Kiss1 in the zebrafish. The autocrine regulation of Kiss1 may activate glutamate co‐release that projects to the vaMR to interact with the 5‐HT system in the MR (Fig. 8). Taken together, our findings indicate that the action of Kiss1/Kiss‐R1 system on 5‐HT neurons in the MR requires the interaction of glutamate as the main excitatory neurotransmitter and to a lesser extent, GABA.
Figure 8.
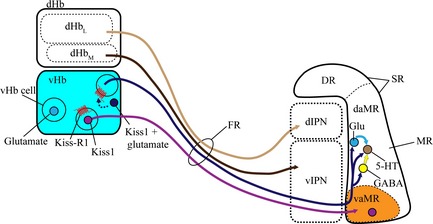
Schematic drawing of hypothetical neural circuit of habenular kisspeptin 1 (Kiss1) neurons. The lateral subnucleus of the dHb projects to the dIPN while the medial subnucleus of the dHb projects to the vIPN. Some cells in the vHb express glutamate (light blue dot). Both Kiss1 (purple dot) and Kiss‐R1 express in the vHb and project down (purple line) to the vaMR, a subregion of the MR [a division of the Superior raphe (SR)], through the fasciculus retroflexus (FR). The presynaptic action of the Kiss1/K‐R1 system causes the release of glutamate in co‐expressing cells (dark blue dot) from the vHb (dark blue line) that potentially regulates the 5‐HT system in the dMR directly or via glutamatergic (light blue line) and GABAergic neurotransmission (yellow line). Modified from Amo et al. (2010, 2014) and Lillesaar et al. (2007).
Author contributions
S.O and I.S.P designed research; F.M.N performed research; S.O and F.M.N analyse data; F.M.N, S.O and I.S.P wrote the paper.
Supporting information
Figure S1. Kiss‐R1‐ir localization and KRBDP2 expression in other regions of the zebrafish brain.
Figure S2. Expression of slc17a6b, gad2 and chat mRNAs in the habenula and raphe regions.
Figure S3. Dual‐fluorescence labeling of Kiss‐R1 (red) in glutamatergic and GABAergic neurons (green) in raphe nuclei.
Figure S4. Co‐expression of slc17a6b and gad2 (red) with 5‐HT (green).
Table S1. Gene abbreviations, the primer sequences, probe size and GenBank accession numbers for probes for DIG‐in situ hybridization.
Acknowledgments and conflict of interest disclosure
We thank Ms. Rachel Shalini Anthonysamy for her technical assistance for confocal imaging. We thank Prof. Hitoshi Okamato (RIKEN, Japan), Dr Christina Lillesaar (Institute of Developmental Genetics, Germany) and Dr Shin‐ichi Higashijima (National Institute of Natural Science, Japan) for generously providing us with the transgenic lines. We also thank Monash University Malaysia for the Higher Degree Research Scholarship to F.M.N. This work is supported by Monash University Malaysia (M‐NEU‐RS‐014), and Malaysian Ministry of Higher Education, FRGS/2/2010/ST/MUSM/03/02 (to S.O.), FRGS/1/2013/SKK01/MUSM/03/2 (to I.S.P.) and FRGS/1/2014/ST03/MUSM/02/1 (to S.O.) and Malaysian Ministry of Science and Technology and Innovation, 02‐02‐10‐SF0161 (to I.S.P.) and 02‐02‐10‐SF0162 (to S.O.). The authors declare no conflict of interest.
All experiments were conducted in compliance with the ARRIVE guidelines.
References
- Aizawa H., Bianco I. H., Hamaoka T., Miyashita T., Uemura O., Concha M. L., Russell C., Wilson S. W. and Okamoto H. (2005) Laterotopic representation of left‐right information onto the dorso‐ventral axis of a zebrafish midbrain target nucleus. Curr. Biol. 15, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H., Amo R. and Okamoto H. (2011) Phylogeny and ontogeny of the habenular structure. Frontiers in Neuroscience 5, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazome Y., Kanda S., Okubo K. and Oka Y. (2010) Functional and evolutionary insights into vertebrate kisspeptin systems from studies of fish brain. J. Fish Biol. 76, 161–182. [DOI] [PubMed] [Google Scholar]
- Amo R., Aizawa H., Takahoko M., Kobayashi M., Takahashi R., Aoki T. and Okamoto H. (2010) Identification of the zebrafish ventral habenula as a homolog of the mammalian lateral habenula. J. Neurosci. 30, 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amo R., Fredes F., Kinoshita M. et al (2014) The habenulo‐raphe serotonergic circuit encodes an aversive expectation value essential for adaptive active avoidance of danger. Neuron 84, 1034–1048. [DOI] [PubMed] [Google Scholar]
- Arai A. C. (2009) The role of kisspeptin and GPR54 in the hippocampus. Peptides 30, 16–25. [DOI] [PubMed] [Google Scholar]
- Behzadi G., Kalen P., Parvopassu F. and Wiklund L. (1990) Afferents to the median raphe nucleus of the rat: retrograde cholera toxin and wheat germ conjugated horseradish peroxidase tracing, and selective D‐[3H]aspartate labelling of possible excitatory amino acid inputs. Neuroscience 37, 77–100. [DOI] [PubMed] [Google Scholar]
- Bianco I. H., Carl M., Russell C., Clarke J. and Wilson S. W. (2008) Brain asymmetry is encoded at the level of axon terminal morphology. Neural. Dev. 3, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biran J., Ben‐Dor S. and Levavi‐Sivan B. (2008) Molecular identification and functional characterization of the kisspeptin/kisspeptin receptor system in lower vertebrates. Biol. Reprod. 79, 776–786. [DOI] [PubMed] [Google Scholar]
- Braitenberg V. and Kemali M. (1970) Exceptions to bilateral symmetry in the epithalamus of lower vertebrates. J. Comp. Neurol. 138, 137–146. [DOI] [PubMed] [Google Scholar]
- Broms J., Antolin‐Fontes B., Tingström A. and Ibañez‐Tallon I. (2015) Conserved expression of the GPR151 receptor in habenular axonal projections of vertebrates. J. Comp. Neurol. 523, 359–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary B. P., Raudsepp T., Frönicke L. and Scherthan H. (1998) Emerging patterns of comparative genome organization in some mammalian species as revealed by Zoo‐FISH. Genome Res. 8, 577–589. [DOI] [PubMed] [Google Scholar]
- Corson J. A. and Erisir A. (2013) Monosynaptic convergence of chorda tympani and glossopharyngeal afferents onto ascending relay neurons in the nucleus of the solitary tract: a high‐resolution confocal and correlative electron microscopy approach. J. Comp. Neurol. 521, 2907–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin J. F. (1996) 5 ‐HT, antidepressant drugs and the psychosocial origins of depression. J. Psychopharmacol. 10, 31–38. [DOI] [PubMed] [Google Scholar]
- DeCarvalho T. N., Subedi A., Rock J., Harfe B. D., Thisse C., Thisse B., Halpern M. E. and Hong E. K. (2014) Neurotransmitter map of the asymmetric dorsal habenular nuclei of zebrafish. Genesis 52, 636–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner K. and Singewald N. (2006) The role of substance P in stress and anxiety responses. Amino Acids 31, 251–272. [DOI] [PubMed] [Google Scholar]
- El Mestikawy S., Wallén‐Mackenzie Å., Fortin G. M., Descarries L. and Trudeau L. (2011) From glutamate co‐release to vesicular synergy: vesicular glutamate transporters. Nat. Rev. Neurosci. 12, 204–216. [DOI] [PubMed] [Google Scholar]
- Escobar S., Servili A., Espigares F. et al (2013) Expression of kisspeptins and kiss receptors suggests a large range of functions for kisspeptin systems in the brain of the European sea bass. PLoS ONE 8, e70177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felip A., Zanuy S., Pineda R., Pinilla L., Carrillo M., Tena‐Sempere M. and Gomez A. (2009) Evidence for two distinct KiSS genes in non‐placental vertebrates that encode kisspeptins with different gonadotropin‐releasing activities in fish and mammals. Mol. Cell. Endocrinol. 312, 61–71. [DOI] [PubMed] [Google Scholar]
- Ferraro G., Montalbano M. E., Sardo P. and La Grutta V. (1996) Lateral habenular influence on dorsal raphe neurons. Brain Res. Bull. 41, 47–52. [DOI] [PubMed] [Google Scholar]
- Filby A. L., van Aerle R., Duitman J. and Tyler C. R. (2008) The kisspeptin/gonadotropin‐releasing hormone pathway and molecular signaling of puberty in fish. Biol. Reprod. 78, 278–289. [DOI] [PubMed] [Google Scholar]
- Freund‐Mercier M. J., Stoeckel M. E. and Klein M. J. (1994) Oxytocin receptors on oxytocin neurones: histoautoradiographic detection in the lactating rat. J. Physiol. 480(Pt 1), 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P. and Lillesaar C. (2012) Probing the diversity of serotonin neurons. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2382–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocho Y., Sakai A., Yanagawa Y., Suzuki H. and Saitow F. (2013) Electrophysiological and pharmacological properties of GABAergic cells in the dorsal raphe nucleus. J. Physiol. Sci. 63, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopurappilly R., Ogawa S. and Parhar I. S. (2013) Functional significance of GnRH and kisspeptin, and their cognate receptors in teleost reproduction. Front. Endocrinol. (Lausanne). 4, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka O. (2010) The habenula: from stress evasion to value‐based decision‐making. Nat. Rev. Neurosci. 11, 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E. K., Santhakumar K., Akitake C. A., Ahn S. J., Thisse C., Thisse B., Wyart C., Mangin J. and Halpern M. E. (2013) Cholinergic left‐right asymmetry in the habenulo‐interpeduncular pathway. Proc. Natl Acad. Sci. 110, 21171–21176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurbin A., Orcel H., Alonso G., Moos F. and Rabie A. (2002) The vasopressin receptors colocalize with vasopressin in the magnocellular neurons of the rat supraoptic nucleus and are modulated by water balance. Endocrinology 143, 456–466. [DOI] [PubMed] [Google Scholar]
- Kalen P., Pritzel M., Nieoullon A. and Wiklund L. (1986) Further evidence for excitatory amino acid transmission in the lateral habenular projection to the rostral raphe nuclei: lesion‐induced decrease of high affinity glutamate uptake. Neurosci. Lett. 68, 35–40. [DOI] [PubMed] [Google Scholar]
- Kanda S., Akazome Y., Mitani Y., Okubo K. and Oka Y. (2013) Neuroanatomical evidence that kisspeptin directly regulates isotocin and vasotocin neurons. PLoS ONE 8, e62776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemali M. and Guglielmotti V. (1977) An electron microscope observation of the right and the two left portions of the habenular nuclei of the frog. J. Comp. Neurol. 176, 133–148. [DOI] [PubMed] [Google Scholar]
- Kitahashi T., Ogawa S. and Parhar I. S. (2009) Cloning and expression of kiss2 in the zebrafish and medaka. Endocrinology 150, 821–831. [DOI] [PubMed] [Google Scholar]
- Lee Y. R., Tsunekawa K., Moon M. J. et al (2009) Molecular evolution of multiple forms of kisspeptins and GPR54 receptors in vertebrates. Endocrinology 150, 2837–2846. [DOI] [PubMed] [Google Scholar]
- Lieschke G. J. and Currie P. D. (2007) Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353–367. [DOI] [PubMed] [Google Scholar]
- Lillesaar C. (2011) The serotonergic system in fish. J. Chem. Neuroanat. 41, 294–308. [DOI] [PubMed] [Google Scholar]
- Lillesaar C., Tannhauser B., Stigloher C., Kremmer E. and Bally‐Cuif L. (2007) The serotonergic phenotype is acquired by converging genetic mechanisms within the zebrafish central nervous system. Dev. Dyn. 236, 1072–1084. [DOI] [PubMed] [Google Scholar]
- Lillesaar C., Stigloher C., Tannhauser B., Wullimann M. F. and Bally‐Cuif L. (2009) Axonal projections originating from raphe serotonergic neurons in the developing and adult zebrafish, Danio rerio, using transgenics to visualize raphe‐specific pet1 expression. J. Comp. Neurol. 512, 158–182. [DOI] [PubMed] [Google Scholar]
- Martin S. C., Heinrich G. and Sandell J. H. (1998) Sequence and expression of glutamic acid decarboxylase isoforms in the developing zebrafish. J. Comp. Neurol. 396, 253–266. [PubMed] [Google Scholar]
- Namba T., Sugimoto Y., Negishi M., Irie A., Ushikubi F., Kakizuka A., Ito S., Ichikawa A. and Narumiya S. (1993) Alternative splicing of C‐terminal tail of prostaglandin E receptor subtype EP3 determines G‐protein specificity. Nature 365, 166–170. [DOI] [PubMed] [Google Scholar]
- Nathan F. M., Ogawa S. and Parhar I. S. (2015) Kisspeptin1 modulates odorant‐evoked fear response via two serotonin receptor subtypes (5‐HT1A and 5‐HT2) in zebrafish. J. Neurochem. 133, 870–878. [DOI] [PubMed] [Google Scholar]
- Ogawa S. and Parhar I. S. (2012) Anatomy of the kisspeptin systems in teleosts. Gen. Comp. Endocrinol. 181, 169–174. [DOI] [PubMed] [Google Scholar]
- Ogawa S., Kitahashi T., Ng K. W. and Parhar I. S. (2010) Autocrine regulation of kiss1 in the habenula of the zebrafish. Proceedings of The 7th International Congress of Neuroendocrinology, ICN2010, Abstract P2–P20.
- Ogawa S., Ng K. W., Ramadasan P. N., Nathan F. M. and Parhar I. S. (2012) Habenular kiss1 neurons modulate the serotonergic system in the brain of zebrafish. Endocrinology 153, 2398–2407. [DOI] [PubMed] [Google Scholar]
- Ogawa S., Nathan F. M. and Parhar I. S. (2014) Habenular kisspeptin modulates fear in the zebrafish. Proc. Natl Acad. Sci. 111, 3841–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Agetsuma M. and Aizawa H. (2012) Genetic dissection of the zebrafish habenula, a possible switching board for selection of behavioral strategy to cope with fear and anxiety. Dev. Neurobiol. 72, 386–394. [DOI] [PubMed] [Google Scholar]
- Onuma T. A. and Duan C. (2012) Duplicated Kiss1 receptor genes in zebrafish: distinct gene expression patterns, different ligand selectivity, and a novel nuclear isoform with transactivating activity. FASEB J. 26, 2941–2950. [DOI] [PubMed] [Google Scholar]
- van den Pol A. N., Gao X. B., Obrietan K., Kilduff T. S. and Belousov A. B. (1998) Presynaptic and postsynaptic actions and modulation of neuroendocrine neurons by a new hypothalamic peptide, hypocretin/orexin. J. Neurosci. 18, 7962–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A. and Anderson K. D. (1990) The patterns of neurotransmitter and neuropeptide co‐occurrence among striatal projection neurons: conclusions based on recent findings. Brain Res. Brain Res. Rev. 15, 251–265. [DOI] [PubMed] [Google Scholar]
- Rink E. and Wullimann M. F. (2001) The teleostean (zebrafish) dopaminergic system ascending to the subpallium (striatum) is located in the basal diencephalon (posterior tuberculum). Brain Res. 889, 316–330. [DOI] [PubMed] [Google Scholar]
- Sato T., Hamaoka T., Aizawa H., Hosoya T. and Okamoto H. (2007) Genetic single‐cell mosaic analysis implicates ephrinB2 reverse signaling in projections from the posterior tectum to the hindbrain in zebrafish. J. Neurosci. 27, 5271–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C., Kimura Y., Hirata H., Suster M. L., Kawakami K. and Higashijima S. (2013) Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development 140, 3927–3931. [DOI] [PubMed] [Google Scholar]
- Servili A., Le Page Y., Leprince J., Caraty A., Escobar S., Parhar I. S., Seong J. Y., Vaudry H. and Kah O. (2011) Organization of two independent kisspeptin systems derived from evolutionary‐ancient kiss genes in the brain of zebrafish. Endocrinology 152, 1527–1540. [DOI] [PubMed] [Google Scholar]
- Shahjahan M., Motohashi E., Doi H. and Ando H. (2010) Elevation of Kiss2 and its receptor gene expression in the brain and pituitary of grass puffer during the spawning season. Gen. Comp. Endocrinol. 169, 48–57. [DOI] [PubMed] [Google Scholar]
- Shutoh F., Ina A., Yoshida S., Konno J. and Hisano S. (2008) Two distinct subtypes of serotonergic fibers classified by co‐expression with vesicular glutamate transporter 3 in rat forebrain. Neurosci. Lett. 432, 132–136. [DOI] [PubMed] [Google Scholar]
- Soga T., Ogawa S., Millar R. P., Sakuma Y. and Parhar I. S. (2005) Localization of the three GnRH types and GnRH receptors in the brain of a cichlid fish: Insights into their neuroendocrine and neuromodulator functions. J. Comp. Neurol. 487, 28–41. [DOI] [PubMed] [Google Scholar]
- Song Y., Duan X., Chen J., Huang W., Zhu Z. and Hu W. (2015) The Distribution of kisspeptin (Kiss)1‐ and Kiss2‐positive neurones and their connections with gonadotrophin‐releasing Hormone‐3 neurones in the zebrafish brain. J. Neuroendocrinol. 27, 198–211. [DOI] [PubMed] [Google Scholar]
- Sutherland R. J. (1982) The dorsal diencephalic conduction system: a review of the anatomy and functions of the habenular complex. Neurosci. Biobehav. Rev. 6, 1–13. [DOI] [PubMed] [Google Scholar]
- Tena‐Sempere M., Felip A., Gómez A., Zanuy S. and Carrillo M. (2012) Comparative insights of the kisspeptin/kisspeptin receptor system: lessons from non‐mammalian vertebrates. Gen. Comp. Endocrinol. 175, 234–243. [DOI] [PubMed] [Google Scholar]
- Ulloa‐Aguirre A., Janovick J. A., Miranda A. L. and Conn P. M. (2006) G‐protein‐coupled receptor trafficking: understanding the chemical basis of health and disease. ACS Chem. Biol. 1, 631–638. [DOI] [PubMed] [Google Scholar]
- Um H. N., Han J. M., Hwang J. I., Hong S. I., Vaudry H. and Seong J. Y. (2010) Molecular coevolution of kisspeptins and their receptors from fish to mammals. Ann. N. Y. Acad. Sci. 1200, 67–74. [DOI] [PubMed] [Google Scholar]
- Valentino R. J., Bey V., Pernar L. and Commons K. G. (2003) Substance P Acts through local circuits within the rat dorsal raphe nucleus to alter serotonergic neuronal activity. J. Neurosci. 23, 7155–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertes R. P., Fortin W. J. and Crane A. M. (1999) Projections of the median raphe nucleus in the rat. J. Comp. Neurol. 407, 555–582. [PubMed] [Google Scholar]
- Wang R. Y. and Aghajanian G. K. (1977) Physiological evidence for habenula as major link between forebrain and midbrain raphe. Science 197, 89–91. [DOI] [PubMed] [Google Scholar]
- Wullimann M. F. and Rink E. (2002) The teleostean forebrain: a comparative and developmental view based on early proliferation, Pax6 activity and catecholaminergic organization. Brain Res. Bull. 57, 363–370. [DOI] [PubMed] [Google Scholar]
- Wulliman M. F., Rupp B. and Reichert H. (1996) Neuroanatomy of the zebrafish brain: a topological atlas. Birkhäuser, Basel. [Google Scholar]
- Yamamoto N. and Ito H. (2008) Visual, lateral line, and auditory ascending pathways to the dorsal telencephalic area through the rostrolateral region of the lateral preglomerular nucleus in cyprinids. J. Comp. Neurol. 508, 615–647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Kiss‐R1‐ir localization and KRBDP2 expression in other regions of the zebrafish brain.
Figure S2. Expression of slc17a6b, gad2 and chat mRNAs in the habenula and raphe regions.
Figure S3. Dual‐fluorescence labeling of Kiss‐R1 (red) in glutamatergic and GABAergic neurons (green) in raphe nuclei.
Figure S4. Co‐expression of slc17a6b and gad2 (red) with 5‐HT (green).
Table S1. Gene abbreviations, the primer sequences, probe size and GenBank accession numbers for probes for DIG‐in situ hybridization.


