Abstract
A premature senescence and death 128 (psd128) mutant was isolated from an ethyl methane sulfonate‐induced rice IR64 mutant bank. The premature senescence phenotype appeared at the six‐leaf stage and the plant died at the early heading stage. psd128 exhibited impaired chloroplast development with significantly reduced photosynthetic ability, chlorophyll and carotenoid contents, root vigor, soluble protein content and increased malonaldehyde content. Furthermore, the expression of senescence‐related genes was significantly altered in psd128. The mutant trait was controlled by a single recessive nuclear gene. Using map‐based strategy, the mutation Oryza sativa cell division cycle 48 (OsCDC48) was isolated and predicted to encode a putative AAA‐type ATPase with 809 amino‐acid residuals. A single base substitution at position C2347T in psd128 resulted in a premature stop codon. Functional complementation could rescue the mutant phenotype. In addition, RNA interference resulted in the premature senescence and death phenotype. OsCDC48 was expressed constitutively in the root, stem, leaf and panicle. Subcellular analysis indicated that OsCDC48:YFP fusion proteins were located both in the cytoplasm and nucleus. OsCDC48 was highly conserved with more than 90% identity in the protein levels among plant species. Our results indicated that the impaired function of OsCDC48 was responsible for the premature senescence and death phenotype.
Keywords: Oryza sativa, premature senescence and death, AAA‐ATPase, OsCDC48
Edited by: Natalia Doudareva, Purdue University, USA
INTRODUCTION
Leaf senescence, the final phase of the development of a plant leaf, is a genetically programmed developmental process and is tightly linked to crop yield and biomass production (Wu et al. 2012). Early occurrence of leaf senescence or premature leaf senescence caused either by environmental stresses or internal genetic factors often lead to a greater loss of the final yield and lower quality of crop grain. In contrast, delayed leaf senescence or so‐called stay green shows potential positive effects on plant yield and survival (Thomas and Howarth 2000; Rivero et al. 2007; Yoo et al. 2007).
Characterization of internal genetic factors mediating leaf senescence is critical to the understanding of its molecular genetic basis. So far, a profile of senescence‐associated genes (SAGs) encoding for transcription factors, enzymes and proteins has been identified in different plant species (Wu et al. 2012; Seo 2014). One of the important enzymes associated with leaf senescence and plant survival is the eukaryotic CDC48 (cell division cycle 48) protein belonging to the AAA‐ATPase family. CDC48 is well known for its wide range of cellular functions including endoplasmic reticulum associated protein degradation (ERAD), DNA replication, cell proliferation and senescence (Ye et al. 2001; Mouysset et al. 2008; Park et al. 2008; Bae et al. 2009; Kuhlbrodt et al. 2011). Its encoding gene CDC48 was first identified in a yeast (Saccharomyces cerevisiae) screen for conditional mutations affecting the cell cycle (Moir et al. 1982). To date, the homologous CDC48 genes have been identified in the yeast, several animal and plant species (Bar‐Nun 2005; Mouysset et al. 2006; Park et al. 2008; Ballar et al. 2011).
The mechanism of CDC48‐regulated apoptosis and leaf senescence or plant survival is complicated and largely unknown. Madeo and colleagues (1997) found that a single base substitution in CDC48 could alter the sensitivity of yeast to cold treatment and lead to cell apoptosis. Similarly, a point mutation in the CDC48 homolog, p97/VCP, causes retarded cellular proliferation and apoptosis of human B‐lymphocytes (Shirogane et al. 1999). In Caenorhabditis elegans, simultaneous depletions of CDC48.1 and CDC48.2 result in the embryonic lethality completely (Yamanaka et al. 2012). Furthermore, RNA interference (RNAi) of CDC48 and its cofactors (Ufd1 and Npl4) inhibits the proliferation of embryonic cells and increases the rate of embryonic lethality in C. elegans (Mouysset et al. 2006; 2008). RNAi also disturbs the disassembly of spindles and ERAD in Xenopus egg extracts, yeast and C. elegans (Cao K et al. 2003; Cheeseman and Desai 2004; Mouysset et al. 2008). Interestingly, overexpression of TbVCP‐1, a CDC48 homolog from Trypanosome could completely abort the cell cycle and lead to apoptosis of most cells within 24 h (Lamb et al. 2001). Although the mechanism of CDC48‐mediated cell death remains to be elucidated, it is believed that the multiple functions of CDC48 mainly depend on its cofactors. For examples, the serine/threonine kinase Pim‐1 is cooperated with CDC48 to prevent apoptosis, and mutations in CDC48 could trigger the cellular apoptosis (Shirogane et al. 1999). More recently, it has been found that CDC48/p97 could modulate the aging and senescence in cooperation with the deubiquitinating enzyme ataxin‐3 in human beings (Kuhlbrodt et al. 2011).
In the present study, we have identified a rice premature senescence and death 128 (psd128) mutant. Here, we present the results of phenotypic and physiological characterization, map‐based isolation and functional validation of the mutation. We show that a single base substitution leading to a premature stop codon in OsCDC48 (Oryza sativa cell division cycle 48) is responsible for the premature senescence and death phenotype in rice.
RESULTS
Phenotypic performance of psd128
The premature senescence and death 128 (originally coded as HM128) mutant was obtained from mutagenesis of cultivar IR64 using ethyl methane sulfonate (EMS). psd128 displayed lower germination rate and slower germination (Figure 1A), retarded growth and development of roots and leaves than those of the wild‐type IR64 (Figure 1B, C; Table S1). The most significant changes between psd128 and IR64 appeared at the 6‐leaf seedling stage under field conditions when the second leaf of psd128 became yellowish with brown spots (Figure 1D). This phenotype first appeared on the leaf tip and spread gradually downward to cover the whole leaf blade. Subsequently, the older leaves turn white and wilted gradually, and the plants died almost completely except a few flag leaves at the early heading stage (Figure 1E, F). DAB (3,3́‐diaminobenzidine tetrachloride) staining indicated that accumulation of H2O2 was presented in and around the brown spots in the spotted leaves of psd128 while the normal green leaves of psd128 and IR64 were clear (Figure 1G). Ultimately, psd128 had shorter panicles and lower seed‐setting rate (Figure 1H), smaller grains with brown spotted hull (Figure 1I), severer chalkiness of milled rice in contrast to those of IR64 (Figure 1J). Except the number of panicles, other major agronomic traits including the plant height and 1000‐grain weight were all significantly reduced in psd128 than those of IR64 (Table 1).
Figure 1.

Phenotypic characteristics of psd128 and the wild‐type IR64 (A) Seed germination of IR64 and psd128. (B) Root growth of IR64 and psd128. (C) Seedling of psd128 and IR64 at the 2‐3 leaf stage. (D) Plant of psd128 and IR64 at the 6 leaf stage. (E) Plant of psd128 and IR64 at the heading stage. (F) Premature senescence leaf of psd128 and IR64 at the tillering stage. (G) Histochemical analysis of IR64 and psd128. 1, IR64; 2, IR64 after DAB staining; 3, psd128; 4, psd128 after DAB staining. (H) Panicle length of IR64 and psd128. (I) Grains of IR64 and psd128. (J) Polished rice of IR64 and psd128.
Table 1.
Agronomic traits of mature plants of psd128 and the wild‐type IR64
| Material | Plant height (cm) | Panicle length (cm) | No. panicle | 1,000‐grain weight (g) | Seed‐setting rate (%) |
|---|---|---|---|---|---|
| IR64 | 103.67 ± 1.53 | 26.53 ± 1.21 | 15.33 ± 1.53 | 25.32 ± 2.48 | 67.10 ± 2.38 |
| psd128 | 64.67 ± 2.52** | 21.53 ± 1.01* | 12.67 ± 0.58 | 17.20 ± 1.33** | 13.15 ± 1.04** |
Values are means ± SD; n = 3;*, significance at P ≤ 0.05; **, high significance at P ≤ 0.01.
Physiological alterations of psd128
To determine the direct cause for the yellowish and whitish leaf phenotype of psd128, we first examined the chlorophyll contents at different leaf developmental stages. Before the initiation of premature senescence there were no significant differences in the contents of chlorophyll a (Chl a), chlorophyll b (Chl b) and carotenoid between psd128 and IR64 at the four‐leaf stage. However, at the six‐leaf stage when the premature senescence phenotype appeared in psd128, the contents of Chl a and carotenoid were significantly lower than those of IR64 while no significant alteration was detected in the content of Chl b between the two genotypes (Table S2). At the maximum tillering stage when the premature senescence phenotype became more severe in the older leaves, the contents of Chl a, Chl b and Chl a/b ratio, except carotenoid in the 2nd upper leaves with slightly yellowish phenotype in psd128 were significantly lower than those of IR64. The contents of Chl a, Chl b, Chl a/b ratio and carotenoid in the 4th upper leaves with apparent senescence in psd128 were all significantly lower than those of IR64 (Table 2). At the early heading stage, the contents of Chl a, Chl b and Chl a/b ratio in the flag leaves were apparently reduced in psd128 in contrast to IR64 while the content of carotenoid was significantly higher than that of IR64. The contents of Chl a, Chl b and Chl a/b ratio except carotenoid in the 2nd upper leaves of psd128 were significantly reduced than those in IR64. The contents of Chl a, Chl b, Chl a/b ratio and carotenoid in the 3rd upper leaves of psd128 were all significantly reduced compared to those of IR64 (Table S3). Our results indicated that psd128 was a chlorophyll and carotenoid‐deficient mutant.
Table 2.
Contents of photosynthetic pigments at the maximal tillering stage
| Pigment | Second upper leaf | Forth upper leaf | ||
|---|---|---|---|---|
| IR64 | psd128 | IR64 | psd128 | |
| Chl a | 4.74 ± 0.06 | 4.02 ± 0.08** | 3.78 ± 0.11 | 1.34 ± 0.16** |
| Chl b | 1.71 ± 0.05 | 1.33 ± 0.01** | 1.48 ± 0.05 | 0.42 ± 0.02** |
| Chl a/b | 2.78 ± 0.05 | 3.02 ± 0.04** | 2.56 ± 0.13 | 3.18 ± 0.17** |
| Carotenoid | 0.74 ± 0.09 | 0.72 ± 0.01 | 0.60 ± 0.04 | 0.33 ± 0.01** |
Chl, chlorophyll; values are means ± SD; n = 3; **, high significance at P ≤ 0.01.
We next examined the photosynthetic parameters and found that there were no significant alterations at the maximal tillering stage in the net photosynthetic rate (Pn), stomatal conductance (Gs), intercelluar CO2 concentration (Ci) and transpiration rate (Tr) between psd128 and IR64 in the 2nd upper leaves without premature senescence (Figure 2A–D). However, Ci of the 3rd upper leaves of psd128 with the mutant phenotype was significantly higher than that of IR64 while no obvious changes were detected in Pn, Gs and Tr between the two genotypes (Figure 2A–D). At the early heading stage, the maximum quantum yield of PSII (Fv/Fm), effective quantum yield of PSII (Y(II)) and electron transport rate (ETR) were all significantly reduced in psd128 compared to those of IR64 in the 2nd upper leaves and flag leaves (Figure 2E–G). The results indicated that psd128 had impaired photosynthetic ability in the premature senescent leaves.
Figure 2.
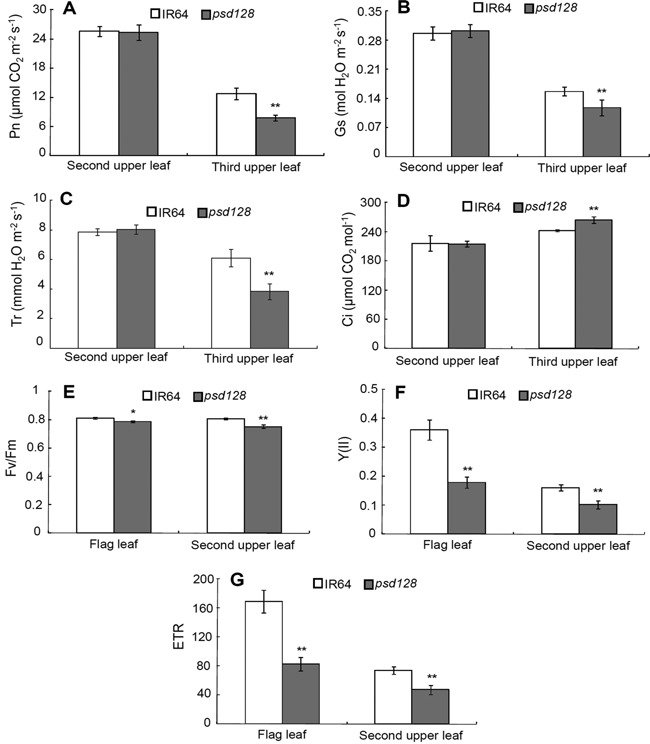
Photosynthetic and chlorophyll fluorescence parameters of IR64 and psd128 (A) Net photosynthetic rate (Pn). (B) stomatal conductance (Gs). (C) transpiration rate (Tr) and (D) intercellular CO2 concentration (Ci) of the 2nd‐upper and 3rd‐upper leaves of IR64 and psd128 at the maximal tillering stage. (E) Maximum photochemical quantum yield of PSII (Fv/Fm). (F) effective photochemical quantum yield of PSII (Y(II)) and (G) electron transport rate (ETR) of the flag and second upper leaves of IR64 and psd128 at the heading stage. Values are means ± SD; n = 3; **, Highly significant at P ≤ 0.01.
We next examined the ultrastructure of chloroplasts using transmission electron microscopy. The results showed that fully developed chloroplasts in mesophyll cells were presented in the normal green leaves of psd128 and IR64 (Figure 3A–D) while a reduced number of chloroplasts, decreased number of thylakoid laminar layers, poor arrangement of grana and increased number of osmiophilic granules were observed in the premature senescent leaves of psd128 (Figure 3E–F), indicating that the mutation significantly affected the development of chloroplasts.
Figure 3.
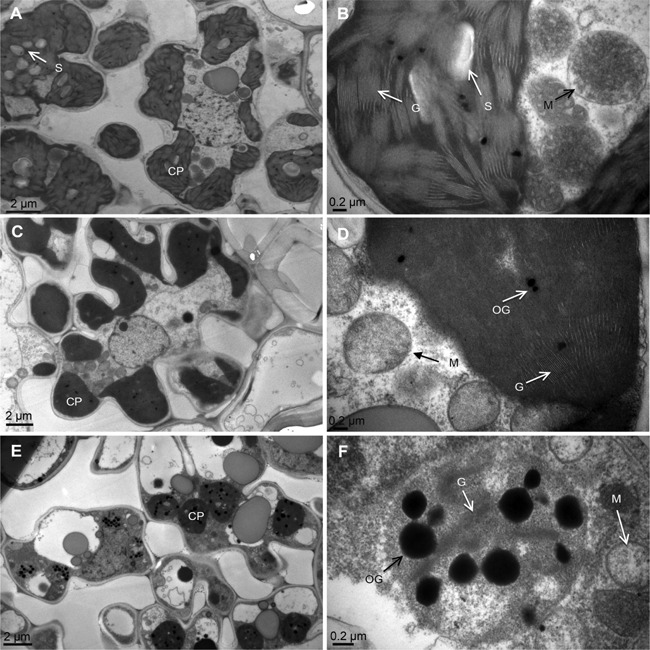
Ultrastructure of the chloroplast (A, B) Ultrastructure of the chloroplast in the wild type IR64. (C, D) Ultrastructure of the chloroplast in the green leaf of psd128. (E, F) Ultrastructure of the chloroplast in the premature senescence leaf of psd128. CP, chloroplast; G, granum; M, mitochondrion; OG, osmiophilic granule; S, starch granule.
A large array of enzymes is associated with plant development and aging. To determine the activity of critical enzymes associated with the plant vigor and scavenging of active radical ions, we first measured the activity of dehydrogenase in the root (Zhang 1992). The result showed that the levels of 2,3,5‐triphenyl tetrazolium chloride (TTC) were all significantly reduced in psd128 than those of IR64 at the four, six‐leaf and maximal tillering stages (Figure 4A), indicating that the root vigor of psd128 was much lower than that of IR64 in the same developmental stages. Our result indicated that the premature senescence also occurred in the mutant root system.
Figure 4.
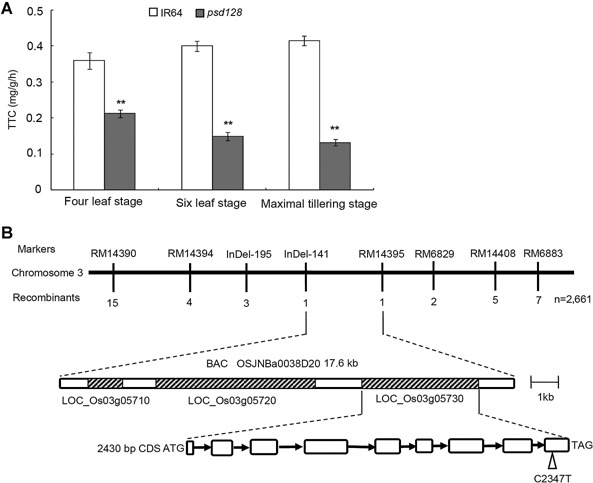
Root activity at different stages and map‐based isolation of OsCDC48 (A) Triphenyl tetrazolium chloride (TTC) contents (mg/g per h) of IR64 and psd128 were determined using 0.5 g fresh roots according to the method described by Zhao et al. (2002) at the four leaf, six leaf and maximal tillering stages. Values are means ± SD; n = 3; **, Highly significant at P ≤ 0.01. (B) A total of 2661 mutant type F2 individuals were used for fine mapping. Open triangle indicates C to T substitution at position 2347 in the 9th exon. Black arrows indicate introns; open boxes indicate exons; molecular markers used for mapping are listed in Table S5.
We then determine the senescence‐related parameters including the enzymatic activities of peroxidase (POD), superoxide dismutase (SOD), catalase (CAT) and ascorbate peroxidase (APX), soluble protein and malonaldehyde (MDA) contents. At the maximal tillering stage, the activities of SOD, POD, APX, soluble protein and MDA contents were similar to those of IR64 except the activity of CAT in the 2nd upper normal green leaves. However, the activities of CAT, POD, SOD and soluble protein content in psd128 were significantly lower than those of IR64 in the 4th upper senescent leaves except the activities of APX, in contrast, the MDA content was significantly higher than that of IR64 (Figure 5A–F). At the heading stage, the activities of SOD was significantly decreased in psd128 than that of IR64 while the activities of CAT, POD, APX and the levels of soluble proteins and MDA were similar to those of IR64 in the 2nd upper leaves without obvious senescent phenotype (Figure 5G–L). The activities of CAT, POD, SOD, APX and the level of soluble proteins were significantly lower than those of IR64 while the content of MDA was significantly higher than that of IR64 in the 3rd upper leaves with severe senescent phenotype (Figure 5G–L). The above results indicated that the activities of CAT, SOD, POD, APX and the level of soluble protein decreased rapidly while the level of MDA increased significantly in psd128 with the development of premature senescence in comparison to those of the wild‐type.
Figure 5.
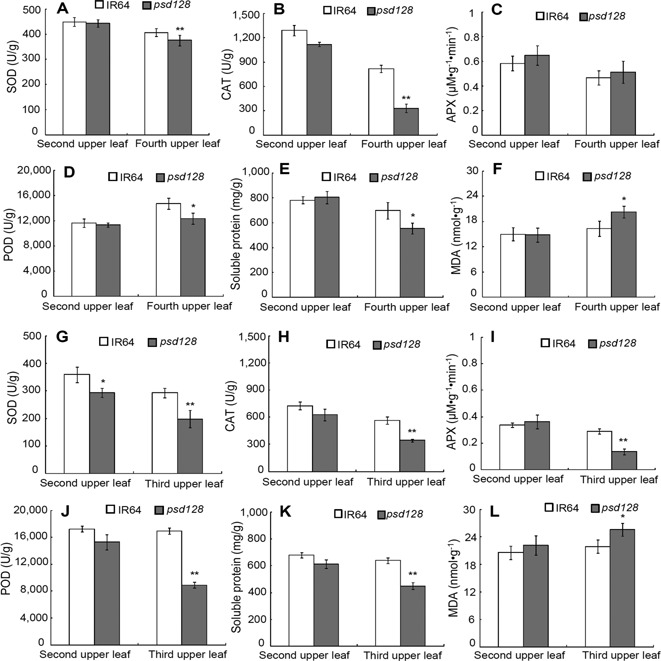
Senescence related‐indices of IR64 and psd128 at different stages (A–F) The enzymatic activities of SOD, CAT, APX and POD, the contents of soluble proteins and MDA from the 2nd and 4th upper leaves at the maximal tillering stage. (G–L) The enzymatic activities of SOD, CAT, APX and POD, the contents of soluble proteins and MDA from the 2nd and 3rd upper leaves at the heading stage. SOD, superoxide dismutase; CAT, catalase; APX, ascorbate peroxidase; POD, peroxidase; MDA, malonaldehyde. Values are means ± SD; n = 3; *, Significance at P ≤ 0.05; **, high significance at P ≤ 0.01.
Map‐based isolation of OsCDC48
To determine the genetic control of the premature senescence and death phenotype, we crossed psd128 with IR64 and Moroberekan, respectively. The F1 plants from both crosses exhibited the normal green phenotype, indicating the mutant phenotype was controlled by recessive gene (s). In the F2 population derived from the cross psd128/PA64, 3424 plants were normal green‐leaved and 1192 plants were premature senescence and death, fitting to a 3:1 ratio (χ 2 = 1.668 < χ 2 0.05 = 3.84). Among the 5690 plants derived from psd128/Moroberekan, 4221 plants were normal green‐leaved and 1469 plants were premature senescence and death, again fitting to the expected 3:1 ratio (χ 2 = 2.027 < χ 2 0.05 = 3.84). The results indicated that the mutation was controlled by a single recessive gene (Table 3). To further confirm this observation, six segregating F3 lines derived from both crosses were planted and phenotyped, again they all fitted to the 3:1 segregation ratios (Table S4). Taken together, the premature senescence and death trait was controlled by a single recessive gene.
Table 3.
Genetic analysis of psd128
| No. F2 individuals | ||||||
|---|---|---|---|---|---|---|
| Cross | F1 | Total | Wild‐type | Mutant‐type | P (3:1) | χ 2 0.05 |
| psd128/PA64 | Normal | 4616 | 3424 | 1192 | 0.20 | 1.67 |
| psd128/Moroberekan | Normal | 5690 | 4221 | 1469 | 0.16 | 2.03 |
To determine the physical location of the mutation, we first used the balk segregant analysis to rapidly link the mutation and SSR markers on chromosome 3, subsequently 2661 mutant‐type individuals (1469 plants derived from psd128/Moroberekan and 1192 plants derived from psd128/PA64) were used for further mapping and delimited the mutation to a 17.6 kb region flanked by InDel‐141 and RM14395 on chromosome 3 (Figure 4B, Table S5). Three open reading frames (LOC_Os03g05710, LOC_Os03g05720, LOC_Os03g05730) were identified in the region based on the annotation in the National Center for Biotechnology Information (NCBI) databank. Sequence analysis indicated that there was no base difference in LOC_Os03g05710 and LOC_Os03g05720 between psd128 and IR64. However, a single base substitution from C to T at position 2347 in the mutant CDS was identified in LOC_Os03g05730 (cell division cycle protein 48, putative, expressed), leading to a premature stop codon TAG. Thus, the locus LOC_Os03g05730, termed as OsCDC48 here afterwards, was considered as the targeted mutation most likely responsible for the mutant phenotype.
To validate the function of OsCDC48, the complementary vector pCAMBIA1300‐OsCDC48 was introduced into the mature embryo‐induced calli from psd128 via Agrobacterium tumefaciens (A. tumefaciens)‐mediated transformation. Five transgenic plants were obtained and all of them exhibited a normal green phenotype as the wild‐type IR64 (Figure 6A–C). Furthermore, the RNAi vector pTCK303‐RNAi was introduced into the mature embryo‐induced calli from cultivar Nipponbare via A. tumefaciens‐mediated transformation. Among a total of six RNAi plants obtained, two plants died at the seedling stage, and the remaining four plants all exhibited the premature senescence and death phenotype similar to that of psd128 (Figure 6D, E). Real‐time PCR analysis indicated that the expression levels of OsCDC48 were significantly reduced in the transgenic RNAi plants compared to those of Nipponbare and IR64 (Figure 6F). Altogether, our results suggested that the point mutation in OsCDC48 was responsible for the premature senescence and death phenotype.
Figure 6.
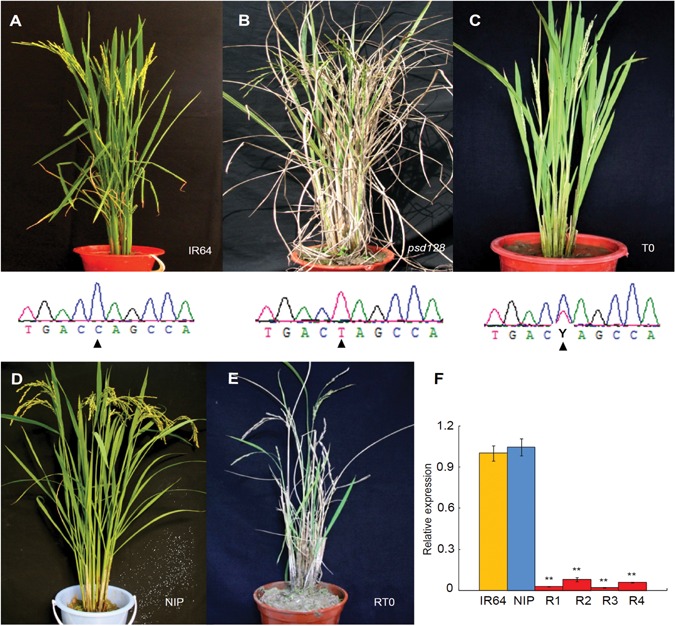
Functional complementation and RNAi (A–C) Complementation of the mutation. (A) IR64 plant with genotype C/C at position 2347 of OsCDC48. (B) psd128 plant with genotype T/T at position 2347 of Oscdc48. (C) T0 transgenic plant with heterozygous genotype Y (C/T) at position 2347 of OsCDC48/Oscdc48. Black triangles indicate the position at 2347. (D–F) RNAi of OsCDC48 and the relative expression of OsCDC48. (D) The receptor cultivar Nipponbare (NIP). (E) T0 RNAi plant (RT0). (F) qRT‐PCR analysis of OsCDC48 in IR64, NIP and R1‐R4 (T0 RNAi plant 1 to plant 4). The values are presented as the mean ± SD from three replications. **, high significance at P ≤ 0.01.
OsCDC48 is constitutively expressed and the mutation affects the expression of senescence‐related genes
To determine the relative abundance of the transcripts, we performed quantitative reverse‐transcription polymerase chain reaction (qRT‐PCR) and found that the expression level of OsCDC48 in the roots was significantly higher than those in the stems, leaves and panicles (Figure 7A). The expression levels of OsCDC48 were similar in the stems, leaves and panicles. Finally, we carried out the promoter assay in transgenic Nipponbare plants and found that GUS was detected in the roots, stems, leaves and panicles/spikelets in consistent to the qRT‐PCR (Figure 7B).
Figure 7.
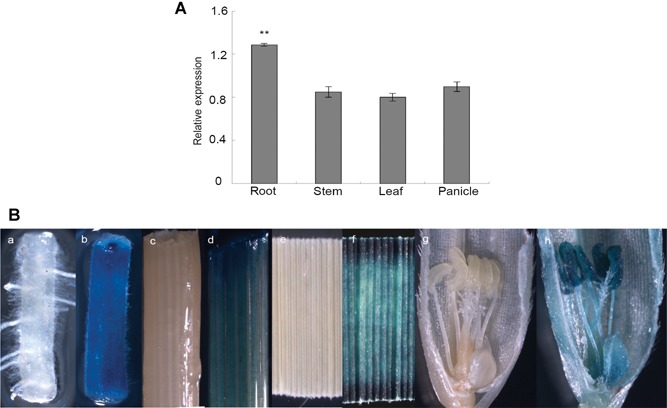
Expression analysis of OsCDC48 (A) qRT‐PCR analysis of the relative expression of OsCDC48. (B) GUS expression analysis of the OsCDC48 promoter. a, c, e, g, wild type control; b, root of pCAMBIA1381Z‐C‐gus transgenic plants; d, stem of pCAMBIA1381Z‐C‐gus transgenic plants; f, leaf of pCAMBIA1381Z‐C‐gus transgenic plants; h, spikelet of pCAMBIA1381Z‐C‐gus transgenic plants. The accession numbers and primer sequences of actin and OsCDC48 are listed in Table S6. **, high significance at P ≤ 0.01.
To determine the effects of the mutation on the expression of senescence‐related genes, we carried qRT‐PCR analysis on six negative regulators (TDC, CAT‐B, OsGS, OsETR2, JARID1C, OsAkɑGal) and a positive regulator of senescence‐related gene (Glu) (Table S6). The expression levels of the six negative regulators in senescent leaves were significantly lower than those of normal leaves of psd128 and IR64, in contrast, the expression level of Glu was significantly higher in senescent leaves (Figure 8). The results indicated that the mutation enhanced the expression of the positive regulators while it inhibited the expression of the negative regulator, and accelerated the overall progression to leaf senescence in psd128.
Figure 8.
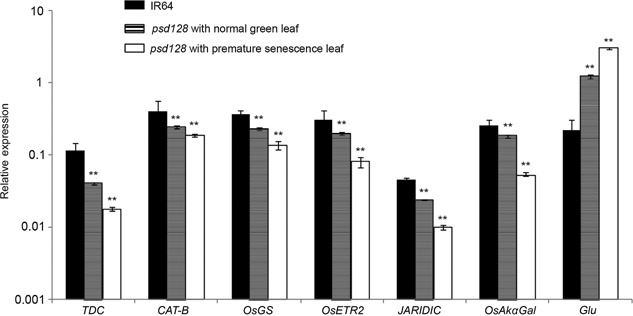
Real‐time PCR analysis of senescence‐related genes in IR64 and psd128 The accession numbers and primer sequences of seven senescence‐related genes (TDC, CAT‐B, OsGS, OsETR2, JAIRDIC, OsAkαGal and Glu) are listed in Table S6. The expression levels were determined by real‐time PCR and normalized to the actin (Os03g0718100). The values are presented as the mean ± SD from three biological replicates. **, highly significance at P ≤ 0.01.
OsCDC48 is located in cytoplasm and nucleus
CDC48 fusion proteins have been located in cytoplasm or/and nucleus in other plant species (Park et al. 2008). To determine its intracellular location in rice, we injected the fusion vector into the tobacco (Nicotiana tabacum) mesophyll cells and found that the OsCDC48:YFP fusion proteins were targeted to the cytoplasm and nucleus as well (Figure 9). This is similar to those of NtCDC48 (Nicotiana tabacum CDC48), AtCDC48 (Arabidopsis thaliana CDC48) and NgCDC48 (Nitotiana glutinosa CDC48) (Feiler et al. 1995; Cheng et al. 2007; Bae et al. 2009). Interestingly, OsCDC48: YFP fusion proteins were also observed in ER‐like structures in the tobacco mesophyll cells in the present study. Whether OsCDC48 is indeed located in ER needs to be validated.
Figure 9.
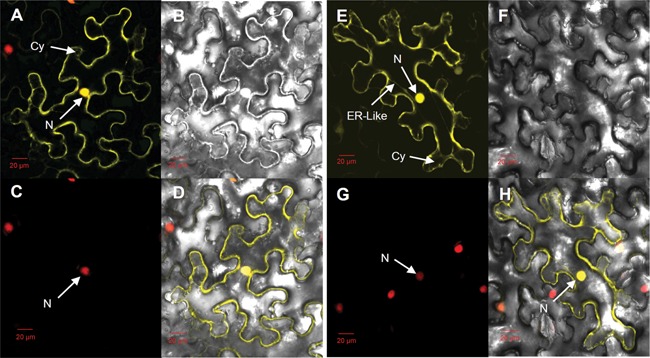
Subcellular localization of OsCDC48 protein in mesophyll cells of Nicotiana tabacum (A) Enhanced yellow fluorescence signal of 35S::YFP. (B) Bright‐field image of YFP. (C) Red fluorescence signal of 35S::RFP:H2B. (D) Merged image of A, B, and C. (E) Enhanced yellow fluorescence signal of OsCDC48:YFP. (F) Bright‐field image of OsCDC48:YFP. (G) Red fluorescence signal of 35S::RFP:H2B. (H) Merged image of E, F, and G. Scale bar = 20 μm. N, nucleus; Cy, cytoplasm; ER‐like, endoplasmic reticulum‐like structure.
Structural and phylogenic analysis of OsCDC48
The cDNA of OsCDC48 consists of 2,915 bp including a 119 bp 5′‐UTR, a 366 bp 3′‐UTR and a 2,430 bp CDS. OsCDC48 has nine exons and eight introns, and encodes a putative AAA‐ATPase with 809 amino acid (aa) residuals. The putative OsCDC48 protein has the typical N terminus (1–199 aa), the C terminus (771–809 aa) and two AAA‐ATPase domains (D1 from 213–466 aa and D2 from 486–771 aa) (Figure 10A). Each of the AAA‐ATPase domains contains the Walker A (magenta color) and Walker B (yellow color) motifs (Figure 10B), indicating OsCDC48 belongs to the family of AAA‐ATPase (Stolz et al. 2011).
Figure 10.
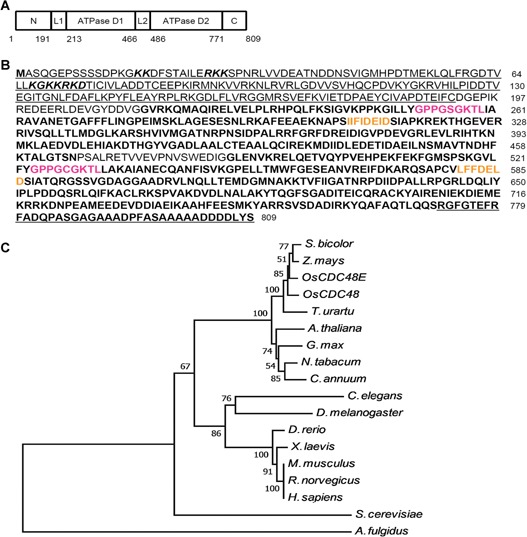
Predicted amino acid sequence of OsCDC48 and dendrogram of CDC48 proteins (A, B) The N‐terminus is underlined (1‐191 aa); two ATPase domains are in bold: D1 (213‐466 aa) and D2 (486‐770 aa); the C‐terminus is in bold and underlined (771‐809 aa); link 1 (DGEPIKREDEERLDEVGYDDVG) is located between the N‐terminus and D1; link 2 (PSALRETVVEVPNVSWEDIG) is located between D1 and D2; two nucleus signal peptides (KK (8×) RKK and KGKKRKD) are in italics in the N‐terminus; the Walker A are in magenta characters; the Walker B are in yellow characters. (C) A total of 18 CDC48 homologs from 17 different species were used for analysis including: OsCDC48 (LOC_Os03g05730), OsCDC48E (Os10g0442600), Arabidopsis thaliana (A. thaliana) (Q9LZF6), Nicotiana tabacum (N. tabacum) (XP_009628175), Glycine max (G. max) (P54774), Sorghum bicolor (S. bicolor) (XP_002465842), Zea mays (Z. mays) (XP_008671992), Triticum urartu (T. urartu) (EMS63012), Capsicum annuum (C. annuum) (Q96372), Archaeoglobus fulgidus (A. fulgidus) (O28972), Caenorhabditis elegans (C. elegans) (P54811), Saccharomyces cerevisiae (S. cerevisiae) (P25694), Danio rerio (D. rerio) (P23787), Drosophila melanogaster (D. melanogaster) (Q7KN62), Mus musculus (M. musculus) (Q01853), Rattus norvegicus (R. norvegicus) (P46462), and Homo sapiens (H. sapiens) (P55072). Distance scale = 0.05.
To construct a phylogenic tree, we searched for the NCBI protein database and identified 18 homozygous CDC48 sequences from 17 species (Figure 10C). Sequence analysis using the neighbor‐joining method showed that these CDC48s were highly conserved with a similarity level ranged from 50%–97% among the living organisms. Especially, the homologous levels of CDC48 among the plant species were as high as 90%–97%, suggesting that the mode of action among CDC48s might be conserved in the organisms. Comparison of the sequence similarity indicated that highly conserved regions were presented in the N‐terminus, D1 and D2 domains with exactly the same in the Walker A and Walker B motifs. The main sequence differences were presented in the C‐terminus although the DDLYS(N) motif was completely identical among a number of CDC48 proteins (Figure 11). In addition, rice has two isoforms of CDC48, OsCDC48 in chromosome 3 and OsCDC48E in chromosome 10. Both of them are highly conserved and share a remarkable high level of similarity at 97% in amino acid residuals. The function of OsCDC48E has yet to be investigated.
Figure 11.
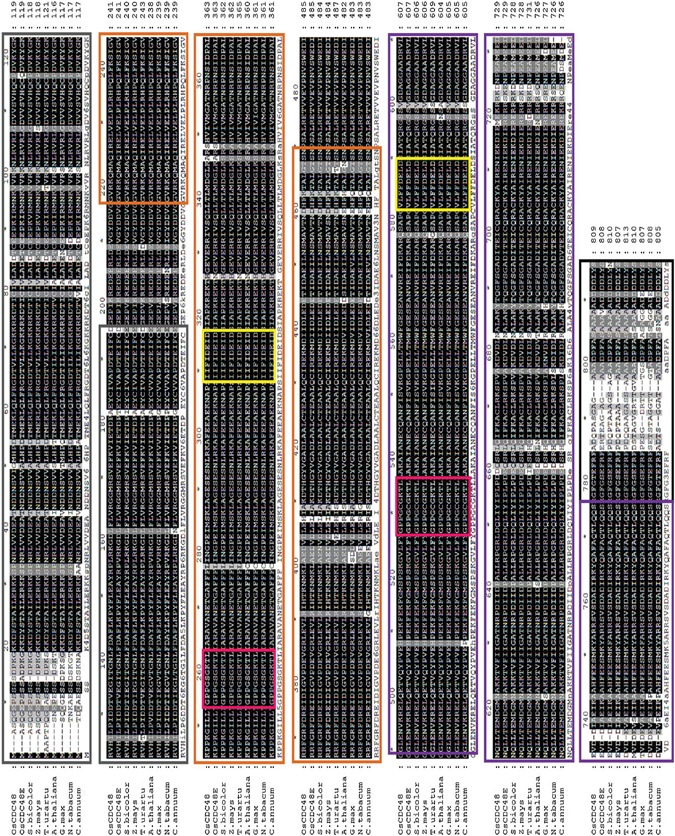
Alignment of OsCDC48 proteins in plants Nine sequences from eight plant species were aligned using ClustalW software. N‐terminus, gray color box; D1, orange color box; D2, purple color box; Walker A, magenta color box; Walker B, yellow color box; C‐terminus, black color box. The accession numbers of the nine sequences are OsCDC48 (LOC_Os03g05730), OsCDC48E (Os10g0442600), A. thaliana (Q9LZF6), N. tabacum (XP_009628175), G. max (P54774), S. bicolor (XP_002465842), Z. mays (XP_008671992), T. urartu (EMS63012) and C. annuum (Q96372).
DISCUSSION
A premature senescence plant often exhibits some important characteristics such as visible color changes, reduction in photosynthesis, degeneration of chloroplasts, degradation of nucleic acids and proteins, and the translocation of macro‐ and micro‐molecules to other parts of the plant, affecting the final grain yield and total biomass production or leading to the death of the senescing plant when the symptom is severe. In the present study, we did not measure the contents of nucleic acids and the remobilization of macro‐ and micro‐molecules in spd128, but we did observe the color changes, reduction of photosynthesis, degeneration of chloroplasts, changes of senescence‐related parameters and lowered performance of agronomic traits. The results were similar to those found in other pre‐senescing mutants in rice although the genes in charge might be different (Li et al. 2005; Jiao et al. 2012).
The premature senescence and death phenotype in psd128 is controlled by a single Mendelian factor OsCDC48 in chromosome 3. Three more premature senescence‐related genes have been identified in the chromosome including psl2 (Zhang et al. 2014), esl5 (Sang et al. 2014) and D343 (Xiao, 2012). All of these genes are located in the different regions in comparison to that of OsCDC48 and none of them have been isolated. In the present study, we isolate OsCDC48 and prove that a single base substitution in the 9th exon at position (C2347T) resulting in a premature stop codon is responsible for the mutant phenotype. To our knowledge, this is the first report on isolation of OsCDC48 encoding a putative AAA‐ATPase conferring premature senescence and death phenotype in rice (Oryza sativa L.).
OsCDC48 is constitutively expressed in the roots, stems, leaves and panicles/spikelets in psd128. The highest expression level of OsCDC48 is observed in the roots and is significantly different from those of stems, leaves and panicles/spikelets in psd128. This is probably due to the root samples containing the meristematic tissue. Similarly, the A. thaliana CDC48A (AtCDC48A) is also constitutively expressed but with the highest level in proliferating cells of the young shoots, roots and flowers (Feiler et al. 1995; Park et al. 2008). To date, most studies of CDC48 proteins in plants have been performed with Arabidopsis. Further experiments are required to prove whether active expression of CDC48 is associated with proliferating cells in other plants including rice.
Many studies have shown that CDC48 proteins are localized to the nucleus and cytoplasm. Specifically, CDC48s are targeted to ER and associated with its well known function in ERAD in the yeasts, nematodes and mammals (Ye et al. 2001; Jarosch et al. 2002; Rabinovich et al. 2002). In addition to its ER location, YFP‐AtCDC48A fusion proteins are targeted to cytoplasm, nuclear envelope, nucleoplasm and nucleolar cavity in A. thaliana. This wild distribution of CDC48 is consistent with its critical roles in cell division, expansion, growth and development (Park et al. 2008). Similarly, the tobacco NtCDC48‐GFP fusion proteins are targeted to the cytoplasm and nucleus, especially to both sides of a cell plate in the phragmoplast zone at anaphase (Feiler et al. 1995; Rancour et al. 2002; Cheng et al. 2007). The results imply that the NtCDC48 may regulate spindle disassembly in the cell division cycle. Recently, a homolog of AtCDC48, AtOM66, was identified and located to the outer mitochondrial membrane. It plays a role in determining the progression to cell death in response to abiotic and biotic stresses (Zhang et al. 2014). In Oryza sativa, the OsCDC48: YFP fusion proteins are located in the cytoplasm and nucleus as well. Furthermore, the fusion proteins are also targeted to the ER‐like structure in the tobacco mesophyll cells. Whether the fusion protein is indeed located in ER and its association with premature senescence in rice has yet to be further validated.
CDC48 is a member of AAA‐ATPase family proteins and regulates ATP hydrolysis to provide energy for the development of a plant. However, the specific roles or pathways in which CDC48 is involved largely depend on its structural sites that interact with its cofactors such as Ufd2, Ufd3 and PNGase (Janiesch et al. 2007). It has been reported that the six amino acid residuals (DDDLYN) in the C‐terminal of CDC48 are necessary for binding to Ufd2 (Baek et al. 2011) while Ufd3 is also able to bind to the C‐terminal of CDC48 through its PUL domain for substrate degradation (Ghislain et al. 1996; Zhao et al. 2009). In addition to Ufd2 and Ufd3, the mouse peptide N‐glycanase (mPNGase) has been reported recently to interact with the C‐terminus of mouse CDC48 in ER‐associated degradation of misfolded glycoproteins (Li et al. 2006). In the rice psd128, a single base substitution causes putatively a truncated OsCDC48 lacking of 38 amino acid residues at the C‐terminus. It seems that the C‐terminal tail of OsCDC48 is unique and irreplaceable for the survival of rice plants especially in the late stage of development although rice has two isoforms of CDC48 with a high degree of identity in the protein level. The role of OsCDC48E remains elusive, and the knockout or site‐specific editing of OsCDC48E using transcription activator‐like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 systems would likely help for the validation of its function (Mahfouz et al. 2014). Nevertheless, identification of cofactors interacting with the C‐terminus of OsCDC48 will be crucial to the elucidation of its function on premature senescence and plant survival. The isolation of OsCDC48 in the present study would facilitate further studies on the mechanism governing for the premature senescence and death phenotype in rice.
MATERIALS AND METHODS
Plant materials
The premature senescence and death 128 (psd128) mutant was isolated from an ethyl methane sulfonate (EMS)‐induced indica rice IR64 mutant bank (Wu et al. 2005). The mutant trait has been stably inherited for more than nine generations under different locations at Lingshui, Hainan and Hangzhou, Zhejiang, China. psd128 was crossed to the parental lines including PA64 and Moroberekan, respectively, to generate F2 populations for genetic and mapping analysis. The wild‐type IR64 and the mutant were grown in the paddy field at the China National Rice Research Institute (CNRRI) in 2012 for the evaluation of agronomic traits including plant height, number of tiller/plant, panicle length, number of filled grains/panicle, seed‐setting rate and 1000‐grain weight. The means from three measurements were used for analysis.
Histochemical analysis
Leaf samples from the wild‐type IR64, and psd128 with and without premature senescence phenotype were collected at the tillering stage. The detection of H2O2 accumulation was carried out using diaminobenzidine (DAB) staining as described by Thordal et al. (1997). For β‐glucuronidase (GUS) assay, a 2406 bp fragment upstream of the start codon covering the promoter of OsCDC48 was amplified from the IR64 genomic DNA using the following primers (OsCDC48‐QF: 5′‐TCCCCCGGGTGGGTTGTTGGGTGTCTCTTCT‐3′ and OsCDC48‐QR: 5′‐CGGAATTCCATTAGAAGCATTGGTTTTGTGAG‐3′). The promoter was then inserted into SmaI and EcoRI double‐digested pCAMBIA1381Z‐C vector to generate a new vector pCAMBIA1381Z‐C‐gus for the transient expression analysis of GUS. pCAMBIA1381Z‐C‐gus was transferred into A. tumefaciens strain EHA105 and introduced into the mature embryo‐induced calli from Nipponbare through A. tumefaciens‐mediated transformation (Hiei et al. 1994). Samples from the roots, leaves, stems and panicles of the transgenic and the wild‐type plants were stained following the method described by Jefferson et al. (1987).
Measurement of photosynthetic pigments and chlorophyll fluorescence
Chlorophyll (Chl) a and b contents were measured at the four‐ and six‐leaf stages in the seedlings, the 2nd upper and 4th upper leaves at the maximal tillering stage, the 2nd, 4th upper and flag leaves at the heading stage as described by Arnon (1949) while the carotenoid contents from the same leaves were determined following the method described by Wellburn (1994). The means from three measurements were used for analysis.
Photosynthetic parameters were measured at 9:00–10:00 am in psd128 and IR64 under field conditions using the portable Li‐6400 (Li‐COR, Lincoln, NB, USA). All the measurements were taken at the saturation irradiance with an incident photosynthetic photo flux density (PPFD) of 1,200 µmol · m−2 · s−1 and an airflow rate at 500 µmol · s−1. These parameters include: net photosynthetic rate (Pn: µmolCO2 · m−2 · s−1), stomatal conductance (Gs: molH2O · m−2 · s−1), intercellular CO2 concentration (Ci: µmolCO2 · mol−1) and transpiration rate (Tr: mmolH2O · m−2 · s−1). Chlorophyll fluorescent parameters were measured using the PAM‐2500 chlorophyll fluorescence system (Heinz Walz, Effeltrich, Germany) under field conditions. Briefly, leaves were dark‐adapted for 30 min, and then continuously illuminated with actinic light at an intensity of 1,100 mmol · m−2 · s−1 for 5 min. The minimum fluorescence (F0) and maximum fluorescence (Fm), effective photochemical quantum yield of PSII (Y(II)) and electron transport rate (ETR) of photosynthetic system II (PSII) were measured under the same conditions. The maximum photochemical quantum yield of PSII, Fv/Fm, was calculated as Fv/Fm = (Fm−F0)/Fm (Kitajima and Butler 1975), and the effective photochemical quantum yield of PSII, Y(II), was calculated as Y(II) = (Fm′−F′)/Fm′ (Genty et al. 1989). The means from three measurements were used for analysis.
Transmission electron microscopy
Leaf sections with premature senescence from psd128, and leaf sections with normal green leaves from psd128 and IR64 at the tillering stage were fixed in 2.5% (v/v) glutaraldehyde (0.2 M phosphate buffer; pH 7.2) for 16 h at 4 °C. After three brief rinses with the phosphate buffer, the samples were treated with 1% (w/v) osmium tetroxide at 4°C for overnight, and then dehydrated through ethanol series (50%, 70%, 85%, 95%, 100% (v/v)). Ethanol was subsequently replaced by a series of Spurr's resin dilutions (25%, 50%, 75%, 100% (v/v)). The samples were embedded in the Spurr's resin for 16 h at 65 °C to thin sectioning. Samples were stained with uranyl acetate and examined with a Tecnai G2 F20 transmission electron microscope at the College of Agriculture and Biotechnology, Zhejiang University.
Assay of enzymatic activity and senescence‐related parameters
All experiments were carried out using the 2nd and 4th upper leaves at the maximal tillering stage, and the 2nd, 4th upper and flag leaves at the heading stage. The activities of CAT and APX were measured following the methods of Zhao et al. (2002). The activity of POD was determined using the method of Chance and Maehly (1995). The activity of SOD was determined using the method of Beauchamp and Fridovich (1971). The content of MDA was measured according to the method of Heath and Packer (1968). The contents of soluble proteins and triphenyl tetrazolium chloride (TTC) were determined as described by Zhao et al. (2002). For the detection of TTC, rice plants were cultured in the nutrient liquid according to the protocol developed by the International Rice Research Institute (Yoshida et al. 1976). For detection of the other remaining parameters, leaf samples were taken from the plants growing in the paddy field at CNRRI. The means from three measurements were used for analysis.
Fine mapping of the mutation
F1 plants derived from the crosses of psd128/PA64 and psd128/Moroberekan, were grown in the paddy field at Lingshui Experimental Station of CNRRI in 2011 to determine if the mutation is dominant or recessive. F2 individuals from both crosses were grown in Hangzhou in the summer of 2011 and were phenotyped for segregation analysis. The individuals with the mutant phenotype from these two populations were collectively used for mapping of the mutation. F3 segregation lines from the two crosses were phenotyped for confirmation of the segregation the next season in Hangzhou in 2012. Bulk segregant analysis was first used to rapidly locate the mutation on a chromosome and a physical linkage map was then constructed using additional markers surrounding the mutation. Equal amount of leaf blades from each of 10 wild‐type and 10 mutant type plants were collected for DNA extraction to form a wild‐type DNA pool and a mutant‐type DNA pool, respectively. DNA of the parents and F2 individuals with the mutant phenotype were extracted following the mini‐preparation method (Lu and Zheng 1992). Simple sequence repeat (SSR) markers were obtained from the website (http://www.gramene.org/) while insertion/deletion (InDel) markers were designed using the Primer 5.0 and DNAStar 5.0 software after comparison of the sequences between the japonica cultivar Nipponbare and the indica cultivar 9311 in the public databases: RGP (http://rgp.dna.affrc.go.jp/E/toppage.html), Gramene (http://gramene.org/genome_ browser/index.html) and the Gene Research Center of the Chinese Academy of Sciences (http://rice.genomics.org.cn/rice/index2.jsp). The primers were synthesized by Sangon Biotech (Shanghai, China). PCR reaction and detection were carried out as described previously (Shi et al. 2009).
Functional complementation and RNAi
For function complementation of the mutation, the wild‐type allele of OsCDC48 was amplified using the forward primer OsCDC48‐F (5′‐TATGACCATGATTACGAATTCTGGTTACTTTTGTTATCTCCCGTT‐3′) and the reverse primer OsCDC48‐R (5′‐ACGACGGCCAGTGCCAAGCTTGTCCAGAATCATTTAGCAAAGCAC‐3′). A 7730 bp genomic DNA covering a 2142 bp promoter region upstream of the 5′UTR, a 4382 bp from the start to the stop codon and a 1183 bp downstream the 3′UTR was amplified and ligated into EcoRI and HindIII double‐digested pCAMBIA1300 to generate a complementary vector pCAMIA1300‐OsCDC48 (Figure S1A). The vector was mobilized to A. tumefaciens strain EHA105 by freeze–thaw method (Dityatkin et al. 1972), and introduced into the mature embryo‐induced calli from psd128 through A. tumefaciens‐mediated transformation (Hiei et al. 1994).
For RNAi of OsCDC48, a 380 bp fragment (position 102–481) from the wild‐type cDNA of OsCDC48 was amplified using the following primers (CDC48‐GSF: 5′‐GGGGTACCACTAGTCGAATCCGCCGACTCCCAT‐3′ and CDC48‐GSR: 5′‐CGGGATCCGAGCTCCACACGCTTGCCGTATTTGACAT‐3′). The fragment was inserted into the SpeI and SacI double‐digested pTCK303 vector to form an intermediate vector pTCK303‐GS. The 380 bp fragment was then inserted into KpnI and BamHI double‐digested pTCK303‐GS to generate the RNAi vector pTCK303‐RNAi (Figure S1B). pTCK303‐RNAi was then transferred to A. tumefaciens strain EHA105 and introduced into the mature embryo‐induced calli from Nipponbare through A. tumefaciens‐mediated transformation (Hiei et al. 1994).
Gene expression analysis
For the quantitative reverse‐transcription PCR (qRT‐PCR) analysis of OsCDC48, the total RNAs were extracted from the roots, leaves, stems and panicles of psd128 and the wild‐type IR64 at the heading stage using the Trizol reagent following the manufacture's instruction (Aidlab Biotechnologies (Beijing), China). Total RNAs from the RNAi transformants, Nipponbare and IR64 plants were extracted from the leaves at the seedling stage using the same method as above. The first‐strand cDNA was synthesized using ReverTra Ace qPCR RT Master Mix according to the manufacturer's protocol (TOYOBO Biotech (Shanghai), China). qRT‐PCR was performed on a Thermal Cycler Dice Real Time System II following the manufacturer's instruction (TaKaRa Biotechnology (Dalian), China). Primers for qRT‐PCR are listed in Table S6. PCR reaction and detection were carried out as described previously (Feng et al. 2013).
For qRT‐PCR analysis of the senescence‐related genes (Table S6), total RNAs were extracted from IR64, psd128 with and without premature senescence phenotype at the maximal tillering stage using the method described as above. PCR was carried out in a Thermal Cycler Dice Real Time System II following the manufacturer's instruction (TaKaRa Biotechnology (Dalian) China). The means from three replications were used for analysis.
Subcelluar localization
A full length coding sequence (CDS) from the wild‐type IR64 was synthesized using the following primers (OsCDC48‐YF: 5′‐GCTCTAGANNACACCCAACAACCCAAAACCT‐3′ and OsCDC48‐YR: 5′‐CGGGATCCNNAGCGGATGCGAAGGGGTCA‐3′). The 2427 bp CDS without the stop codon was double‐digested with XbaI and BamHI, and then ligated into XbaI and BamHI double‐digested vector pCAMBIA1300‐YFP containing the yellow fluorescent protein (YFP) reporter gene to form a new construct pCDC48‐YFP which was then introduced into the A. tumefaciens strain EHA105 by the freeze‐thaw method (Dityatkin et al. 1972), and transformed into the mesophyll cells of Nicotiana tabacum containing the red fluorescence signal in its nucleus (35S::RFP:H2B) through injection. Observation was made under a confocal fluorescence microscope Zeiss LSM 780 (Carl Zeiss AG, Germany).
Phylogenetic analysis
BLAST analysis was performed on the NCBI website (http://www.ncbi.nlm.nih.gov/) to search for homologs of OsCDC48. A total of 18 sequences from 17 species were identified (Figure 11). The sequences were aligned using ClustalW software and the neighbor‐joining tree was generated using the Poisson correction method in MEGA 6.0 software (Tamura et al. 2007). Bootstrap replication with 1,000 times was used for a statistical support for the nodes in the phylogenetic tree.
Accession Numbers
Sequence data from this article can be found in GenBank database (http://www.ncbi.nlm.nih.gov/) under the following accession numbers: Genomic DNA of OsCDC48 (KP718614); CDS of OsCDC48 (KP718613).
AUTHOR CONTRIBUTIONS
Q.H. and Y.S. performed most of the research and Q.H. drafted the manuscript. X.Z. and B. F. carried out marker analysis. H.W. and X.X. carried out trasformation experiments. L.S. X.L. and D.G. performed some field experiments. Q.H. and J.W. designed the experiment. J.W. supervised the study and revised the manuscript.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Figure S1. Vectors for functional complementation and RNAi of OsCDC48. (A) Functional complementation vector pCAMBIA1300‐OsCDC48. Kan, kanamycin resistance gene; Hyg, hygromycin resistance gene; 35S, CaMV35S promoter. (B) RNAi vector pTCK303‐RNAi. Black arrow, 380 bp cDNA fragment of the wild type OsCDC48; open arrow, CaMV35S promoter; open box, Intron
Table S1. Seed germination, root length and seedling height
Table S2. Contents of photosynthetic pigments at the seedling stage
Table S3. Contents of photosynthetic pigments at the heading stage
Table S4. Segregation of F3 lines
Table S5. Molecular markers used for mapping of the mutation
Table S6. List of genes used for real‐time PCR analysis
ACKNOWLEDGMENTS
This work was supported by the National High Technology Research and Development Program of China (2014AA10A603 and 2012AA101102).
Huang QN, Shi YF, Zhang XB, Song LX, Feng BH, Wang HM, Xu X, Li XH, Guo D, Wu JL ( 2015) Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J Integr Plant Biol 58: 12–28
Available online on Jun. 4, 2015 at www.wileyonlinelibrary.com/journal/jipb
REFERENCES
- Arnon DI ( 1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris . Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae H, Choi SM, Yang SW, Pai HS, Kim WT ( 2009) Suppression of the ER‐localized AAA ATPase NgCDC48 inhibits tobacco growth and development. Mol Cells 28: 57–65 [DOI] [PubMed] [Google Scholar]
- Baek GH, Kim I, Rao H ( 2011) The Cdc48 ATPase modulates the interaction between two proteolytic factors Ufd2 and Rad23. Proc Natl Acad Sci USA 16: 13558–13563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballar P, Pabuccuoglu A, Kose FA ( 2011) Different p97/VCP complexes function in retrotranslocation step of mammalian ER‐associated degradation (ERAD). Int J Biochem Cell Bio 43: 613–621 [DOI] [PubMed] [Google Scholar]
- Bar‐Nun S ( 2005) The role of p97/Cdc48p in endoplasmic reticulum‐associated degradation: From the immune system to yeast. Cur Top Microbiol Immunol 300: 95–125 [DOI] [PubMed] [Google Scholar]
- Beauchamp C, Fridovich I ( 1971) Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal Biochem 44: 276–287 [DOI] [PubMed] [Google Scholar]
- Cao K, Nakajima R, Meyer HH, Zheng Y ( 2003) The AAA‐ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115: 355–367 [DOI] [PubMed] [Google Scholar]
- Chance B, Maehly AC ( 1995) Assay of catalases and peroxidases. Methods Enzymol 2: 764–775 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A ( 2004) Cell division: AAA‐tacking the mitotic spindle. Curr Biol 14: R70–72 [DOI] [PubMed] [Google Scholar]
- Cheng ZL, Yu Y, Liu LN, Xia GX ( 2007) Isolation, characterization and functional analysis of a cdc48 homologue from tobacco BY‐2 cells. Pro Nat Sci 2: 156–162 [Google Scholar]
- Dityatkin SY, Lisovskaya KV, Panzhava NN, Iliashenko BN ( 1972) Frozen‐thawed bacteria as recipients of isolated coliphage DNA. Biochim Biophys Acta 281: 319–323 [DOI] [PubMed] [Google Scholar]
- Feiler HS, Desprez T, Santoni V, Kronenberger J, Caboche M, Traas J ( 1995) The higher plant Arabidopsis thaliana encodes a functional CDC48 homologue which is highly expressed in dividing and expanding cells. EMBO J 14: 5626–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng BH, Yang Y, Shi YF, Shen HC, Wang HM, Huang QN, Xu X, Lü XG, Wu JL ( 2013) Characterization and genetic analysis of a novel rice spotted‐leaf mutant HM47 with broad‐spectrum resistance to Xanthomonas oryzae pv. oryzae . J Integr Plant Biol 55: 473–483 [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR ( 1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A ( 1996) Cdc48p interacts with ufd3p, a WD repeat protein required for ubiquitin‐mediated proteolysis in Saccharomyces cerevisiae . EMBO J 15: 4884–4899 [PMC free article] [PubMed] [Google Scholar]
- Heath RL, Packer L ( 1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125: 189–198 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T ( 1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmüller H, Cassata G, Krause S, Hoppe T ( 2007) The ubiquitin‐selective chaperone CDC‐48/p97 links myosin assembly to human myopathy. Nat Cell Biol 9: 379–390 [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T ( 2002) Protein dislocation from the ER requires polyubiquitination and the AAA‐ATPase Cdc48. Nat Cell Biol 4: 134–139 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW ( 1987) GUS fusions: Beta‐glucuronidase as a sensitive and versatile gene fusionmarker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao BB, Wang JJ, Zhu XD, Zeng LJ, Li Q, He ZH ( 2012) A novel protein RLS1 with NB‐ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol Plant 5: 205–217 [DOI] [PubMed] [Google Scholar]
- Kitajima M, Butler WL ( 1975) Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim Biophys Acta 376: 105–115 [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Janiesch PC, Kevei É, Segref A, Barikbin R, Hoppe T ( 2011) The Machado‐Joseph disease deubiquitylase ATX‐3 couples longevity and proteostasis. Nat Cell Biol 13: 273–281 [DOI] [PubMed] [Google Scholar]
- Lamb JR, Fu V, Wirtz E, Bangs JD ( 2001) Functional analysis of the trypanosomal AAA protein TbVCP with trans‐dominant ATP hydrolysis mutants. J Biol Chem 276: 21512–21520 [DOI] [PubMed] [Google Scholar]
- Li FZ, Jin SH, Hu GC, Fu YP, Si HM, Jiang DA, Sun ZX ( 2005) Isolation and physiological characteristics of a premature senescence mutant in rice (Oryza sativa L.). J Zhejiang Univ Sci B 6: 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhao G, Zhou X, Schindelin H, Lennarz WJ ( 2006) The AAA ATPase p97 links peptide N‐glycanase to the endoplasmic reticulum‐associated E3 ligase autocrine motility factor receptor. Proc Natl Acad Sci USA 103: 8348–8353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Zheng KL ( 1992) A simple method for isolation of rice DNA. Chin J Rice Sci 6: 47–48 (in Chinese) [Google Scholar]
- Madeo F, Fröhlich E, Fröhlich KU ( 1997) A yeast mutant showing diagnostic makers of early and late apoptosis. J Cell Biol 139: 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Piatek A, Stewart CN Jr ( 2014) Genome engineering via TALENs and CRISPR/Cas9 systems: Challenges and perspectives. Plant Biotechnol J 12: 1006–1014 [DOI] [PubMed] [Google Scholar]
- Moir D, Stewart SE, Osmond BC, Botstein D ( 1982) Cold‐sensitive cell‐division‐cycle mutants of yeast: Isolation, properties, and pseudoreversion studies. Genetics 100: 547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouysset J, Deichsel A, Moser S, Hoege C, Hyman AA, Gartner A, Hoppe T ( 2008) Cell cycle progression requires the CDC‐48UFD‐1/NPL‐4 complex for efficient DNA replication. Proc Natl Acad Sci USA 105: 12879–12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouysset J, Kähler C, Hoppe T ( 2006) A conserved role of Caenorhabditis elegans CDC‐48 in ER‐associated protein degradation. J Struct Biol 156: 41–49 [DOI] [PubMed] [Google Scholar]
- Park S, Rancour DM, Bednarek SY ( 2008) In planta analysis of the cell cycle‐dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiol 148: 246–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Fröhlich KU, Diamant N, Bar‐Nun S ( 2002) AAA‐ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum‐associated protein degradation. Mol Cell Biol 22: 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour DM, Dickey CE, Park S, Bednarek SY ( 2002) Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol 130: 1241–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E ( 2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104: 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang XC, Xu FF, Zhu XY, Xing YD, He PL, Zhang CW, Yang ZL, He GH ( 2014) Identification and gene fine mapping of early senescence leaf mutant esl5 in Oryza sativa . Acta Agron Sin 40: 1182–1189 (in Chinese with an English abstract) [Google Scholar]
- Seo PJ ( 2014) Recent advances in plant membrane‐bound transcription factor research: Emphasis on intracellular movement. J Integr Plant Biol 56: 334–342 [DOI] [PubMed] [Google Scholar]
- Shi YF, Chen J, Liu WQ, Huang QN, Shen B, Leung H, Wu JL ( 2009) Genetic analysis and gene mapping of a new rolled‐leaf mutant in rice (Oryza sativa L.). Sci China C Life Sci 52: 885–890 [DOI] [PubMed] [Google Scholar]
- Shirogane T, FUkada T, Muller JM, Shima DT, Hibi M, Hirano T ( 1999) Synergistic roles for Pim‐1 and c‐Myc in STAT3‐mediated cell cycle progression and antiapoptosis. Immunity 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Stolz A, Hilt W, Buchberger A, Wolf DH ( 2011) Cdc48: A power machine in protein degradation. Trends Biochem Sci 36: 515–523 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S ( 2007) MEG A4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thomas H, Howarth CJ ( 2000) Five ways to stay green. J Exp Bot 51: 329–337 [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen H, Zhang ZG, Wei YD, Collinge DB ( 1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley‐powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Wellburn AR ( 1994) The spectral determination of chlorophyll a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Plant Physiol 144: 307–313 [Google Scholar]
- Wu JL, Wu C, Lei C, Baraoidan M, Bordeos A, Madamba MR, Ramos‐Pamplona M, Mauleon R, Portugal A, Ulat VJ, Bruskiewich R, Wang G, Leach J, Khush G, Leung H ( 2005) Chemical and irradiation induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol Biol 59: 85–97 [DOI] [PubMed] [Google Scholar]
- Wu XY, Kuai BK, Jia JZ, Jing HC ( 2012) Regulation of leaf senescence and crop genetic improvement. J Integr Plant Biol 54: 936–952 [DOI] [PubMed] [Google Scholar]
- Xiao R ( 2012) Genetic analysis and gene mapping of two rice premature senescence mutants. Sichuan Agricultural University (in Chinese with an English abstract)
- Yamanaka K, Sasagawa Y, Ogura T ( 2012) Recent advances in p97/VCP/Cdc48 cellular functions. Biochim Biophys Acta 1823: 130–137 [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA ( 2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
- Yoo SC, Cho SH, Zhang H, Paik HC, Lee CH, Li J, Yoo JH, Lee BW, Koh HJ, Seo HS, Paek NC ( 2007) Quantitative trait loci associated with functional stay‐green SNU‐SG1 in rice. Mol Cells 24: 83–94 [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock J, Gomez KA ( 1976) Laboratory Manual for Physiological Studies of Rice. The International Rice Research Institute (Third edition), Manila, Philippines
- Zhang B, Van Aken O, Thatcher L, De Clercq I, Duncan O, Law SR, Murcha MW, van der Merwe M, Seifi HS, Carrie C, Cazzonelli C, Radomiljac J, Höfte M, Singh KB, Van Breusegem F, Whelan J ( 2014) The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana . Plant J 80: 709–727 [DOI] [PubMed] [Google Scholar]
- Zhang T, Sen YY, Zheng JM, Cheng ZJ, Jiang KF, Yang L, Cao YJ, You SM, Wan JM, Zheng JK ( 2014) Genetic analysis and fine mapping of a premature leaf senescence mutant in rice (Oryza sativa L.). Acta Agron Sin 40: 2070–2080 (in Chinese with an English abstract) [Google Scholar]
- Zhang XZ ( 1992) Methods for Crop Physiology. Beijing, Agriculture Press; (in Chinese) [Google Scholar]
- Zhao G, Li G, Schindelin H, Lennarz WJ ( 2009) An armadillo motif in Ufd3 interacts with cdc48 and is involved in ubiquitin homeostasis and protein degradation. Proc Natl Acad Sci USA 106: 16197–16202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SJ, Shi GA, Dong XC ( 2002) Laboratory Guides for Plant Physiology. Chinese Agricultural Science and Technology Press (in Chinese)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Figure S1. Vectors for functional complementation and RNAi of OsCDC48. (A) Functional complementation vector pCAMBIA1300‐OsCDC48. Kan, kanamycin resistance gene; Hyg, hygromycin resistance gene; 35S, CaMV35S promoter. (B) RNAi vector pTCK303‐RNAi. Black arrow, 380 bp cDNA fragment of the wild type OsCDC48; open arrow, CaMV35S promoter; open box, Intron
Table S1. Seed germination, root length and seedling height
Table S2. Contents of photosynthetic pigments at the seedling stage
Table S3. Contents of photosynthetic pigments at the heading stage
Table S4. Segregation of F3 lines
Table S5. Molecular markers used for mapping of the mutation
Table S6. List of genes used for real‐time PCR analysis


