Abstract
Glutathione reductase (GR) catalyzes the reduction of glutathione disulfide (GSSG) to reduced glutathione (GSH) and participates in the ascorbate‐glutathione cycle, which scavenges H2O2. Here, we report that chloroplastic/mitochondrial GR2 is an important regulator of leaf senescence. Seed development of the homozygous gr2 knockout mutant was blocked at the globular stage. Therefore, to investigate the function of GR2 in leaf senescence, we generated transgenic Arabidopsis plants with decreased GR2 using RNAi. The GR2 RNAi plants displayed early onset of age‐dependent and dark‐ and H2O2‐induced leaf senescence, which was accompanied by the induction of the senescence‐related marker genes SAG12 and SAG13. Furthermore, transcriptome analysis revealed that genes related to leaf senescence, oxidative stress, and phytohormone pathways were upregulated directly before senescence in RNAi plants. In addition, H2O2 accumulated to higher levels in RNAi plants than in wild‐type plants and the levels of H2O2 peaked in RNAi plants directly before the early onset of leaf senescence. RNAi plants showed a greater decrease in GSH/GSSG levels than wild‐type plants during leaf development. Our results suggest that GR2 plays an important role in leaf senescence by modulating H2O2 and glutathione signaling in Arabidopsis.
Keywords: Arabidopsis thaliana, ascorbate‐glutathione cycle, glutathione reductase2, hydrogen peroxide, leaf senescence
Edited by: Chun‐Ming Liu, Institute of Botany, CAS, China
INTRODUCTION
Glutathione (γ‐l‐glutamyl‐l‐cysteinylglycine) is a multifunctional metabolite in plants. It is an important regulator of gene expression, cell signaling, the cell cycle, plant development, and cell death, and plays essential roles in processes such as glyoxylase and formaldehyde metabolism, sulfur assimilation, and the Calvin‐Benson and Krebs cycles (Noctor et al. 2012). Glutathione exists in two forms: a reduced form (GSH) and an oxidized form (GSSG). Under normal growth conditions, more than 90% of the glutathione in plant cells is in the reduced form (Noctor et al. 2002). As most physiological functions of glutathione in plants are attributed to its reduced form (Alscher 1989), normal growth and development depend on the presence of a high proportion of GSH. GSH synthesis in chloroplasts and cytosols and reduction of GSSG to GSH by glutathione reductase (GR, EC 1.6.4.2) maintain high proportions of GSH (Noctor and Foyer 1998; Asada 1999).
GSSG is reduced to GSH by GR in the ascorbate‐glutathione cycle, an important reactive oxygen species (ROS)‐scavenging pathway present in almost all cellular compartments in plants (Mittler 2002). In this cycle, ascorbate peroxidase reduces H2O2 to water and concurrently oxidizes ascorbate to monodehydroascorbate. The reduction of monodehydroascorbate occurs via the catalytic action of monodehydroascorbate reductase, which uses NAD(P)H as an electron donor. Monodehydroascorbate can also disproportionate to dehydroascorbate and ascorbate. Dehydroascorbate can then be reduced to ascorbate by dehydroascorbate reductase, an enzyme that uses GSH as an electron donor, resulting in the formation of GSSG. GSSG in turn is re‐reduced to GSH by GR, in a reaction that uses reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) as an electron donor (Asada 1999; Foyer and Noctor 2005). Thus, GR maintains the cellular glutathione redox status and thereby protects plants against oxidative stress (Mittler 2002; Noctor et al. 2012).
Subcellular fractionation studies detected GR activities in the chloroplast, cytosol, mitochondria, and peroxisome in many plant species (Edwards et al. 1990; Jiménez et al. 1998; Kataya and Reumann 2010). GR activities differ both in different cellular compartments and in different species. In Pisum sativum (pea) and Nicotiana tabacum (tobacco) plants, most GR activity is found in the chloroplast. For example, in pea plants, about 77% of the total GR activity is in the chloroplast and only about 20% and 3% is in the cytosol and mitochondria, respectively (Edwards et al. 1990). In tobacco plants, about 70% of total GR activity is in the chloroplast (Ding et al. 2009). Two genes, GR1 (At3g24170) and GR2 (At3g54660), encode GR in Arabidopsis. The subcellular distribution of GR activities in Arabidopsis differs from that in pea and tobacco plants, with GR1 activity localizing to the cytosol and peroxisomes (Marty et al. 2009; Kataya and Reumann 2010) and GR2 to chloroplasts and mitochondria (Creissen et al. 1995; Chew et al. 2003; Yu et al. 2013). GR1 accounts for 40% to 65% of the total GR activity in Arabidopsis leaves (Marty et al. 2009; Mhamdi et al. 2010; Yu et al. 2013).
The possible roles of GR in different cellular compartments have been investigated extensively. Elevated levels of cytosolic GR activity in transgenic tobacco plants have no effect on the size of the glutathione pool and the reduction status of glutathione under optimal or oxidative conditions (Foyer et al. 1991). In Arabidopsis, GR1 does not affect plant development under normal growth conditions (Marty et al. 2009). However, GR1 plays an essential role in H2O2 metabolism and signaling (Mhamdi et al. 2010). By contrast, chloroplastic GR improves resistance to abiotic stresses. Transgenic tobacco, poplar, and Gossypium hirsutum (cotton) plants with enhanced chloroplastic GR activities had increased resistance to oxidative stress caused by ozone, paraquat, high light, or chilling stresses (Aono et al. 1993; Broadbent et al. 1995; Foyer et al. 1995; Payton et al. 2001; Kornyeyev et al. 2003). By contrast, tobacco plants with decreased chloroplastic GR activity were more sensitive than the wild type to oxidative stress caused by paraquat or chilling stresses (Aono et al. 1995; Ding et al. 2012, 2009). Moreover, mutation of Arabidopsis GR2 is lethal in early embryo development (Tzafrir et al. 2004; Marty et al. 2009), suggesting that GR2 is crucial for several aspects of plant development, even under normal growth conditions. Indeed, recent work showed that GR2 is essential for root apical meristem maintenance in Arabidopsis, as it regulates the glutathione redox status under normal growth conditions (Yu et al. 2013).
To date, the functions of GR2 in leaf development remain unclear. Here, we report that Arabidopsis GR2 regulates leaf senescence and embryo development. GR2 RNAi plants showed early onset of age‐dependent and dark‐ and H2O2‐induced leaf senescence, accumulated high levels of H2O2, and exhibited enhanced sensitivity to H2O2. Transcriptome analysis revealed that the expression of genes related to oxidative stress, leaf senescence, and phytohormone pathways was upregulated in GR2 RNAi plants. These results demonstrate that GR2 plays an important role in leaf senescence by modulating H2O2 and glutathione signaling in Arabidopsis.
RESULTS
GR2 is essential for embryo development
Previous work suggested that GR2 deletion is embryonic lethal (Tzafrir et al. 2004). To investigate how GR2 deletion affects embryo development, we examined the gr2 mutant (Salk_040170), which contains a T‐DNA insertion in exon 9 of At3g54660 (Figure S1A). We found no homozygous gr2 mutant plants and heterozygous gr2 plants showed a wild‐type phenotype. Whereas the developing siliques of wild‐type plants contained green seeds, those of heterozygous gr2 plants contained both green and white seeds (Figure S1B). In mature siliques, the seeds of the gr2 mutant were small and shriveled and could not germinate (Figure S1C). About one‐quarter of the embryos were aborted, which is consistent with the trait being controlled by a single, recessive allele. To identify which stages of embryogenesis were arrested in the gr2 mutant, we examined the embryos in the cleared seeds of a developmental series of siliques. We found that development of the homozygous gr2 seeds was blocked at the globular stage, whereas embryos in wild‐type and heterozygous gr2 seeds underwent normal development (Figure S1D).
To confirm that GR2 was responsible for the embryo‐lethal phenotype of the gr2 mutant, we introduced full‐length GR2 under the control of the 35S promoter into heterozygous gr2 plants. The complemented plants showed normal embryonic development, as observed in wild‐type plants (Figure S1B, C). Thus, this indicates that the mutation in GR2 is responsible for the embryo‐lethal phenotype of gr2.
GR2 is highly expressed in mature and senescent leaves
GR2 is expressed throughout the seedling and is widely expressed in various organs and tissues in Arabidopsis, including roots, leaves, flowers, and siliques (Yu et al. 2013). We examined the expression of GR2 in developing young leaves, mature leaves, and senescent leaves, using the senescence‐associated gene (SAG) SAG13 as a senescence marker (Schippers et al. 2008). Reverse transcription‐polymerase chain reaction (RT‐PCR), GUS staining, and quantitative real‐time PCR analyses showed that GR2 was expressed throughout leaf development, but was highly expressed in mature and senescent leaves (Figure 1), suggesting that GR2 might be involved in leaf development.
Figure 1.
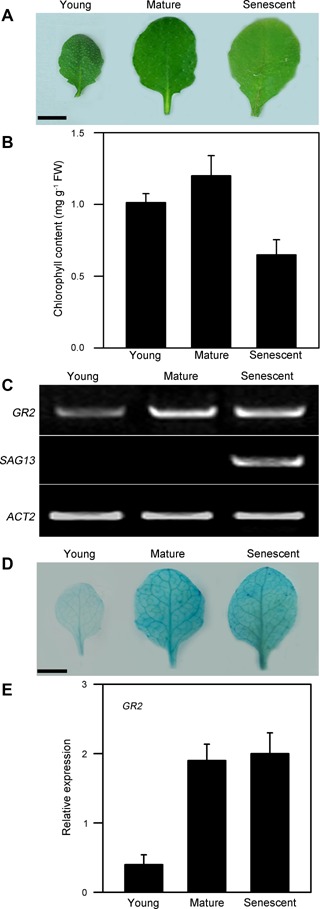
GR2 expression in young, mature, and senescent leaves in wild‐type plants (A) Morphologies of young, mature, and senescent leaves. Bars = 5 mm. (B) Chlorophyll contents in young, mature, and senescent leaves. (C) Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of GR2 and SAG13 expressions in young, mature, and senescent leaves. ACT2 was used as the loading control. (D) GR2pro:GUS expression in young, mature, and senescent leaves. Bars = 5 mm. (E) Transcript levels of GR2 were determined by real‐time PCR using ACT2 as an internal control. The fourth rosette leaves of wild‐type plants grown in soil were used. The young, mature, and senescent leaves were analyzed at 10, 35, and 55 d after emergence, respectively. The values are means ± SD of three independent experiments.
Generation of transgenic plants with decreased GR2 expression
To investigate the functions of GR2 in leaf development, we generated transgenic Arabidopsis plants with decreased levels of GR2 using RNAi technology. We selected three independent RNAi lines (igr2‐7, igr2‐9, and igr2‐14) with greatly reduced GR2 mRNA levels and used the T4 progeny of these lines in subsequent analyses. Using 3‐week‐old seedlings, we compared the GR2 mRNA levels, GR2 protein levels, and GR activity between wild‐type and RNAi plants (Figure 2). GR2 mRNA levels were significantly reduced in the three RNAi lines (Figure 2A, B). Quantitative real‐time PCR showed that the level of GR2 transcript was about 50% in igr2‐7 plants and about 10% in igr2‐9 and igr2‐14 compared to that in wild‐type plants (Figure 2A, B). Immunoblot analysis using antiserum specific for Arabidopsis GR2 identified a protein band of about 58 kDa. The GR2 protein level in igr2‐7, igr2‐9, and igr2‐14 plants decreased to about 50, 10, and 10% of that in wild‐type plants, respectively (Figure 2C). The total GR activity in the leaves of igr2‐7, igr2‐9, and igr2‐14 plants decreased to 70, 39, and 40% of that in wild‐type plants, respectively (Figure 2D). GR1 was reported to account for 40%–65% of the total GR activity in Arabidopsis leaves (Marty et al. 2009; Mhamdi et al. 2010). Thus, GR2 activity is greatly inhibited in igr2‐9 and igr2‐14 plants.
Figure 2.
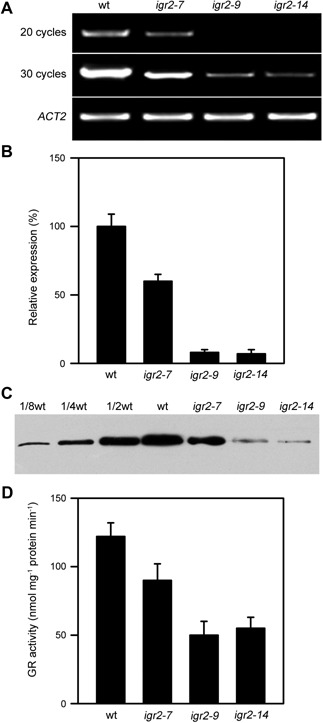
The levels of GR2 mRNA, GR2 protein, and total glutathione reductase (GR) activity in wild‐type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants (A) GR2 mRNA levels determined by RT‐PCR. (B) GR2 mRNA levels analyzed by real‐time PCR. (C) GR2 protein levels. (D) Total GR activity. Three‐week‐old seedlings were used to compare GR2 mRNA levels, GR2 protein levels, and total GR activity between wt and RNAi plants. Specific primers used are listed in Table S1. The values shown are means ± SD of three independent experiments.
Decreased GR2 promotes early leaf senescence
Whereas the igr2‐7 plants showed no aberrant phenotype, the igr2‐9 and igr2‐14 plants were smaller than the wild type (Figure 3A). The leaf area of igr2‐9 and igr2‐14 plants was about 40% less than that of wild‐type plants at 30 d after seed germination (Figure 3B). Early leaf senescence symptoms were observed at the flowering stage in igr2‐9 and igr2‐14 plants, but not in the wild‐type and igr2‐7 plants (Figure 3C). These results prompted us to investigate how GR2 influences leaf senescence. We harvested the fourth rosette leaves of wild‐type and RNAi plants at the indicated time points. We observed early onset of leaf senescence in igr2‐9 and igr2‐14 plants (Figure 4A). Leaf senescence was further characterized by examining well‐established physiological parameters for leaf senescence, including chlorophyll content, maximal efficiency of PSII photochemistry (Fv/Fm), malondialdehyde (MDA) content, and membrane ion leakage (Lim et al. 2007). Chlorophyll contents started to decrease at 45 d in wild‐type plants but at 35 d in igr2‐9 and igr2‐14 plants. Moreover, igr2‐9 and igr2‐14 plants showed a greater decrease in chlorophyll levels than did the wild‐type plants (Figure 4B). Similar results were observed for Fv/Fm in wild‐type and igr2‐9 and igr2‐14 plants (Figure 4C). The MDA content started to increase at 40 d in wild‐type plants, but already at 30 d in the igr2‐9 and igr2‐14 plants, and the increase was much greater in igr2‐9 and igr2‐14 plants than in wild‐type plants (Figure 4D). The patterns for membrane ion leakage in wild‐type and igr2‐9 and igr2‐14 plants were similar to those observed for MDA content (Figure 4E).
Figure 3.
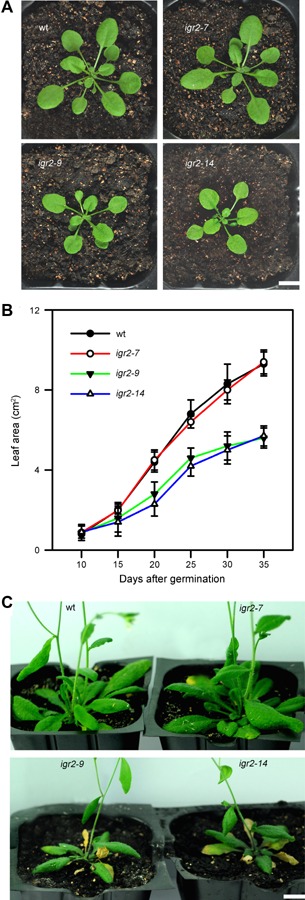
The phenotypes and growth kinetics of wild type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants (A) Three‐week‐old plants. Bars = 1 cm. (B) Growth kinetics of wt and RNAi plants. The total leaf area was determined at the indicated days after seed germination. The values shown are means ± SD of three independent experiments. (C) Eight‐week‐old plants. Bars = 1 cm.
Figure 4.
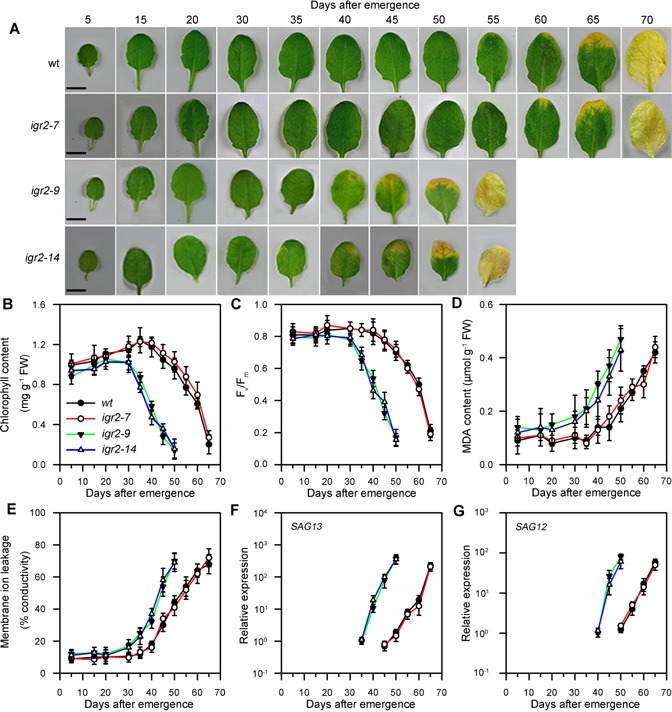
Age‐dependent senescence in wild‐type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants (A) Morphologies of leaves. Bars = 5 mm. (B, C, D, and E) Measurements of chlorophyll content, Fv/Fm, malondialdehyde (MDA) content, and membrane ion leakage. (F, G) Expression of the senescence‐related marker genes, SAG12 and SAG13. Transcript levels were determined by real‐time PCR using ACT2 as an internal control. For SAG13, the relative transcript levels detected at day 35 in igr2‐9 plants were assigned a value of 1 after normalization to ACT2 transcript levels. For SAG12, the relative transcript levels detected at day 40 in the igr2‐9 plant were assigned a value of 1 after normalization to ACT2 transcript levels. The fourth rosette leaves from wt and RNAi plants grown in soil were sampled at the indicated days after emergence. The values are means ± SD of three independent experiments.
To further characterize early leaf senescence in igr2‐9 and igr2‐14 plants, we examined the expression of two established senescence marker genes, SAG12 and SAG13 (Figure 4F, G). SAG13 expression, which is associated with oxidative stress and senescence (Weaver et al. 1998), was detected starting at day 35 in igr2‐9 and igr2‐14 plants, but only starting at day 45 in wild‐type plants. SAG13 expression in igr2‐9 and igr2‐14 plants was about 200‐fold higher at day 50 than in wild‐type plants (Figure 4F). Expression of SAG12, a hallmark of age‐induced senescence (Weaver et al. 1998), was detected starting at day 40 in igr2‐9 and igr2‐14 plants, but starting at day 50 in wild‐type plants. At day 55, SAG12 expression was about 80‐fold higher in igr2‐9 and igr2‐14 plants than in the wild type (Figure 4G).
Decreased GR2 leads to a change in glutathione redox status during leaf development
Glutathione reductase regulates the redox status of glutathione pools, indicated as the GSH/GSSG ratio, by catalyzing the reduction of GSSG. To investigate how decreased GR2 affects the GSH/GSSG ratio during leaf development, we measured the glutathione pools in wild‐type and RNAi plants (Figure 5). We found that total glutathione (GSH + GSSG), GSH, and GSSG contents increased gradually in young leaves during leaf development, peaked in mature leaves, and then decreased considerably in senescent leaves in wild‐type and RNAi plants (Figure 5A–C). However, total glutathione, GSH, and GSSG contents peaked much earlier in igr2‐9 and igr2‐14 plants than in wild‐type plants. Total glutathione and GSH contents peaked at 20 d after leaf emergence in igr2‐9 and igr2‐14 plants, but at 35 d in wild‐type plants. GSSG content peaked at 35 d in igr2‐9 and igr2‐14 plants, but at 45 d in wild‐type plants. In addition, the increase in total glutathione, GSH, and GSSG contents in young leaves was much greater in igr2‐9 and igr2‐14 plants than in wild‐type plants. By contrast, the decrease in GSH and GSSG contents in senescent leaves was much greater in igr2‐9 and igr2‐14 plants than in wild‐type plants. The GSH/GSSG ratio decreased with leaf development in wild‐type and RNAi plants and the decrease was much greater in igr2‐9 and igr2‐14 plants than in wild‐type plants. Moreover, the GSH/GSSG ratio was much lower in igr2‐9 and igr2‐14 plants than in wild‐type plants (Figure 5D). Similar changes in glutathione content during leaf senescence have been reported in Arabidopsis and Triticum aestivum (wheat) (Pavet et al. 2005; Li et al. 2014).
Figure 5.
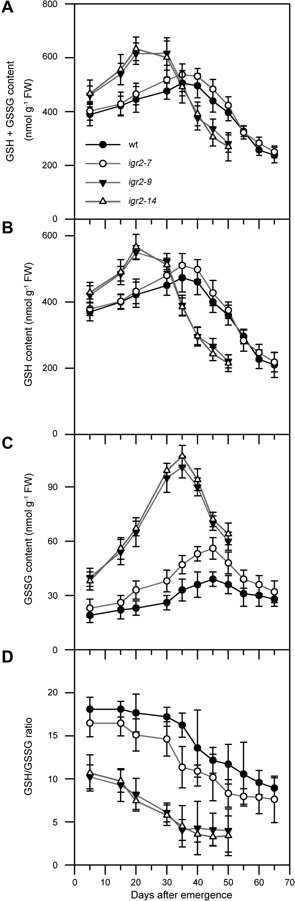
Changes in glutathione pool size in wild‐type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants during leaf development (A) GSH + GSSG content. (B) GSH content. (C) GSSG content. (D) GSH/GSSG ratio. The fourth rosette leaves from wt and RNAi plants grown in soil were harvested at the indicated days after emergence and used for glutathione extraction. Six independent biological replicates were performed. The values are means ± SD (n = 6).
Decreased GR2 results in elevated levels of H2O2
The reaction catalyzed by GR is part of the ascorbate‐glutathione cycle, which scavenges H2O2 (Asada 1999). Therefore, we monitored the levels of reactive oxygen species (ROS), including H2O2, superoxide (O2 •−), and singlet oxygen (1O2), during leaf development in wild‐type and RNAi plants (Figure S2). DAB (3,3′‐diaminobenzidine tetrachloride) staining showed that the level of H2O2 increased slightly with leaf development and peaked at around 40 d after leaf emergence in wild‐type plants, whereas the level of H2O2 peaked at 30 d and then decreased slightly in igr2‐9 and igr2‐14 plants. Furthermore, the level of H2O2 was greater in igr2‐9 and igr2‐14 plants than in wild‐type plants (Figure S2A). NBT (4‐nitroblue tetrazolium chloride) staining and Singlet Oxygen Sensor Green (SOSG) fluorescence analysis showed that the levels of O2 •− and 1O2 remained low in young leaves and increased after leaf maturation in wild‐type and GR2 RNAi plants. There was no difference in the levels of O2 •− and 1O2 in young or senescent leaves between wild‐type and igr2‐9 and igr2‐14 plants (Figure S2B, C).
We then quantified the level of H2O2 using an improved spectrophotometric assay (Figure 6). The H2O2 contents both in young and senescent leaves were higher in igr2‐9 and igr2‐14 plants than in wild‐type plants. The H2O2 content increased rapidly in the young leaves of igr2‐9 and igr2‐14 plants and peaked at 30 d after leaf emergence. However, in the young leaves of wild‐type plants, the H2O2 content increased slowly, peaked at 40 d, and then decreased slightly. Taken together, these results show that H2O2 accumulates to higher levels in the leaves of igr2‐9 and igr2‐14 plants than in wild‐type plants during development.
Figure 6.
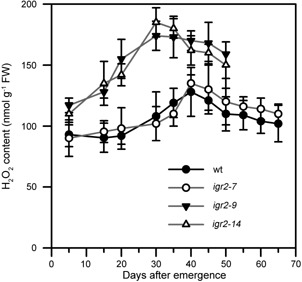
H2O2 levels in wild‐type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants during leaf development The fourth rosette leaves of wt and RNAi plants grown in soil were harvested at the indicated days after emergence and used for H2O2 measurements. The values are means ± SD of four to six independent experiments.
Decreased GR2 leads to accelerated darkness‐ and H2O2‐induced leaf senescence
H2O2 accumulation has been proposed to function as a signal in senescence in Arabidopsis (Foyer and Noctor 2005; Zentgraf and Hemleben 2008; Smykowski et al. 2010). Thus, the early leaf senescence observed in igr2‐9 and igr2‐14 plants may be associated with elevated levels of H2O2. To further examine this possibility, we investigated the response of wild‐type and RNAi plants to a 3‐d exposure to elevated H2O2 (10 mM) (Figure 7). To quantify the leaf senescence induced by elevated H2O2, we examined the four physiological parameters and two senescence marker genes described above in the fourth rosette leaves at 20 d after emergence. Indeed, we detected signs of senescence in the leaves of igr2‐9 and igr2‐14 plants (Figure 7A). Elevated H2O2 induced a decrease in chlorophyll and Fv/Fm and an increase in MDA content and membrane ion leakage in wild‐type and RNAi plants. However, compared to wild‐type plants, igr2‐9 and igr2‐14 plants showed a greater decrease in chlorophyll content and Fv/Fm and a greater increase in MDA content and membrane ion leakage (Figure 7B). In addition, the expression of SAG13 and SAG12 was higher in igr2‐9 and igr2‐14 plants than in wild‐type plants after exposure to elevated levels of H2O2 (Figure 7C). These results indicate that the leaves of igr2‐9 and igr2‐14 plants are more sensitive to H2O2‐inducible senescence than are those of wild‐type plants.
Figure 7.
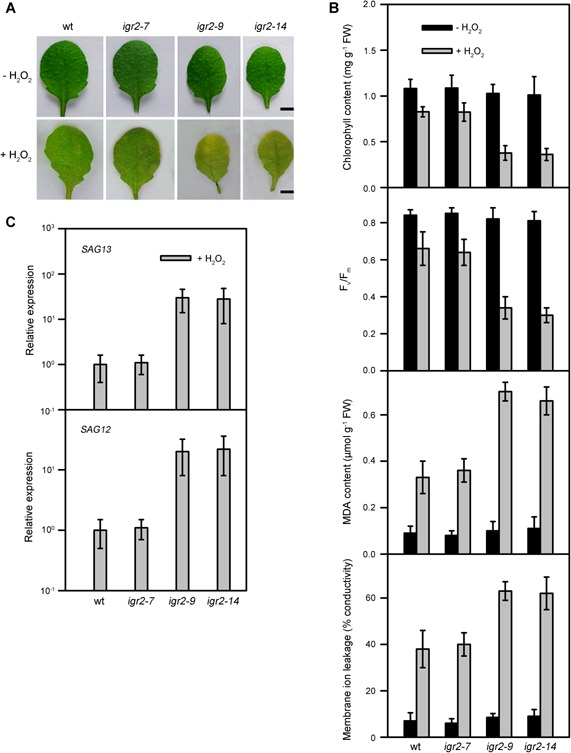
Effects of 10 mM H2O2 on detached leaves from wild‐type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants (A) Morphologies of leaves. Bars = 5 mm. (B) Chlorophyll content, Fv/Fm, MDA content, and membrane ion leakage. (C) Expression of the senescence‐related marker genes, SAG12 and SAG13. Transcript levels were determined by real‐time PCR using ACT2 as an internal control. For SAG13, the relative transcript levels detected in wt after H2O2 treatments were assigned a value of 1 after normalization to the ACT2 transcript levels. For SAG12, the relative transcript levels detected in wt after H2O2 treatments were assigned a value of 1 after normalization to the ACT2 transcript levels. SAG12 and SAG13 expression was not detected in wt and RNAi plants in the absence of the H2O2 treatment. The fourth rosette leaves at 20 d after emergence were used in the experiments. The values are means ± SD of three independent experiments.
Continuous darkness can induce leaf senescence in whole plants and detached leaves (Lin and Wu 2004; Chen et al. 2011). Therefore, we next examined whether GR2 is also involved in darkness‐induced leaf senescence (Figure 8). After dark treatment, the leaves of igr2‐9 and igr2‐14 plants were more yellow than those of wild‐type plants (Figure 8A). Darkness resulted in a decrease in chlorophyll contents and Fv/Fm, an increase in MDA contents, and membrane leakage in wild‐type and RNAi plants. However, there was a greater decrease in chlorophyll content and Fv/Fm and a greater increase in MDA content and membrane leakage in igr2‐9 and igr2‐14 plants than in wild‐type plants (Figure 8B). We also measured SAG12 and SAG13 mRNA levels by real‐time PCR and found that these genes were induced earlier and to a greater extent in leaves of igr2‐9 and igr2‐14 plants than those in wild‐type plants (Figure 8C). In addition, H2O2 accumulated to greater levels in igr2‐9 and igr2‐14 plants than in wild‐type plants during dark treatments (Figure 8D). These results suggest that decreased GR2 also promotes darkness‐induced leaf senescence.
Figure 8.
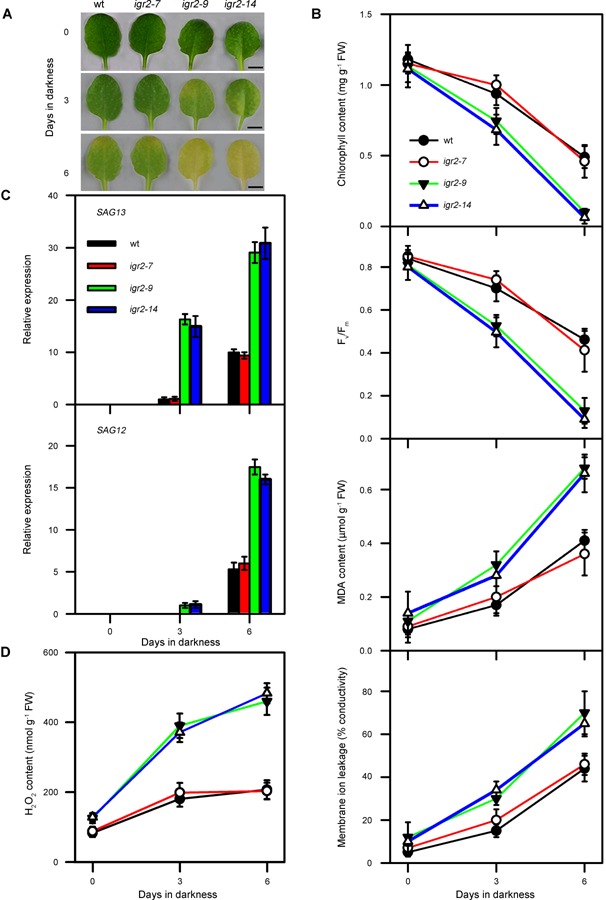
Effects of darkness on detached leaves from wild‐type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants (A) Morphologies of leaves. Bars = 5 mm. (B) Chlorophyll content, Fv/Fm, MDA content, and membrane ion leakage. The values are means ± SD of three independent experiments. (C) Expression of the senescence‐related marker genes, SAG12 and SAG13. Transcript levels were determined by real‐time PCR using ACT2 as an internal control. For SAG13, the relative transcript levels detected in wt after 3 d of dark treatment were assigned a value of 1 after normalization to ACT2 transcript levels. For SAG12, the relative transcript levels detected in the igr2‐9 after 3 d dark treatments were assigned a value of 1 after normalization to the ACT2 transcript levels. The values are means ± SD of three independent experiments. (D) H2O2 levels. The values are means ± SD of four to six independent experiments. The fourth rosette leaves at 20 d after emergence were used for dark treatments.
We further investigated whether GR2 expression is induced by elevated H2O2 exposure and continuous darkness. Detached leaves of wild‐type plants (3‐week‐old seedlings) were treated with 10 mM H2O2 and continuous darkness. GR2 expression was induced 6 h after exposure to H2O2 and levels of expression remained high for 3 d in the presence of H2O2 (Figure S3A). In addition, GR2 expression was induced 12 h after exposure to continuous darkness and levels remained elevated until 5 d after exposure to continuous darkness (Figure S3B). These data suggest that GR2 regulates the leaf senescence induced both by elevated H2O2 and continuous darkness.
Decreased GR2 induces the expression of genes related to oxidative stress, senescence, and phytohormones
To examine the function of GR2, we conducted microarray experiments that compared the transcriptome profiles of the fourth leaves (at 20 d after emergence) of wild‐type and igr2‐9 plants. At this age, there was no visible sign of leaf senescence. We selected genes that were differentially expressed (with a ≥2‐fold change cut‐off) in wild‐type and igr2‐9 plants for GO analysis. We conducted a Fisher's test to calculate the significance of the percentage distribution of GO annotations for comparison with those in the whole genome (Figure 9). We identified 494 genes that were upregulated and 629 genes that were downregulated genes in igr2‐9 compared with wild‐type plants (Table S2). Genes upregulated by decreased GR2 expression had GO annotations highly associated with “response to stress” and “response to abiotic or biotic stimulus”, with a P‐value of 2.21×10−15 and 1.63×10−9, respectively (Figure 9A), suggesting that stress‐responsive genes are overrepresented in igr2‐9 plants. Furthermore, genes upregulated by decreased GR2 expression were highly related to “response to oxidative stress”, “defense response”, and “leaf senescence”, with P‐values of 2.05×10−30, 1.64×10−7, and 2.24×10−4, respectively (Figure 9B). In addition, genes upregulated by decreased GR2 expression were highly related to “salicylic acid and jasmonic acid mediated signaling pathways” and to “responses to ethylene, jasmonic acid, salicylic acid, and abscisic acid stimulus” (Figure 9B). These results suggest that GR2 regulates the response to oxidative stress, leaf senescence, and phytohormone pathways. Further analysis indicated that several groups of genes involved in “response to high light”, “response to heat”, “response to wounding”, and “response to water deprivation” were significantly overrepresented in igr2‐9 plants as compared to the whole genome. However, GR2 may not be involved in all types of stress response, such as cold stress since genes involved in cold stress were not affected in igr2‐9 plants (Figure 9B).
Figure 9.
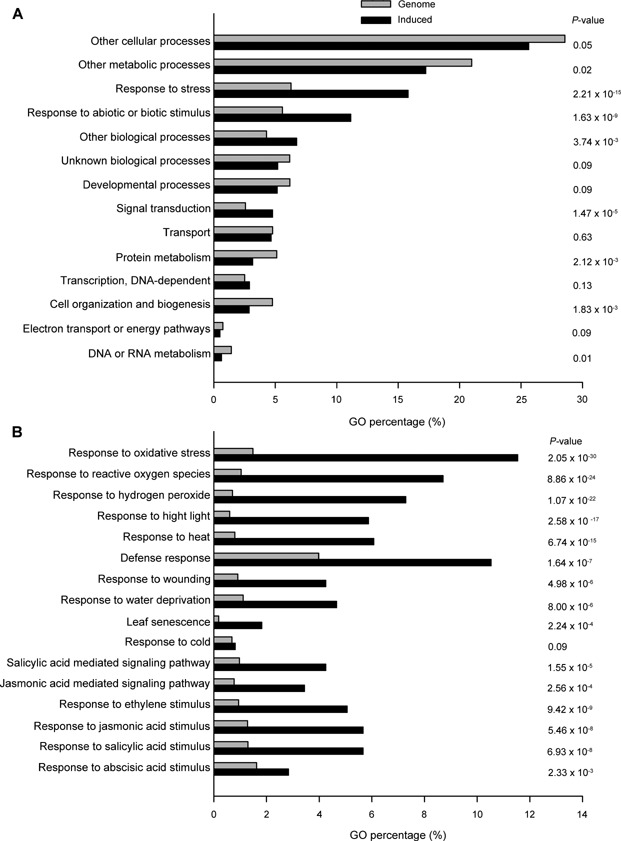
Global analysis of gene expression profiles in the igr2‐9 line (A) Expression profiles of genes up‐regulated in the igr2‐9 based on the gene ontology (GO) annotation (biological process) in TAIR. (B) Analysis of genes induced in igr2‐9 by specific GO terms. The fourth rosette leaves (at 20 d after emergence) were used for the microarray analysis. P‐values in A and B were calculated by Fisher's test to compare the percentage distribution of GO annotation from genes induced by decreased GR2 and the whole genome.
To confirm that genes identified as being differentially expressed in the microarray analysis were indeed up‐ or downregulated in igr2‐9 plants, we examined the expression of selected genes by real‐time PCR (Table 1). Senescence‐related genes, such as NAC053, WRKY53, SEN1, and NAC2, were about 6‐, 10‐, 7‐, and 7‐fold more strongly expressed, respectively, in igr2‐9 plants than in the wild type. The expression of oxidative stress‐related genes, such as GRXS13, CDSP32, and APX2, and several defense‐related genes, such as SOT12, PR1, and HYS1, were upregulated in igr2‐9 plants. In addition, the expression of phytohormone‐related genes, such as ERF1, PDF1.2A, ERF13, WRKY50, and EDL3, were upregulated in igr2‐9 plants. Therefore, these results suggest that decreased GR2 may induce the expression of oxidative stress‐, senescence‐, and phytohormone‐related genes before senescence.
Table 1.
Genes related to senescence, oxidative stress, defense, and phytohormones are upregulated in the fourth rosette leaves at 20 d after emergence in the igr2‐9 line
| AGI number | Description | Fold changea | RERb |
|---|---|---|---|
| Senescence | |||
| AT3G10500 | NAC053 | 2.52 | 5.92 ± 0.4 |
| AT4G23810 | WRKY transcription factor 53 (WRKY53) | 2.27 | 9.83 ± 1.7 |
| AT4G35770 | Senescence 1 (SEN1) | 2.12 | 7.35 ± 1.2 |
| AT5G39610 | NAC2 | 2.17 | 7.29 ± 1.9 |
| Oxidative stress | |||
| AT1G02930 | Glutathione transferase | 2.00 | 6.03 ± 1.7 |
| AT1G03850 | Glutaredoxin (GRXS13) | 2.72 | 5.46 ± 0.9 |
| AT1G76080 | Thioredoxin (CDSP32) | 3.64 | 9.64 ± 1.7 |
| AT3G09640 | L‐ascorbate peroxidase (APX2) | 2.98 | 5.18 ± 1.9 |
| AT4G16270 | Peroxidase superfamily protein | 2.84 | 8.15 ± 1.2 |
| AT5G39580 | Peroxidase superfamily protein | 2.97 | 16.42 ± 1.8 |
| Defense response | |||
| AT2G03760 | Brassinosteroid sulfotransferase (SOT12) | 2.63 | 7.22 ± 1.4 |
| AT2G14610 | Pathogenesis‐related gene1 (PR1) | 2.97 | 7.34 ± 0.9 |
| AT3G26830 | Phytoalexin deficient (PAD3) | 2.25 | 5.66 ± 1.4 |
| AT4G12470 | Azelaic acid induced 1 (AZI1) | 2.62 | 7.65 ± 1.1 |
| AT5G46050 | Peptide transporter PTR3‐A (PTR3) | 2.27 | 8.05 ± 1.5 |
| AT5G64930 | Regulator of pathogenesis‐related (HYS1) | 2.66 | 6.00 ± 1.7 |
| AT3G57240 | Beta‐1,3‐glucanase 3 (BG3) | 4.58 | 7.92 ± 1.2 |
| Phytohormone pathway | |||
| AT4G11890 | Protein kinase family protein (ARCK1) | 2.07 | 4.66 ± 0.4 |
| AT2G13810 | AGD2‐like defense response protein 1 (ALD1) | 2.66 | 5.50 ± 0.9 |
| AT3G23240 | Ethylene response factor1 (ERF1) | 3.77 | 10.42 ± 0.9 |
| AT5G44420 | Plant defensin (PDF1.2A) | 3.34 | 7.92 ± 1.2 |
| AT2G44840 | Ethylene‐responsive element binding factor 13 (ERF13) | 2.21 | 5.72 ± 1.3 |
| AT2G19190 | Senescence‐induced receptor‐like serine/threonine‐protein kinase (FRK1) | 2.72 | 6.50 ± 0.9 |
| AT2G35980 | Late embryogenesis abundant hydroxyproline‐rich glycoprotein (YLS9) | 2.32 | 5.40 ± 1.0 |
| AT5G26170 | Putative WRKY transcription factor 50 (WRKY50) | 3.62 | 7.80 ± 1.5 |
| AT4G27410 | NAC domain‐containing protein 72 (RD26) | 2.33 | 5.40 ± 1.2 |
| AT4G17490 | Ethylene response factor subfamily B‐3 of ERF/AP2 transcription factor family (ERF6) | 2.54 | 6.00 ± 1.1 |
| AT2G32020 | GCN5‐related N‐acetyltransferase‐like protein | 3.53 | 8.37 ± 1.5 |
| AT3G63060 | F‐box protein (EDL3) | 2.03 | 6.22 ± 0.8 |
Fold change is the mean of three independent experiments with the ratio of raw signal values in igr2‐9 and wild‐type plants.
Gene expression levels were quantified by real‐time PCR and the relative expression rate (RER) was compared between igr2‐9 and wild‐type plants. Data represent means ± SD (n = 3).
DISCUSSION
Glutathione reductase reduces GSSG to GSH in the ascorbate‐glutathione cycle, which scavenges H2O2 (Foyer and Noctor 2009). Arabidopsis GR1 is localized to the cytosol and peroxisomes. GR1 is not required for growth in optimal conditions but plays an important role when intracellular H2O2 production is increased and during pathogen challenge (Marty et al. 2009; Kataya and Reumann 2010; Mhamdi et al. 2010). Arabidopsis GR2 localizes to chloroplasts and mitochondria (Creissen et al. 1995; Chew et al. 2003). Many studies have shown that chloroplastic GR plays an important role in protecting plants against environmental and oxidative stresses (Aono et al. 1993, 1995; Foyer et al. 1995; Broadbent et al. 1995; Payton et al. 2001; Kornyeyev et al. 2003; Ding et al. 2012, 2009). Moreover, GR2 plays important roles in several aspects of plant development, since mutation of GR2 is embryo lethal (Tzafrir et al. 2004; Marty et al. 2009) and GR2 is essential for root apical meristem maintenance (Yu et al. 2013). Our results confirmed that GR2 is essential for embryo development (Figure S1). The gsh1 mutation, which disrupts glutathione synthesis, was also shown to be embryo lethal, but seed development of the homozygous gsh1 mutant was blocked at the torpedo stage (Cairns et al. 2006). This suggests that a deficiency in GR2 leads to embryo lethality through a different mechanism from deficiency in GSH synthesis. We showed that a reduction in GR2 results in early onset of age‐dependent and dark‐ and H2O2‐induced leaf senescence (Figures 3, 4, 7 and 8). Thus, GR2 is an important regulator of leaf senescence in Arabidopsis. However, the expression pattern of GR2 did not affect the onset of age‐dependent leaf senescence in the wild type, since GR2 is already highly expressed in mature leaves (Figure 1). The finding that growth of GR2 RNAi seedlings was retarded (Figure 3B) suggests that GR2 may promote growth.
How does GR2 regulate leaf senescence? Our results showed that large reductions in GR2 increased H2O2 accumulation in developing young leaves, mature leaves, and senescent leaves (Figure 6), indicating that GR2 is important for maintaining H2O2 homeostasis during leaf development in Arabidopsis. However, H2O2 accumulation varies throughout leaf development in GR2 RNAi plants. The level of H2O2 increased significantly in the developing young leaves of GR2 RNAi plants, peaking at 30 d after leaf emergence and then declining gradually (Figure 6). Leaf senescence commenced directly after H2O2 levels peaked in GR2 RNAi plants, as reflected by a decrease in chlorophyll content and Fv/Fm, an increase in MDA content and membrane ion leakage, and an induction of the senescence marker genes, SAG12 and SAG13 (Figure 4, 6). In addition, we observed that the leaves of GR2 RNAi plants turned yellow more rapidly than did those of wild‐type plants following exposure to elevated levels of H2O2. The changes in the physiological parameters for leaf senescence further confirmed that GR2 RNAi plants underwent early onset of leaf senescence in response to elevated H2O2 levels (Figure 7). Furthermore, the endogenous levels of H2O2 were higher in detached leaves from GR2 RNAi plants than in those from the wild‐type plants when senescence‐inducing dark treatment was applied (Figure 8D). As expected, compared with wild‐type leaves, the leaves of GR2 RNAi plants showed an early senescence phenotype in response to darkness treatment, which was accompanied by a greater decrease in chlorophyll content and Fv/Fm and a greater increase in MDA content and membrane ion leakage as well as a greater increase in the expression of SAG12 and SAG13 (Figure 8A, B). Moreover, GR2 was highly expressed in wild‐type senescent leaves (Figure 1). Taken together, these results suggest that early onset leaf senescence in GR2 RNAi plants is associated with H2O2 accumulation.
Was early onset of leaf senescence in GR2 RNAi plants due to physiochemical damage to the cell caused by elevated H2O2 or, alternatively, could elevated H2O2 levels in GR2 RNAi plants act as a signal that activates the gene expression pathways that regulate senescence? ROS are proposed to play an essential role in leaf senescence. Indeed, previous studies showed that ROS accumulate during leaf senescence (Navabpour et al. [Link]; Khanna‐Chopra 2012) and high levels of ROS are present in the chloroplasts of ageing plants (Munné and Alegre 2002). In addition, several SAGs are known to be induced in response to oxidative stress (Miller et al. 1999; Navabpour et al. [Link]). The connection between oxidative stress and leaf senescence is further supported by the finding that mutants with delayed leaf senescence, such as ore1, ore3/ein2, ore9, and gigantea, are resistant to oxidative stress (Kurepa et al. 1998; Woo et al. 2004), while mutants with accelerated leaf senescence, such as old5, aaf‐OX, and Cdf1, are associated with disturbed cellular redox balance or hypersensitivity to oxidative stress (Schippers et al. 2008; Chen et al. 2011; Cui et al. 2013). Recent genetic evidence suggests that altered ROS levels likely act as signals that are implemented in the developmental program of the leaf, causing early onset of senescence (Foyer and Noctor 2005; Queval et al. 2007; Zentgraf and Hemleben 2008; Smykowski et al. 2010). In this study, we observed an early leaf senescence phenotype in GR2 RNAi plants after the leaves were exposed to high levels of H2O2 (Figure 7). We also observed an early leaf senescence phenotype in GR2 RNAi plants after a senescence‐inducing dark treatment, which caused H2O2 to accumulate (Figure 8). In addition, we showed that considerable levels of H2O2 had already accumulated in the young leaves of GR2 RNAi plants and that H2O2 peaked in leaves 30 d after emergence (Figure 6). At this stage, however, an apparent senescence phenotype was not visible and there were no significant differences in the physiological parameters for leaf senescence between GR2 RNAi and wild‐type plants (Figure 4). More importantly, the results from the microarray analysis show that genes involved in “leaf senescence” and “oxidative stress” were overrepresented in the leaves of GR2 RNAi plants at 20 d after leaf emergence (Figure 9), suggesting that the senescence‐ and stress‐related genes were already induced at the developmental stage before senescence.
To investigate how GR2 regulates leaf senescence, we compared the transcriptome of the igr2‐9 line with that of wild‐type Arabidopsis at different developmental stages before the onset of leaf senescence (Breeze et al. 2011) (Figure S4 and Table S3). We analyzed the upregulated genes of transcriptome datasets from the report by Breeze et al. (2011) using GO::TermFinder as described in this study. We showed that the expression of genes related to response to oxidative stress”, “response to ROS”, “defense response”, and “leaf senescence” is significantly overrepresented during the early stages of leaf senescence in wild‐type Arabidopsis, similar to the finding for igr2‐9. Breeze et al. (2011) showed that ROS response genes are induced early in leaf senescence. Most of these ROS response genes differ from those induced in igr2‐9; 51 ROS response genes were upregulated 29 d after sowing (Breeze et al. 2011) and 41 ROS response genes were upregulated in igr2‐9, but only 13 overlapped. However, these overlapping genes include some important senescence‐related genes, such as NAC2/ORE1/ANAC092 and SAG21 (Table S3). We investigated if elevated H2O2 induced the expression of H2O2 marker genes, such as glycosyltransferase family 61 protein, heat shock protein 17.6A, GSTU9, PAD3, WRKY53, and GSTU24, in igr2‐9 plants (Vanderauwera et al. 2005; Gadjev et al. 2006; Queval et al. 2007; Sewelama et al. 2014). Indeed, we observed that these H2O2 marker genes were upregulated in the leaves of igr2‐9 plants at 20 d after emergence (Table 2). We also observed the induction of WRKY53, NAC2, and SEN1 in igr2‐9 plants before senescence (Table 1). WRKY53, SEN1, and NAC2 are marker genes for leaf senescence. It has been shown that WRKY53 and its regulators are induced by ROS (Zentgraf et al. 2010). WRKY53 positively regulates developmental senescence and wrky53 knock‐out mutants exhibit a delayed senescence phenotype. WRKY53 regulates senescence‐associated gene expression and acts at an upstream position in the WRKY signaling cascade (Miao et al. 2004). SEN1 expression is associated with Arabidopsis leaf senescence and SEN1 is induced by oxidative stresses (Oh et al. 1996; Woo et al. 2004). The NAC2/ORE1/ANAC092 loss‐of‐function mutant shows a delayed leaf‐senescence syndrome, whereas overexpression of AtNAC2 induces early senescence (Oh et al. 1997; Kim et al. 2009; Balazadeh et al. [Link]). Taken together, these results suggest that the elevated accumulation of H2O2 in the leaves of GR2 RNAi plants acts a signal mediator for early leaf senescence.
Table 2.
Induction of H2O2 marker transcripts in the fourth rosette leaves at 20 d after emergence in the igr2‐9 line
| AGI number | Description | Fold changea | RERb |
|---|---|---|---|
| AT3G10320 | Glycosyltransferase family 61 protein | 2.20 | 5.81 ± 1.7 |
| AT5G12030 | Heat shock protein 17.6A | 1.84 | 3.22 ± 0.8 |
| AT5G62480 | Glutathione S‐transferase tau 9 (GSTU9) | 4.25 | 8.43 ± 2.0 |
| AT3G26830 | Cytochrome P450 71B15 (PAD3) | 2.25 | 6.70 ± 1.3 |
| AT4G23810 | WRKY transcription factor 53 (WRKY53) | 2.27 | 5.97 ± 1.6 |
| AT1G17170 | Glutathione S‐transferase TAU 24 (GSTU24) | 1.66 | 4.01 ± 1.8 |
Fold change is the mean of three independent experiments with the ratio of raw signal values in igr2‐9 and wild‐type plants.
Gene expression levels were quantified by real‐time PCR and the relative expression rate (RER) was compared between igr2‐9 and wild‐type plants. Data represent means ± SD (n = 3).
H2O2 can be produced in different cellular compartments, such as chloroplasts, mitochondria, peroxisomes, and the cytoplasm. H2O2 pools in different cell compartments may have different effects on leaf senescence. Chloroplasts are thought to be the main target of age‐associated oxidative stress in plants (Munńe‐Bosch and Alegre 2002) and play a principal role in the regulation of leaf senescence (Zapata et al. 2005; Martínez et al. 2008). Peroxisomes and the ROS generated in these organelles play a central role in natural and dark‐induced senescence in pea plants (del Río et al. 1998). Downregulation of CAT2 in peroxisomes, which results in accumulation of H2O2 activates leaf senescence (Zimmermann et al. 2006; Smykowski et al. 2010). H2O2 produced in peroxisomes upregulates genes involved in protein repair responses, whereas H2O2 produced in chloroplasts induces early signaling responses, by upregulating the expression of transcription factors and biosynthetic genes involved in the production of secondary signaling messengers (Sewelam et al. 2014). Cytoplasmic H2O2 also induces leaf senescence, and it appears to have a greater role in senescence signaling than does peroxisomal H2O2 (del Río et al. 1998; Bieker et al. 2012). GR2 localizes to both chloroplasts and mitochondria (Creissen et al. 1995; Chew et al. 2003). Since most of the GR activity is in the chloroplast (Edwards et al. 1990; Ding et al. 2009), it is proposed that accumulated H2O2 in GR2 RNAi plants resulted mainly from chloroplasts. Arabidopsis plants overexpressing glycolate oxidase in the chloroplast (GO5) represent an excellent system in which to study the action of H2O2 as a signal molecule in chloroplasts (Fahnenstich et al. 2008). Recently, Sewelam et al. (2014) investigated H2O2 signaling from the chloroplast using GO5 plants. They found that genes related to “oxidative stress”, defense response”, “leaf senescence”, and “phytohormone pathways” were overrepresented in GO5 plants after 8 h of H2O2 production (Figure S5). They also found that H2O2 from chloroplasts induces early signaling responses, upregulating the expression of many transcription factors (Sewelam et al. 2014). Indeed, we observed that the expression of many transcription factors was induced in the igr2‐9 line (Table S2). NAC and WRKY transcription factors were shown to be the two largest groups of transcription factors differentially expressed in the senescence transcriptome (Guo et al. 2004). Some of these transcription factors have been shown to be central regulators of senescence in wheat and Arabidopsis (Miao et al. 2004; Guo and Gan 2006; Uauy et al. 2006; Balazadeh et al. [Link], 2011; Breeze et al. 2011; Yang et al. 2011). Many NAC and WRKY transcription factors were induced in igr2‐9 and GO5 plants (Table S4). Thus, H2O2 pools in the chloroplasts of GR2 RNAi plants may regulate leaf senescence. GR2 also localizes to mitochondria. The decreased GR2 in GR2 RNAi plants may cause H2O2 to accumulate in mitochondria. Our results show that leaf senescence was accelerated in RNAi plants under dark conditions (Figure 8). Therefore, H2O2 in mitochondria and possibly also in peroxisomes may promote dark‐induced leaf senescence in GR2 RNAi plants (del Río et al. 1998).
Recent studies demonstrated that glutathione status is central to the regulation of cell signaling under optimal and stress conditions (Noctor et al. 2012, 2013; Mhamdi et al. 2013; Schnaubelt et al. 2015). Is the early leaf senescence in GR2 RNAi plants associated with changes in glutathione status? Studies of the cat2 knockout mutant under photorespiratory conditions showed that glutathione status plays an important role in the H2O2‐dependent induction of salicylic acid and jasmonic acid signaling pathways (Mhamdi et al. 2013, 2010; Queval et al. 2012; Han et al. [Link], [Link]). To explore the role of changes in glutathione status in inducing leaf senescence in GR2 RNAi plants, we compared the transcriptome datasets of gr1, cad2, rml1, and cat2 plants with those of igr2‐9 plants. We found that the expression of eight genes involved in jasmonic acid signaling was repressed in the gr1 mutant, whereas that of seven genes involved in jasmonic acid signaling was upregulated under long‐day conditions (Mhamdi et al. 2010; Table S5). The expression of 34 and 58 genes involved in jasmonic acid signaling was repressed and up‐regulated, respectively, in the cad2 mutant, which has impaired glutathione synthesis, under long‐day conditions (Han et al. [Link]; Table S6). Due to the limited number of genes upregulated in the gr1 and cad2 mutants (Tables S5, S6), we were unable to identify genes associated with specific cellular processes by GO::TermFinder analysis. These results demonstrate that glutathione interacts with the jasmonic acid pathway. Studies of the cat2 gr1 and cat2 cad2 double mutants showed that glutathione status affects the ability of H2O2 to activate the pathogenesis‐related jasmonic acid and salicylic acid signaling pathways (Han et al. [Link], [Link]; Mhamdi et al. 2013, 2010). Transcriptome analyses of the cat2 mutant, which has a modified glutathione status due to increased levels of H2O2, and the rml1 mutant, which has severely impaired glutathione synthesis, showed that genes involved in the jasmonic acid, salicylic acid, ethylene, and abscisic acid pathways as well as pathogenesis‐related genes were highly upregulated in both cat2 and rml1 (Mhamdi et al. 2010; Schnaubelt et al. 2015) (Figures S6, S7, Tables S7–9). Our results also showed that genes involved in jasmonic acid, salicylic acid, ethylene, and abscisic acid pathways as well as pathogenesis‐related genes were overrepresented in igr2‐9 (Figure 9, Tables S7‐9). Thus, our results suggest that the glutathione status of GR2 RNAi plants may affect leaf senescence by modulating phytohormone pathways and the expression of pathogenesis‐related genes (Figure 5).
Glutathione is present in almost all cellular compartments, including chloroplasts, mitochondria, peroxisomes, and the cytosol (Mitter 2002). Oxidative stress may drive characteristic changes in the amounts of glutathione in different sub‐cellular compartments. Increased intracellular H2O2 availability preferentially drives glutathione accumulation in vacuoles and chloroplasts (Queval et al. 2011). We observed a significant increase in H2O2 levels and glutathione accumulation in the leaves of igr2‐9 and igr2‐14 plants than in those of the wild type before a leaf age of 30 d after emergence (Figures 5, 6). Whether such an increase in H2O2 levels drives the changes in glutathione accumulation in different cellular compartments in igr2‐9 and igr2‐14 plants is not clear. It has been reported that about 70% of total GR activity is in the chloroplast (Edwards et al. 1990) and that decreased chloroplastic GR activity results in significant accumulation of H2O2 in chloroplasts (Ding et al. 2009). In addition, increased H2O2 drives a substantial increase in GSSG accumulation in the chloroplast (Queval et al. 2011). Therefore, we expect that there should be a significant increase in GSSG accumulation in the chloroplast of GR2 RNAi plants when H2O2 is increased. GSSG accumulation in chloroplasts could reflect oxidation of GSH already present in the chloroplast in GR2 RNAi plants with increased levels of endogenous H2O2 and have important consequences for redox regulation in this compartment. This redox regulation as a signaling mechanism in the chloroplast plays an important role in regulating leaf senescence as discussed above. GR2 also localizes to mitochondria and thus H2O2 should also accumulate in mitochondria in GR2 RNAi plants. Therefore, it is most likely that there is an accumulation of glutathione in mitochondria in GR2 RNAi plants. Whether and how glutathione accumulation is affected in other subcellular compartments, such as vacuoles, peroxisomes, nuclei, and the cytosol, in GR2 RNAi plants remains to be determined.
In this study, we showed that decreased GR2 resulted in early onset of age‐dependent and dark‐ and H2O2‐induced senescence in Arabidopsis. Previous studies showed that lines overexpressing chloroplastic GR had a normal phenotype under normal growth conditions in poplar trees, cotton, and tobacco plants, but displayed enhanced resistance to photoinhibition (including chilling‐induced photoinhibition) and oxidative stress (Aono et al. 1993; Broadbent et al. 1995; Foyer et al. 1995; Payton et al. 2001; Kornyeyev et al. 2003). These studies suggest that overexpression of chloroplastic GR may protect plants against various environmental stresses. Thus, we propose that Arabidopsis lines overexpressing GR2 display a delayed senescence phenotype under environmental stress conditions, but a normal phenotype under normal growth conditions.
In conclusion, we showed that decreased GR2 led to early onset of age‐dependent and dark‐ and H2O2‐induced leaf senescence. Our results suggest that the elevated levels of H2O2 and modified glutathione status in GR2 RNAi plants act as signals that mediate early leaf senescence. Thus, GR2 is an important regulator of leaf senescence in Arabidopsis.
MATERIALS AND METHODS
Plant materials and growth conditions
All of RNAi lines and mutants used in this study were derived from the wild‐type Arabidopsis thaliana Columbia (Col‐0) ecotype and cultivated in growth chambers with a photosynthetic photon flux density of 100 µmol m−2 s−1, a relative humidity of 75%–80%, a temperature of 22 ± 1°C and a photoperiod of 16/8 h light/dark. The T‐DNA insertion gr2 mutant (SALK_040170) was obtained from the Arabidopsis Biological Resource Center (ABRC) and PCR was used to confirm homozygosity of gr2 with the primers GR2LP and GR2RP (see Table S1). For rapid PCR screening of mutants and RNAi plants, genomic DNA was extracted for analysis as described in our previous study (Zhong et al. 2013). Leaf areas were measured using an LI‐3000 A Portable Area Meter (LI‐Cor).
Analyses of H2O2‐ and dark‐induced senescence
For H2O2‐induced senescence, the fourth rosette leaves at 20 d after emergence were incubated in 3 mM MES buffer (pH 5.8) in the presence of 10 mM H2O2 for 3 d. For dark‐induced senescence, the fourth rosette leaves at 20 d after emergence were incubated in 3 mM MES buffer (pH 5.8) and then transferred to complete darkness for periods of up to 6 d.
GR activity assay and quantification of GSH content
Glutathione reductase activity was determined by monitoring the rate of GSH formation at 412 nm, as described in our previous study (Ding et al. 2009). The in vivo glutathione level was quantified by reverse‐phase HPLC after derivatization with monobromobimane, according to established procedures (Strohm et al. 1995).
Physiological assays for leaf senescence
The fourth rosette leaves of individual plants were used to determine physiological parameters for leaf senescence, including chlorophyll content, Fv/Fm, MDA content, and membrane ion leakage. Chlorophyll content and Fv/Fm were measured according to our previous study (Ding et al. 2012). MDA content was measured according to Hodges et al. (1999). Membrane ion leakage was analyzed by monitoring electrolytes released from leaves using a bench‐top conductivity meter (CON500, CLEAN Instruments), according to Chen et al. (2011).
Detection and measurement of ROS in leaves
Total leaf H2O2 content was determined by spectrometric assay according to the improved method of Veljovic‐Jovanovic et al. (2002). Leaves were ground to a fine powder in liquid nitrogen and the powder was extracted in 1 M HClO4 and 5% insoluble PVP. The homogenate was centrifuged at 12,000 g for 10 min at room temperature and the supernatant was neutralized with 5 M K2CO3 to pH 5.6 in the presence of 50 µL 0.3 M phosphate buffer (pH 5.6). The homogenate was centrifuged at 12,000 g for 1 min at room temperature to remove KClO4. The sample was incubated prior to assay for 10 min with 1 U ascorbate oxidase (Sigma, Watford, UK) to oxidize ascorbate and thereby eliminate the interference of ascorbate. The reaction mixture consisted of 0.1 M phosphate buffer (pH 6.5); 3.3 mM 3‐(dimethylamino) benzoic acid; 0.07 mM 3‐methyl‐2‐benzothiazolinone hydrazone; and 50 ng horseradish peroxidase (Sigma, Watford, UK). The reaction was initiated by the addition of an aliquot of the sample. The absorbance change at 590 nm was monitored at 25°C. To eliminate lipid peroxide interference, a parallel aliquot for each sample was incubated for 10 min with 1 U catalase (Sigma, Watford, UK) to catalyze H2O2. The lipid peroxide content was then determined using the same procedure as described above and subtracted from each sample. For each assay, the H2O2 content in the extract was quantified relative to an internal standard.
For in situ detection of H2O2, detached leaves were vacuum‐infiltrated with 1 mg mL−1 3,3′‐diaminobenzidine (DAB) solution (pH 3.8) for 5 min and incubated in the dark at room temperature for 6 h (Thordal‐Christensen et al. 1997). For in situ detection of O2 •−, detached leaves were vacuum‐infiltrated with 1 mg mL−1 nitroblue tetrazolium (NBT) solution in 10 mM potassium phosphate buffer (pH 7.8) for 5 min and incubated for 20 min in darkness at room temperature (Kawai‐Yamada et al. 2004). Stained leaves were cleared by boiling in acetic acid/glycerol/ethanol (1:1:3, v/v/v) solution before photographs were taken. Imaging of singlet oxygen production was performed as described by Flors et al. (2006). Briefly, detached leaves were immersed in 10 mM SOSG (Invitrogen) and 50 mM phosphate buffer (pH 7.5) for 2 h in darkness and then transferred to light conditions for 4 h. Following excitation of SOSG by UV light, fluorescence images were acquired with a charge‐coupled device camera (Olympus) with a Green Fluorescent Protein A (GFPA) interference filter in the objective. Fluorescence intensity was determined using ImageJ software (http://rsb.info.nih.gov/ij/) and the background was subtracted from the readings.
Microscopy of developing seeds
Developing seeds were analyzed by microscopy (Leica DMRBE) according to Meinke (1994). Seeds were removed from the siliques of wild‐type and heterozygous gr2 plants and cleared in Hoyer's solution (3.75 g of gum arabic, 50 g of chloral hydrate, and 2.5 mL of glycerol in 15 mL of water). Cleared seeds were then observed by light microscopy (Leica DMRBE) using differential interference contrast.
Antiserum production
For the production of polyclonal antibodies against GR2, the nucleotide sequence encoding a specific part of GR2 was amplified from cDNA (for primers used, see Table S1). The resulting DNA fragment was fused in frame with the N‐terminal His affinity tag of pET28a, and the resulting plasmids were transformed into Escherichia coli strain BL21 (DE3). The fusion proteins were purified on a nickel‐nitrilotriacetic acid agarose resin matrix and polyclonal antibody was raised in rabbit using the purified antigen. In immunoblot analyses, the dilution ratio for the antibody against GR2 was 1:2000.
Immunoblot analysis
Total leaf proteins were separated using 15% SDS polyacrylamide gels containing 6 M urea. After electrophoresis, the proteins were transferred electrophoretically to polyvinylidene difluoride membranes (Amersham Biosciences, Waukesha, Wisconsin, USA), probed with specific primary antibodies, and visualized using the enhanced chemiluminescence method (Ding et al. 2012).
GR2 promoter construction and GUS staining
GR2pro:GUS was generated by amplifying the 2‐kb sequence upstream of the GR2 translation start sites and subcloning the fragment into the pCAMBIA 1381Z binary vector (for primers used, GR2pro1 and GR2pro2, see Table S1). Plant samples were incubated in staining solution (50 mM sodium phosphate buffer, pH 7.2, 0.2% Triton X‐100, 10 mM potassium ferrocyanide, 10 mM potassium ferricyanide, and 1 mM 5‐bromo‐4‐chloro‐3‐indolyl‐b‐D‐glucuronic acid, cyclohexylammonium salt) at 37°C overnight. Samples were then washed in 70% ethanol before photographs were taken (Zhong et al. 2013).
RNAi and complementation of the gr2 mutant
To obtain RNAi plants, the partial coding region for GR2 (at3g54660) was amplified with igr2 sense and igr2 antisense primers (Table S1) and cloned into the pKANNIBAL vector between the XbaI‐HindIII sites in the sense orientation and the XhoI‐EcoRI sites in the antisense orientation. The construct generated in pKANNIBAL was subcloned as a Notl fragment into pART27. The resultant construct was transformed into the Agrobacterium tumefaciens GV3101 strain and introduced into Arabidopsis plants (Ding et al. 2009). For complementation of gr2, the cDNA containing the coding region of GR2 was amplified by PCR with atgr2 primers (Table S1) and subcloned into the plant expression vector pCAMBIA1301 under the control of P35S. The resulting plasmid was introduced into heterozygous gr2 plants.
RNA isolation and real‐time PCR
Total RNA was isolated using TRIZOL reagent (Sigma–Aldrich) according to the manufacturer's instructions. Next, 500 ng of total RNA was used as template to generate the first‐strand cDNA in a 100 µL reaction with the Superscript II cDNA synthesis system (Invitrogen), according to the manufacturer's instructions. The resulting cDNA samples were used for real‐time PCR using M×3000P (Stratagene) with SYBR Green I (Invitrogen), according to the manufacturers' protocols. Primer pairs for real‐time PCR were designed with the open‐source PCR primer design program DNAMAN version 1.1.10 around an intron to obtain a PCR product of 200 to 400 bp. The primer sequences are available in Table S1. The presence of a single PCR product was verified by melt‐curve analysis and gel electrophoresis.
Microarray analysis
Microarray analysis was conducted using commercial oligonucleotide microarrays (Genechip Arabidopsis ATH1 genome arrays, Affymetrix). Three biological replicates were used for both wild‐type and igr2‐9 plants. Each replicate (200 mg) was based on the fourth leaves with an age of 20 d after emergence. Standard Affymetrix protocols were followed throughout. The data were analyzed using GeneSpring GX 11.00. A change in signal of at least twofold compared with the wild type was considered meaningful, if present in all three biological replicates. The distribution of gene ontology (GO) annotations of genes upregulated in igr2‐9 plants was processed using the web‐based program at TAIR (http://www.arabidopsis.org). GO‐specific terms were investigated using GO::TermFinder (Boyle et al. 2004). P‐values were calculated using Fisher's test (Agresti 1992). Statistical analysis was carried out using Student's t‐test. A significant difference between the control and experimental groups was considered with P<0.01.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Figure S1. Gene structure of At3g54660 and microscopy analysis of developing embryos in the siliques of wild‐type and heterozygous gr2 plants
Figure S2. ROS levels in wild‐type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants during leaf development
Figure S3. Expression of GR2 is induced by elevated H2O2 levels and continuous darkness
Figure S4. Analysis of genes upregulated during the early stages of leaf development in Arabidopsis by specific GO terms
Figure S5. Analysis of genes specifically upregulated after 8 h of H2O2 production in leaf chloroplasts in Arabidopsis overexpressing glycolate oxidase in the chloroplast
Figure S6. Analysis of genes up‐regulated in cat2 following induction of glutathione oxidation after transfer from high CO2 to ambient air
Figure S7. Analysis of genes upregulated in shoots of the 7‐day‐old rootmeristemless1‐1 (rml1‐1) mutant
Table S1. List of primers used in this study
Table S2. Genes found to be significantly (P<0.01) upregulated (≥2, A) or downregulated (≤2, B) in igr2‐9 in a microarray analysis
Table S3. Comparative analysis of the genes enriched in specific GO terms between the transcriptomes of leaves of igr2‐9 and wild‐type Arabidopsis at 29 d after sowing (DAS)
Table S4. List of NAC and WRKY transcription factors induced in the leaves of igr2‐9 and GO5 (with overexpressed glycolate oxidase in the chloroplast) plants
Table S5. Genes induced in the leaves of gr1 plants grown under long‐day conditions
Table S6. Genes induced in the leaves of cad2 plants grown under long‐day conditions
Table S7. Comparative analysis of the upregulated genes enriched in specific GO terms between the transcriptomes of leaves of igr2‐9 and cat2 plants grown under short‐day (SD) conditions
Table S8. Comparative analysis of the upregulated genes enriched in specific GO terms between the transcriptomes of leaves of igr2‐9 and cat2 plants grown under long‐day (LD) conditions
Table S9. Comparative analysis of the upregulated genes enriched in specific GO terms between the transcriptomes of leaves of igr2‐9 and rml1 plants
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (30970218) and the State Key Basic Research and Development Plan of China (2015CB150105). We thank the ABRC for the seed stocks.
Ding S, Wang L, Yang Z, Lu Q, Wen X, Lu C ( 2015) Decreased glutathione reductase2 leads to early leaf senescence in Arabidopsis . J Integr Plant Biol 58: 29–47
Available online on Jun. 1, 2015 at www.wileyonlinelibrary.com/journal/jipb
REFERENCES
- Agresti A ( 1992) A survey of exact inference for contingency tables. Stat Sci 7: 131–153 [Google Scholar]
- Alscher RG ( 1989) Biosynthesis and antioxidant function of glutathione in plants. Physiol Plant 77: 457–464 [Google Scholar]
- Aono M, Kubo A, Saji H, Tanaka KK, Kondo N ( 1993) Enhanced tolerance to photooxidative stress of transgenic Nicotiana tabacum with chloroplastic glutathione reductase activity. Plant Cell Physiol 34: 129–135 [Google Scholar]
- Aono M, Saji H, Fujiyama K, Sugita M, Kondo N, Tanaka K ( 1995) Decrease in activity of glutathione reductase enhanced paraquat sensitivity in transgenic Nicotiana tabacum . Plant Physiol 107: 645–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K ( 1999) The water‐water cycle in chloroplasts: Scavenging active oxygen species and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Hamad S, Allu AD, Matallana‐Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller‐Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt‐promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Bieker S, Riester L, Stahl M, Franzaring J, Zentgraf U ( 2012) Senescence‐specific alteration of hydrogen peroxide levels in Arabidopsis thaliana and oilseed rape spring variety Brassica napus L. cv. Mozart. J Integr Plant Biol 54: 540–554 [DOI] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G ( 2004) GO::TermFinder‐Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, Zhang C, Morris K, Jenner C, Jackson S, Thomas B, Tabrett A, Legaie R, Moore JD, Wild DL, Ott S, Rand D, Beynon J, Denby K, Mead A, Buchanan‐Wollaston V ( 2011) High‐resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent P, Creissen GP, Kular B, Wellburn AR, Mullineaux PM ( 1995) Oxidative stress responses in transgenic tobacco containing altered levels of glutathione reductase activity. Plant J 8: 247–255 [Google Scholar]
- Buchanan‐Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ ( 2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation‐induced senescence in Arabidopsis . Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Cairns NG, Pasternak M, Wachter A, Cobbett CS, Meyer AJ ( 2006) Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol 141: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GH, Liu CP, Chen SCG, Wang LC ( 2011) Role of ARABIDOPSIS A‐FIFTEEN in regulating leaf senescence involves response to reactive oxygen species and is dependent on ETHYLENE INSENSITIVE2 . J Exp Bot 62: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew O, Whelan J, Millar AH ( 2003) Molecular definition of the ascorbate‐glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278: 46869–46877 [DOI] [PubMed] [Google Scholar]
- Creissen GP, Reynolds H, Xue Y, Mullineaux PM ( 1995) Simultaneous targeting of pea glutathione reductase and of a bacterial fusion protein to chloroplasts and mitochondria in transgenic tobacco. Plant J 8: 167–175 [DOI] [PubMed] [Google Scholar]
- Cui MH, Ok SH, Yoo KS, Jung KW, Yoo SD, Shin JS ( 2013) An Arabidopsis cell growth defect factor‐related protein CRS, promotes plant senescence by increasing the production of hydrogen peroxide. Plant Cell Physiol 54: 155–167 [DOI] [PubMed] [Google Scholar]
- del Rio LA, Pastori GM, Palma JM, Sandalio LM, Sevilla F, Corpas FJ, Jimenez A, Lopez‐Huertas E, Hernandez JA ( 1998) The activated oxygen role of peroxisomes in senescence. Plant Physiol 116: 1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SH, Lei M, Lu QT, Zhang AH, Yin Y, Wen XG, Zhang LX, Lu CM ( 2012) Enhanced sensitivity and characterization of photosystem II in transgenic tobacco plants with decreased chloroplast glutathione reductase under chilling stress. Biochim Biophys Acta 1817: 1979–1991 [DOI] [PubMed] [Google Scholar]
- Ding SH, Lu QT, Zhang Y, Yang ZP, Wen XG, Zhang LX, Lu CM ( 2009) Enhanced sensitivity to oxidative stress in transgenic tobacco plants with decreased glutathione reductase activity leads to a decrease in ascorbate pool and ascorbate redox state. Plant Mol Biol 69: 577–592 [DOI] [PubMed] [Google Scholar]
- Edwards EA, Rawsthorne S, Mullineaux PM ( 1990) Subcellular distribution of multiple forms of glutathione reductase in leaves of pea (Pisum sativum L.). Planta 180: 278–284 [DOI] [PubMed] [Google Scholar]
- Fahnenstich H, Scarpeci TE, Valle EM, Flügge UI, Maurino VG ( 2008) Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol 148: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors C, Fryer MJ, Waring J, Reeder B, Bechtold U, Mullineaux PM, Nonell S, Wilson MT, Baker NR ( 2006) Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J Exp Bot 57: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Lelandais M, Galap C, Kunert KJ ( 1991) Effects of elevated cytosolic glutathione reductase activity on the cellular glutathione pool and photosynthesis in leaves under normal and stress conditions. Plant Physiol 97: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G ( 2005) Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G ( 2009) Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid Redox Signal 11: 861–905 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Souriau N, Perret S, Lelandais M, Kunert KJ, Pruvost C, Jouanin L ( 1995) Overexpression of the glutathione reductase but not glutathione synthetase leads to increases in antioxidant capacity and resistance to photoinhibition in poplar trees. Plant Physiol 109: 1047–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Breusegem FV ( 2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis . Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S ( 2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27: 521–549 [Google Scholar]
- Guo Y, Gan S ( 2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G (2013a) Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid Redox Signal 18: 2106–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Mhamdi A, Chaouch S, Noctor G (2013b) Regulation of basal and oxidative stress‐triggered jasmonic acid‐related gene expression by glutathione. Plant Cell Environ 36: 1135–1146 [DOI] [PubMed] [Google Scholar]
- Hodges DM, De Long JM, Forney CF, Prange RK ( 1999) Improving the thiobarbituric acid‐reactive‐substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 270: 604–611 [DOI] [PubMed] [Google Scholar]
- Jiménez A, Hernandez JA, Pastori G, del Rio LA, Sevilla F ( 1998) Role of the ascorbate‐glutathione cycle of mitochondria and peroxisomes in the senescence of pea leaves. Plant Physiol 118: 1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataya ARA, Reumann S ( 2010) Arabidopsis glutathione reductase1 is dually targeted to peroxisomes and the cytosol. Plant Signal Behav 5: 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai‐Yamada M, Ohori Y, Uchimiya H ( 2004) Dissection of Arabidopsis Bax inhibitor‐1 suppressing Bax‐, hydrogen peroxide‐, and salicylic acid‐induced cell death. Plant Cell 16: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna‐Chopra R ( 2012) Leaf senescence and abiotic stresses share reactive oxygen species‐mediated chloroplast degradation. Protoplasma 249: 469–481 [DOI] [PubMed] [Google Scholar]
- Kim J, Woo R, Kim J, Lim P, Lee I, Choi S, Hwang D, Nam H ( 2009) Trifurcate feed‐forward regulation of age‐dependent cell death involving miR164 in Arabidopsis . Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kornyeyev D, Logan BA, Payton P, Allen RD, Holaday AS ( 2003) Elevated chloroplastic glutathione reducatase activities decrease chilling‐induced photoinhibition by increasing rates of photochemistry, but not thermal energy dissipation, in transgenic cotton. Funct Plant Biol 30: 101–110 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Smalle J, Va M, Montagu N, Inzé D ( 1998) Oxidative stress tolerance and longevity in Arabidopsis: The late‐flowering mutant gigantea is tolerant to paraquat. Plant J 14: 759–764 [DOI] [PubMed] [Google Scholar]
- Li H, Wang G, Liu S, An Q, Zheng Q, Li B, Li Z ( 2014) Comparative changes in the antioxidant system in the flag leaf of early and normally senescing near‐isogenic lines of wheat (Triticum aestivum L.). Plant Cell Rep 33: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Lim P, Kim H, Nam H ( 2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lin J, Wu S ( 2004) Molecular events in senescing Arabidopsis leaves. Plant J 39: 612–628 [DOI] [PubMed] [Google Scholar]
- Martínez DE, Costa ML, Guiamet JJ ( 2008) Senescence‐associated degradation of chloroplast proteins inside and outside the organelle. Plant Biol 10: 15–22 [DOI] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtza M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld JP, Hell R ( 2009) The NADPH‐dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis . Proc Natl Acad Sci USA 106: 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW ( 1994) Seed development in Arabidopsis thaliana In: Meyerowitz EM, Somerville CR, eds. Arabidopsis. Cold spring Harbor Laboratory Press, New York: pp. 253–259 [Google Scholar]
- Mhamdi A, Han Y, Noctor G ( 2013) Glutathione‐dependent phytohormone responses: Teasing apart signaling and antioxidant functions. Plant Signal Behav 8: e24181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat L, Saindrenan P, Gouia H, Issakidis‐Bourguet E, Renou JP, Noctor G ( 2010) Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol 153: 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U ( 2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis . Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Miller JD, Arteca RN, Pell EJ ( 1999) Senescence‐associated gene expression during ozone‐induced leaf senescence in Arabidopsis . Plant Physiol 120: 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R ( 2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Munné‐Bosch S, Alegre L ( 2002) Plant ageing increases oxidative stress in chloroplasts. Planta 214: 608–615 [DOI] [PubMed] [Google Scholar]
- Navabpour S, Morris K, Allen R, Harrison E, A‐H‐Mackerness S, Buchanan‐Wollaston V (2003) Expression of senescence‐enhanced genes in response to oxidative stress. J Exp Bot 54: 2285–2292 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH ( 1998) Ascorbate and glutathione: Keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Noctor G, Gomez L, Vanacker H, Foyer CH ( 2002) Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J Exp Bot 53: 1283–1304 [DOI] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez‐Garcia B, Queval G, Foyer CH ( 2012) Glutathione functions in plants: An integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Queval G, Foyer CH ( 2013) Regulating the redox gatekeeper: Vacuolar sequestration puts glutathione disulfide in its place. Plant Physiol 163: 665–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Lee S, Chung I, Lee C, Nam H ( 1996) A senescence‐associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Oh S, Park J, Lee G, Paek K, Park S, Nam H ( 1997) Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana . Plant J 12: 527–535 [DOI] [PubMed] [Google Scholar]
- Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J, Alvarez ME, Foyer CH ( 2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis . Plant Physiol 139: 1291–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton P, Webb R, Kornyeyev D, Allen R, Holaday AS ( 2001) Protecting cotton photosynthesis during moderate chilling at high light intensity by increasing chloroplastic antioxidant enzyme activity. J Exp Bot 52: 2345–2354 [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis‐Bourguet E, Hoeberichts FA, Vandorpe M, Gakiére B, Vanacker H, Miginiac‐Maslow M, Breusegem FV, Noctor G ( 2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of day length‐dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2‐induced cell death. Plant J 52: 640–657 [DOI] [PubMed] [Google Scholar]
- Queval G, Jaillard D, Zechmann B, Noctor G ( 2011) Increased intracellular H2O2 availability preferentially drives glutathione accumulation in vacuoles and chloroplasts. Plant Cell Environ 34: 21–32 [DOI] [PubMed] [Google Scholar]
- Queval G, Neukermans J, Vanderauwera S, Van Breusegem F, Noctor G ( 2012) Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in Arabidopsis . Plant Cell Environ 35: 374–387 [DOI] [PubMed] [Google Scholar]
- Schippers JHM, Nunes‐Nesi A, Apetrei R, Hille J, Fernie AR, Dijkwel PP ( 2008) The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20: 2909–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaubelt D, Queval G, Dong Y, Diaz‐Vivancos P, Makgopa ME, Howell G, Simone AD, Bai J, Hannah MA, Foyer CH ( 2015) Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol in Arabidopsis thaliana . Plant Cell Environ 38: 266–279 [DOI] [PubMed] [Google Scholar]
- Sewelama N, Jaspert N, Kelen KVD, Tognetti VB, Schmitz J, Frerigmanne H, Stahl E, Zeier J, Frank Van Breusegem FV, Maurino VG ( 2014) Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol Plant 7: 1191–1210 [DOI] [PubMed] [Google Scholar]
- Smykowski A, Zimmermann P, Zentgraf U ( 2010) G‐box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis thaliana . Plant Physiol 153: 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Strohm M, Jouanin L, Kunert KJ, Pruvost C, Polle A, Foyer CH, Rennenberg H ( 1995) Regulation of glutathione synthesis in leaves of transgenic poplar (Populus tremula X P. alba) overexpressing glutathione synthetase. Plant J 7: 141–145 [Google Scholar]
- Thordal‐Christensen H, Zhang Z, Wei Y, Collinge DB ( 1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley‐powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tzafrir I, Pena‐Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, Meinke D ( 2004) Identification of genes required for embryo development in Arabidopsis . Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J ( 2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeel S, Langebartels C, Gruissem W, Inzé D, Breusegem FV ( 2005) Genome‐wide analysis of hydrogen peroxide‐regulated gene expression in Arabidopsis reveals a high light‐induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljovic‐Jovanovic S, Noctor G, Foyer CH ( 2002) Are leaf hydrogen peroxide concentrations commonly overestimated? The potential influence of artefactual interference by tissue phenolics and ascorbate. Plant Physiol Biochem 40: 501–507 [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM ( 1998) A comparison of the expression patterns of several senescence‐associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 445–469 [DOI] [PubMed] [Google Scholar]
- Woo H, Kim J, Nam H, Lim P ( 2004) The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant Cell Physiol 45: 923–932 [DOI] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM ( 2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Pasternak T, Eiblmeier M, Ditengou F, Kochersperger P, Sun JQ, Wang H, Rennenberg H, Teale W, Paponov I, Zhou WK, Li CY, Li XG, Palme K ( 2013) Plastid‐localized glutathione reductase2‐regulated glutathione redox status is essential for Arabidopsis root apical meristem maintenance. Plant Cell 25: 4451–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata JM, Guéra A, Esteban‐Carrasco A, Martín M, Sabater B ( 2005) Chloroplasts regulate leaf senescence: Delayed senescence in transgenic ndhF‐defective tobacco. Cell Death Differ 12: 1277–1284 [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Hemleben V ( 2008) Molecular cell biology: Are reactive oxygen species regulators of leaf senescence? Prog Bot 69: 117–138 [Google Scholar]
- Zentgraf U, Laun T, Miao Y ( 2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana . Eur J Cell Biol 89: 133–137 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Heinlein C, Orendi G, Zentgraf U ( 2006) Senescence‐specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ 29: 1049–1060 [DOI] [PubMed] [Google Scholar]
- Zhong LL, Zhou W, Wang HJ, Ding SH, Lu QT, Wen XG, Peng LW, Zhang LX, Lu CM ( 2013) Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25: 2925–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Figure S1. Gene structure of At3g54660 and microscopy analysis of developing embryos in the siliques of wild‐type and heterozygous gr2 plants
Figure S2. ROS levels in wild‐type (wt) and GR2 RNAi (igr2‐7, igr2‐9, and igr2‐14) plants during leaf development
Figure S3. Expression of GR2 is induced by elevated H2O2 levels and continuous darkness
Figure S4. Analysis of genes upregulated during the early stages of leaf development in Arabidopsis by specific GO terms
Figure S5. Analysis of genes specifically upregulated after 8 h of H2O2 production in leaf chloroplasts in Arabidopsis overexpressing glycolate oxidase in the chloroplast
Figure S6. Analysis of genes up‐regulated in cat2 following induction of glutathione oxidation after transfer from high CO2 to ambient air
Figure S7. Analysis of genes upregulated in shoots of the 7‐day‐old rootmeristemless1‐1 (rml1‐1) mutant
Table S1. List of primers used in this study
Table S2. Genes found to be significantly (P<0.01) upregulated (≥2, A) or downregulated (≤2, B) in igr2‐9 in a microarray analysis
Table S3. Comparative analysis of the genes enriched in specific GO terms between the transcriptomes of leaves of igr2‐9 and wild‐type Arabidopsis at 29 d after sowing (DAS)
Table S4. List of NAC and WRKY transcription factors induced in the leaves of igr2‐9 and GO5 (with overexpressed glycolate oxidase in the chloroplast) plants
Table S5. Genes induced in the leaves of gr1 plants grown under long‐day conditions
Table S6. Genes induced in the leaves of cad2 plants grown under long‐day conditions
Table S7. Comparative analysis of the upregulated genes enriched in specific GO terms between the transcriptomes of leaves of igr2‐9 and cat2 plants grown under short‐day (SD) conditions
Table S8. Comparative analysis of the upregulated genes enriched in specific GO terms between the transcriptomes of leaves of igr2‐9 and cat2 plants grown under long‐day (LD) conditions
Table S9. Comparative analysis of the upregulated genes enriched in specific GO terms between the transcriptomes of leaves of igr2‐9 and rml1 plants


