Abstract
Whole genome sequencing of bacteria has recently emerged as a cost-effective and convenient approach for addressing many microbiological questions. Here we review the current status of clinical microbiology and how it has already begun to be transformed by the use of next-generation sequencing. We focus on three essential tasks: identifying the species of an isolate, testing its properties such as resistance to antibiotics and virulence, and monitoring the emergence and spread of bacterial pathogens. The application of next-generation sequencing will soon be sufficiently fast, accurate and cheap to be used in routine clinical microbiology practice, where it could replace many complex current techniques with a single, more efficient workflow.
Introduction
Clinical microbiology is a discipline focussed on rapidly characterizing pathogen samples in order to direct the management of individual infected patients (diagnostic microbiology) and monitor the epidemiology of infectious disease (public health microbiology). Applications to epidemiology include the detection of outbreaks, changing trends in infection and the emergence of new threats. Ongoing developments in DNA sequencing technologies are likely to affect the diagnosis and monitoring of all pathogens including viruses, bacteria, fungi and parasites, but for this review we focus on bacterial pathogens to illustrate the likely changes arising from the adoption of routine whole-genome sequencing.
Bacterial pathogens account for much of the worldwide burden of infection. For patients with bacterial infections, the crucial steps are to grow an isolate from a specimen, identify its species, determine its pathogenic potential and test its susceptibility to antimicrobial drugs. Together this information facilitates the specific and rational treatment of patients. For public health purposes, knowledge also needs to be gained about the relatedness of the pathogen to other strains of the same species to investigate transmission routes and enable the recognition of outbreaks1. Each of the steps in this process of characterising the pathogen depends on many specialized, species-specific methodologies that have been developed over decades. These require the extensive knowledge-base of clinical microbiologists who apply labour-intensive, complex and often slow techniques to yield the relevant information. This multi-step process takes from days (for the isolation by culture, species identification and susceptibility testing for rapidly growing bacteria, such as Escherichia coli) to months (for slow growing bacteria such as Mycobacterium tuberculosis, or to produce full typing for any pathogen) (Figure 1).
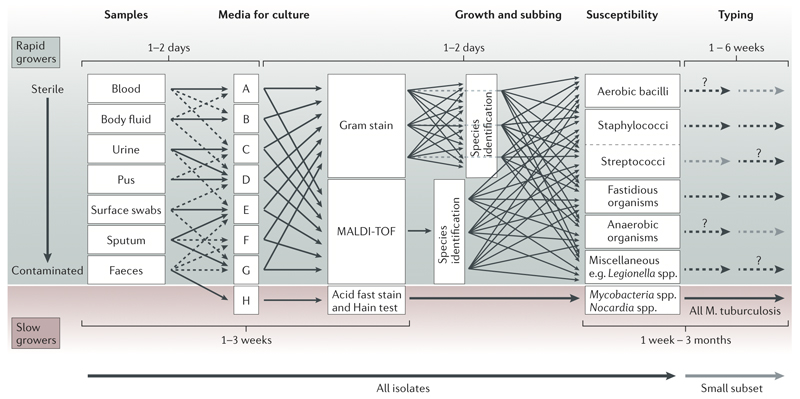
Figure 1. Principles of current processing of bacterial pathogens.
Schematic representation of the current workflow for processing samples for bacterial pathogens is presented, with high complexity and a typical timescale of a few weeks to a few months. The schematic is an approximation that highlights the principle steps in the workflow; it is not intended to be a comprehensive or precise description. Samples that are likely to be normally sterile are often cultured on rich medium that will support the grown of any culturable organism. Those from samples contaminated with colonising flora present a challenge for growing the infecting pathogen. Many types of culture media (referred to as selective media) are used to favour the growth of the suspected pathogen; this approach is particularly important for culturing pathogens from faeces. Boxes A to H arbitrarily represents the many different media for culture. The medium H represents a medium designed for growing mycobacteria that have specific growth requirements.
Once an organism is growing, the morphological appearance and density of growth are properties that need specialist knowledge for deciding whether it is likely to be pathogenic. The likely pathogens are then processed through a complex pathway that has many contingencies to determine species and antimicrobial susceptibility. Broadly, there are two approaches. One approach uses MALDI-TOF for species identification prior to setting up susceptibility testing. The other uses Gram staining followed by biochemical testing to determine species; susceptibility testing is often set up simultaneously with doing biochemical tests. Categorisation of pathogens into groups of species is needed to choose the appropriate susceptibility testing panel. Lastly, depending on the species and perceived likelihood of an outbreak, a small subset of isolates may be chosen for further investigation using a wide range of typing tests often only provided by reference laboratories. The dashed lines and question marks are positioned arbitrarily to indicate that the further investigation is varied and happens only in a small number of cases.
Ideally, all the information necessary for both individual treatment and public health protection would be gained in a single step. In principle, the genome sequence of an isolate contains all, or nearly all, the information required to direct treatment and inform public health measures. Indeed it is becoming clear …that rapid, inexpensive genome sequencing (Box 1) holds the potential to replace many complex multi-faceted procedures used to characterise a pathogen after it has been isolated by culture2, 3. However, there are substantial challenges to be overcome and success will depend on development of the genomic knowledge and analytical methods required to correctly extract and interpret this information. Indeed, application of new sequencing technologies will be highly disruptive and we predict that it will take many years to fully transform clinical microbiology laboratories. Ultimately, deployment will critically require substantial validation of genotypic prediction of phenotype, particularly for antimicrobial resistance; this work is yet to be done. In this review, we provide a brief overview of current practice and then we outline the potential of sequencing technology to deliver the following key diagnostic information in the clinical laboratory after culture of an isolate: identification of species, antimicrobial resistance, presence of virulence determinants and strain typing to detect outbreaks and support surveillance.
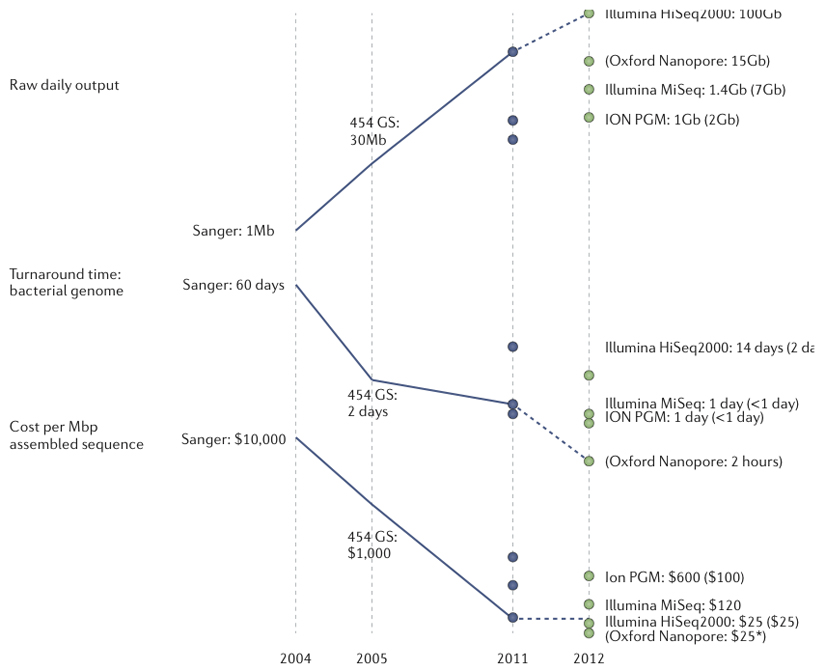
Box 1 Sequencing platforms for clinical microbiology.
Released in 2005 with reads of ~110b, the first next-generation sequencers, from Roche-454, could sequence bacterial genomes in a single run 60. Initial applications were focused on diversity discovery. Later versions of the 454 platform have increased read length (~500b) to approach that of Sanger sequencing but at much lower cost, and so have retained a role in producing high-contiguity assemblies of bacterial genomes.
Initially launched in 2006 with short (36b) reads, Illumina Genome Analyzers have captured the bulk of the sequencing market for both microbiology and larger-organisms. With incrementally increasing capacity and read-length, the current standard configuration (at the end of 2011) delivers ~300Gb raw data per 8-lane flowcell in the form of 100b, paired reads. Tagging each sample with its own 6-8b index sequence allows at least 96 samples to be sequenced simultaneously in each lane. This approach makes the Illumina HiSeq platform useful and cost-effective for large bacterial sample collections.
It is clear that for most uses in microbiology, fast, compact bench-top machines will be preferred to the large, very high capacity machines designed for human sequencing. Two such platforms, the Ion PGM and the Illumina MiSeq, both of which use established chemistries that both involve library preparation and amplification as the first steps in sequencing, are becoming popular among microbiologists61. In a new platform from Oxford Nanopore Technologies, which is slated for commercial release in 201262, the sequence of a single DNA molecule passing through a protein nanopore under the control of a processive enzyme is measured as fluctuations in electrical current across a lipid membrane. According to the company, data is collected in real time at around 200-400 bases per second, and they expect up to 1000 bases per second in the future. These data are translated to sequence information in real time using on-board electronics. The company have said that chips are configured to read 2000 or 8000 pores simultaneously and that reads can be up to tens of kb in length. Because it reads native DNA, the Oxford Nanopore technology is anticipated to work with relatively crude samples and low DNA concentrations. The company plans two machines: the scalable ‘GridION’ in which multiple sequencing units (each with a projected output of ~2Gb of data an hour) can be combined in parallel; and the single-use, USB-connected MinION, with a projected hourly capacity of ~150Mb. If per-base accuracy can be improved to current next-generation standards the long reads will enable complete genomes to be generated in minutes with either machine. This new technology is the first in a new breed of similarly designed platforms which are likely to produce dramatic improvement is sequencing technology. The figure shows the development of sequencing technologies relevant to microbiology, highlighting the continuing increases in throughput and speed, and reductions in costs.
Current clinical microbiology
The principles behind diagnostic bacteriology have changed little over the past 50 years. Most of the output from a microbiological laboratory is dependent on isolating a viable organism. Over a century of experimentation has led to the development of a wide repertoire of methods for isolating culturable bacterial pathogens. After culture, diagnostic characterisation depends on a wide range of testing pathways (Figure 1), many aspects of which are species-specific4–6. Complexity and a lack of automation prevent the rapid return of the complete diagnostic information about a bacterial isolate.
The cardinal steps in processing a sample are isolation of a pathogen, determination of species, testing of antimicrobial susceptibility and virulence, and, in specific settings, intra-species typing. The first three steps are critical for optimal management of an infected patient and the last step is valuable for identifying outbreaks and surveillance.
Culture of pathogen
The aim of culture is to investigate the microbial composition of a sample, identify colonies that deserve further attention, and to produce sufficient mass of pure organisms for subsequent use. Although most bacterial diseases are caused by ~20 species (Table 1), up to 1000 other species may sometimes cause disease6. The majority of these pathogens can be grown in appropriate culture media (using a wide variety of methods), but a minority (< 10%) of infecting bacterial pathogens are believed to be non-culturable or difficult to grow; for these species diagnosis currently depends on serological, antigen and nucleic acid amplification tests.
Table 1. The bacterial pathogens reported by the Oxford University Hospitals Trust microbiology laboratory in the last 15 years.
The 15 year output of isolates by the Oxford University Hospitals Trust microbiology laboratory are shown as an example of the frequency of pathogens isolated by a large service with comprehensive diagnostic throughput. Of 751134 isolates cultured, 557581 (74%) were categorised into 301 species using routine phenotypic methods. 158,157150 (21%) were characterised to genus or other grouping (71 categories) (e.g. Pseudomonas spp. or coagulase negative staphylococci, respectively). 36403 (5%) were isolated but not characterised beyond the gram stain (not shown). On a global scale the proportions of species may differ by country. For example M. tuberculosis will be a major component of laboratory activity in communities with high prevalence whereas Oxford has a very low incidence of TB.
| Examples of difficult to culture species | |
| Species | Species |
| Chlamidia trachomatis | Bartonella henselae |
| Chlamydophila pneumoniae | Bartonella elizabethae |
| Chlamydophila psitticae | Ehrlichia ewingii |
| Mycoplasma pneumoniae | Ehrlichia chaffeensis |
| Ureaplasma urealyticum | Anaplasma phagocytophilum |
| Treponema pallidum | Rickettsia conorii |
| Borrelia burgdorferi | Orientia tsutsugamushi |
| borrelia recurrentis | Rickettsia prowazekii |
| Leptospira interrogans | Rickettsia typhi |
| Coxiella burnettei | Rickettsia rickettsii |
| Mycobacterium leprae | Rickettsia akari |
Culture is complex and contingent on the origin of the sample. Samples from usually-sterile sites (such as cerebrospinal fluid) and bacterially contaminated samples (such as faeces) represent opposite extremes. For sterile sites, a full report of all organisms present is possible, although not all may be clinically relevant. For highly contaminated samples, isolation of pathogens requires selective media, assisted by, for example, inspection of colony morphology and gram staining. Educated guesses about likely pathogens alter the choice of protocol, and the growth time before further analysis can vary from hours to weeks. A full description of culture methodology is beyond the scope of this review and is available in from extensive literature, for example, a Clinical Microbiology textbook5.
Species identification
Knowing the species of an isolate is often vital to make effective clinical decisions. Determining species is directly informative about pathogenic potential, and allows differentiation of infecting pathogens from non-infecting ‘contaminating pathogens’. A typical example is that Staphylococcus aureus isolated from a blood culture (rather than a skin swab sample) has a high probability of being an infecting pathogen whereas Staphylococcus epidermidis would likely be a contaminating isolate. Currently, species identification is first based on gram staining, colony growth and morphology, rapid biochemical reactions and ancillary tests. These take up to 24 hours for organisms that require extensive biochemical panels (for example, using the Vitek system commercialised by BioMerieux or the BD Phoenix system from Beckton Dickinson). 16S rRNA sequencing is increasingly used in ambiguous cases, but this has a number of drawbacks7 including the fact that it usually takes a further two days8.
Recently, matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry has yielded rapid species identification by analysing the biomolecules present in pure suspensions of any isolate and comparing the results with known profiles9, 10. The cost of a MALDI-TOF mass spectrometer is high (several hundred thousand dollars) but the running cost is low11 (around one dollar per isolate) and results can be obtained in minutes9 so this approach is rapidly being adopted for routine use. Therefore, MALDI-TOF represents an attractive alternative to traditional methods by increasing the speed of identification of species12 and highlights the potential of new technology combined with sophisticated software and databases to simplify and improve an important aspect of diagnostic microbiology. However, questions remain about the level of resolution it can achieve9, 13 , for example to distinguish between the closely related species M. bovis and M. tuberculosis14. In addition, no further information about the isolate, such as antibiotic susceptibility or virulence, is yielded by MALDI TOF.
Testing for antibiotic resistance
Determining the antimicrobial resistance properties of an isolate is possibly the single most important procedure for managing bacterial infectious disease at the individual patient level. This is largely because falsely recording an organism as susceptible to an antibiotic represents a serious risk to the infected patient if they are treated with an ineffective antibiotic. Current knowledge of susceptibility testing is vast, complex and embodied in guidelines and various textbooks15. The phenotypic methods for susceptibility testing are almost exclusively based on inhibition of growth of the bacteria when exposed to the test antimicrobial.
Infectious diseases practice is critically dependent on confidence in this system of testing. However, the sensitivity and specificity of a particular method of testing is based on comparison to in vitro ‘gold standard’ susceptibility testing systems (such as the micro-dilution method15) which are regarded as surrogates for clinical outcome15. Consequently, even with phenotypic testing the in vivo (that is, clinical) susceptibility of an isolate is not known with complete certainty15. Clinicians have come to accept this uncertainty in clinical decision making, and indeed they are familiar with treating some pathogens without testing since for some organisms there are no accurate tests available.
Advantageously, phenotypic testing yields information not only on those agents to which an organism is resistant, but also those to which it is susceptible, which is of direct clinical value. However, no single pathway for resistance and/or susceptibility testing exists. Tests are grouped by species, which adds to complexity and time taken for thorough testing. The tests are subject to many assumptions about degree of susceptibility based on minimum inhibitory concentration (MIC) and require selection of a ‘breakpoint‘ for each antibiotic, that is a MIC level above which the isolate is deemed resistant to therapy. These breakpoints are chosen based on diverse but imperfect factors such as the distribution of MICs, chemical concentration, mutual interactions between host and drug, animal models and clinical treatment experience. Consequently, there is considerable debate on how to set the breakpoints, and these are not always agreed across countries and organizations. The effect of susceptibility testing on clinical response to infection is difficult to study given the multiple factors that influence patient outcome, so that the sensitivity and specificity for determining resistance or susceptibility of phenotypic tests are often poorly measured. In addition, phenotypic testing has proven unreliable in some well-described situations. For example, the emergence of quinolone resistance in Salmonella enterica serovar Typhi that was not detected by routine phenotypic testing – these isolates were falsely found to be susceptible16. This failure has since been found to be caused by the emergence of a new resistance mechanism and new testing recommendations have been formulated17. Furthermore, complete testing can take days for rapid growers such as E. coli and S. aureus and even months for slow growers such as M. tuberculosis.
At present, the presence or absence of resistance genes is used in a few situations to direct early treatment of patients. For example, detection of the mecA gene determines whether an isolate of Staphylococcus aureus is meticilin susceptible or resistant18, which in turn is associated with increased mortality19. Another example is the Hain test which uses DNA hybridisation of primers unique to a limited number of common resistance determinants to predict resistance of M. tuberculosis isolates to a few key anti-mycobacterial drugs8. This has gained credibility and wide use and is a good proof of principle example for the future of using DNA sequence to predict resistance.
Detecting virulence determinants
Identifying virulence determinants is rarely a priority in treating individual patients. There are on the other hand a few examples where this is critical. For example, in Corynebacterium diphtheriae infections detecting the presence of toxin is crucial to administering anti-toxin to the patient4, 5. Similarly, determining whether a strain of Clostridium difficile is toxin producing or not is crucial to diagnose whether C. difficile is pathogenic and what treatment is required. Historically, most virulence determinants have been detected using bio-assays (e.g. detection of botulinum toxin) or serotyping (e.g. presence of pneumococcal capsule). Increasingly, detection of virulence factors is based on detecting the bacterial sequences encoding virulence factors using PCR (for example for factors such as the C. difficile toxin B)11. These tests are rarely included in the repertoire of routine laboratories and are usually performed by reference laboratories. For public health purposes, virulence determinants such as capsule type is important, particularly for species where capsule based vaccines are in wide use4, 5, e.g. Haemophilus influenzae type b, Streptococcus pneumoniae and Neisseria meningitidis.
Outbreak detection and surveillance
Pathogen surveillance and outbreak detection is mostly informal and reactive. The isolates chosen for investigation of relatedness to identify outbreaks have been dependent on extemporary choices, often based on loosely defined epidemiological criteria at the level of the routine clinical laboratory or infection control team. Consequently, many outbreaks are likely missed. The typing used for identifying epidemic transmission can take months to complete as most typing schemes are species specific, depend on many variables and only a handful of laboratories in the whole world perform routine typing. Methods commonly used now include PCR (e.g. Multiple-Locus Variable Number of Tandem Repeats Analysis)9, restriction fragment length polymorphisms (e.g. pulsed field gel electrophoresis: PFGE)20 or fractional sequencing (e.g. multi-locus sequence typing: MLST)21. Through substantial investment in monitoring and reference facilities, turn-around time for these methods can be reduced down to a few days. However, because most locations do not benefit from such facilities, typing typically contributes little to the immediate control of an outbreak.
Potential of genome sequencing
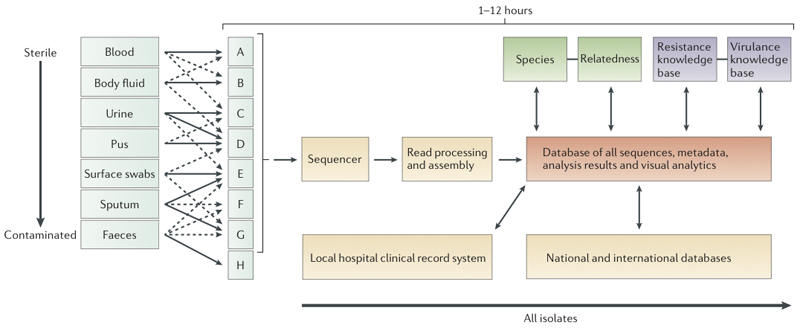
The major advantage of whole genome sequencing is to yield all the available DNA information content on isolates in a single rapid step following culture (sequencing without culture will be discussed in the Future directions section). In principle, the result contains all the data currently used for diagnostic and typing needs, even though it is not always yet known how to interpret this data. However, the genome also includes vast amounts of additional data presently unavailable from routine processing, thus opening the prospect for large-scale research into pathogen genotype-phenotype associations from routinely collected data. The hurdles to implementing whole genome sequencing in clinical and public health laboratories are substantial, as widespread adoption would require incorporating the knowledge from over a century of characterising pathogens - presently delivered by a skilled workforce - into an entirely new framework of mainly computer-driven genome processing (Figure 2). This would require a radical shift to a new operational paradigm for routine laboratories. In addition, a new understanding of genotype-to-phenotype relationships needs to be established, evaluated and deployed in parallel with current routine methods, which will require a major effort leading to gradual replacement of present day methodologies over many years.
Figure 2. Hypothetical workflow based on whole genome sequencing.
Schematic representation of the workflow anticipated after adoption of whole genome sequencing, with an expected timescale that could fit within a single day. The culture steps would be the same as currently used in a routine microbiology laboratory. Some types of sample might be directly sequenced (see ‘future directions’, not shown here). Once a sample or likely pathogen is ready for sequencing, DNA will be extracted. This procedure is becoming simpler, as the input required for successful sequencing is reducing; it is now possible to use as little as 5 ng and to purify this in <30 minutes. For current bench-top machines it can take as little as 2 hours to prepare the DNA for sequencing, and new platforms (Box 1) could enable sequencing without preparation. Therefore, bacterial genome sequencing in hours and possibly even minutes is a realistic prospect.
After sequencing, the main processes for yielding information will be computational. The development of software and databases is a major challenge to overcome before a pathogen sequencing can be deployed in clinical microbiology. Automated sequence assembly algorithms will be necessary for processing the raw sequence data (Box 1). This assembled sequence would then be analysed by modular software to determine species, relationship to other isolates of the same species, antimicrobial resistance profile and virulence gene content. Results of this analysis will be reported through hospital information systems. All the results will also be used for outbreak detection and infectious diseases surveillance. These developments will require new large database and other informatics technology and will take time to develop. In particular, it will need ‘intelligent systems’ which will incorporate elements of machine learning to enable automatic updating of key knowledge bases for species identification, antimicrobial resistance determination and virulence detection. Formal evaluation of such a solution will also need robust testing to ensure it performs at least as well as current methods.
Crucially, the translation of sequence technology into new practices in clinical microbiology is facilitated by genetic features of bacteria. Compared to eukaryotic genomes, bacterial genomes are much smaller (2-6 Megabases) and bacteria usually possess a single haploid chromosome (though a few possess two haploid chromosomes). On the other hand, they are much more diverse than eukaryotic species, partly because about 10% to 40% of the genome may consist of dispensable sequences which are not shared in all members of a same species22. Many of these dispensable elements are also mobile, for example episomal structures such as plasmids. The plasmids and other mobile elements often encode antibiotic resistance and even virulence determinants and, as such, are highly relevant to clinical microbiology.
Species identification
As highlighted above, identification of species is a crucial initial step in managing infectious diseases and tracking pathogens. Currently, taxonomic approaches are based on keeping a type strain collection as a gold standard (with the exception of MALDI-TOF which can use a set of references for each species). Using whole genome sequencing, this could be replaced by a ‘type sequence’. That is, species would be taxonomically defined by their sequence and the ‘type sequence’ would constitute a reference point against which to compare sequence data from other isolates. The relationship of the species to all previously sequenced organisms can be determined using phylogenetic analysis (Box 3).
Box 3 Phylogenetic analysis.
In order to identify the species of an isolate or to investigate whether it is part of an outbreak, it is often useful to construct a phylogeny illustrating how the isolate is related to other strains. To do so, Bayesian phylogenetics is an attractive alternative method to classical non-statistical phylogenetic techniques78. The most popular software for Bayesian phylogenetic inference is BEAST79. A key advantage of the Bayesian method is that assumptions are made explicitly, and can be relaxed or tested. Many such extensions are implemented in BEAST, for example to account for differences in sampling dates80, non-constant population sizes81, non-constant molecular clocks82 and geographical origins of the individuals83. Bayesian phylogenetic methods can be slow for genome-scale data, but parallel computing approaches can help with this issue83.
Recombination in bacteria may be frequent, occur at rates that vary among lineages, and have effects on sequence diversification (these effects may often be larger than those of mutation)78, 84. Ignoring the effect of recombination can therefore impair phylogenetic reconstruction85. Furthermore, understanding the recombination process itself is often informative about ecological28 or pathological86 properties of bacterial lineages. An intuitive approach is to detect recombinant fragments and account for them during phylogenetic reconstruction87. It is possible to do this formally by expanding Bayesian phylogenetic methods to include a model of recombination, for example as implemented in ClonalFrame86 and ClonalOrigin78.
A phylogenetic tree is not a direct reflection of transmission events88, but it can still be informative about the way they occurred89. In this context, an important first step is to estimate the molecular clock (the rate of molecular substitution) in order to re-scale the tree in units of time. Such a clock rate can be estimated from longitudinal samples from a single infected individual90, 91; it can be estimated jointly with the phylogeny in BEAST48 or it can be estimated from the reconstructed tree by exploiting the correlation between tree root-to-tip distances and year of isolation32, 87. Such estimates are only reliable if the range of sampling dates is significant (typically at least 10 years) compared to the time to the most recent common ancestor. The table contains estimates of molecular clock rates for a variety of bacterial pathogens. Although these vary substantially, they are all of the order of one mutation per year per genome. Once a molecular clock is estimated, the common ancestors on the phylogenetic tree can be dated, so that epidemiological interpretations of microevolution become possible, which are in turn informative about patterns of transmission at a larger scale.
| Pathogen | Mutations per site per year | Mutations per genome per year | References |
|---|---|---|---|
| Staphylococcus aureus | 3.0x10-6 | 8.4 | 43,48 |
| Clostridium difficile | 5.3x10-7 | 2.3 | a |
| Mycobacterium tuberculosis | 1.1x10-7 | 0.5 | 90 |
| Streptococcus pneumoniae | 1.6x10-6 | 3.5 | 87 |
| Helicobacter pylori | 1.9x10-5 | 30.4 | 91 |
| Vibrio cholerae | 8.3x10-7 | 3.3 | 32 |
| Escherichia coli | 2.26x10−7 | 1.1 | 92 |
Didelot, X. et al. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. submitted (2012)
A ribosomal multilocus sequence typing (rMLST) scheme has recently been proposed23 that relies of the sequences of 53 genes encoding ribosomal proteins, which are present in all bacteria. Acquiring the sequences of such a large number of genes is best done by first sequencing the whole genome and then extracting individual genes, for example using BLAST24. The BIGSdb database system is an integrated platform that enables users to find many genes in many genomes using BLAST and to record the results for future use25. More than 1,900 bacterial genomes from 452 bacterial genera have been analysed using the rMLST scheme23. Any newly sequenced genome can easily be added to the database, have its ribosomal genes extracted and its phylogenetic relationships with other genomes assessed. In a separate effort, a new method has recently been developed that allows the automatic in silico application to any genome sequence of the MLST schemes of 66 distinct species based on hundreds of genes, thus potentially revealing both the species to which the genome belongs and its sequence type within the relevant MLST scheme26.
With further development, these comparative approaches could reach the level required to replicate current species identification procedures with high precision. As this is progressively being achieved, our definitions of bacterial species will probably need refining to reflect new accumulated knowledge based on sequence comparison. Indeed it has already been shown that sequence data, even at the level of fractional sequencing (e.g. MLST), is robust at differentiating Streptococcus pneumoniae or Campylobacter jejuni from closely related species27, 28. On the other hand, it has also revealed that some named species do not represent monophyletic units of diversity, for example in the case of Bacillus cereus and B. thuringiensis20. While the increased statistical power of having the whole genomic sequence data considerably improves the precision of such analysis for differentiating all species, it will probably also reveal more ambiguity at the boundaries of currently defined species than has already been recognised from fractional sequencing28, 29. Such findings are likely to give impetus to a reconsideration of the notion of bacterial species, eventually leading to great simplification and clarity to the early steps in diagnostic clinical microbiology. For example, a genomic criterion for species definition has been proposed whereby two isolates belong to the same species if their average nucleotide identity is at least 95%22 and this was shown to closely replicate current definitions based on DNA-DNA hybridization tests30.
Several challenges remain to be overcome before routine species identification via whole genome sequencing can become a reality for most pathogens. This includes achieving a turn-around time approaching hours for sequencing and analysing the isolate data. This will depend on new rapid sequencing (Box 1), new assembly techniques (Box 2), new phylogenetic techniques (Box 3) and developing software and databases able to store very large numbers of genomes (Figure 2). Software packages will need to be user-friendly and yield clinically meaningful results. Quality control procedures will need to be developed as well as criteria for run success, software validation, and proficiency testing for laboratories. Prior to deployment as a diagnostic system, a detailed clinical evaluation will be needed including a comparison with currently used methods.
Box 2 Assembly and alignment techniques.
High-throughput sequencing techniques produce many short (30 to 100bp) overlapping reads from the target genome. The first task of any analysis is therefore to assemble these reads into larger parts of the genome 63. A first approach to do so is called “reference based assembly” and consists of comparing the reads to a previously sequenced “reference genome” in order to determine where they fit. Maq64 and STAMPY65 are two popular software packages to perform such assembly. Once reads have been mapped to the reference genome, positions that differ can be found, for example using SAMtool66. A first obvious drawback of this approach is that any element absent from the reference genome will not be assembled. A second difficulty is that the ability to map reads accurately to the reference genome decreases with the genetic distance between target and reference genomes. A closely related reference genome is therefore needed to accurately assemble the target genome. Furthermore, when several genomes are assembled using the same reference, the genomes more closely related to the reference will be better assembled, which can introduce significant biases is downstream analysis.
For these reasons, there is growing interest in assembling genomes in a reference-free manner, a task often called “de-novo assembly” and performed for example by the programs Newbler67 or Velvet68. With new high-throughput sequencing techniques that can produce longer reads (for example, the Pacific Biosciences platform and, in the future, the Oxford Nanopore platform), individual reads contain larger overlapping regions so that it is easier to see how they fit with each other along the target genome. De-novo assemblies do not have the two difficulties described above for reference based assembly: the whole of the genome is assembled, and the quality of the assembly does not depend on the choice of reference. De-novo assembly however suffers from the fact that it results in tens or hundreds of contigs representing different segments of the genome. Further assembly of these contigs into a complete genome is typically made impossible by the presence of repetitive elements, for which reads from separate elements can have high levels of homology.
When genomes are assembled de novo, they need to be aligned before they can be compared. Alignment of bacterial de novo genome assemblies is complicated by rearrangements that have destroyed the colinearity of the genomes69. The computer package Mauve has been designed to align whole genomes, accounting for rearrangements70, 71. However it is limited in the number of genomes it can align simultaneously (in our experience up to 20-40 genomes, depending on their diversity). A solution is to align the genomes in a pairwise fashion to a reference, but this raises the same difficulties as described above for reference-based assembly.
An alternative that is useful for most practical purposes is to take a gene-by-gene approach. For example genes can be retrieved from genomes using BLAST24. This gene-querying approach is useful when genes of interest are known in advance, for example when performing species identification based on 16S rDNA72, or assessing the presence of known genetic markers of resistance73 or virulence74. A full description of genetic content of the genomes may however require automatic annotation. This can be performed by Glimmer75 or one of the several pipelines based on this program, such as xBASE76 or DIYA77.
Testing for antibiotic resistance
In principle, it should be possible to predict resistance phenotypes by identifying genetic determinants of antimicrobial resistance and thus enable rapid antibiotic treatment decision making. Currently there are a few examples (including from S. aureus31, Vibrio cholerae32 and Burkholderia dolosa33) in which genetic determinants of antimicrobial resistance identified from whole genome data are consistent with recorded variation in phenotype. This early data suggests that a sequence-based approach holds substantial promise. Indeed, a few methods for predicting antibiotic resistance from genetic rather than phenotypic data are already widely used: for example, the detection by PCR of mecA, which confers methicillin resistance in S. aureus34, and sequences known to encode resistance to isoniazid, rifampicin, ethambutol, aminoglycosides, capreomycin and fluoroqinolones in M. tuberculosis (known as the MTBDR35). In principle, whole genome data could improve these tests, as computational querying of the sequence may be more sensitive than using PCR primers and it would be easier to search for more determinants.
Several challenges need to be overcome to achieve clinical adoption of whole genome sequencing in resistance prediction. First, a comprehensive set of genetic determinants of antimicrobial resistance would need to be identified for each species. Such genetic determinants include: genes whose presence confers resistance (such as TEM β-lactamase36); point mutations in essential genes (such as in rpoB, which confers rifampicin resistance37); and changes in expression of genes (for example reversion in the mutant operator sequences of E. coli ampC which leads to an increase in β-lactamase expression38). Importantly, even where resistance determinants are well characterized, others may be revealed by further research39. Furthermore, new mechanisms of antimicrobial resistance arise all too frequently: recent examples include quinolone resistance in S. typhi16, NDM β-lactamase in Enterobacteriaciae40 and multi-resistance in Neisseria gonorrhoeae41. Therefore, compiling a list of genetic determinants of resistance would be an on-going task.
The sequence details of these determinants would need to be incorporated in a database that is kept up-to-date (to include novel resistance determinants) and allows international data exchange, via for example CDC Atlanta and ECDC Stockholm. Such a database would also facilitate the identification and reporting of trends in resistance and new acquisition of resistance genes from other species. Predictions about resistance and susceptibility from sequence data need to be accurate: falsely inferring susceptibility where the isolate is resistant represents a substantial risk to the patient. Therefore, performance needs to be established to high degrees of confidence in robust and well powered clinical studies before deployment in a regulated environment. For example, in the UK this would require Clinical Pathology Accreditation and in the USA this would require approval from the Federal Food and Drug Administration.
Therefore, although sequence data has the potential to support fast and cheap identification of resistance, we envisage a two-pronged approach that combines on-going comparison of clinical outcome data with genetic data and phenotypic resistance screening. For example, on-going phenotypic testing will be needed to identify new resistance and to keep the proposed database up-to-date.
Detecting virulence determinants
The genetic basis of many recognised virulence phenotypes is known and yet our understanding of virulence factors is incomplete. The genome sequence of an isolate could yield information on all the known virulence factors in one step and create the opportunity for the discovery of new virulence factors through association studies that link the isolate genomic data with patient disease manifestation and outcome data. One early example of a finding from such an association study was the discovery of a prophage associated with whether Neisseria meningitidis causes meningitis42. Another example is the finding that non-synonymous mutations in specific genes in S. aureus occurred just before the development of invasive disease43 .
More recently, whole genome sequencing of isolates from major outbreaks has demonstrated the potential for identifying recognised virulence genes and pathogenicity gene clusters and for providing new understanding of virulence factors. For example, the recent analysis of whole genome data from E. coli O 10444, 45 illustrated the speed and precision of whole genome sequencing. Draft sequencing took three days using the IonTorrent PGM27and the first assembly was released two days later28, 29. Within a week of data becoming available, the strain was shown to be a novel E. coli O104:H4 variant that had acquired a prophage encoding Shiga toxin 2 and additional virulence and antibiotic-resistance determinants45. Similarly, sequencing of isolates from the 2010 Haitian Vibrio cholerae outbreak was claimed to be achievable in less than a day using the PacBio system34 and sequence analysis allowed the detection and characterization of a toxin encoded by the CTX phage46.
Similar to the situation for antimicrobial resistance, identifying virulence determinants from analysis of whole genomic sequences is at early stage and substantial challenges need to be overcome before implementing this approach in a routine service environment. In particular, it requires the development of a database that includes all known virulence determinants and can incorporate new determinants. New software is needed to analyse genome sequences for the presence and absence of known virulence determinants as well as conducting on-going association studies as described for antimicrobial resistance. The requirement for high sensitivity is generally lower for identifying virulence factors than for antimicrobial resistance, as identifying virulence only has major clinical consequences in a few cases.
Outbreak detection and surveillance
Genome sequences potentially provide a high resolution, accurate and reproducible means for relating organisms. For example, sequencing the genomes of a diverse collection of Chlamydia trachomatis isolates has demonstrated the limitations of current clinical typing techniques for identifying phylogenetic relationships47. Compelling examples of the effectiveness of whole genome analyses for unravelling the origins and dispersal of pathogens at regional and global scales have recently been published. This approach was used to investigate the emergence and global dispersal of ST239 isolates of meticillin resistant S. aureus48. In another example, the emergence of serotype 19A pneumococcal capsular variants, following the introduction in the USA of a pneumococcal vaccine, was documented and its spread tracked across the USA49. A comparative study of 154 whole genomes of Vibrio cholerae enabled the history of pandemic cholera over the last fifty years to be compiled, revealing that the seventh and current cholera pandemic has comprised three successive, partially overlapping waves with strong geographical and temporal structure32. In M. leprae, genome sequencing of isolates from 50 patients and 33 wild armadillos showed that these animals represent a major source of zoonotic transmission of leprosy in the southern United States50. In a previous study, the spread of M. leprae was shown to follow human migration and historical trade routes51. Finally, a comparison of 17 whole genomes and SNP typing in 286 globally representative isolates established strong geographical clustering in Y. pestis compatible with a Chinese origin for the Black Death pandemic52.
Early reports also strongly suggest that using sequencing to detect outbreaks that include person-to-person transmission within communities and hospitals is a major benefit to health-care; this has been recently illustrated for S. aureus and C. difficile using rapid bench-top sequencing53, 54. A report on using whole genome sequencing to study a TB outbreak on Vancouver Island55 suggested that genealogical analysis of whole genomic sequences could be a major advance for TB contact tracing, compared to the current cumbersome approaches. The current approaches depend heavily on identifying transmission networks through interviews, supplemented by M. tuberculosis-specific multiple locus Variable Number of Tandem Repeats (MIRU VNTR) typing56, which is less discriminatory than whole genome sequencing. Similar observations have been reported for a subset of MRSA isolates cultured from a hospital in Thailand, suggesting that phylogenetic analysis could be used to infer local hospital transmission48. The previously discussed studies of V. cholerae46 and shigatoxin-producing E. coli O 10444, 45 indicate that sequencing can also rapidly provide a clear understanding of the origins of a local outbreak.
Whole genome sequencing is becoming the method of choice in research settings for monitoring pathogens over long time courses and wide geographical scales, as well as for identifying outbreaks. Sequence data gathered for diagnostic purposes can be accumulated for pathogen surveillance, outbreak detection and evolutionary studies. In principle, detection of an outbreak could occur as early as the first secondary case. Consequently, deployment of sequencing technology for diagnostic purposes in local laboratories would also meet the needs for surveillance, as long as the genome sequences can be linked with the epidemiological information. To be fully useful, data would have to be shared locally, nationally and internationally: new integrated approaches to jointly store epidemiological and genomic data are under development57. It can be expected that national reference laboratories will adopt whole genome sequencing as a single technology for typing all pathogens - replacing many species-specific typing methods - even if this is not done in the near future in routine diagnostic laboratories. A number of agencies including the Public Health England (Health Protection Agency), England, UK, are exploring adoption of whole genomic sequencing, initially to supplement current methods for typing high value pathogens with the intention of implementing this approach more widely as the preferred typing method for outbreak investigation and pathogen surveillance.
Future directions
Clinical microbiology is on the threshold of incorporating genome sequencing into routine practice. Although this review focuses on the promise of this technology for bacterial pathogens, there is also rapid progress towards its adoption for viral, fungal and parasitic pathogen diagnostics and surveillance. The potential advantages of sequencing as a primary technology, and the requirement for robust evaluation, have been set out in this review.
It is likely that commercial developments based on sequencing technologies will focus on steps in current processing of cultured isolates that are discrete, high-cost and high value. An example where adoption may occur soon is in the analysis of mycobacterial cultures. Whole genome sequence is likely to soon provide, at a lower cost, all of the information provided currently by the MTBDR test(s)35 and also more details about species identification and resistance determinants. Similarly, sequencing could yield, at little additional cost, more definitive typing information than MIRU VNTR testing. As discussed above, another setting where adoption of whole genome sequencing has already started is the investigation of putative outbreaks of major pathogens.
In this Review, we have focused on cases in which the pathogen has been cultured, but there is also potential for sequencing without culturing, that is, to sequence the entire DNA in a sample (e.g. pus, cerebrospinal fluid, sputum). Such a metagenomics approach has been used to define the microbiomes of diverse samples and environments58, 59. Approaches such as bioinformatically masking the human sequences then assembling pathogen genomes de novo, or mapping reads to a reference genome from the hypothesized pathogen, are likely to be useful, subject to the availability of sufficient data to overcome the relatively low proportion of pathogen DNA in a clinical sample. In samples where pathogen cell counts are very low, such as M. tuberculosis present among many other organisms in sputum or the blood of a bacteraemic patient with 1-100 acterial colony forming units /ml, recovering complete bacterial genome sequences may depend on very cheap, fast sequencing or enhanced methods to deplete background material. New very fast single-molecule long-read sequencing approaches (Box 1) should make it possible to sequence at great depth and low cost.
Adopting whole pathogen sequencing would require major changes in the organisation, skill mix and infrastructure of diagnostic laboratories and would therefore be disruptive, even in if the main use of sequencing were after culture of the pathogen. Areas for focus will be strengthening competence in bioinformatics and software development. Advances are required in databases, efficient software and algorithms for analysis, software that automatically updates knowledge-bases, and sophisticated links between pathogen genomics databases and patient clinical record systems. To ensure the benefits are accessible to the wider community, especially where a number of providers (commercial or otherwise) are developing systems, information needs to be shared in line with agreed standards. The opportunities for global surveillance of infectious diseases are vast, but political resolve is required to enable the sharing of sequence and meta-data on a global scale.
Supplementary Material
Online summary.
Whole genome sequencing of bacterial isolates is becoming more and more widespread, paving the way for a transformation of many current procedures in clinical microbiology.
Identifying the species of an isolate is currently a very complex laboratory process. A few methods have already been proposed for doing this based on the genome sequence, which could result in a re-evaluation of the bacterial species concept.
Testing antibiotics resistance properties is often crucial to determine appropriate treatment. Since resistance is encoded by specific genes, this susceptibility assessment could be performed in silico based on the genome sequence.
The same is true about determining virulence properties of a strain, with the difference that correlations between genotype and phenotype is often more complex (involving several genes) than for resistance. Association mapping techniques can however be used to detect such complex correlations, leading to a better understanding of pathogenicity.
Several studies have already demonstrated the great potential of whole genome sequencing in epidemiological investigations. These have so far been performed after the course of an outbreak, but with improving technology could be carried on an on-going basis to detect epidemiological risks as they arise and react accordingly.
Bacteria culturing is a pre-requirement even for whole-genome sequencing as currently performed. This represents an important bottleneck since some bacteria are slow-growing while others can not be cultured, but metagenomics approaches could provide a solution to this long-standing issue.
Acknowledgements
DWC and TAEP are part funded by the NIHR Oxford Biomedical Research Centre and are NIHR Senior Investigators. XD, DJW and RB are funded by the UK CRC. We thank Dr John Paul for helpful advice and suggestions.
Glossary
- Escherichia coli
A common inhabitant of the guts of many animals, but some strains can cause serious food poisoning as reminded by the 2011 outbreak in Germany.
- Mycobacterium tuberculosis
The causative agent of tuberculosis (TB), it infects about a third of the human population and claims over a million lives per year, making it the most deadly bacterial pathogens of humans.
- Staphylococcus aureus
Found as a harmless colonizer of the skin of around 20% of the human population, it can cause life-threatening symptoms and be resistant to some antibiotics (eg Methicilin Resistant Staphylococcus aureus aka MRSA)
- Staphylococcus epidermidis
A normal part of the human skin flora, it can become pathogenic if introduced into deeper tissues following surgery.
- Mycobacterium bovis
The causative agent of bovine TB, it is a close relative of Mycobacterium tuberculosis and can occasionally cause tuberculosis in humans as well.
- Salmonella Typhi
All Salmonella cause disease, but the Typhi lineage is the main causative agent of typhoid fever, which claim hundreds of thousands of lives per annum.
- Haemophilus influenzae
Responsible for a wide range of clinical diseases (but not the flu as originally thought and the name might still suggest) especially in young children, it was the first free living organism to have its genome completely sequenced.
- Streptococcus pneumoniae
A major cause pneumonia, it can also cause a variety of other severe conditions, and has recently developed resistance to some antibiotics. It causes around a million deaths per year, mostly in children.
- Neisseria meningitidis
A commensal inhabitant of the nasopharynx in up to a quarter of the human population, it occasionally get into the blood resulting in over a hundred thousand deaths per year through meningitis and septicaemia.
- Clostridium difficile
A leading cause of diarrhoea and more severe conditions, especially in the elderly following disruption of the normal gut flora through the use of antibiotics.
- Corynebacterium diphtheriae
The causative agent of diphtheria, a respiratory illness which has been mostly eradicated in developed countries through vaccination but has resurged in recent years in Eastern Europe.
- Bacillus cereus and thuringiensis
These bacteria live predominantly in the soil, but can occasionally infect humans, causing food poisoning with symptoms of vomiting and diarrhoea.
- Campylobacter jejuni
A natural colonizer of the digestive tracts of many birds and cattle, it is typically transmitted to humans by ingestion of contaminated food and results in severe diarrheal diseases.
- Vibrio cholerae
The agent of cholera is transmitted via contaminated waters, and can cause death through dehydration. It caused millions of deaths in Europe in the 19th century, but has since mostly disappeared from industrialised countries. It still claims over a hundred thousand lives per annum in developping countries.
- Chlamydia trachomatis
The cause of over a hundred million sexually transmitted infections annually, as well as trachoma, an infection of the eye that can result in blindness.
- Mycobacterium leprae
The causative agent of leprosy, which has affected humanity for thousands of years but is now almost eradicated.
Biographies
Author biographies
Xavier Didelot
Xavier Didelot received his D.Phil. in statistical genetics from the University of Oxford in 2007. He was a research fellow at the University of Warwick for three years before moving back to Oxford to work on the Modernising Medical Microbiology (MMM) project. He is perhaps best known as the author of ClonalFrame, software that reconstructs the genealogy of a sample of bacteria while accounting for the confounding effect of recombination.
Rory Bowden
Rory Bowden received his PhD in Molecular Virology from the University of Cambridge, with post-doctoral training in virus population genetics and evolution at Oxford and Glasgow. His recent research has been in practical aspects of the genomics of a wide range of organisms from bacteria to chimpanzees. He has recently been appointed Deputy Head of High-Throughput Genomics at the Wellcome Trust Centre for Human Genetics at the University of Oxford.
Daniel Wilson
Daniel J. Wilson read biological sciences before completing a D.Phil. in pathogen population genetics at the University of Oxford in 2005. He was a postdoctoral research associate at Lancaster University before moving to the University of Chicago as a postdoctoral research scholar. In 2010 he returned to the University of Oxford as a research fellow in pathogen population genomics. His laboratory focuses on understanding the evolution and epidemiology of bacterial and viral pathogens such as Staphylococcus aureus and norovirus through whole genome analysis.
Tim Peto
Tim Peto read for a DPhil and qualified in Medicine at the University of Oxford. He is an infectious diseases physician and Professor of Medicine at the Oxford University Hospitals and a NIHR Senior Investigator. As a clinical epidemiologist and clinical trialist for over 20 years, he has been overseeing key large scale trials in HIV, malaria and TB. In the last 5 years, he has been studying the use of whole genome sequencing for tracking common pathogens in both hospitals and the community.
Derrick Crook
Derrick Crook qualified in Medicine at the University of Witwatersrand South Africa, studied at the London School of Tropical Medicine and trained in the Department of Medicine, University of Virginia and the Department of Infectious Diseases, New England Medical Centre, Boston. He is a clinical microbiologist, infectious diseases physician, Professor of Microbiology at the Oxford University Hospitals and a NIHR Senior Investigator. He is the infection control doctor for all the Oxford hospitals. His research encompasses diagnostics, epidemiology, new sequencing and informatics technologies aimed at improving management of infectious diseases.
Footnotes
Weblinks
The website of the Modernising Medical Microbiology project is accessible at http://www.modmedmicro.ac.uk/
References
- 1.Burlage RS. Principles of public health microbiology. Jones & Bartlett Learning; Sudbury, MA: 2012. [Google Scholar]
- 2.Relman DA. Microbial genomics and infectious diseases. N Engl J Med. 2011;365:347–57. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkhill J, Wren BW. Bacterial epidemiology and biology - lessons from genome sequencing. Genome Biol. 2011;12:230. doi: 10.1186/gb-2011-12-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandell GL, Bennett JE, Dolin R. Mandell, Douglas, and Bennett's principles and practice of infectious diseases. Churchill Livingstone/Elsevier; Philadelphia, PA: 2010. [Google Scholar]
- 5.Murray PR, Rosenthal KS, Pfaller MA. Medical microbiology. Mosby/Elsevier; Philadelphia: 2009. [Google Scholar]
- 6.Warrell DA, Cox TM, Firth JD. Oxford textbook of medicine. Oxford University Press; Oxford; New York: 2010. [Google Scholar]
- 7.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–4. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarridge JE., 3rd Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–62. doi: 10.1128/CMR.17.4.840-862.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seng P, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–51. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 10.van Veen SQ, Claas EC, Kuijper EJ. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol. 2010;48:900–7. doi: 10.1128/JCM.02071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherkaoui A, et al. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol. 2010;48:1169–75. doi: 10.1128/JCM.01881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaillot O, et al. Cost-effectiveness of switch to matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine bacterial identification. J Clin Microbiol. 2011;49:4412. doi: 10.1128/JCM.05429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson LG, Drake SK, Murray PR. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010;48:444–7. doi: 10.1128/JCM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shitikov E, et al. Mass spectrometry based methods for the discrimination and typing of mycobacteria. Infect Genet Evol. 2012;12:838–45. doi: 10.1016/j.meegid.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Lorian V. Antibiotics in laboratory medicine. Lippincott Williams & Wilkins; Philadelphia, PA: 2005. [Google Scholar]
- 16.Wain J, et al. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis. 1997;25:1404–10. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 17.Cavaco LM, Hasman H, Xia S, Aarestrup FM. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob Agents Chemother. 2009;53:603–8. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bode LG, van Wunnik P, Vaessen N, Savelkoul PH, Smeets LC. Rapid detection of methicillin-resistant Staphylococcus aureus in screening samples by relative quantification between the mecA gene and the SA442 gene. J Microbiol Methods. 2012;89:129–32. doi: 10.1016/j.mimet.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 19.Cosgrove SE, et al. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003;36:53–9. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 20.Priest FG, Barker M, Baillie LW, Holmes EC, Maiden MC. Population structure and evolution of the Bacillus cereus group. J Bacteriol. 2004;186:7959–70. doi: 10.1128/JB.186.23.7959-7970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–88. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 22.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 2005;102:2567–72. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolley KA, et al. Ribosomal Multi-Locus Sequence Typing: universal characterisation of bacteria from domain to strain. Microbiology. 2012 doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen MV, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–61. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothberg JM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–52. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 28.Suerbaum S. No tech gaps in E. coli outbreak. Nature. 2011;476:33. doi: 10.1038/476033a. [DOI] [PubMed] [Google Scholar]
- 29.Mellmann A, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 31.McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS ONE. 2011;6:e24301. doi: 10.1371/journal.pone.0024301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011 doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korlach J, et al. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 2010;472:431–55. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- 35.MTBDR G. Hain Lifescience; Nehren, Germany: [Google Scholar]
- 36.Livermore DM. beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–84. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boehme CC, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caroff N, Espaze E, Gautreau D, Richet H, Reynaud A. Analysis of the effects of -42 and -32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing ampC. J Antimicrob Chemother. 2000;45:783–8. doi: 10.1093/jac/45.6.783. [DOI] [PubMed] [Google Scholar]
- 39.Devasia R, et al. High proportion of fluoroquinolone-resistant Mycobacterium tuberculosis isolates with novel gyrase polymorphisms and a gyrA region associated with fluoroquinolone susceptibility. J Clin Microbiol. 2012;50:1390–6. doi: 10.1128/JCM.05286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–62. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 41.Bolan GA, Sparling PF, Wasserheit JN. The emerging threat of untreatable gonococcal infection. N Engl J Med. 2012;366:485–7. doi: 10.1056/NEJMp1112456. [DOI] [PubMed] [Google Scholar]
- 42.Bille E, et al. A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med. 2005;201:1905–13. doi: 10.1084/jem.20050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young BC, et al. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A. 2012;109:4550–5. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohde H, et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med. 2011;365:718–24. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 45.Rasko DA, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–17. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chin CS, et al. The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris SR, et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 2012;44:413–9. doi: 10.1038/ng.2214. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golubchik T, et al. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet. 2012 doi: 10.1038/ng.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Truman RW, et al. Probable zoonotic leprosy in the southern United States. N Engl J Med. 2011;364:1626–33. doi: 10.1056/NEJMoa1010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monot M, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41:1282–9. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- 52.Morelli G, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42:1140–3. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koser CU, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012;366:2267–75. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eyre DW, et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open. 2012;2 doi: 10.1136/bmjopen-2012-001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gardy JL, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–9. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 56.Cardoso Oelemann M, et al. The forest behind the tree: phylogenetic exploration of a dominant Mycobacterium tuberculosis strain lineage from a high tuberculosis burden country. PLoS ONE. 2011;6:e18256. doi: 10.1371/journal.pone.0018256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aanensen DM, Huntley DM, Feil EJ, al-Own F, Spratt BG. EpiCollect: linking smartphones to web applications for epidemiology, ecology and community data collection. PLoS One. 2009;4:e6968. doi: 10.1371/journal.pone.0006968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuczynski J, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2012;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:562. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 62.Check Hayden E. Nature. 2012 [Google Scholar]
- 63.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12:443–51. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–9. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–30. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darling AE, Miklos I, Ragan MA. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 2008;4:e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yarza P, et al. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–50. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 73.Liu B, Pop M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37:D443–7. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu H-J, Wang AHJ, Jennings MP. Discovery of virulence factors of pathogenic bacteria. Current Opinion in Chemical Biology. 2008;12:93–101. doi: 10.1016/j.cbpa.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 75.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaudhuri RR, et al. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 2008;36:D543–6. doi: 10.1093/nar/gkm928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stewart AC, Osborne B, Read TD. DIYA: a bacterial annotation pipeline for any genomics lab. Bioinformatics. 2009;25:962–3. doi: 10.1093/bioinformatics/btp097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Didelot X. Bacterial Population Genetics in Infectious Disease 37-60. John Wiley & Sons, Inc; 2010. [Google Scholar]
- 79.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rodrigo AG, et al. Coalescent estimates of HIV-1 generation time in vivo. Proc Natl Acad Sci U S A. 1999;96:2187–91. doi: 10.1073/pnas.96.5.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–92. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 82.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 85.Schierup MH, Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–91. doi: 10.1093/genetics/156.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Didelot X, Achtman M, Parkhill J, Thomson NR, Falush D. A bimodal pattern of relatedness between the Salmonella Paratyphi A and Typhi genomes: convergence or divergence by homologous recombination? Genome Res. 2007;17:61–8. doi: 10.1101/gr.5512906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–4. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10:540–50. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cottam EM, et al. Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007. PLoS Pathog. 2008;4:e1000050. doi: 10.1371/journal.ppat.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ford CB, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43:482–6. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kennemann L, et al. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci U S A. 2011;108:5033–8. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reeves PR, et al. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS ONE. 2011;6:e26907. doi: 10.1371/journal.pone.0026907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Highlighted References
- •.Konstantinidis, K.T. & Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102, 2567-72 (2005).; [First description of a computation criteria to define bacterial species based on whole-genome sequencing.] [Google Scholar]
- •.Jolley, K.A. & Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010).; [A database system for whole genomes that provides a smooth transition for users from working with MLST to working with genomes.] [Google Scholar]
- •.Bille, E. et al. A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med 201, 1905-13 (2005).; [First example of an association mapping study to determine virulence factors in Neisseria meningitidis.] [Google Scholar]
- •.Young, B.C. et al. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A 109, 4550-5 (2012).; [A detailed investigation of Staphylococcus aureus within-host genomic diversification in time revealing a probable evolution towards increased virulence.] [Google Scholar]
- •.Rasko, D.A. et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365, 709-17 (2011).; [Epidemiological investigation based on whole-genome sequencing for the 2011 German outbreak of Escherichia coli.] [Google Scholar]
- •.Chin, C.S. et al. The origin of the Haitian cholera outbreak strain. N Engl J Med 364, 33-42 (2011).; [A study of the origin of the on-going Haitian outbreak of Vibrio cholerae based on whole-genome comparison with other strains.] [Google Scholar]
- •.Harris, S.R. et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet 44, 413-9, S1 (2012).; [An example of how current typing techniques can be misleading compared to whole-genome sequencing.] [Google Scholar]
- •.Harris, S.R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469-74 (2010).; [One of the first studies to illustrate the great potential of whole-genome sequencing to reconstruct person-to-person transmission pathways within a hospital.] [Google Scholar]
- •.Golubchik, T. et al. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet (2012).; [An example of the great evolutionary potential of highly recombinogenic bacteria to escape epidemiological interventions.] [Google Scholar]
- •.Eyre, D.W. et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2 (2012).; [A demonstration of the usefulness of benchtop sequencing to answer epidemiological questions in near real-time.] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.