Abstract
Introduction
Palmitate, the typical end product released from fatty acid synthase, is of interest to many researchers performing metabolomics. Although palmitate can be readily detected by using mass spectrometry, many metabolomic platforms involve the use of plastic consumables that introduce a competing background signal of palmitate.
Objectives
The goal of this study was to quantify palmitate contamination in metabolomics and isotope tracer studies and to examine the reliability of approaches for reducing error.
Methods
We measured the quantitative error introduced by palmitate contamination from 4 vendors of plastic consumables used in combination with several different extraction solvents.
Results
The background palmitate signal was as much as sixfold higher than the biological palmitate signal from 4 million 3T3-L1 cells. Importantly, the palmitate contamination signal was highly variable between plastic consumables (even within the same lot) and therefore could not be accurately removed by subtracting the background as measured from a blank. In addition to affecting relative and absolute quantitation, the palmitate background signal from disposable plastics also led to the underestimation of labeled palmitate in isotope tracer experiments.
Conclusion
When measuring palmitate standard solutions, the best results were obtained when glass vials and glass pipettes were used. However, much of the palmitate background signal could be eliminated by pre-rinsing plastic vials and plastic pipette tips with methanol prior to sample introduction. For isotope tracer studies, error could also be minimized by estimating palmitate enrichment from palmitoylcarnitine, which does not have a competing contamination signal from plastic consumables.
Keywords: Isotope labeling, Lipidomics, Palmitate, Plastic contamination, Stearate
1 Introduction
Palmitate, a saturated fatty acid containing 16 carbons, is built from acetyl-CoA precursors as the major end product released from fatty acid synthase. Other fatty acids of varying length and unsaturation are derived from palmitate via elongation, oxidation, and/or desaturation reactions. Together, these fatty acids serve as building blocks for complex lipids that are used for storing energy, signaling, and structural components of cellular membranes.
Given its central position in metabolism, analysis of palmitate can provide important insight into a cell's biochemical state. Palmitate labeling has been used to assess reductive glutamine metabolism, acetyl-CoA labeling, fatty acid uptake, and redox homeostasis (Jiang et al. 2016; Kamphorst et al. 2014; Metallo et al. 2012; Yao et al. 2016). To investigate carbon sources that support lipid synthesis in rapidly proliferating cells, various labeled nutrients have been provided to cells and palmitate enrichment evaluated. All of these studies are similar in that their success relies upon accurately measuring the levels of labeled and unlabeled palmitate.
Lipidomic platforms are optimized to quantify hydrophobic molecules such as palmitate (Gross and Han 2011). The challenge of comprehensive metabolomic platforms is that they target both hydrophobic and hydrophilic molecules. Unlike lipidomic platforms, metabolomic platforms often use plastic microcentrifuge tubes and plastic pipette tips with organic solvents to solubilize lipids. This workflow is convenient for profiling tricarboxylic acid cycle and glycolytic intermediates, but the use of plastic consumables and organic solvents introduces the risk of sample contamination with palmitate and stearate (Tumanov et al. 2015). Fatty acids are often used as slip agents and lubricants during plastic production and therefore can be manufacturing residues (Lee et al. 2012; Olivieri et al. 2012). While it has been demonstrated that metabolomic analysis of biological samples with plastic consumables enables the detection of a palmitate signal, the quantitative reliability of the measurement has not been carefully assessed (Ivanisevic et al. 2013; Kurczy et al. 2015).
Here we show that conventional metabolomic platforms using plastic consumables can overestimate the level of unlabeled palmitate by 0.2–1.15 nmol per sample. When we performed an experiment with 4 million 3T3-L1 cells, this contamination signal led to the overestimation of unlabeled palmitate by as much as sixfold. We found that palmitate contamination could be significantly minimized by washing plastic consumables with methanol prior to the introduction of sample.
2 Materials and methods
2.1 Palmitate quantitation
A stock solution was made by dissolving 3.29 μmol of U-13C palmitate (Cambridge Isotopes) in 50.0 mL of LC-grade methanol (Sigma Aldrich) and stored at −20 °C in an LC-grade glass vial (Supelco). As an internal control, various aliquots of the stock solution were mixed with extraction solvents prior to sample introduction. Extracted samples were separated with a Luna aminopropyl column (3 μm, 150 × 1.0 mm I.D., Phenomenex) and analyzed by an Agilent 6540 QTOF as previously described (Mahieu et al. 2015). A hydrophilic interaction liquid chromatography separation was used instead of a reversed-phase separation to minimize carry over. Absolute concentrations of palmitate were determined by calculating the ratio of the fully unlabeled peak of samples to the fully labeled peak of the internal standard.
2.2 Plasticware and glassware
We tested polypropylene microcentrifuge tubes from three vendors (Eppendorf, Fisherbrand, and VWR) and polypropylene 1000 μL pipette tips from three vendors (Fisherbrand, VWR, and Thermo) to evaluate palmitate and stearate contamination. We compared these results to identical experiments performed with borosilicate glass culture tubes (VWR) and borosilicate glass serological pipettes (Fisherbrand and VWR). We note that in experiments performed with plastic microcentrifuge tubes and plastic pipettes, all other sample-handling steps involved glass unless otherwise indicated.
2.3 Quantifying the contamination from plasticware
We first aimed to quantify the amount of palmitate detected in “blank” preparations. Here we define a blank as a sample that goes through the extraction procedure, but no biological material is ever introduced. We tested select steps of the extraction procedure separately. First, we measured the palmitate contamination signal arising from pipetting 0.50 mL of LC-grade methanol repetitively with a single plastic pipette tip 30 times. All steps of this experiment other than pipetting involved glass. Second, we measured the palmitate contamination signal from vortexing and sonication. We tested three LC-grade extraction solvents: water (Milli-Q), acetonitrile (Sigma), and methanol. A 1.00 mL aliquot of each was added to plastic microcentrifuge tubes and then each tube was vortexed for 30 s before being bath sonicated for 30 min. All other steps of the experiment were performed with glass. Finally, we tested the effects of storing methanol in plastic microcentrifuge tubes for various periods of time, which often occurs in metabolite-extraction protocols. We transferred 1.00 mL of methanol to Fisherbrand microcentrifuge tubes and then stored them at −20 °C for 24 h, 48 h, or 120 h. After the incubation period, samples were transferred to glass vials and analyzed by liquid chromatography/mass spectrometry (LC/MS). All other steps of the experiment involved glass. For comparison, we transferred 1.00 mL of methanol to glass vials stored at −20 °C for 120 h and analyzed them by LC/MS.
2.4 Quantifying error in measurements of palmitate stock solutions
Stock solutions were prepared by conjugating palmitate to bovine serum albumin (BSA) as previously described (BSA:palmitate, 1:6 molar ratio) (Yao et al. 2016). Briefly, 2.00 mM palmitate (Sigma) was conjugated to 0.333 mM fatty acid free BSA (Sigma) in 150 mM NaCl solution at 37 °C for 1 h. The resulting BSA-palmitate solution was serial diluted with water to 100, 40.0, 20.0, 10.0, and 0 μM (water only). Aliquots of 50.0 μL of each BSA-palmitate solution were added to 150 μL of methanol: acetonitrile (2:1) extraction solvent. The mixtures were vortexed for 30 s, bath sonicated for 30 min, incubated at −20 °C for 2 h, centrifuged, and the supernatant was analyzed by LC/MS. Quantitation was accomplished by using an internal U-13C palmitate standard as described above. While pipetting 50 and 100 μL involved the use of a 200 μL plastic pipette tip (VWR) in both the plasticware and glassware experiments, we found that pipetting such small aliquots one time introduced minimal contamination (see Results).
2.5 Quantifying palmitate levels in FBS and 3T3-L1 cells
Fetal bovine serum (FBS, Gibco) was extracted by using the same method as for the BSA-palmitate solution. Four million fibroblasts (3T3-L1) were extracted with glassware by using a modified version of the previously described protocol (Ivanisevic et al. 2013). Here, instead of drying samples by using a vacuum concentrator as in the referenced protocol, we dried samples under nitrogen gas. Palmitate concentrations were determined by using an internal U-13C palmitate standard. Data were fit to a standard curve generated from glass extractions to account for error resulting from contamination and inefficient extraction.
2.6 Steady-state isotopic labeling and ISA
3T3-L1 cells were cultured in U-13C glucose for 48 h. Cells were washed with phosphate-buffer saline (PBS) twice prior to washing with water. Cells were then removed from the flask with a plastic cell scraper. The use of a plastic cell scraper with water minimized palmitate contamination. Half of the cells were transferred to cold methanol stored in a plastic microcentrifuge tube and the other half were transferred to cold methanol stored in a glass tube. Prior to LC/MS analysis, each sample was extracted by using either plasticware or glassware, respectively, as described above. Palmitoylcarnitine detection was accomplished in positive-ion mode with a reversed-phase C18 separation (Zorbax, 5 μm, 150 × 0.5 mm I.D, Agilent Technologies) as previously reported (Nikolskiy et al. 2013). The isotopologue distribution of palmitate and palmitoylcarnitine was corrected for the contribution of naturally occurring isotopes by IsoCor (Millard et al. 2012). Isotopomer spectral analysis (ISA) was performed by using the convISA algorithm (Tredwell and Keun 2015).
3 Results and discussion
The use of plasticware can introduce a competing background signal of palmitate and stearate that interferes with the ability to accurately measure endogenous palmitate and stearate in biological samples. These contaminating background signals lead to the overestimation of absolute concentration and the underestimation of isotope labeling in isotope tracer experiments. The latter is particularly relevant to palmitate labeling, which is commonly used to assess de novo lipid synthesis. Given that metabolomic platforms often use plastic microcentrifuge tubes and plastic pipette tips, the objective of this study was to quantify the error resulting from contamination. Although we quantified palmitate and stearate contamination by using LC/MS here, it is important to note that our results are not technology specific. We expect similar errors to be introduced from the use of plastic consumables in GC/MS- and NMR-based platforms.
3.1 Measuring contamination from plastic pipette tips
We repetitively pipetted LC-grade methanol 30 times with Fisherbrand, VWR, and Thermo Scientific plastic pipette tips. We then quantified the amount of palmitate and stearate in the methanol by using LC/MS. In this experiment, only the pipette tips were plastic. All other steps involved glass supplies. Additionally, we tested our column and mass spectrometer prior to each analysis to ensure that there was no palmitate or stearate signal due to carry over or any other unidentified source. The results show that each plastic pipette tip introduces considerable palmitate and stearate contamination that is comparable among the vendors tested (Table 1).
Table 1.
Quantifying palmitate and stearate contamination from 1 mL of methanol that was repeatedly pipetted 30 times with plastic pipette tips from the indicated vendors
| Fisherbrand | Thermo scientific | VWR | |
|---|---|---|---|
| Palmitate (nmol) | 0.09 ± 0.03 | 0.10 ± 0.02 | 0.3 ± 0.2 |
| Stearate (nmol) | 0.11 ± 0.02 | 0.11 ± 0.03 | 0.5 ± 0.3 |
Data are presented as mean ± SD (n = 3)
3.2 Measuring contamination from plastic microcentrifuge tubes
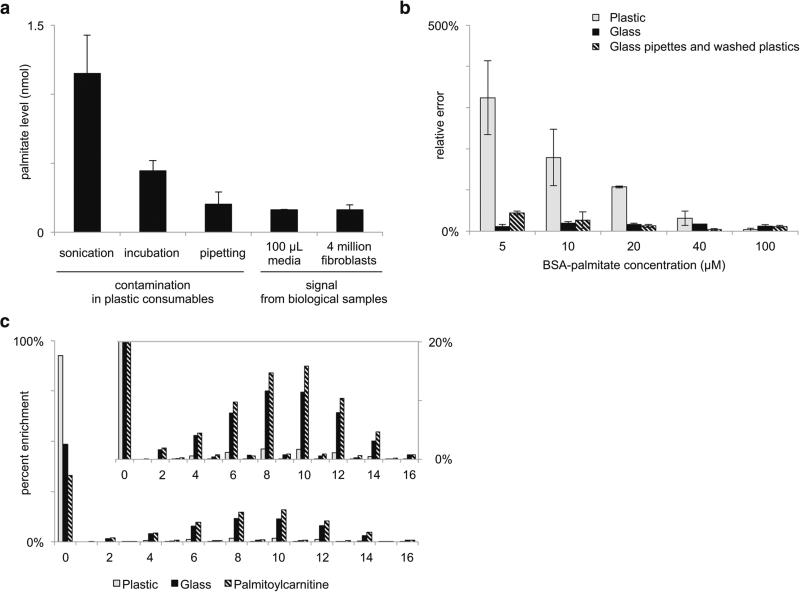
To evaluate the palmitate and stearate contamination introduced by plastic microcentrifuge tubes, we transferred 1.00 mL aliquots of water, acetonitrile, or methanol to Eppendorf, Fisherbrand, and VWR plastic microcentrifuge tubes. These solvents are commonly used in metabolomic workflows. After vortexing and bath sonicating, we then measured the amount of palmitate and stearate detected from each solvent. All other steps of the experiment involved glass, and we tested the column and mass spectrometer to ensure that there was no background palmitate or stearate signal due to carry over or any other unidentified source. Palmitate and stearate signals were not detected in the water preparations. In contrast, we detected significant signals for palmitate and stearate in acetonitrile and methanol preparations. Most likely these signal intensities are due to the different solubility of palmitate and stearate in each solvent (Stillwell 2013). Contamination signals from methanol preparations were generally about twice as large as those from acetonitrile preparations (Table 2). Results were comparable among the vendors tested. As shown in Fig. 1a, the intensity of the palmitate contamination signal from different sample-preparation techniques can be as high as sixfold greater than the intensity of the biological palmitate signal from 100 μL of 10 % FBS media or from 4 million extracted 3T3-L1 cells.
Table 2.
Quantifying palmitate and stearate contamination from 1 mL of the indicated solvents after they were vortexed and bath sonicated in plastic microcentrifuge tubes from each of the vendors listed
| Palmitate or stearate detected in solvents after sonication (nmol) |
|||
|---|---|---|---|
| Water | Acetonitrile | Methanol | |
| Palmitate: Eppendorf | not detectable | 0.15 ± 0.08 | 0.4 ± 0.2 |
| Palmitate: Fisherbrand | not detectable | 0.67 ± 0.08 | 1.2 ± 0.2 |
| Palmitate: VWR | not detectable | 0.26 ± 0.03 | 0.40 ± 0.08 |
| Stearate: Eppendorf | not detectable | 0.10 ± 0.07 | 0.6 ± 0.2 |
| Stearate: Fisherbrand | not detectable | 1.8 ± 0.1 | 3.4 ± 0.3 |
| Stearate: VWR | not detectable | 0.38 ± 0.02 | 0.8 ± 0.1 |
Data are presented as mean ± SD (n = 3)
Fig. 1.
Palmitate contamination from plasticware affects quantitative analysis. a Nanomoles of palmitate detected in: methanol from plastic microcentrifuge tubes that were bath sonicated, methanol incubated in plastic microcentrifuge tubes (no sonication), methanol aspirated and dispensed from plastic pipette tips, 100 μL of 10 % FBS media extracted with glassware, or 4 million 3T3-L1 cells extracted with glassware. b Palmitate standard solutions of the indicated concentration were extracted with: plastic microcentrifuge tubes and plastic pipettes, methanol-rinsed plastic microcentrifuge tubes and glass pipettes, or all glassware. Relative error is shown as the absolute value of the difference between the measured concentration and the actual concentration. Data are represented as mean ± SD (n = 3 replicates for each vendor listed in Materials and Methods). c Isotope labeling pattern of palmitate from 3T3-L1 cells enriched with U-13C glucose for 48 h. Data are from 3T3-L1 cells extracted with plastic or glassware as indicated. The palmitate labeling pattern inferred from palmitoylcarnitine was not significantly different between the plastic- and glass-extracted samples, and the displayed data represent an average
3.3 Quantitative error due to plastic contamination
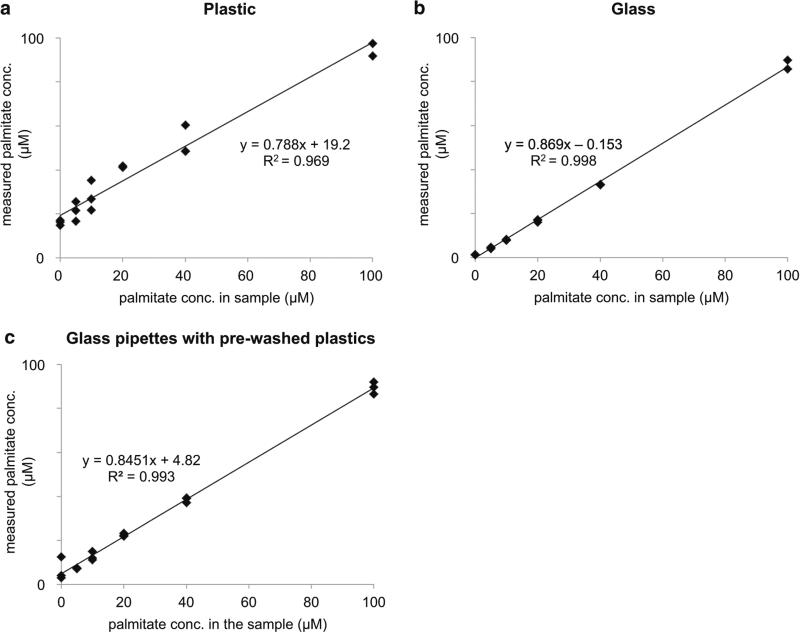
We next made stock solutions of known palmitate concentration by conjugation with BSA, performed a conventional metabolite extraction, and then quantified the concentration of palmitate in the samples. Each sample was analyzed by using either plastic pipette tips and plastic microcentrifuge tubes or glassware. The percentage of relative error from each experiment is shown in Fig. 1b. As expected, the error was largest for plastic-based extractions when the standard solution of palmitate was the lowest concentration. At higher concentrations, the amount of palmitate from contamination is small relative to the concentration of the standard and therefore the percent error decreases. In contrast, the percent error from glass experiments was small and remained relatively constant for all concentrations. We note that the percent error from glass appears to be larger than the percent error from plastic for the 100 μM standard solution. We suspect that this is due to the incomplete recovery of standard palmitate during extraction, which is partially balanced by the gain of palmitate from contamination when plastic consumables are used. We also found that the precision between replicates was significantly lower when the standard solutions were extracted with plastic compared to glassware (Fig. 2). These data show that the amount of palmitate contamination varies between plastic consumables, even though the plastic consumables were from the same lot number.
Fig. 2.
Standard curves of measured palmitate concentration versus actual palmitate concentration for samples extracted with plastic microcentrifuge tubes and plastic pipettes (a), all glassware (b), or plastic microcentrifuge tubes rinsed with methanol and glass pipettes (c). Note the difference in y-intercepts and R2 values
3.4 Contamination leads to the overestimation of unlabeled palmitate
Measuring the labeling pattern of palmitate (or the mass isotopomer distribution) after the introduction of a 13C-labeled substrate provides information about acetyl-CoA labeling and the contribution of de novo lipid synthesis to the total intracellular palmitate pool. The latter analysis has proven valuable to understanding the metabolism of rapidly dividing cells, but it relies on accurate quantitation of mass isotopologues. Palmitate contamination from plastic consumables distorts the palmitate labeling pattern and results in an underestimation of isotopic enrichment. When ISA is applied, this leads to an underestimation of de novo lipid synthesis.
Here we cultured 3T3-L1 cells with U-13C glucose for 48 h and then extracted them with plastic or glass pipettes and tubes prior to LC/MS analysis. We analyzed the palmitate labeling patterns by using the convISA algorithm implemented in MATLAB. The use of plastic compared to glassware led to a twofold increase in the M + 0 peak and a resulting eightfold decrease in the calculated contribution of de novo lipid synthesis, g(48 h) (Fig. 1c and Table 3). After correcting for naturally occurring isotopes, palmitate contamination only contributes to the M + 0 peak. Thus, the calculation of acetyl-CoA labeling is not affected as shown by the D values in Table 3.
Table 3.
ISA values for palmitate (PA) and palmitoylcarnitine from 3T3-L1 cells labeled with U-13C glucose for 48 h and extracted with either plasticware or glassware
| ISA | PA (plastic) | PA (glass) | Palmitoylcarnitine |
|---|---|---|---|
| D | 0.55 | 0.55 | 0.56 |
| g(48 h) | 0.07 | 0.51 | 0.65 |
3.5 Minimizing error from plastics to improve palmitate measurements
The best method to remove plastic contamination is to use glassware in all steps of the sample preparation involving organic solvents. For some laboratories, however, this may be challenging because not all equipment used in preparing metabolomic samples is compatible with glass (e.g., centrifuges, vacuum concentrators, Pipetman pipettes). The complication of subtracting the palmitate contamination signal measured in a blank from the data of experimental samples is that the palmitate contamination signal is variable from plastic disposable to plastic disposable (see Fig. 2 and note that these data are for plastic pieces from the same lot number). Interestingly, we found that the exposure time of plastic consumables to methanol did not significantly change the amount of palmitate and stearate contamination detected (Table 4). Thus, we explored whether plastic consumables could be washed with methanol prior to sample introduction to reduce palmitate and stearate contamination. When we extracted our standard palmitate solutions by using glass pipettes and plastic microcentrifuge tubes that had been pre-washed with methanol, the relative error was reduced sevenfold for the 5 μM palmitate solution and not statistically different from glass for all other palmitate solutions (Fig. 1b). Additionally, the standard deviation of palmitate measurements was reduced (Fig. 2). These results suggest that washing plastic consumables with methanol prior to sample introduction is an effective method to minimize error associated with palmitate and stearate contamination.
Table 4.
Quantifying palmitate and stearate contamination from 1 mL of methanol stored in a plastic microcentrifuge tube at −20 °C for the indicated time
| Plastic |
Glass | |||
|---|---|---|---|---|
| 24 h | 48 h | 5 d | 5 d | |
| Palmitate (nmol) | 0.8 ± 0.1 | 0.45 ± 0.04 | 0.46 ± 0.08 | 0.002 ± 0.008 |
| Stearate (nmol) | 3.2 ± 0.3 | 2.5 ± 0.2 | 2.8 ± 0.2 | 0.01 ± 0.01 |
Data are presented as mean ± SD (n = 3)
For isotope tracer experiments, another possible method to minimize error associated with plastic contamination is to infer palmitate labeling from other lipids incorporating palmitate. Palmitoylcarnitine is a reasonable choice because it is relatively easy to detect and it does not have a competing contamination signal from plastic consumables. In 3T3-L1 cells labeled with U-13C glucose for 48 h, the palmitate labeling pattern measured from cells extracted with glass is consistent with the labeling pattern inferred from palmitoylcarnitine (Fig. 1c). Notably, because there is no competing contamination signal, the labeling pattern of palmitoylcarnitine does not change between 3T3-L1 cells extracted with glass and 3T3-L1 cells extracted with plasticware. When palmitate and palmitoylcarnitine were considered from glass-extracted 3T3-L1 cells, ISA provided similar values for D and g(48 h) (Table 3). The deviation in g(48 h) may be due to palmitate contamination in glass-extracted samples or some preferential channeling of glucose-derived palmitate to palmitoylcarnitine (Glatz et al. 2010; Li et al. 2015).
4 Concluding remarks
The use of plastic pipettes and plastic microcentrifuge tubes introduces palmitate contamination that is as high as sixfold larger than the palmitate measured from 4 million extracted 3T3-L1 cells. This error prevents accurate quantitation of palmitate in profiling and isotope tracer experiments. The plastic contamination signal varies between consumables in the same lot, making it challenging to remove by simple subtraction using a blank sample. Rinsing plastic microcentrifuge tubes with methanol prior to the introduction of sample reduced palmitate contamination substantially. For isotope tracer experiments, error could also be minimized by inferring palmitate enrichment from palmitoylcarnitine, which does not have a competing plastic contamination signal. However, the best results were obtained when extractions were performed with glassware.
Acknowledgments
This work was supported by funding from the National Institutes of Health grants R01 ES022181 (GJP), R21 CA191097 (GJP), and R01 HL118639-03 (RWG) as well as grants from the Alfred P. Sloan Foundation (GJP), the Camille & Henry Dreyfus Foundation (GJP), and the Pew Scholars Program in the Biomedical Sciences (GJP).
References
- Glatz JF, Luiken JJ, Bonen A. Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiological Reviews. 2010;90:367–417. doi: 10.1152/physrev.00003.2009. doi:10.1152/physrev.00003.2009. [DOI] [PubMed] [Google Scholar]
- Gross RW, Han X. Lipidomics at the interface of structure and function in systems biology. Chemistry & Biology. 2011;18:284–291. doi: 10.1016/j.chembiol.2011.01.014. doi:10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisevic J, et al. Toward ‘omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Analytical Chemistry. 2013;85:6876–6884. doi: 10.1021/ac401140h. doi:10.1021/ac401140h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. doi:10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Chung MK, Fan J, Rabinowitz JD. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2014;2:23. doi: 10.1186/2049-3002-2-23. doi:10.1186/2049-3002-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurczy ME, Ivanisevic J, Johnson CH, Uritboonthai W, Hoang L, Fang M, et al. Determining conserved metabolic biomarkers from a million database queries. Bioinformatics. 2015;31:3721–3724. doi: 10.1093/bioinformatics/btv475. doi:10.1093/bioinformatics/btv475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TW, Tumanov S, Villas-Boas SG, Montgomery JM, Birch NP. Chemicals eluting from disposable plastic syringes and syringe filters alter neurite growth, axogenesis and the microtubule cytoskeleton in cultured hippocampal neurons. Journal of Neurochemistry. 2015;133(1):53–65. doi: 10.1111/jnc.13009. [DOI] [PubMed] [Google Scholar]
- Li Q, Deng S, Ibarra RA, Anderson VE, Brunengraber H, Zhang GF. Multiple mass isotopomer tracing of acetyl-CoA metabolism in Langendorff-perfused rat hearts: channeling of acetyl-CoA from pyruvate dehydrogenase to carnitine acetyltransferase. Journal of Biological Chemistry. 2015;290:8121–8132. doi: 10.1074/jbc.M114.631549. doi:10.1074/jbc.M114.631549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieu NG, Spalding J, Patti GJ. Warpgroup: increased Precision of metabolomic data processing by consensus integration bound analysis. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv564. doi:10.1093/bioinformatics/btv564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. doi:10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard P, Letisse F, Sokol S, Portais JC. IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics. 2012;28:1294–1296. doi: 10.1093/bioinformatics/bts127. doi:10.1093/bioinformatics/bts127. [DOI] [PubMed] [Google Scholar]
- Nikolskiy I, Mahieu NG, Chen YJ, Tautenhahn R, Patti GJ. An untargeted metabolomic workflow to improve structural characterization of metabolites. Analytical Chemistry. 2013;85:7713–7719. doi: 10.1021/ac400751j. doi:10.1021/ac400751j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri A, Degenhardt OS, McDonald GR, Narang D, Paulsen IM, Kozuska JL, Holt A. On the disruption of biochemical and biological assays by chemicals leaching from disposable laboratory plasticware. Canadian Journal of Physiology and Pharmacology. 2012;90:697–703. doi: 10.1139/y2012-049. doi:10.1139/y2012-049. [DOI] [PubMed] [Google Scholar]
- Stillwell W. Chapter 4—membrane lipids: fatty acids an introduction to biological membranes. Elsevier; San Diego: 2013. pp. 43–56. [Google Scholar]
- Tredwell GD, Keun HC. convISA: a simple, convoluted method for isotopomer spectral analysis of fatty acids and cholesterol. Metabolic Engineering. 2015;32:125–132. doi: 10.1016/j.ymben.2015.09.008. doi:10.1016/j.ymben.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Tumanov S, Bulusu V, Kamphorst JJ. Analysis of fatty acid metabolism using stable isotope tracers and mass spec-trometry. Methods in Enzymology. 2015;561:197–217. doi: 10.1016/bs.mie.2015.05.017. doi:10.1016/ bs.mie.2015.05.017. [DOI] [PubMed] [Google Scholar]
- Yao CH, Fowle-Grider R, Mahieu NG, Liu GY, Chen YJ, Wang R, et al. Exogenous fatty acids are the preferred source of membrane lipids in proliferating fibroblasts. Cell Chemical Biology. 2016;23:483–493. doi: 10.1016/j.chembiol.2016.03.007. doi:10.1016/j.chembiol.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]