Abstract
RORα is a member of the nuclear receptor (NR) superfamily and analysis of the (global) RORα-deficient mouse model revealed this NR has a role in glycemic control and fat deposition. Therefore, we generated an adipose-specific RORα ‘gain of function’ mouse model under the control of the fatty acid binding protein 4 (FABP4) promoter to elucidate the function of RORα in adipose tissue. The Tg-FABP4-RORα4 mice demonstrated a shift in fat distribution to non-adipose tissues when challenged with a high fat diet (HFD). Specifically, we observed a subcutaneous lipodystrophy, accompanied by hepatomegaly (fatty liver/mild portal fibrosis) and splenomegaly; in a background of decreased weight gain and total body fat after HFD. Moreover, we observed significantly higher fasting blood glucose and impaired clearance of glucose in Tg-FABP4-RORα4 mice. Genome wide expression and qPCR profiling analysis identified: (i) subcutaneous adipose specific decreases in the expression of genes involved in fatty acid biosynthesis, lipid droplet expansion and glycemic control, and (ii) the fibrosis pathway as the most significant pathway [including dysregulation of the collagen/extracellular matrix (ECM) pathways] in subcutaneous adipose and liver. The pathology presented in the Tg-FABP4-RORα4 mice is reminiscent of human metabolic disease (associated with aberrant ECM expression) highlighting the therapeutic potential of this NR.
Keywords: RORα, Subcutaneous adipose, Obesity, Hepatomegaly, Fibrosis, Collagen
Highlights
-
•
Adipose-specific expression of RORα is associated with subcutaneous lipodystrophy and hepatomegaly with fibrosis.
-
•
The phenotype is associated with impaired glycemic control and decreased weight gain on a high fat diet.
-
•
Gene expression profiling reveals significant dysregulation of extra cellular matrix signaling.
We have generated a ‘gain of function’ animal model with the nuclear hormone receptor RORα4 to understand the function of this protein in fat. Over expression of the RORα4 gene, was associated with fat deposition in non-adipose tissues on a high fat diet. Moreover, we observed a decrease in fat tissue (located under the skin) accompanied by enlargement of the liver and spleen. In addition, over expression of this receptor was associated with impaired glycemic control. The pathology in this animal model is reminiscent of metabolic disease in humans, highlighting the therapeutic potential of pharmacologically manipulating this nuclear receptor.
1. Introduction
Obesity and diabetes are worldwide health burdens. The incidence of obesity has doubled since 1980 with over 1.9 billion adults overweight in 2014, and globally > 400 million people have type 2 diabetes. Unfortunately, existing anti-obesity and anti-diabetic therapeutic display insufficient efficacy and adverse effects (Aguiree et al., 2013). Therefore, there is a global need for novel therapeutic targets that regulate excessive adiposity and glycemic pathophysiology to treat those populations challenged by adverse clinical outcomes. The nuclear hormone receptor (NR) superfamily is comprised of hormone-dependent transcription factors that translate physiological signals into gene regulation to control metabolism in an organ-specific manner. In the context of metabolic disease, dysfunctional NR signaling results in dyslipidemia, diabetes, and obesity. There are 48 NRs in humans, all implicated in disease, although not all with identified natural ligands; these NRs are denoted as orphans. The NR, RORα, has been implicated in the regulation of glucose and fat homeostasis, but has historically belonged to the orphan class of NRs. However, interest in this specific receptor has been stimulated by the rapid emergence of small molecule agonist and inverse agonists (Smith and Muscat, 2006, Marciano et al., 2014).
Several studies suggest that RORα is a constitutively active receptor. The constitutive recruitment of coactivators such as p300, glutamate receptor interacting protein 1 and steroid receptor coactivator 1 (reviewed in Fitzsimmons et al., 2012) by RORα in the absence of exogenous ligands supports a state of constitutive activation of RORα by a common endogenous ligand (such as cholesterol metabolites and intermediates) (reviewed in Fitzsimmons et al., 2012, Marciano et al., 2014). For example, crystallographic and mass spectrometry studies showed that the RORα-LBD is constitutively bound to endogenous ligands including fatty acids, cholesterol metabolites and all trans retinoic acid (reviewed in Marciano et al., 2014). Furthermore, the basal activity of RORs is highlighted by the observation that natural inverse agonists are exploited (for example ursolic acid) in RORγ reporter assays to identify agonists in reporter assays (Santori, 2015, Chang et al., 2016). Recently, it was confirmed that oxygenated sterols (natural LXR ligands and agonists) function as native ligands, but as inverse agonists for both RORα and RORγ (reviewed in Marciano et al., 2014, Kojetin and Burris, 2014). Moreover, the crosstalk between the (oxysterol activated NR) LXR and RORα/γ signaling pathways is underscored by our recent study demonstrating decreased expression of the cholesterol 25-hydroxylase mRNA (encodes the enzyme that produces the LXR agonist 25-hydroxycholesterol, 25-HC) and aberrant phagocytosis in the Rorα-deficient staggerer mice (sg/sg) (Tuong et al., 2013). New studies (Tuong et al., 2016) implicate Rorα in regulating lipid storage in macrophages in a process modulated by 25-HC levels.
The biological significance of RORα in the regulation of important metabolic pathways is underscored by studies on the (global) Rorα-deficient staggerer (sg/sg) mouse model. For example, analysis of the global Rorα knockout model has revealed that Rorα regulates (i) adiposity (Lau et al., 2004, Lau et al., 2008), (ii) resistance to diet-induced obesity and hepatic steatosis (Lau et al., 2008), (iii) thermogenesis and browning/beiging of subcutaneous adipose tissue (SAT) (Lau et al., 2015), (iv) insulin sensitivity and signaling (Lau et al., 2011), (v) inflammation and phagocytosis (Tuong et al., 2013) and (vi) lipid droplet homeostasis (Tuong et al., 2016).
Specifically, the Rorα-deficient sg/sg mice display increased AKT signaling in skeletal muscle (Lau et al., 2011), improved glucose tolerance and insulin sensitivity. The lean phenotype in sg/sg mice is associated with reduced serum triglyceride and cholesterol levels (Lau et al., 2008, Lau et al., 2015, Kang et al., 2011, Mamontova et al., 1998). In addition, decreased adiposity is associated with an increased metabolic rate and cold tolerance in Rorα-deficient sg/sg mice. This phenotype involves browning/beiging of SAT, increased uncoupling protein 1 (Ucp1) expression (mRNA and protein) and thermogenic gene expression (Lau et al., 2015), and significantly increased expression of the (cell-fate controlling) histone-lysine N-methyltransferase 1 (Ehmt1), which stabilizes the Prdm16 transcriptional complex. However, the significance of RORα in the regulation of adipose physiology remains unclear as it is difficult to dissect the contribution of this widely expressed receptor from the complex interactions that give rise to the lean phenotype. Several in vitro studies have suggested that RORα transcriptional activity acts to suppress adipocyte differentiation. Embryonic fibroblasts from Rorα-deficient sg/sg mice displayed enhanced differentiation into functional adipocytes (Duez et al., 2009) and in 3T3-L1 cells RORα constrained differentiation via increased expression during late adipogenesis (Okada et al., 2009). However, these authors also report a similar differentiation potentiality in pre-adipocytes sourced from homozygous sg/sg mice as their heterozygous sg/+ counterparts. It is evident that an in vivo investigation of RORα function - specifically in adipose tissue (i.e. an organ/tissue specific mouse model) that can account for the developmental, metabolic and compartmental context, is warranted. Therefore, we generated an adipose-specific RORα ‘gain of function’ transgenic mouse model in order to further understand the adipose-specific function of RORα in (i) lipid deposition, (ii) glucose tolerance and insulin sensitivity, (iii) obesity, and (iv) gene regulation. This is highly significant within the context of obesity as the capacity to expand the number of adipocyte cells to accommodate increased lipid storage requirements is a key determinant of the degree of metabolic dysfunction that accompanies increased adiposity.
Our studies indicate that (heterozygous, Tg-FABP4-RORα4 tg/+) mice with adipose-specific RORα4 expression have impaired glucose tolerance, decreased SAT, and hepatomegaly on a high fat diet, a phenotype often associated with obesity (Abdennour et al., 2014, Porter et al., 2009, Tam et al., 2012). RNA-seq, targeted qPCR and canonical pathway analysis suggests that adipose-specific RORα4 phenotype is associated with differential regulation of the fibrosis pathway in adipose and hepatic tissue. For example, genes that encode the extracellular matrix (ECM) collagen proteins are down-regulated in the SAT, but increased in tg/+ hepatic tissue. This is in accord with the role of collagen production in adipose development and physiology, as well as adipose plasticity to suit metabolic demands and changes (Mariman and Wang, 2010) and ectopic/aberrant fat deposition (Aikio et al., 2014).
2. Materials and Methods
2.1. Transgene Plasmid Generation
hRORα4 was amplified by PCR from pSG5-RZR (courtesy of Dr. OA MacDougald, University of Michigan Medical School, USA (Becker-Andre et al., 1993)) using a forward primer containing a SalI cut site: AGCGTCGACCATGATGTATTTTGTGATCG and a reverse primer containing a NotI cut site: AATGCGGCCGCTTTACCCATCAATTTG. After SalI/NotI double digestion, RZR was subcloned into SalI NotI double digested-pCRII-FABP4 promoter vector (Longo et al., 2004) to generate FABP4-RORα4 construct and verified by sequencing, prior to pronuclear injection.
2.2. Animals and Tissue Collection
Generation of Tg-FABP4-RORα4 mice was performed by Transgenic Animal Services Queensland. Pronuclear injection was performed in C57BL/6/CBA mixed background mice. Founders were identified by genotyping and FABP4-RORα4 positive transgenic mouse lines were selected for phenotypic analysis after > 5 generations of backcrossing to C57BL/6 background. 16 and 32 week old WT and FABP4-RORα4 transgenic littermates (Tg-FABP4-RORα4) were obtained from crossing heterozygous tg/+ Tg-FABP4-RORα4 transgenic breeders with C57BL/6 mice. Homozygous tg/tg Tg-FABP4-RORα4 mice were generated by crossing heterozygous Tg-FABP4-RORα4 males and females. All animals were housed in the Queensland Bioscience Precinct Vivarium (UQ) with a 12 h light-dark cycle. The high fat diet used in this study is as described in (Pearen et al., 2013). Animals were weaned at 4 weeks of age and were fed the standard chow diet ad libitum (which contains 4.6% total fat). In contrast, the high fat diet (SF03-002 Fat Modified Rodent Diet; very high fat modification of AIN93G) used in the study contains 36% fat. Both diets were acquired from Specialty Feeds (Glen Forrest, Western Australia). Experimental mice were weighed weekly. For tissue collection, mice were fasted overnight in a new food-free holding cage and subsequently euthanized. Tissues were collected and immediately snap-frozen in liquid nitrogen and then stored at − 80 °C. All aspects of animal experimentation were approved by The University of Queensland Animal Ethics Committee.
2.3. Intraperitoneal Glucose Tolerance Test and Insulin Tolerance Test
Blood glucose measurements were obtained from the tail vein of 6 h fasted animals (14–16 weeks old or 22 week old mice on high fat diet) following glucose or insulin challenge, using a blood glucose testing system (Accu-chek Performa; Roche Diagnostics, Castle Hill, NSW, Australia) as described (Raichur et al., 2010). Glucose was administered to each mouse at a dose of 2 g/kg and insulin was given at 1.0 U/kg.
2.4. Insulin Enzyme-linked Immunosorbent Assay (ELISA)
The ALPCO Mouse ultrasensitive Insulin ELISA assay kit was used for the quantitative determination of insulin plasma from 6 h or overnight fasted mice. All procedures were performed according to manufacturer's instructions.
2.5. Protein Extraction and Immunoblot Analysis
Protein extraction from adipose tissue was previously described (Lau et al., 2015) with modifications. Inguinal white adipose tissues were homogenized in 1 mM EDTA, 10 mM Tris, and 0.25 M sucrose (pH 7.5) with 1xComplete protease inhibitor and 1xPHOS-STOP (Roche Diagnostics, Mannheim, Germany). Infranatant and pellet were separated from the top layer of fat cake after centrifugation. Detergent was then added to a final concentration of 1% Triton X-100, 1% NP-40, and 0.1% SDS for the infranatant (cytosolic proteins) and pellet (nuclear and membrane proteins) separately, incubated for 30 min and sheared eight times with an insulin syringe. The pellet was sheared multiple times using P200 pipette tips and insulin syringe. Protein concentration was measured with BCA reagent (Thermoscientific; Pierce, Rockford, IL). Immunoblot analysis was performed as described previously except that the membranes were blocked in 5% skim milk and probed with anti-RORα (ab60134 1:1000; Abcam), anti-AKT (#9272, 1:1000; Cell Signaling Technology, Danvers, MA), pAKT (ser473) (#4058; 1:1000; Cell Signaling Technology), anti-TBP (sc-204; 1:1000, Santa Cruz Biotechnology), or anti-α-tubulin (#2144, 1:1000; Cell Signaling Technology).
2.6. Histology
Hematoxylin and eosin (H&E) staining was performed as described (Lau et al., 2015), on paraffin-embedded tissues fixed in 10% buffered formalin (Sigma-Aldrich). Accustain® Trichrome Stains (Masson) (Sigma-Aldrich) was used for staining collagen fibers in the liver according to manufacturer's instructions. To assess macrophage infiltration into adipose, immunohistochemistry was performed on deparaffinized and re-hydrated sections. Enzymatic antigen retrieval was performed using Proteinase K (20 μg/mL) in TE buffer (50 mM Tris, 1 mM EDTA, pH 8.0) for 3 min at room temperature and rinsed with PBS. Macrophages were detected using a specific primary antibody for F4/80 (rat anti-mouse, Abcam, ab6640) at 4 °C overnight. This was followed by 0.3% H2O2 treatment to reduce endogenous peroxidase activity, and staining with species-matched horseradish peroxidase-conjugated secondary antibody (goat anti-rat antibody, Santa Cruz Biotechnology) at room temperature for 1 h. All blocking steps were performed in 0.5% BSA/PBS in the presence of 10% species-matched anti-sera. Sections were then counterstained using Mayer's hematoxylin and mounted using permanent mounting media. Sections were examined using an Olympus BX-51 microscope with a DP-70 digital camera and DP controller imaging software (Olympus Imaging Systems, PA, USA).
2.7. Blood Lipid Profiling and Measurement of Hepatic Triglyceride Concentration
Measurements of total cholesterol, triglyceride, and non-esterified fatty acid in heparinized plasma samples were analyzed by Clinical Pathology Laboratory, School of Veterinary Science, The University of Queensland (Gatton, Queensland, Australia).
2.8. Nuclear Magnetic Resonance (NMR) and Dual-energy X-ray Absorptiometry (DEXA) Scans
NMR scanning was performed using a Bruker MiniSpec LF50 (Ettlingen, Germany). Total body percentage fat and percentage lean mass values were extracted from the analysis. DEXA scanning was performed on frozen liver tissues harvested from euthanized overnight-fasted mice (32 weeks of age) and measured for lean mass and fat mass using a PIXImus Densitometer (GE Lunar, Madison, WI).
2.9. Genotyping and Tissue Detection of Transgene Expression
Genotyping of mice from Tg-FABP4-RORα4 transgenic lines was performed on toe/tail/ear puncture samples using QuickExtract™ DNA Extraction Solution (Epicentre Biotechnologies, Madison, Wisconsin, USA) according to manufacturer's protocol. For confirmation of transgene expression, RNA was extracted from tissues and cDNA synthesis was performed as described (Pearen et al., 2009, Myers et al., 2009). SYBR primers were designed using Primer Express (Applied Biosystems, Foster City, CA) for the following: mouse Rorα forward CAATGCCACCTACTCCTGTCC and reverse GCCAGGCATTTCTGCAGC for real-time endogenous expression and genotyping, and human RORα reverse CTACGGCAAGGCATTTCTGTAAT for real-time ectopic expression and genotyping. The same forward primer was used for both real-time endogenous and ectopic expression.
2.10. RNA Extraction and Purification and cDNA Synthesis
Total RNA was extracted using TRI-Reagent (Sigma-Aldrich, St. Louis, MO) and RNA purification was performed using the RNeasy mini kit (Qiagen, Clifton Hill, Victoria, Australia) according to the manufacturers' instructions. Complementary DNA (cDNA) was synthesized from 0.5–2 μg of purified total RNA using Superscript III Reverse Transcriptase (Invitrogen) and random hexameric primers according to the manufacturer's instructions.
2.11. qPCR Analysis
Relative expression of genes was determined using the Applied Biosystems (ABI) ViiA™ 7 Real-Time PCR System (ABI, Singapore) as previously described (Lau et al., 2008, Tuong et al., 2013). Relative gene expression was analyzed by qPCR using either TaqMan Gene Expression Assays (ABI, Foster City, CA), or TaqMan low-density arrays (TLDAs) as described (Pearen et al., 2013), or primers designed for use with SYBR master mix. Assay on demand primers for TaqMan qPCR used are described (Lau et al., 2004, Lau et al., 2015) and include the following: total RORα (mouse + human; Mm00443103_m1), Tle3 (Mm00437097_m1), Ehmt1 (Mm00553234_m1), Prdm16 (Mm00712556_m1), Ucp1 (Mm01244861_m1), Cpt2 (Mm00487202_m1), Pnpla2 (Mm00503040_m1), Adrb3 (Mm00442669_m1), Ndufb5 (Mm00452592_m1), Dio2 (Mm00515664_m1), and Pparα (Mm00440939_m1). The TaqMan primers for the housekeeping genes are as follows: 18s (catalog number: 4319413E) and Gusb (Mm00446953_m1). Primers for SYBR assays include the following: Cidea, CAAACCATGACCGAAGTAGCC and AACCAGGCCAGTTGTGATGAC; Acot11, GATCATGGCTTGGATGGAGAA and GGCCTCGGAAATGGAACAT; Errα, CTCTGGCTACCACTACGGTGTG and AGCTGTACTCGATGCTCCCCT; Mcpt1 ATCATGTATCGCCGCAAACT and CCATCTGGTAGGAGCACATGG. Primers for Col18a1 that detects long, medium short, and all isoforms were as published (Aikio et al., 2014). The SYBR primer sets used as endogenous controls are as follows: Gusb, GTGAGCAACGCCAAATATGATG and TCCAAATGCCCATAGTCATGATAC; 18s, GATCCATTGGAGGGCAAGTCT and CCAAGATCCAACTACGAGCTTTTT; Rplp0, AGATGCAGCAGATCCGCA and GTTCTTGCCCATCAGCACC.
2.12. RNA-seq Analysis and CIBERSORT
Library preparation and sequencing were performed by the IMB Sequencing Facility at the University of Queensland. Total RNA sample libraries were generated using the Illumina TruSeq Stranded mRNA LT sample preparation kit (Illumina, Part no. RS-122-2101 and RS-122-2102), according to the standard manufacturer's protocol (Part no. 15031047 Rev. E October 2013). The mRNA denaturation and elution was performed with 0.1 μg to 0.2 μg of total RNA (depending on amount of sample available) prior to a heat fragmentation step aimed at producing libraries with an insert size between 120 and 200 bp. cDNA was then synthesized from the enriched and fragmented RNA using SuperScript III Reverse Transcriptase (Invitrogen, Catalog no. 18064014) and random primers. The resulting cDNA was converted into double stranded DNA in the presence of dUTP to prevent subsequent amplification of the second strand and thus maintain the strandedness of the library. Following 3′ adenylation and adaptor ligation, libraries were subjected to 15 cycles of PCR to produce RNA-seq libraries ready for sequencing. Prior to sequencing, RNA-seq libraries were qualified via the Agilent Bioanalyzer with the High Sensitivity DNA kit (Integrated Sciences, Part no. 4067-4626). Quantification of libraries for clustering was performed using the KAPA Library Quantification Kit - Illumina/Universal (KAPA Biosystems, Part no. KK4824) in combination with the Life Technologies Viia 7 real-time PCR instrument. Sequencing was performed using the Illumina NextSeq500 (NextSeq control software v1.2/Real Time Analysis v2.1) platform. The library pool was diluted and denatured according to the standard NextSeq500 protocol and sequencing was carried out to generate single-end 76 bp reads using a 75 cycle NextSeq500 High Output reagent Kit (Catalog # FC-404-1005). Reads were aligned to GRCm38.p2 (Mus musculus) using STAR (Dobin et al., 2013), and read counts for each gene in the Ensembl annotation were generated using htseq-count in the HTSeq python package (Anders et al., 2015) and the GENCODE annotation (Harrow et al., 2012). Differential gene expression was detected using the DESeq2 (Love et al., 2014) packages in R. To quantify the relative expression levels of distinct cell types from complex tissues, we utilized the CIBERSORT (Newman et al., 2015). Variance stabilized transformed read count was generated using the DESeq2 package (Love et al., 2014) and used as input data for CIBERSORT. The LM22 signature was selected as a default parameter and CIBERSORT was run with 100 permutation. Although the tool and the embedded LM22 signature dataset were not designed for interrogating RNA-seq data, the authors note that “the linearity assumptions made by our method are likely to hold, as previously suggested”. Enrichment scores were transferred into the stack format as published (Newman et al., 2015).
2.13. Ingenuity Pathway Analysis
The canonical pathways and functional analyses were generated through the use of IPA (Ingenuity Systems) from the differential expression gene data set acquired from RNA-seq analysis. In the context of the identification of activation or inhibition of pathways, the z-score algorithm matches the direction of gene changes with expected activated/inhibited state in the canonical pathways (orange indicates predicted activation/positive z-score and blue indicates predicted inhibition/negative z-score).
2.14. Database for Annotation, Visualization and Integrated Discovery (DAVID) Analysis
The entire gene list of significantly regulated genes identified from the RNA-seq analysis was uploaded onto the web documentation of DAVID. Included in the documentation is a modified Fisher exact probability for ‘over-represented’ or most relevant biological terms associated with the given gene list where P < 0.05 was considered significant (Huang da et al., 2009). Data was extracted after DAVID annotates, organizes and condenses the data into meaningful biological functions that are associated with the gene list provided.
2.15. Statistical Analyses
Statistical analyses for all other analyses not included in the RNA-seq workflow were performed using GraphPad Prism version 5.0 (GraphPad Software, San Diego, USA). In general, significance was calculated using unpaired two-tailed Student's t-tests or two-way ANOVAs with Bonferroni's post-tests where applicable. Correlation analysis was tested in an assumption-free model and the R2 value reports the Pearson correlation coefficient. The associated P-value tests whether the correlation effect between groups could be due to random sampling. A small P-value allows for the rejection of the null hypothesis that the correlation is due to random sampling.
3. Results
3.1. Transgenic Overexpression of RORα4 in Adipose Tissue
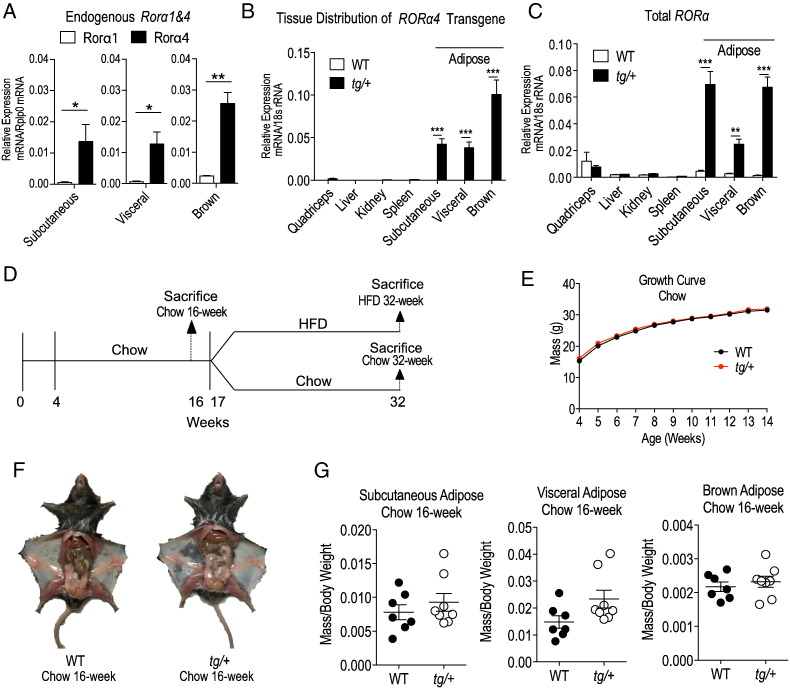
Our previous reports investigating the (global) Rorα-deficient staggerer sg/sg mouse model, and a transgenic muscle-specific line expressing dominant negative RORα indicated that this NR regulates glycemic control and Akt2 signaling. Further studies in the sg/sg mouse model indicated this NR regulated fat deposition, and responses to dietary challenges. We were interested in further investigating the specific in vivo functional role(s) of RORα signaling in adipose tissue, a major peripheral tissue involved in energy storage. Therefore, we pursued targeted adipose-specific expression of RORα in transgenic mice. We selected the RORα4 isoform for transgenic expression, because qPCR analysis of subcutaneous, visceral and brown adipose tissues from wild-type (WT) C57BL/6 mice clearly demonstrated that of the two isoforms expressed in rodents (RORα1 and α4), RORα4 was the most abundantly expressed isoform in the three adipose tissue depots (Fig. 1A).
Fig. 1.
Human RORα4 transgene tissue specificity, growth curve and tissue weights of WT versus Tg-FABP4-RORα4 mice on normal chow. (A–C) RNA was extracted from selected tissues (kidney, quadriceps muscle, liver, SAT (inguinal fat), visceral adipose (epididymal fat) and brown adipose (interscapular fat), and examined for relative gene expression of (A) endogenous (mouse) Rorα1 and Rorα4, (B) the transgene (human RORα4) and (C) total RORα (mouse and human) compared to the endogenous control (18s) using real-time PCR (n = 3 littermate pairs in duplicate experiments). Statistical analyses were performed using unpaired two-tailed Student's t-test for (A) and two-way ANOVAs with Bonferroni's post-test applied for (B–C) where *P < 0.05; **P < 0.01; ***P < 0.001. (D) Schematic timeline of diet regime employed. (E) Body weight (in grams) of male WT (black) and tg/+ (red) mice (n = 18 littermate pairs) measured weekly up to 14 weeks of age. Statistical analyses were performed using two-way ANOVAs with Bonferroni's post-test applied but results were not significant. (F) Representative images of dissected WT and tg/+ littermate pair (chow diet). (G) Mean relative mass/total weight ± S.E.M. of adipose tissues dissected from WT and tg/+ mice (n = 7 littermate pairs). Statistical analyses were performed on each tissue using unpaired two-tailed Student's t-test was performed where *P < 0.05.
RORα is a constitutively active NR, and we confirmed the (transcriptional) activity of native RORα4 by examining its ability to transactivate a well-characterized RORα dependent reporter gene (mPCP2tkluc) (Supplementary Fig. 1A). We produced transgenic mice that selectively express a transgene encoding a native (human) RORα4 expressing construct driven by fatty acid binding protein 4 (FABP4) promoter (also known as adipocyte protein 2) to achieve adipose-specific expression. To validate the selective expression of the transgene in adipose tissues, RNA from major organ/tissues (skeletal muscle, liver, kidney and spleen), including inguinal, epididymal and interscapular fat tissue representing the subcutaneous, visceral and brown adipose depots, was extracted from male WT and transgenic [heterozygous (tg/+) Tg-FABP4-RORα4] mice, and ectopic RORα4 transgene expression was examined via quantitative real-time PCR (qPCR) relative to 18s rRNA (Fig. 1B). The heterozygous transgenic mice selectively, predominantly, and abundantly expressed the ectopic human transgene (Fig. 1B), resulting in significantly increased total (i.e. endogenous mouse and ectopic human) RORα expression (Fig. 1C) in adipose tissues from tg/+ mice relative to WT littermate pairs.
As expected, significantly lower transgene expression was observed in the non-adipose tissues (quadriceps muscle, liver, spleen, and kidney) compared to the adipose tissues. For example, ectopic RORα4 expression was > 50- and > 500-fold greater in SAT, relative to spleen and liver, respectively. Western blot analysis was performed to validate RORα overexpression using the proteins from the nuclear fractions obtained from inguinal adipose tissues of WT and heterozygote tg/+ mice. We showed that total RORα protein is expressed at a higher level (by ~ 8-fold) in heterozygote transgenic mice (tg/+) relative to WT siblings in n = 2 littermate pairs (Supplementary Fig. 1B and C). Thus using the published FABP4 promoter (He et al., 2003) for driving adipose expression, we have successfully generated an adipose-specific RORα transgenic mouse line (Tg-FABP4-RORα4).
3.2. Adipose-specific RORα4 Transgene Expression Decreases Weight Gain and Subcutaneous Adiposity
We hypothesized that RORα overexpression in adipose would perturb energy storage and homeostasis. Hence, we compared mice on normal chow and high fat diets (HFD), and examined growth, organ weights, fat deposition etc., after an initial phase where normal chow diet (< 10% of total calories from fat) was fed ad libitum for 16 weeks, and a second phase where the diet was switched to an energy-dense HFD (~ 40% of energy from fat) for 14–16 weeks (Fig. 1D). The control cohort remained on chow for the entire duration.
During the growth phase (4–14 weeks of age), comparable weight gain was observed between transgenic (FABP4-RORα4) heterozygous tg/+ (Fig. 1E) and homozygous tg/tg mice (Supplementary Fig. 2B) compared to WT littermates on a normal chow diet. We detected no dramatic morphological abnormalities and the tissue weights (normalized to total body weight) of the three adipose depots examined were similar (from 16 week old mice) in WT, tg/+ (Fig. 1F and G) or tg/tg littermates (Supplementary Fig. 2C), and did not display any significant differences. We conclude that the FABP4-RORα4 transgene did not overtly effect the growth or fat deposition in the three adipose depots of tg/+ or tg/tg littermate mice maintained on normal chow diet.
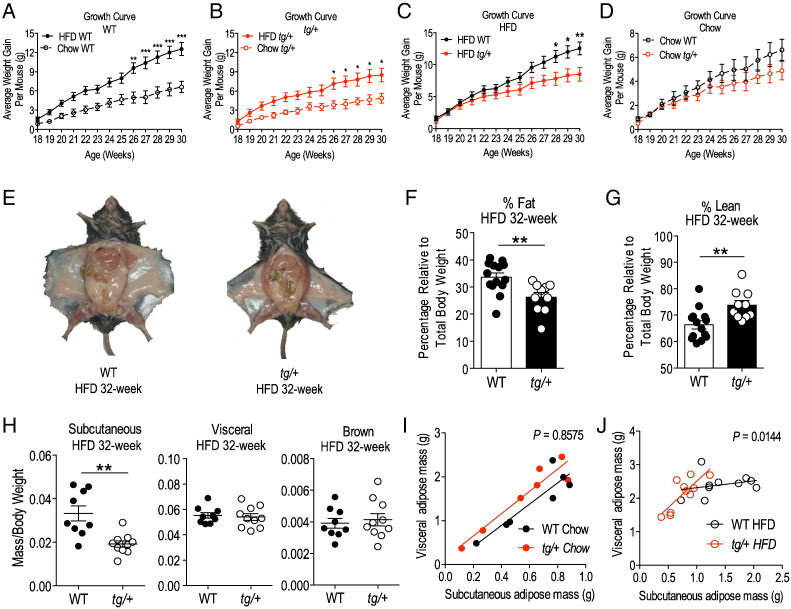
The heterozygous transgenic mice were subsequently placed on high fat diet (HFD) and examined from 17 to 32 weeks of age (relative to chow diet fed mice as controls). After 8 weeks on HFD, both WT and heterozygous tg/+ mice displayed significant increases in weight gain (by ~ 50%) when compared to their respective chow-fed controls (chow-fed for the same duration) (Fig. 2A and B). However, between HFD-fed WT and tg/+ littermate pairs, we observed a marked and statistically significant divergence in weight gain from week 28 (approximately 10 weeks into the diet) and by week 30, tg/+ mice averaged 4 g less in weight than WT littermates (Fig. 2C). In corresponding sets of mice on a normal chow diet, for the same duration, the tg/+ mice trended toward less weight gain compared to WT littermates (Fig. 2D), however, this observation did not attain significance. Thus transgenic mice appear to be leaner on a HFD. Gross anatomical examination of frontal views of mice after dissection revealed a striking decrease in subcutaneous fat deposition in the HFD-fed tg/+ mice relative to WT littermates (Fig. 2E). The difference in adiposity for the HFD-fed tg/+ mice is corroborated by an independent measurement of total body fat and lean mass percentage using nuclear magnetic resonance (NMR) scanning. This NMR analysis was performed prior to dissection on non-fasted, live, HFD-fed mice, and showed a significant ~ 25% decrease in percentage total body fat of tg/+ mice relative to WT littermate pairs (with a corresponding increase in percentage total lean mass) (Fig. 2F and G). Adipose depot weight analysis confirmed a significant ~ 40% decrease in the tissue weight of the SAT from HFD fed tg/+ mice relative to the corresponding tissue from WT littermate pairs (Fig. 2H). However, there were no such respective differences in weights of other adipose tissues examined [visceral (epididymal) adipose tissue and brown (interscapular) adipose tissue] (Fig. 2H). This significantly decreased subcutaneous depot mass in the tg/+ relative to WT littermates on the HFD was further confirmed by examining the correlation between the visceral (epididymal) vs. subcutaneous (inguinal) fat mass from WT and heterozygous tg/+ littermates on chow (Fig. 2I) and HFD (Fig. 2J), respectively. We fitted the data points using correlation analysis and it demonstrated the attenuated capacity of the tg/+ subcutaneous adipose depot to expand in the face of a HFD challenge, in contrast to the WT littermate (Fig. 2J). If the overall slopes were identical during the HFD challenge, there is a ~ 1.4% chance of randomly choosing data points that will fit identical slopes, concluding that the differences between the slopes are significant. In contrast, mice fed a chow diet in parallel did not present with any observable (and/or significant differences; ~ 86% chance of randomly choosing data points that will fit the current slope) in fat deposition in WT and tg/+ littermates on the chow diet (Fig. 2I and Supplementary Fig. 1C and D). This type of correlation analysis has been employed previously to characterize depot specific adipose expansion (Jeffery et al., 2016).
Fig. 2.
Growth curve of WT and tg/+ Tg-FABP4-RORα4 mice on HFD and measurement of adiposity. (A–D) Average weight gain ± S.E.M of WT (black) and tg/+ (red) mice. Growth curves plotted to compare HFD and chow diet in (A) WT mice and (B) tg/+ mice separately. (C) HFD growth curve on tg/+ mice relative to WT (solid circles, n = 10 littermate pairs) with commencement of HFD at week 17. (D) Parallel growth curve data of WT and tg/+ mice on chow diet (hollow circles, n = 7 littermate pairs). Statistical analyses were performed using two-way ANOVAs with Bonferroni's post-test applied where *P < 0.05; **P < 0.01; ***P < 0.001. Representative images of (E) dissected WT and tg/+ littermate pair at the end of the HFD study (32-week). (F–G) NMR scans were performed on non-fasted live WT and tg/+ littermate pairs (HFD) and represented as the mean ± S.E.M. (F) percentage total body fat and (G) percentage lean mass relative to total body weight from n = 8 littermate pairs of mice. (H) Mean relative mass/total weight ± S.E.M. of adipose tissues dissected from WT and tg/+ littermate pairs after HFD (n = 9 littermate pairs). Statistical analysis was performed using unpaired two-tailed Student's t-test where **P < 0.05. (I–J) Assumption-free correlation analysis of visceral and SAT mass in (I) chow 32-week study (n = 7 littermate pairs) and (J) HFD study (n = 10 littermate pairs). The Pearson correlation R2 value reports the fraction of “shared” variance while the P value tests the null hypothesis that there is no linear trend between the groups, where a small P value indicates a statistical significant linear trend.
In summary, our initial observations suggested that adipose-specific RORα4 overexpression prevented overall weight gain by limiting prevents subcutaneous fat expansion and deposition on a high fat diet. The potentially selective effect of the FABP4-RORα4 transgene on SAT fat deposition and expansion, i.e. a selective SAT lipodystrophy, is of particular interest in the light of pre-clinical studies indicating removal of SAT induces glucose intolerance (Gentile et al., 2015), and that intra-abdominal subcutaneous fat transplantation reverses glucose intolerance induced by an energy-dense diet (Hocking et al., 2015).
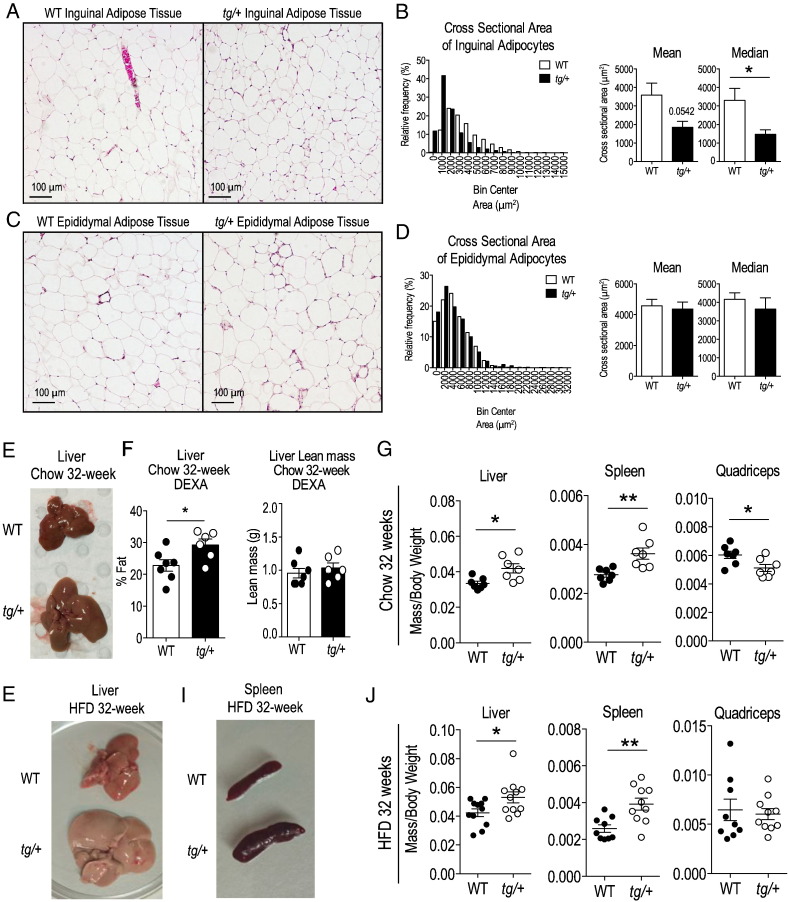
3.3. Adipocytes From tg/+ Subcutaneous Fat Tissue Are significantly Decreased in Median Size
We further characterized HFD-induced fat deposition in different types of adipose tissues at a cellular level, by measuring the cross-sectional area of adipocytes in hematoxylin and eosin (H&E)-stained sections of SAT from WT and heterozygous tg/+ mice. Staining revealed that adipocytes from the SAT fat pad in tg/+ mice were smaller relative to respective cells in WT littermate tissues (Fig. 3A). This difference was confirmed by quantification, with tg/+ mice displaying a higher proportion of adipocytes with smaller cell area and a significant decrease (~ 50%) in median area relative to WT littermates (Fig. 3B). This phenotype was not observed in visceral adipose tissue, rather epididymal adipocytes from WT and tg/+ mice had a similar cross-sectional area (Fig. 3C and D). The predictable hyperplasia in response to high fat diet feeding observed in adipocytes from the visceral compartment indicated that tg/+ mice did not have a generalized lipid storage defect, rather that excess lipids were being directed away from SAT depots.
Fig. 3.
H&E staining of adipose tissue and weights of non-adipose tissues in HFD-fed WT and tg/+ Tg-FABP4-RORα4 mice. (A) Representative images of WT and tg/+ SAT stained by H&E. (B) Quantification of cross sectional area of ~ 300 inguinal adipocytes from n = 4 littermate pairs of WT and tg/+ mice is shown as a histogram of the relative frequency of adipocyte sizes with bin intervals of 1000 μm2 and mean and median area measurements. (C) Representative images of WT and tg/+ visceral epididymal adipose tissues stained by H&E. (D) Quantification of cross sectional area of ~ 300 epididymal adipocytes from n = 4 littermate pairs of WT and tg/+ mice is shown as a histogram of the relative frequency of adipocyte sizes with bin intervals of 2000 μm2 and mean and median area measurements. Significance was calculated using unpaired two-tailed Student's t-test where *P < 0.05. (E) Representative image of liver dissected from chow-fed 32-week old WT and tg/+ mice. (F) DEXA scanning of dissected livers from chow-fed WT and tg/+ mice (n = 7 littermate pairs). (G) Mean relative mass/total weight ± S.E.M. of non-adipose tissues (liver, spleen and quadriceps muscle) dissected from chow-fed 32-week old WT and tg/+ littermate pairs (n = 7 littermate pairs). (H–I) Representative images of (H) liver and (I) spleen dissected from HFD-fed WT and tg/+ mice. (J) Mean relative mass/total weight ± S.E.M. of non-adipose tissues (liver, spleen and quadriceps muscle) dissected from WT and tg/+ littermate pairs after HFD (n = 9 littermate pairs). Statistical significance was calculated using unpaired two-tailed Student's t-test for where *P < 0.05; **P < 0.01.
3.4. Transgenic Adipose Specific RORα4 Expression Leads to Hepatomegaly and Splenomegaly
The selective decrease in inguinal (i.e. subcutaneous) adiposity after HFD in Tg-FABP4-RORα4 transgenic mice was somewhat unexpected as previous studies from our group and others reported decreased total adiposity in the homozygous (global) Rorα-deficient sg/sg mouse line on both normal chow and HFD (Lau et al., 2008, Lau et al., 2015, Kang et al., 2011). However, in the chow-fed study comparing non-adipose tissues between WT and tg/+ mice, we noted that 32 week old tg/+ mice displayed a modest decrease in relative quadriceps muscle mass (~ 15%) and a pronounced enlargement of the liver and spleen (both ~ 30% increased relative weight, Fig. 3E and G). Dual-energy X-ray absorptiometry (DEXA) scanning of livers from this cohort of chow-fed mice, confirmed significantly increased fat composition (by ~ 30%) in tg/+ livers compared to those of WT littermates (in contrast to no changes in the liver lean mass composition) (Fig. 3F).
An even more striking hepatomegaly and splenomegaly was observed in HFD fed tg/+ mice (Fig. 3H–J). Quantification confirmed the significant weight increase of the liver and spleen in tg/+ mice on HFD relative to WT littermates (n = 9–10) (Fig. 3J). Notably, quadriceps muscles were not significantly different in weight between WT and tg/+ mice on a HFD (Fig. 3J).
Collectively, results from the extended chow study point to an age-dependent effect of transgenic FABP4-RORα4 overexpression on hepatic and splenic lipid loading. Due to the lipodystrophy observed in the SAT and the hepatomegaly after HFD, we focused our subsequent attention on the contribution of these tissues to the overall phenotype of the transgenic model.
It has been shown that SAT modulates liver triglyceride accumulation (Hocking et al., 2015). Moreover, failure to sustain or expand adequate subcutaneous fat storage adversely impacts glucose tolerance and contributes to ectopic fat accumulation in non-adipose organs such as the liver, increasing susceptibility to inflammatory stress and cancer (Gentile et al., 2015, Hocking et al., 2015, Wree et al., 2011). To gain a histological perspective of the changes in fat deposition of tg/+ livers, we performed H&E staining on deparaffinized liver sections from HFD-fed WT and tg/+ mice to identify significant morphological differences. We observed a striking increase in vacuole-like lipid bodies in tg/+ liver relative to WT littermates (Supplementary Fig. 3A), suggesting increased lipid storage in the tg/+ livers could be involved in hepatomegaly (Fig. 3E and H).
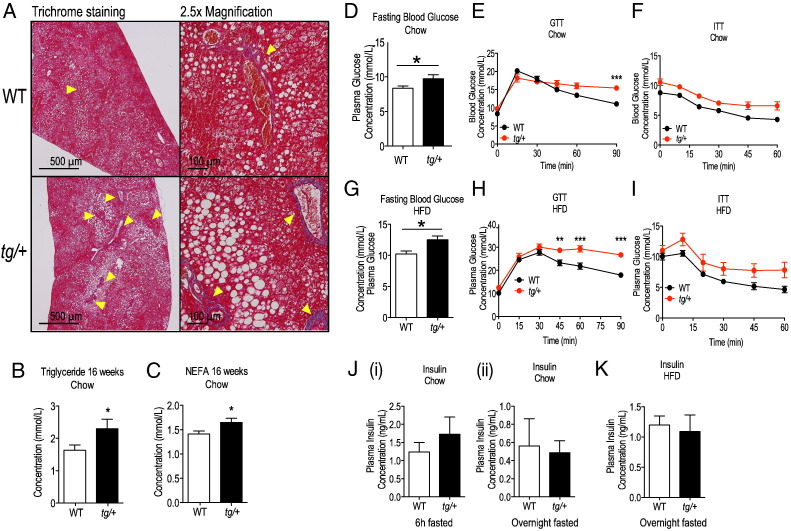
In the liver, steatosis can be accompanied by fibrosis. To check for development of fibrosis in the tg/+ livers, we performed Masson's trichrome staining to visualize collagen accumulation, a hallmark of fibrosis. In both WT and tg/+ livers of mice (fed on a HFD for 14 weeks) we observed signs of portal vein collagen accumulation with no signs of central fibrosis occurring in both strains of mice (Fig. 4A). A modest increase in collagen-positive portal veins/sites in transgenic liver was present, consistent with early, mild portal fibrosis (Fig. 4A).
Fig. 4.
Trichrome staining of HFD livers and glycemic challenge of WT and Tg-FABP4-RORα4 mice. (A) Formalin-fixed and paraffinized liver tissues of HFD-fed WT and tg/+ mice were sectioned at 0.7 μm-step intervals and stained for (A) collagen using Masson's trichrome staining. Red denotes cytoplasmic staining and the yellow arrowheads indicate collagen staining (blue). Representative images acquired from n = 4 littermate pairs are shown. (B and C) Analysis of mean ± S.E.M. fasting plasma (B) triglyceride and (C) NEFAs from 16 weeks chow-fed WT and tg/+ mice (n = 6 littermate pairs). Significance was calculated using unpaired two-tailed Student's t-test where *P < 0.05. (D) Fasting blood glucose levels from 6 h-fasted WT (white) and tg/+ (black) (n = 10 littermate pairs) mice in the chow study (16 weeks old). (E and F) Blood glucose was measured at various times after intra-peritoneal administration of (E) glucose or (F) insulin (t = 0) on 6 h-fasted WT (black) and tg/+ (red) mice (16 week chow, n = 10 littermate pairs). (G) Fasting blood glucose levels from 6 h-fasted WT (white) and tg/+ (black) mice (n = 5 littermate pairs) in the HFD study. (H and I) Blood glucose was measured at various times after intra-peritoneal administration of (H) glucose or (I) insulin (t = 0) on 6 h-fasted WT (black) and tg/+ (red) mice in the HFD study (n = 5 littermate pairs). Statistical analyses were performed using unpaired two-tailed Student's t-test for panels D and G and two-way ANOVAs with Bonferroni's post-test applied for panels E, F, H and I where *P < 0.05; **P < 0.01;***P < 0.001. (J) Plasma insulin levels of (i) 6 h-fasted (n = 5 littermate pairs) and (ii) overnight fasted WT and tg/+ littermates (n = 8 littermate pairs) from the 32-week chow study. Statistical analysis was performed using unpaired two-tailed Student's t-test but comparisons were not significant. (K) Plasma insulin levels of overnight fasted WT and tg/+ mice in the HFD study (n = 8 littermate pairs). Statistical analysis was performed using unpaired two-tailed Student's t-test but comparisons were not significant.
In summary, we observed the development of hepatomegaly and splenomegaly with FABP4-RORα4 transgene overexpression on chow diet. Upon HFD challenge, transgenic mice displayed decreased total body weight associated with a specific decrease in subcutaneous adiposity, and hepatomegaly was further exacerbated. DEXA and histological analyses of transgenic livers showed increased lipid accumulation. This was accompanied by significantly increased levels of circulating triglycerides and NEFA's in the Tg/+ mice (relative to WT littermates) on the chow diet (Fig. 4B and C); however, the blood lipid profiles on the HFD were not significantly different (data not shown). Interestingly, transgenic livers appear to be susceptible to develop mild portal fibrosis, warranting more rigorous characterization of this phenotype. Thus the effect of HFD challenge was to alter the energy storage response in the tg/+ mice from the traditional lipid depots (adipose tissues) to non-conventional sites such as the liver, imposing additional stress on these organs. This indicates a poor prognosis for the tg/+ mice because disorders in adipose fat storage function can lead to increased liver steatosis, glucose intolerance and cancer (Wree et al., 2011). Consequently, we investigated the association between aberrant energy storage and glycemic control in the FABP4-RORα4 transgenic mouse model.
3.5. Glucose Intolerance in Tg-FABP4-RORα4 Mice
Previous studies on the global Rorα-deficient staggerer (sg/sg) mutant mice revealed overall improved glucose tolerance and increased insulin sensitivity (Lau et al., 2011). Hence we examined the effect of adipose-specific RORα transgene expression on these parameters.
Compared to their WT counterparts, both tg/+ (heterozygous) (Fig. 4D) and tg/tg (homozygous) (Supplementary Fig. 2D) Tg-FABP4-RORα4 transgenic mice fed on normal chow diets exhibited elevated blood glucose levels after a 6 h fast. We further examined systemic glucose metabolism and insulin sensitivity by performing intraperitoneal glucose tolerance tests (GTTs) and insulin tolerance tests (ITTs). Although blood glucose levels in tg/+ mice peaked at a similar level to WT littermate mice, glucose clearance was impaired at the later timepoint – remaining ~ 40% higher than WT littermates at the end of the time course (Fig. 4E). Interestingly, glucose clearance in the homozygous tg/tg mice (that we could only obtain in limited number) was markedly impaired with a significant increase in peak blood glucose levels (~ 30%) after glucose challenge and sustained higher blood glucose levels were recorded throughout the time course (Supplementary Fig. 2E). The blood glucose level in tg/tg mice was ~ 65% higher than WT littermates at the end of the time course (Supplementary Fig. 2E). In contrast, WT, tg/+ (Fig. 4F) and tg/tg (Supplementary Fig. 2F) mice showed similar responsiveness to intraperitoneal insulin, indicating normal insulin action in the transgenic mice. Furthermore, plasma insulin levels in 6 h and overnight fasted tg/+ mice were not significantly different than the comparable levels in WT mice [Fig. 4J(i and ii)]. Preliminary intraperitoneal pyruvate tolerance testing also revealed no significant differences between WT and tg/+ littermates (data not shown), allowing us to exclude alterations in hepatic gluconeogenesis as a contributor to the impaired glucose clearance observed during IP glucose challenge.
In the HFD study, tg/+ mice also displayed significantly higher fasting blood glucose levels (~ 20% higher) relative to WT littermates (Fig. 4G). After intraperitoneal administration of glucose, both WT and tg/+ mice showed similar peak blood glucose levels (~ 30% increase compared to those on chow diet) and the impairment in glucose clearance (Fig. 4H). However, this was more pronounced compared to data from the chow study in Fig. 4E (blood glucose level in HFD tg/+ mice was ~ 50% higher than WT littermates at the end of the time course). While tg/+ mice did appear to be less sensitive to insulin administration, this trend did not attain significance (Fig. 4I). Fasted plasma insulin levels were also not significantly different between WT and tg/+ mice (Fig. 4K. Therefore, the impairment to glucose tolerance in Tg-FABP4-RORα4 mice appears to be independent of insulin action.
In summary, these results demonstrated a dose-dependent negative effect of adipose-RORα expression on whole body glucose clearance – an effect that was amplified in the context of high fat diet. It is intriguing to speculate whether the SAT lipodystrophy that is observed in this model contributes to compromised glucose tolerance as other studies have indicated it is essential for the maintenance of glycemic control under both normal and high fat dietary conditions (Gentile et al., 2015, Hocking et al., 2015).
3.6. Insights Into the Molecular Pathways Underlying the SAT Lipodystrophy and the Hepatomegaly Phenotype in the Tg-FABP4-RORα4 Mice
In an effort to gain molecular insights into the SAT lipodystrophy and hepatomegaly, accompanied by glucose intolerance (in the absence of changes in whole body insulin sensitivity, we undertook broad-scale (qPCR and RNA-seq) transcriptional profiling of SAT and the liver in order to identify changes in underlying molecular pathways.
3.6.1. Subcutaneous Adipose Gene Expression in Tg-FABP4-RORα4 Mice
Our strategy to identify the gene expression patterns underlying the selective SAT lipodystrophy involved a bifurcated approach, that investigated: (i) specific differences in subcutaneous relative to visceral adipose in the transgenic mouse, and (ii) differences in SAT in the transgenic relative to WT mice.
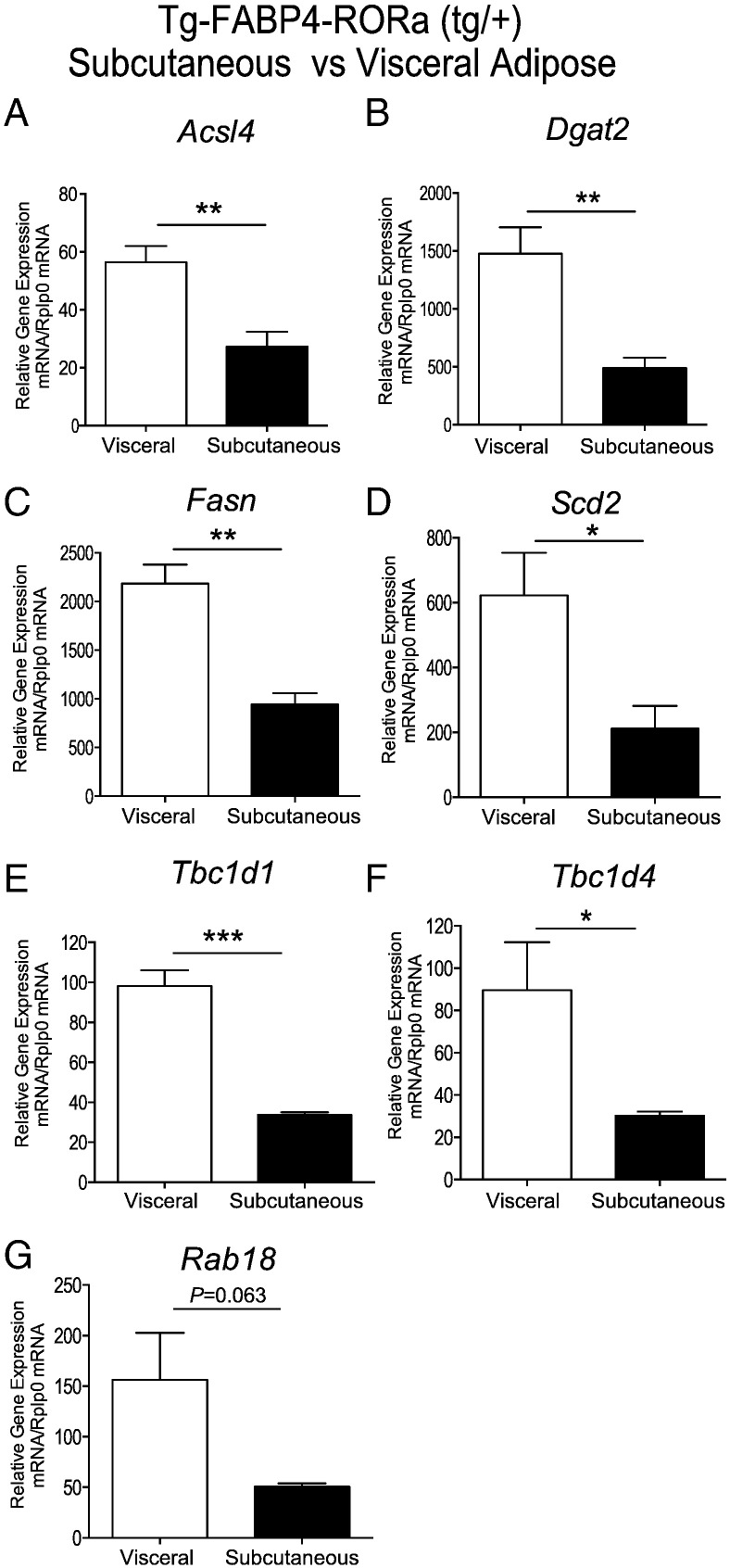
Initially, we examined the differential expression of genes in subcutaneous relative to visceral adipose (by qPCR) in the transgenic model that may influence lipid homeostasis and glycemic control. This analysis identified significantly decreased expression of several critical genes that drive fatty acid biosynthesis, and lipid droplet expansion including Acs14, Dgat2, Fasn and Scd2 in SAT relative to visceral adipose tissue from the Tg-FABP4-RORα4 (Fig. 5A–D). Moreover, we observed significantly decreased expression of the Rab-GTPase-activating proteins (Rab-GAPs), Tbc1d1 and AS160/Tbc1d4 (Fig. 5E and F) in the Tg-FABP4-RORα4 SAT. Decreased expression of Rab GTPase-activating proteins has been reported to correlate with decreased glucose tolerance in an intraperitoneal GTT assay, aberrant glucose uptake and dysfunctional carbohydrate and energy homeostasis (Hargett et al., 2016, Chadt et al., 2015). Furthermore, we observed decreased Rab18 expression (that failed to attain significance, P = 0.06), which has been associated with impaired fat storage (Pulido et al., 2011), and aberrant processing of lipids in adipose tissue (Pulido et al., 2013) (Fig. 5F). In summary, this analysis identified the differential expression of several critical genes in SAT (relative to visceral adipose) that provided some insights into the subcutaneous lipodystrophic and glucose intolerant phenotype in the Tg-FABP4-RORα4.
Fig. 5.
Taqman low density array (TLDA) analysis of mRNA expression from SAT relative to visceral adipose tissue of Tg-FABP4-RORα4 mice on a high fat diet. Relative gene expression (normalized against Rplp0) of (A) Acsl4, (B) Dgat2, (C) Fasn, (D) Scd2, (E) Tbc1d1, (F) Tbc1d4 and (G) Rab18. TLDAs performed as described (Pearen et al., 2012, Pearen et al., 2013). Statistical analysis was performed on data acquired from n = 4 littermate pairs using unpaired two-tailed Student's t-test where *P < 0.05; **P < 0.01; ***P < 0.001.
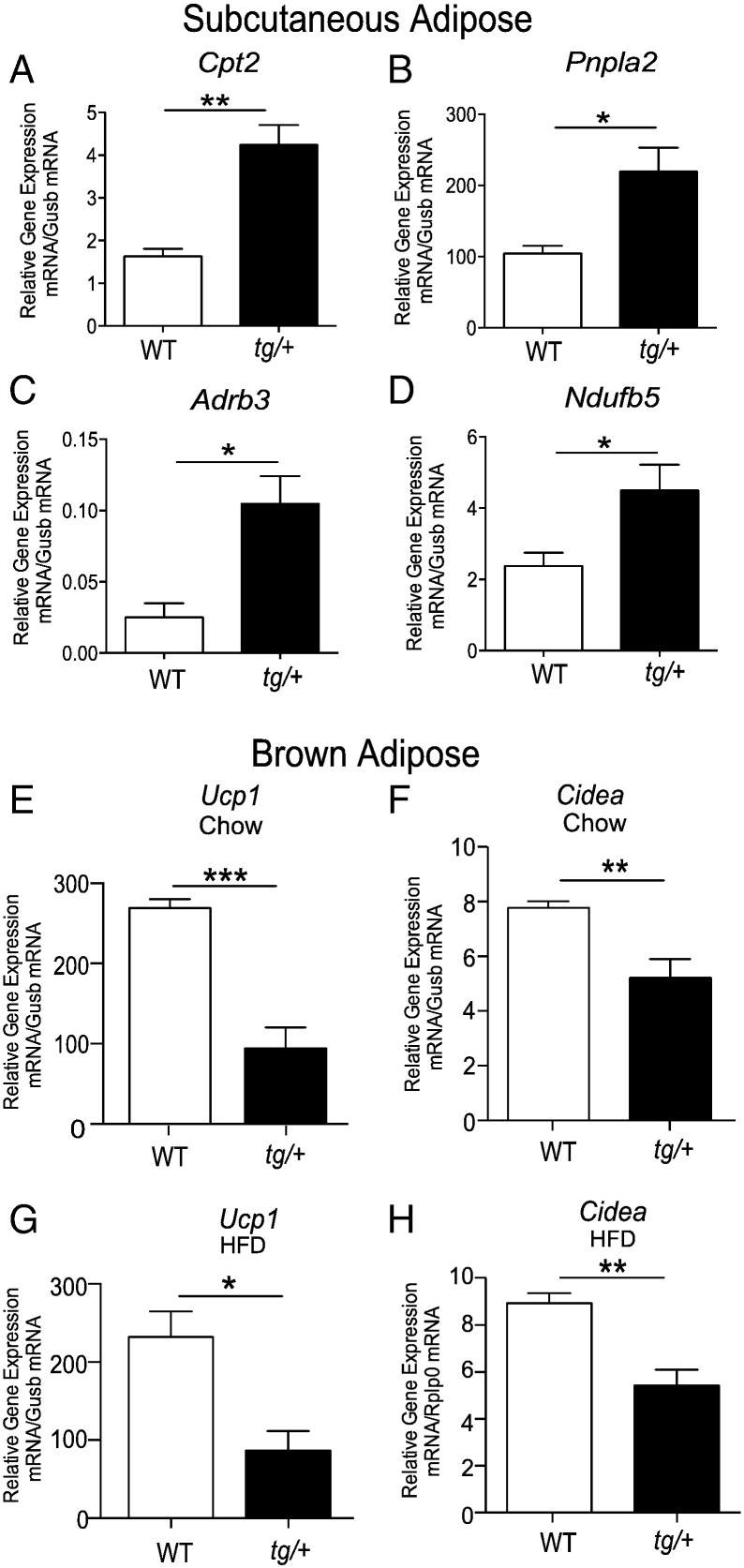
To gain further insights we examined the differential expression of several metabolic genes that may contribute to decreased SAT deposition/expansion (on HFD) in the Tg-FABP4-RORα4 mouse model, relative to WT littermates. We uncovered several gene changes that are consistent with elevated lipid mobilization and catabolism in the SAT. For example, we identified significant up-regulation of the gene encoding carnitine palmitoyltransferase 2 (Cpt2) in the adipose tissue of heterozygote tg/+ mice (Fig. 6A). Cpt2 catalyzes rate-limiting steps in fatty acid uptake and oxidation by mitochondria (Bonnefont et al., 2004, Jogl et al., 2004). In addition, we also observed significant up-regulation of patatin-like phospholipase domain containing 2 [Pnpla2, also known as adipose triglyceride lipase (ATGL)] in tg/+ SAT (Fig. 6B). PNPLA2/ATGL is the rate-limiting enzyme that catalyzes the hydrolysis of triglycerides and plays a key role in lipid droplet degradation (Smirnova et al., 2006). Moreover, we observed increased expression of the mRNAs encoding the β3-adrenergic receptor (Adrb3) (Fig. 6C) in concordance with increased lipolysis and fatty acid utilization (Ghorbani et al., 1997, Granneman et al., 2005, Mottillo et al., 2010). Furthermore, the gene that encodes one of the critical NADH dehydrogenases that are part of the mitochondrial complex I machinery (Ndufb5) was significantly up-regulated in the SAT of Tg-FABP4-RORα4 tg/+ mice (Fig. 6D). Differential and increased expression of Cpt2, Pnplas/ATGL, Adrb3, and Ndufb5 is entirely concordant with increased lipid mobilization and utilization in the SAT from Tg-FABP4-RORα4 tg/+ mice, and would contribute to decreased fat deposition/expansion in this depot by mice that overexpress RORα4 in adipose tissue.
Fig. 6.
Taqman low density array (TLDA) analysis of mRNA expression from subcutaneous adipose and brown adipose tissue from Tg-FABP4-RORα4 relative to WT mice. Relative gene expression of (A–D) Cpt2, Pnpla2, Adrb3, Ndufb5, respectively in SAT from HFD fed 32-week tg/+ Tg-FABP4-RORα4 and WT littermates (from n = 4). TLDAs performed as described (Pearen et al., 2012, Pearen et al., 2013). and presented as relative gene expression (normalized against Gusb). Relative gene expression of Ucp1 and Cidea, respectively in brown adipose tissue from chow (E–F) and high fat diet (G–H) fed 32-week tg/+ Tg-FABP4-RORα4 and WT littermates (n = 4). Quantitative PCR was performed on RNA fractionated from interscapular brown adipose tissue, and measured Ucp1 and Cidea expression (using Taqman and SYBR assays), and presented as relative gene expression (normalized against Gusb, and Rplp0, respectively) in samples from 32-week chow (E–F) and high fat diet fed (G–H) Tg-FABP4-RORα4 and WT littermates (from n = 5). Statistical analysis was performed using unpaired two-tailed Student's t-test where *P < 0.05; **P < 0.01; ***P < 0.001.
We subsequently investigated gene expression associated with adaptive thermogenesis in the brown adipose tissue from the Tg-FABP4-RORα4 transgenic mouse model, relative to WT littermates. Previously, we identified that decreased adiposity in the Rorα-deficient sg/sg mice was associated with differential expression of the hierarchical adipose transcriptional regulators (Ehmt1, Tle3, Prdm16), increased Ucp1 expression, and elevated expression of genes associated with the induction of the thermogenic program (Lau et al., 2015). Therefore, we performed qPCR analysis on selected key thermogenic regulatory genes from chow-fed Tg-FABP4-RORα4 tg/+ mice, including Ehmt1, Tle3, Prdm16, Ucp1 and the accompanying genes associated with thermogenesis (e.g. Cidea, Acot11, Dio2, Errα, Pparα, and Mcpt1). We observed no significant differences in the mRNA expression of the transcriptional regulators, Ehmt1, Tle3 and Prdm16 in brown adipose tissue (Supplementary Fig. 4), but we observed a significant decrease in the expression of the mRNAs encoding Ucp1 (by ~ 60–70%) and Cidea (by ~ 30–40%) in the brown adipose from Tg-FABP4-RORα4 tg/+ mice, relative to WT littermates on the chow and high fat diets, respectively (Fig. 6E and F vs. Fig. 6G and H, respectively). However, we did not observe any significant differences in the expression of the other genes involved in the thermogenic program in the brown adipose from Tg-FABP4-RORα4 tg/+ mice, relative to WT littermates on the chow and high fat diets, respectively (Supplementary Fig. 4). The decreased expression of Ucp1, is consistent with increased Ucp1 mRNA expression in the Rorα-deficient sg/sg mouse model, and provides further evidence for the association between RORα signaling and Ucp1 mRNA expression. Moreover, decreased Cidea expression correlates with increased lipolysis, and decreased triglyceride deposition/lipid droplet formation (Puri et al., 2008). Overall, the lipodystrophic SAT phenotype in the Tg-FABP4-RORα4 tg/+ mice is in accord with gene expression changes that affect lipid homeostasis.
3.6.2. RNA-seq and Pathway Analysis
We subsequently performed genome wide RNA-seq expression profiling analysis on RNA collected from SAT and hepatic tissue to gain deeper insights into the underlying molecular pathways. The analysis identified > 2400 genes/mRNA (total, after Benjamini and Hochberg correction) that were differentially expressed in the SAT in a significant manner. We could also identify that RORα was significantly up-regulated by ~ 10-fold (P < 6.08 × 10− 89) in tg/+ SAT (Supplementary Table 1, highlighted in gray). In contrast, only ~ 70 genes (total, after Benjamini and Hochberg correction) in the liver were differentially expressed in a significant manner (n = 3 littermate pairs). The top 50 up- and down-regulated genes in SAT are listed in Supplementary Table 1 and the up- and down-regulated genes in the liver are listed in Supplementary Table 1.
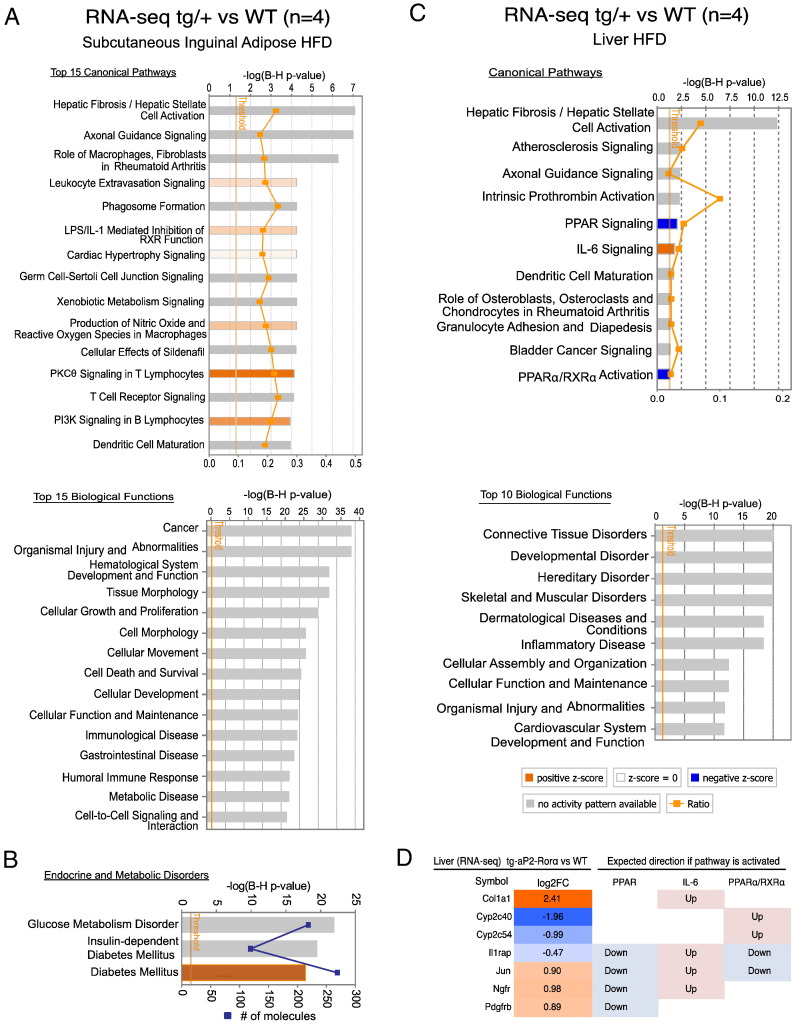
Ingenuity pathway analysis was performed to identify canonical pathways and biological functions that were significantly enriched with differentially expressed genes from the RNA-seq analysis. This was performed with a cut-off value of ≥±1.3 fold-change (~ 1800 genes after cut-off). Pathway analysis on SAT identified significant enrichment of differentially expressed genes involved in ~ 140 canonical pathways in Tg-FABP4-RORα4 tg/+ mice. The analysis revealed that the differentially expressed genes enrich approximately ~ 20% of each pathway (Fig. 7A). The top 15 significantly enriched canonical pathways and biological functions are shown in Fig. 7A. These include (the most highly ranked pathway) hepatic fibrosis (P < 8.28 × 10− 8). Interestingly, the pathway analysis also identified significant enrichment of functions associated with endocrine disorders and metabolic diseases, corresponding to glucose metabolism disorder (P < 5.62 × 10− 24), insulin-dependent diabetes mellitus (P < 2.39 × 10− 21) and diabetes mellitus (P < 1.44 × 10− 19) and further predicts that diabetes mellitus is increased (z-score = 2.303) (Fig. 7B). Interestingly, 10 out of the top 15 significantly enriched canonical pathways are associated with immune signaling and inflammatory response. In summary, this is in accord with the phenotypes of impaired glucose clearance observed and highlights increased susceptibility in developing diabetes mellitus at the transcript level with adipose-specific RORα expression.
Fig. 7.
RNA-seq and ingenuity pathway analysis of HFD inguinal adipose tissue and liver. RNA-seq was performed on RNA isolated from overnight fasted WT and Tg-FABP4-RORα4 (heterozygous) tg/+ mice that were in the HFD study (n = 4 littermate pairs). (A) The top 15 canonical pathways and biological functions enriched by differentially expressed genes from the RNA-seq analysis comparing WT and tg/+ inguinal adipose tissues (HFD) highlighted by ingenuity pathway analysis. Ratio indicates the percentage of differentially expressed genes per any given pathway and P-value threshold is set at P < 0.05 derived after Benjamini and Hochberg (B–H) multiple testing correction. (B) Significantly enriched metabolic functions. Orange bars indicate positive z-score (predicted activation) and gray bars indicate no prediction of activation/inhibition state. (C) Significantly enriched canonical pathways and top 10 biological functions associated with differentially expressed genes in tg/+ liver (HFD). (D) Gene expression changes in liver tissues of Tg-FABP4-RORα4 vs. WT (n = 4 littermate pairs) comparing to expected expression direction of PPAR, IL-6 and PPARα/RXRα signaling activation.
Ingenuity pathway analysis of the liver is limited, identifying only 11 canonical pathways as significantly enriched with the list of differentially expressed genes from RNA-seq analysis (~ 70 genes) between WT and Tg-FABP4-RORα4 tg/+ hepatic tissue (Fig. 7C). Notably, hepatic fibrosis was the most significant pathway in tg/+ liver (that displayed hepatomegaly) as discussed previously. The pathway analysis and heatmap depicted in Fig. 7D, also predicts that pathways associated with PPAR signaling (P < 8.47 × 10− 3) and PPARα activation (P < 4.69 × 10− 2) are inhibited while IL-6 signaling pathway (P < 1.52 × 10− 2) is activated (Fig. 7C/D). PPARs are well-known NRs that play a central role in fatty acid metabolism (in particular lipid catabolism). IL-6 is a pro-inflammatory cytokine and has been linked to development of hepatic steatosis and hepatic insulin resistance (Sabio et al., 2008). These observations in the hepatic tissue are in accord with increased lipid storage or possible tissue damage/fibrosis in Tg-FABP4-RORα4 tg/+ livers. Moreover, functional DAVID pathway analysis of the liver (Supplementary Fig. 3B) revealed significant enrichment of functions that correspond to organization of collagen fibrils (P < 3.59 × 10− 13) and inflammation of organ (P < 2.07 × 10− 09).
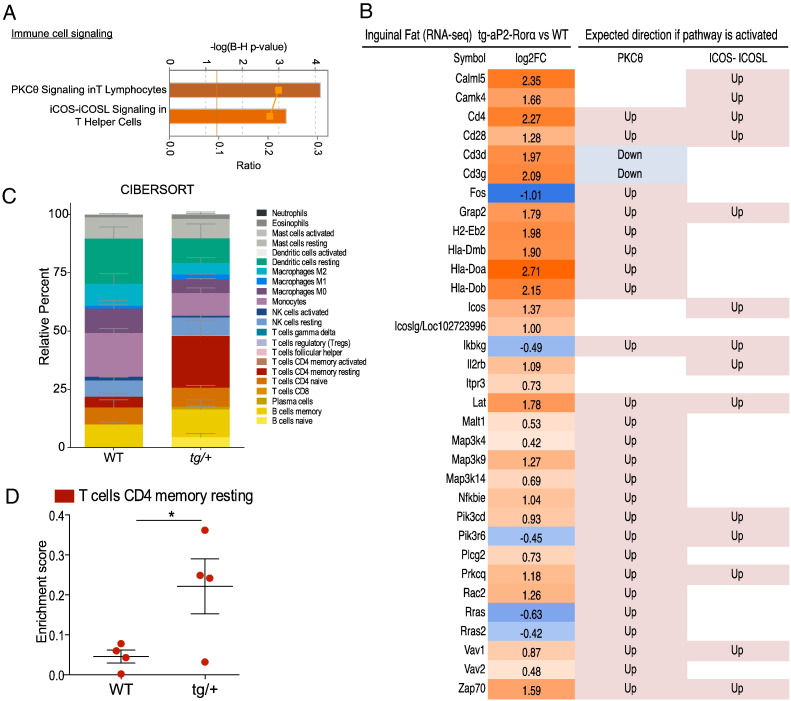
3.6.3. RORα4 Expression and Immunity in SAT
The association between RORα4 expression and immunity in adipose tissue is underscored by the pathway analysis identification of significant activation of T lymphocytes signaling pathways, including PKC-θ signaling in T lymphocytes (~ 30% of pathway enriched, z-score = 2.502, P < 7.22 × 10− 05) and inducible T-cell co-stimulator (ICOS)-ICOS-ligand (ICOSL) signaling in T helper (Th) cells (~ 22% of pathway enriched, z-score = 2.673, P < 6.28 × 10− 4) (Fig. 8A-immune cell signaling). The two significantly enriched T-cell signaling pathways comprised of several overlapping genes suggest that inflammatory signaling may be activated in the SAT of Tg-FABP4-RORα4 tg/+ mice as a result of RORα4 overexpression. The heatmap in Fig. 8B details the fold changes of the genes involved in the two pathways accompanied with expected direction of gene changes when the immune response pathways are activated. Thus, the gene changes identified in the RNA-seq/pathway analysis of the SAT, highlight potential changes associated with T-cell signaling and PKC-θ signaling in tg/+ SAT. Recently, there have been several high profile publications utilizing analytical tools to allow for the discrimination of cell subsets and cell composition in complex tissues from their gene expression profiles (Newman et al., 2015, Qi et al., 2014). We utilized the CIBERSORT gene expression deconvolution analytical tool (Newman et al., 2015) on our RNA-seq data and plotted the predicted relative frequency of the immune cell subtypes contained in the LM22 signature into a stacked format. CIBERSORT was able to identify a significant increase in CD4 T-cell signature (red) in the SAT from 3/4 of the tg/+ Tg-FABP4-RORα4 mice relative to WT littermates (Fig. 8C and D).
Fig. 8.
Activation of T-cell signaling and CIBERSORT analysis. (A) Significantly enriched immune cell signaling functions in HFD tg/+ SAT. Orange bars indicate positive z-score (predicted activation from ingenuity pathway analysis). (B) The heatmap presents differentially expressed in SAT from Tg-FABP4-RORα4 vs. WT (n = 4 littermate pairs). Fold changes were shown in log2 scale with expected expression direction of PKC-θ and ICOS-ICOSL pathways (activation). (C–D) CIBERSORT analysis of SAT RNA-seq data for the quantification of relative levels (percentage) of 22 immune cell subtypes (LM22 gene signature). Relative percentages are plotted in a stacked format in (C). The data specific to memory resting CD4 T cells listed in C (red module) was extracted and an unpaired two-tailed Student's t-test was performed to calculate for significance where *P < 0.05.
3.6.4. RORα4 Expression, Fibrosis and Extracellular Matrix Remodeling in SAT and Liver Tissue
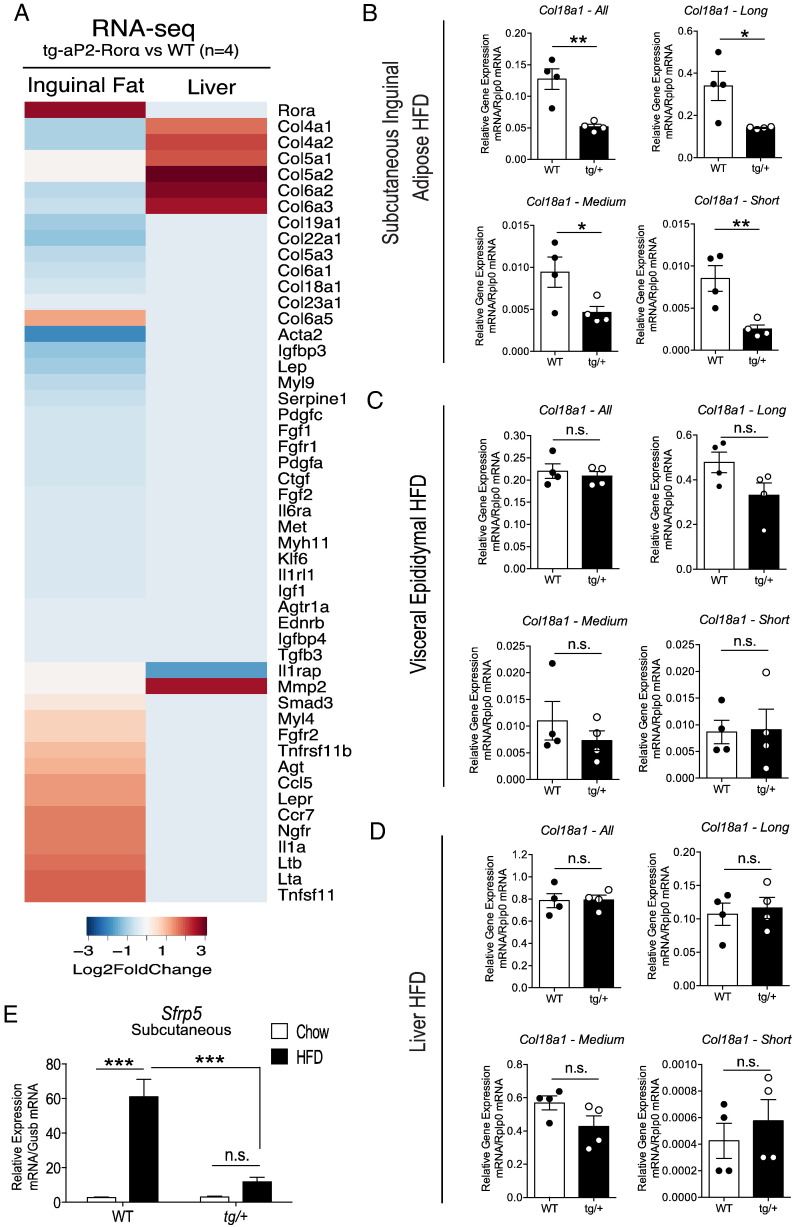
Interestingly, the most significant canonical pathway in both Tg-FABP4-RORα4 tg/+ SAT (P < 8.28 × 10− 8) and hepatic tissue (P < 5.11 × 10− 13) is hepatic fibrosis (Fig. 7A and C). Although, there is a contrast in the regulation of the genes between the two tissues, this is in accordance with the phenotype. For example, many of the genes that encode the extracellular matrix (ECM) collagen proteins are down-regulated in the Tg-FABP4-RORα4 tg/+ SAT, but increased in hepatic tissue (Fig. 9A). This is highlighted by the differential expression of collagens 4, 5, 6, 18, 19 and 23 (Fig. 9A), indicating ECM directed molecular mechanisms are affecting fat deposition/energy storage in adipose and liver. Collagen production and regulation is important for adipose development and physiology, as well as adipose plasticity to suit metabolic demands and changes (Mariman and Wang, 2010). In addition ECM reprogramming is necessary for adipose growth and expansion on energy dense diets. In contrast, collagen production in liver is associated with liver fibrosis. The role of collagens in controlling adiposity is highlighted by loss of collagen XVIII (18) (significantly down-regulated in Tg-FABP4-RORα4 tg/+ SAT RNA-seq data) that results in reduced adiposity, ectopic deposition of fat in the liver and hypertriglyceridemia (Aikio et al., 2014). This effect was attributed to reduced fat storage capacity due to perturbations in ECM remodeling and adipocyte development associated with aberrant Wnt/β-catenin signaling (Aikio et al., 2014). Therefore, we used qPCR to validate and demonstrate the (~ 2–3-fold) significantly decreased expression of the Col18a1 long, medium and short isoforms in SAT (Fig. 9B). In contrast, no significant differences in expression were observed in visceral adipose tissue and hepatic tissue (Fig. 9C and D). The recognized association between aberrant ECM and Wnt/β-catenin signaling in dysfunctional fat deposition was underscored by the RNA-seq analysis in SAT, that identified significant down-regulation of the mRNA encoding secreted frizzled-related protein 5 (Sfrp5) by ~ 5-fold (P < 8.55 × 10− 56) (Supplemental Table 1, highlighted in gray). Sfrp5 is a Wnt antagonist that is typically upregulated in adipose during diet-induced obesity. Quantitative PCR analysis further highlighted the attenuated response of Sfrp5 mRNA expression to a high fat diet challenge in the Tg-FABP4-RORα4 relative to WT mice, and the significantly decreased expression of Sfrp5 mRNA expression in the SAT from Tg-FABP4-RORα4 relative to wild type on the high fat diet (Fig. 9E). Overall, the difference in Sfrp5 induction in Tg-FABP4-RORα4 tg/+ mice supports the findings of reduced weight gain associated with specific reduction of SAT fat deposition/expansion in HFD-fed Tg-FABP4-RORα4 tg/+ mice, and further suggests that the ability to expand the adipose tissue may be altered in these mice.
Fig. 9.
Gene expression changes in the hepatic fibrosis pathway and ECM regulation. (A) Heatmap of differentially expressed genes associated with hepatic fibrosis. Data was extracted from STAR-DESeq2 output (RNA-seq pipeline). Each column represents log2 fold-changes comparing SAT and liver tissues of Tg-FABP4-RORα4 vs. WT (n = 4 littermate pairs). Relative gene expression of Col18a1 in (B) SAT, (C) visceral adipose tissue, and (D) liver, respectively from tg/+ Tg-FABP4-RORα4 and WT littermates in the HFD study (n = 4–5 littermate pairs). Quantitative PCR (utilizing SYBR primers) was performed on RNA fractionated from the tissues and presented as relative gene expression normalized against Rplp0. Statistical analysis was performed using unpaired two-tailed Student's t-test where *P < 0.05; **P < 0.01; ***P < 0.001. (E) Relative gene expression of Sfrp5 in inguinal SAT from tg/+ Tg-FABP4-RORα4 and WT mice in the HFD study (n = 4 littermate pairs). Quantitative PCR was performed using TaqMan assays and presented as relative gene expression (normalized against Gusb). Statistical analysis was performed using a two-way ANOVA with Bonferroni's post-test applied where ***P < 0.001; n.s. denotes non-significant.
In summary, adipose specific transgenic RORα4 expression results in the differential expression of collagen genes (in SAT and liver) that are critical for ECM remodeling, and aberrant Wnt signaling - both of which are the underlying factors in altered fat deposition in adipose and hepatic tissues. This is in accord with the very recent study demonstrating the fat microenvironment controls depot specific expansion and tissue mass on obesogenic diets (Jeffery et al., 2016), and obesogenic studies on Wnt signaling (Mori et al., 2012, Gutierrez-Vidal et al., 2015). Furthermore, the two gene expression profiling approaches (qPCR and RNA-seq) identified differential expression of many genes involved in lipid, and glucose homeostasis coupled to ECM remodeling, that are in accord with hepatic fibrosis as the most significantly enriched pathway in Tg-FABP4-RORα4 tg/+ SAT and liver. This provides molecular evidence supporting our observations of reduced subcutaneous adiposity, impaired glucose tolerance, ectopic fat accumulation and liver fibrosis. This phenotype highlights important crosstalk between the adipose tissue and the liver, under the modulation of adipose-specific ROR alpha expression.
4. Discussion
The sterol-sensing ‘orphan’ NR, RORα, is expressed in a number of metabolically active tissues and cell types, including the adipose tissue, muscle, resident tissue macrophages and lymphocytes (Bookout et al., 2006). This NR has been implicated in the regulation of lipid homeostasis and responses to energy dense diets. These factors, and the significance of lipid distribution in adipose depots and lean tissue suggested a potentially important role for RORα in obesity and inflammation. Adipose tissue is important for the regulation of energy homeostasis, in part due to its role in storing triglyceride and secreting many endocrine signaling proteins (Scherer, 2006). The local and systemic effects of overexpressing RORα in the adipose tissues of Tg-FABP4-RORα4 mice were examined herein.
Perhaps the most striking result from this study was that Tg-FABP4-RORα4 mice demonstrated a remarkable shift in energy storage and fat distribution from the SAT to non-adipose tissues when challenged with a high fat diet (HFD). Specifically, we observed a subcutaneous lipodystrophy associated with pronounced hepatomegaly and splenomegaly. These changes were also accompanied with decreased overall weight gain and decreased total body fat percentage after HFD. Decreased adiposity and reduced weight gain were most probably a reflection of decreased fat deposition in the subcutaneous depot. Moreover, we also observed significantly higher fasting blood glucose levels and impaired clearance of intra-peritoneally injected glucose in chow-fed Tg-FABP4-RORα4 mice. The glucose clearance impairment was further exacerbated when Tg-FABP4-RORα4 mice were placed on the HFD challenge. However, there were no apparent differences in insulin sensitivity, or circulating insulin levels. Histological analysis confirmed markedly increased accumulation of lipid bodies and signs of portal fibrosis throughout the liver derived from Tg-FABP4-RORα4 mice. In comparison, chow-fed Tg-FABP4-RORα4 mice also presented with splenomegaly and mild hepatomegaly at the end of the study, but do not display changes to the adipose tissues and overall growth. This suggests that the Tg-FABP4-RORα4 mice do not adapt appropriately to the physiological challenges of an energy-dense diet in the context of lipid storage and expansion of adipose tissue, but rather are associated with aberrant energy storage and ectopic fat deposition in other (lean mass) organs.
We utilized two approaches to understand the molecular mechanisms underscoring the observed phenotype of subcutaneous lipodystrophy with accompanying hepatic lipid accumulation and impaired glucose tolerance: targeted qPCR profiling of critical metabolic genes and RNA-seq deep sequencing coupled with pathway analyses.
Focused qPCR analysis of critical metabolic genes in the transgenic SAT (relative to the visceral adipose tissue) identified the selective and significantly decreased expression of several important genes that drive fatty acid biosynthesis, and lipid droplet expansion including Acs14, Dgat2, Fasn and Scd2 in SAT from the Tg-FABP4-RORα4 line. Furthermore, we identified decreased expression of Rab 18 and the Rab-GAPs, Tbc1d1 and AS160/Tbc1d4. This pattern of expression in adipose tissue has been associated with impaired glycemic control, energy homeostasis, and fat storage (Hargett et al., 2016, Chadt et al., 2015, Pulido et al., 2013). The subcutaneous specific differential expression of several critical genes provided some insights into the subcutaneous lipodystrophic and glucose intolerant phenotype in the Tg-FABP4-RORα4.
Moreover, additional qPCR profiling revealed the SAT lipodystrophy in the Tg-FABP4-RORα4 mouse model is also associated with increased lipid mobilization and catabolism (relative to WT littermates). For example, we identified increased expression of the genes encoding Cpt2, Pnpla2/ATGL and Adrb3 (Bonnefont et al., 2004, Jogl et al., 2004, Smirnova et al., 2006, Ghorbani et al., 1997, Granneman et al., 2005, Mottillo et al., 2010). This phenotype was coupled with decreased expression of the (adaptive) thermogenic driver, Ucp1, and Cidea (a marker of increased lipolysis) in brown adipose tissue. This was in accord with increased Ucp1 and Cidea expression in the brown adipose from RORa deficient staggerer mice.
RNA-seq and pathway analysis identified differential expression of many genes involved in lipid localization and transport, glucose metabolism disorder and diabetes in Tg-FABP4-RORα4 SAT. The analysis revealed fibrosis as the most significant pathway in both the SAT and liver. Interestingly, these two tissues display dysfunctional and aberrant (and often reciprocal) regulation of mRNAs encoding ECM collagen proteins; collagen transcripts were down-regulated in the SAT and inversely up-regulated in the liver. Collagen (5, 6 and 18) production and regulation has been implicated in adipose development and physiology, as well as adipose plasticity to suit metabolic demands and changes (Mariman and Wang, 2010). Aberrant regulation and expression of the extracellular matrix has been demonstrated to drive perturbed adipose reprogramming during obesity and ectopic fat accumulation (Tam et al., 2012, Aikio et al., 2014), ECM-guided molecular mechanisms control the adipogenic pathways and adipocyte size. Specifically, decreased collagen expression in the SAT is in accord with decreased adipocyte size, decreased fat deposition and ectopic fat accumulation (Aikio et al., 2014). In this context we observed the selective decrease in the expression of the mRNAs encoding the short, medium and long forms of Col18a1 in SAT, but not in visceral adipose tissue or liver.
Furthermore, in this context we identified the significantly decreased expression of (the Wnt antagonist) SFRP5. Down regulation of Wnt signaling and gene expression are associated with adipogenic differentiation (triglyceride storage), and Wnt3A driven dedifferentiation of adipocytes decreases the medium and long forms of Col18a1 (Aikio et al., 2014). In contrast, SFRP5 is normally induced during differentiation to attenuate Wnt signaling (Christodoulides et al., 2009) and increased during diet-induced obesity (Koza et al., 2006, Lagathu et al., 2009, Mori et al., 2012, Okada et al., 2009). Positive Wnt/β-catenin signaling inhibits adipogenesis and Sfrp5 is strongly induced during adipocyte differentiation to counteract it (Christodoulides et al., 2009). Consequently, Sfrp5-deficient mice are resistant to obesity (Mori et al., 2012). Interestingly, reduced hepatic SFRP5 (mRNA and protein) expression in morbidly obese women is associated with fatty liver disease (Gutierrez-Vidal et al., 2015). Overall, the difference in Sfrp5 induction in Tg-FABP4-RORα4 tg/+ mice supports the findings of reduced weight gain associated with specific reduction of SAT fat deposition/expansion in HFD-fed Tg-FABP4-RORα4 tg/+ mice, and further suggests that the ability to expand the adipose tissue may be altered in these mice.
Conversely, ECM accumulation in the liver is a hallmark of hepatic fibrosis and is associated with hepatic stellate cell activation due to inflammatory signaling (Bataller and Brenner, 2005). ECM reprogramming is necessary for adipose growth and expansion on energy dense diets, and fibrosis in fat tissue is associated with obesity in humans. In children, the presence of collagen in fat is associated with adipocyte size, body mass index and M2 phenotype macrophages, providing further evidence of the association between ECM remodeling and innate immunity (Tam et al., 2012). Interestingly, loss of collagen 18 (significantly down-regulated by ~ 2–3-fold in Tg-FABP4-RORα4 SAT) results in reduced adiposity, ectopic deposition of fat in the liver and hypertriglyceridemia (Aikio et al., 2014). The phenotype was attributed to reduce fat storage capacity as a result of perturbations in adipocyte development. Interestingly, subcutaneous lipodystrophy, liver steatosis and glucose intolerance are observed in humans with PPAR γ mutations Clearly both are NR dependent, and the metabolic phenotypes have clear parallels (Savage et al., 2003).
Another interesting feature revealed by the RNA-seq analysis suggests increased T-cell involvement/recruitment in the SAT of Tg-FABP4-RORα4 mice. While RORα is known to regulate inflammation and influence the development of specific lymphocyte populations, for example T helper 17 cells and group 2 innate-like lymphocytes (Halim et al., 2012, Mjosberg et al., 2012), the biological significance of the increased lymphocyte infiltrate in this mouse model and its relation to the phenotypes remains to be elucidated.
Overall, our collective data suggests that RORα overexpression in the SAT inhibits adipose plasticity, reducing fat deposition and expansion in the tissue, in addition to a shift in lipid homeostasis toward increased lipolysis and mobilization to secondary organs. This potentially perpetuates increased inflammatory signaling and hepatic stellate cell activation in the liver, activating the fibrotic program. The evident adverse effects on the liver are possibly a compensatory mechanism for adipose dysfunction. Pathway analysis of the liver provides support for this hypothesis, predicting inhibition of PPARα signaling and activation of IL-6 signaling.
In humans, there is a positive correlation between greater amounts of lower-body (gluteo-femoral in particular) SAT depots and protection against glucose intolerance and insulin resistance, dyslipidemia and atherosclerosis (reviewed in Manolopoulos et al., 2010). In this context, our data is in line with the view that subcutaneous fat serves as a protective metabolic sink for excess energy and loss of this depot/protection leads to ectopic fat accumulation and impaired glucose clearance. This severely hinders normal tissue function and perpetuates considerable amounts of stress in these organs. Moreover, recent studies report metabolic benefits acquired after SAT transplantation into the intra-abdominal cavity in mice, effectively conferring protection against HFD-induced glucose intolerance and hepatic lipid loading (Hocking et al., 2015, Konrad et al., 2007, Tran et al., 2008). For example, mice implanted intra-abdominally with SAT, but not epididymal visceral tissues, were protected against HFD-induced glucose intolerance. These mice were also protected against hepatic triacylglycerol accumulation and inflammation after HFD. However, the underlying mechanism remains obscure as there were no differences in weight gain, glucose uptake by other tissues (including the skeletal muscle), or plasma adipokine concentrations (Hocking et al., 2015).
In light of the metabolic benefits conferred by expansion of SAT and transplantation of SAT into intra-abdominal cavity, it is clear that maintenance (or even supplementation) of SAT integrity offers protection against glucose intolerance and lipid imbalance during metabolic disease. Failure to sustain or expand adequate subcutaneous fat storage adversely impacts glucose tolerance and contributes to ectopic fat accumulation in non-adipose organs such as the liver, increasing susceptibility to inflammatory stress and cancer (Gentile et al., 2015, Hocking et al., 2015, Wree et al., 2011). The pathology presented thus far in the Tg-FABP4-RORα4 transgenic mouse model is reminiscent of metabolic dysfunction, such as those of childhood and adolescent obesity. Obese youths typically present with (i) decreased subcutaneous adiposity, (ii) adipose tissue dysfunction accompanied with macrophage, dendritic cells and T-cells recruitment and pro-inflammatory signaling, (iii) increased hepatic lipid deposition/steatosis and muscle lipid deposition, and (iv) impaired glucose tolerance and insulin sensitivity (Corgosinho et al., 2012, Aguilar et al., 2013, Rigamonti et al., 2013, Burgert et al., 2006, Santoro et al., 2013).
Finally, in the context of this adipose specific FABP4 transgenic (over expression) mouse model phenotype, it is essential to discuss the caveats of this animal model. In the last five years several reviews have highlighted the advantages and disadvantages of the different promoters (for example FABP4 vs. adiponectin) for adipose specific gain and loss of function studies including the following: (i) physiological implications of leaky expression in non-adipose tissues and over expression and (ii) ectopic gene position and copy number (Wang et al., 2010, Kang et al., 2014, Jeffery et al., 2014, Wang et al., 2014, Lee et al., 2013). Recently it has been demonstrated that the FABP4 and adiponectin Cre produced very similar phenotypes in side by side comparisons (Kim et al., 2016). However, these qualifications we discussed above need to be considered in the interpretation of any transgenic over expression model.
In conclusion, this animal model study suggests that the NR, RORα4, has a critical regulatory role in the phenotype associated with decreased subcutaneous fat deposition, fatty liver and impaired glucose tolerance. Thus, establishing a role for RORα in these processes will potentially allow us to pharmacological exploit its regulation for treatment of these conditions.
Author Contributions
Z.K.T., and R.F., analyzed the cellular and metabolic phenotype of the transgenic mice. Z.K.T., S-C.M.W., and P.L. performed RNA extractions, Taqman low density array and qPCR analysis. T.G.O. analyzed RNA seq data, performed IPA canonical and pathway analyses, cibersort and bioinformatic characterization. P.L. constructed the transgenic expression vector and constructed the transgenic FABP4-RORα4 line. F.S and G.T. performed experiments to analyze body composition and utilization of MRI and DEXA. Z.K.T., S-C.M.W., T.G.O. and P.L. prepared and edited the figures. G.E.O.M. conceived and supervised the project, was involved in experimental analysis and interpretation. Z.K.T., R.F., and G.E.O.M. wrote the manuscript. All the authors were involved in the editing and proofreading of the manuscript.
Conflicts of Interest
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Carol Wicking for generously providing the trichrome staining kit.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.08.027.
Contributor Information
Rebecca Fitzsimmons, Email: r.fitzsimmons@imb.uq.edu.au.
George E.O. Muscat, Email: g.muscat@imb.uq.edu.au.
Appendix A. Supplementary Data
Supplementary material.
References
- Abdennour M., Reggio S., Le Naour G., Liu Y., Poitou C., Aron-Wisnewsky J., Charlotte F., Bouillot J.L., Torcivia A., Sasso M., Miette V., Zucker J.D., Bedossa P., Tordjman J., Clement K. Association of adipose tissue and liver fibrosis with tissue stiffness in morbid obesity: links with diabetes and BMI loss after gastric bypass. J. Clin. Endocrinol. Metab. 2014;99:898–907. doi: 10.1210/jc.2013-3253. [DOI] [PubMed] [Google Scholar]
- Aguilar M.J., Gonzalez-Jimenez E., Antelo A., Perona J.S. Insulin resistance and inflammation markers: correlations in obese adolescents. J. Clin. Nurs. 2013;22:2002–2010. doi: 10.1111/jocn.12034. [DOI] [PubMed] [Google Scholar]
- Aguiree F., Brown A., Cho N.H., Dahlquist G., Dodd S., Dunning T., Hirst M., Hwang C., Magliano D., Patterson C. 2013. IDF Diabetes Atlas. [Google Scholar]
- Aikio M., Elamaa H., Vicente D., Izzi V., Kaur I., Seppinen L., Speedy H.E., Kaminska D., Kuusisto S., Sormunen R., Heljasvaara R., Jones E.L., Muilu M., Jauhiainen M., Pihlajamaki J., Savolainen M.J., Shoulders C.C., Pihlajaniemi T. Specific collagen XVIII isoforms promote adipose tissue accrual via mechanisms determining adipocyte number and affect fat deposition. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3043–E3052. doi: 10.1073/pnas.1405879111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataller R., Brenner D.A. Liver fibrosis. J. Clin. Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Andre M., Andre E., Delamarter J.F. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem. Biophys. Res. Commun. 1993;194:1371–1379. doi: 10.1006/bbrc.1993.1976. [DOI] [PubMed] [Google Scholar]
- Bonnefont J.P., Djouadi F., Prip-Buus C., Gobin S., Munnich A., Bastin J. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol. Asp. Med. 2004;25:495–520. doi: 10.1016/j.mam.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Bookout A.L., Jeong Y., Downes M., Yu R.T., Evans R.M., Mangelsdorf D.J. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgert T.S., Taksali S.E., Dziura J., Goodman T.R., Yeckel C.W., Papademetris X., Constable R.T., Weiss R., Tamborlane W.V., Savoye M., Seyal A.A., Caprio S. Alanine aminotransferase levels and fatty liver in childhood obesity: associations with insulin resistance, adiponectin, and visceral fat. J. Clin. Endocrinol. Metab. 2006;91:4287–4294. doi: 10.1210/jc.2006-1010. [DOI] [PubMed] [Google Scholar]
- Chadt A., Immisch A., De Wendt C., Springer C., Zhou Z., Stermann T., Holman G.D., Loffing-Cueni D., Loffing J., Joost H.G., Al-Hasani H. Deletion of both Rab-GTPase-activating proteins TBC14KO and TBC1D4 in mice eliminates insulin- and AICAR-stimulated glucose transport. Diabetes. 2015;64:746–759. doi: 10.2337/db14-0368. (2015, Diabetes, 64, 1492) [DOI] [PubMed] [Google Scholar]
- Chang M.R., Dharmarajan V., Doebelin C., Garcia-Ordonez R.D., Novick S.J., Kuruvilla D.S., Kamenecka T.M., Griffin P.R. Synthetic RORgammat agonists enhance protective immunity. ACS Chem. Biol. 2016;11:1012–1018. doi: 10.1021/acschembio.5b00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulides C., Lagathu C., Sethi J.K., Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol. Metab. 2009;20:16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corgosinho F.C., De Piano A., Sanches P.L., Campos R.M., Silva P.L., Carnier J., Oyama L.M., Tock L., Tufik S., De Mello M.T., Damaso A.R. The role of PAI-1 and adiponectin on the inflammatory state and energy balance in obese adolescents with metabolic syndrome. Inflammation. 2012;35:944–951. doi: 10.1007/s10753-011-9397-2. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duez H., Duhem C., Laitinen S., Patole P.S., Abdelkarim M., Bois-Joyeux B., Danan J.L., Staels B. Inhibition of adipocyte differentiation by RORalpha. FEBS Lett. 2009;583:2031–2036. doi: 10.1016/j.febslet.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons R.L., Lau P., Muscat G.E. Retinoid-related orphan receptor alpha and the regulation of lipid homeostasis. J. Steroid Biochem. Mol. Biol. 2012;130:159–168. doi: 10.1016/j.jsbmb.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Gentile C.L., Weir T.L., Cox-York K.A., Wei Y., Wang D., Reese L., Moran G., Estrada A., Mulligan C., Pagliassotti M.J., Foster M.T. The role of visceral and subcutaneous adipose tissue fatty acid composition in liver pathophysiology associated with NAFLD. Adipocyte. 2015;4:101–112. doi: 10.4161/21623945.2014.978662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani M., Claus T.H., Himms-Hagen J. Hypertrophy of brown adipocytes in brown and white adipose tissues and reversal of diet-induced obesity in rats treated with a beta3-adrenoceptor agonist. Biochem. Pharmacol. 1997;54:121–131. doi: 10.1016/s0006-2952(97)00162-7. [DOI] [PubMed] [Google Scholar]
- Granneman J.G., Li P., Zhu Z., Lu Y. Metabolic and cellular plasticity in white adipose tissue I: effects of beta3-adrenergic receptor activation. Am. J. Physiol. Endocrinol. Metab. 2005;289:E608–E616. doi: 10.1152/ajpendo.00009.2005. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Vidal R., Vega-Badillo J., Reyes-Fermin L.M., Hernandez-Perez H.A., Sanchez-Munoz F., Lopez-Alvarez G.S., Larrieta-Carrasco E., Fernandez-Silva I., Mendez-Sanchez N., Tovar A.R., Villamil-Ramirez H., Mejia-Dominguez A.M., Villarreal-Molina T., Hernandez-Pando R., Campos-Perez F., Aguilar-Salinas C.A., Canizales-Quinteros S. SFRP5 hepatic expression is associated with non-alcoholic liver disease in morbidly obese women. Ann. Hepatol. 2015;14(5):666–674. [PubMed] [Google Scholar]
- Halim T.Y., Maclaren A., Romanish M.T., Gold M.J., Mcnagny K.M., Takei F. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012;37:463–474. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Hargett S.R., Walker N.N., Keller S.R. Rab GAPs AS160 and Tbc1d1 play nonredundant roles in the regulation of glucose and energy homeostasis in mice. Am. J. Physiol. Endocrinol. Metab. 2016;310:E276–E288. doi: 10.1152/ajpendo.00342.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., Barnes I., Bignell A., Boychenko V., Hunt T., Kay M., Mukherjee G., Rajan J., Despacio-Reyes G., Saunders G., Steward C., Harte R., Lin M., Howald C., Tanzer A., Derrien T., Chrast J., Walters N., Balasubramanian S., Pei B., Tress M., Rodriguez J.M., Ezkurdia I., Van Baren J., Brent M., Haussler D., Kellis M., Valencia A., Reymond A., Gerstein M., Guigo R., Hubbard T.J. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Barak Y., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking S.L., Stewart R.L., Brandon A.E., Suryana E., Stuart E., Baldwin E.M., Kolumam G.A., Modrusan Z., Junutula J.R., Gunton J.E., Medynskyj M., Blaber S.P., Karsten E., Herbert B.R., James D.E., Cooney G.J., Swarbrick M.M. Subcutaneous fat transplantation alleviates diet-induced glucose intolerance and inflammation in mice. Diabetologia. 2015;58:1587–1600. doi: 10.1007/s00125-015-3583-y. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jeffery E., Berry R., Church C.D., Yu S., Shook B.A., Horsley V., Rosen E.D., Rodeheffer M.S. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte. 2014;3:206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E., Wing A., Holtrup B., Sebo Z., Kaplan J.L., Saavedra-Pena R., Church C.D., Colman L., Berry R., Rodeheffer M.S. The adipose tissue microenvironment regulates depot-specific adipogenesis in obesity. Cell Metab. 2016 doi: 10.1016/j.cmet.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogl G., Hsiao Y.S., Tong L. Structure and function of carnitine acyltransferases. Ann. N. Y. Acad. Sci. 2004;1033:17–29. doi: 10.1196/annals.1320.002. [DOI] [PubMed] [Google Scholar]
- Kang H.S., Okamoto K., Takeda Y., Beak J.Y., Gerrish K., Bortner C.D., Degraff L.M., Wada T., Xie W., Jetten A.M. Transcriptional profiling reveals a role for RORalpha in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol. Genomics. 2011;43:818–828. doi: 10.1152/physiolgenomics.00206.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Kong X., Rosen E.D. Adipocyte-specific transgenic and knockout models. Methods Enzymol. 2014;537:1–16. doi: 10.1016/B978-0-12-411619-1.00001-X. [DOI] [PubMed] [Google Scholar]
- Kim S.J., Tang T., Abbott M., Viscarra J.A., Wang Y., Sul H.S. AMPK phosphorylates desnutrin/ATGL and hormone-sensitive lipase to regulate lipolysis and fatty acid oxidation within adipose tissue. Mol. Cell. Biol. 2016;36:1961–1976. doi: 10.1128/MCB.00244-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojetin D.J., Burris T.P. REV-ERB and ROR nuclear receptors as drug targets. Nat. Rev. Drug Discov. 2014;13:197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad D., Rudich A., Schoenle E.J. Improved glucose tolerance in mice receiving intraperitoneal transplantation of normal fat tissue. Diabetologia. 2007;50:833–839. doi: 10.1007/s00125-007-0596-1. [DOI] [PubMed] [Google Scholar]
- Koza R.A., Nikonova L., Hogan J., Rim J.S., Mendoza T., Faulk C., Skaf J., Kozak L.P. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagathu C., Christodoulides C., Virtue S., Cawthorn W.P., Franzin C., Kimber W.A., Nora E.D., Campbell M., Medina-Gomez G., Cheyette B.N., Vidal-Puig A.J., Sethi J.K. Dact1, a nutritionally regulated preadipocyte gene, controls adipogenesis by coordinating the Wnt/beta-catenin signaling network. Diabetes. 2009;58:609–619. doi: 10.2337/db08-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P., Nixon S.J., Parton R.G., Muscat G.E. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J. Biol. Chem. 2004;279:36828–36840. doi: 10.1074/jbc.M404927200. [DOI] [PubMed] [Google Scholar]
- Lau P., Fitzsimmons R.L., Raichur S., Wang S.C., Lechtken A., Muscat G.E. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- Lau P., Fitzsimmons R.L., Pearen M.A., Watt M.J., Muscat G.E. Homozygous staggerer (sg/sg) mice display improved insulin sensitivity and enhanced glucose uptake in skeletal muscle. Diabetologia. 2011;54:1169–1180. doi: 10.1007/s00125-011-2046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P., Tuong Z.K., Wang S.C., Fitzsimmons R.L., Goode J.M., Thomas G.P., Cowin G.J., Pearen M.A., Mardon K., Stow J.L., Muscat G.E. Roralpha deficiency and decreased adiposity are associated with induction of thermogenic gene expression in subcutaneous white adipose and brown adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2015;308:E159–E171. doi: 10.1152/ajpendo.00056.2014. [DOI] [PubMed] [Google Scholar]
- Lee K.Y., Russell S.J., Ussar S., Boucher J., Vernochet C., Mori M.A., Smyth G., Rourk M., Cederquist C., Rosen E.D., Kahn B.B., Kahn C.R. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo K.A., Wright W.S., Kang S., Gerin I., Chiang S.H., Lucas P.C., Opp M.R., Macdougald O.A. Wnt10b inhibits development of white and brown adipose tissues. J. Biol. Chem. 2004;279:35503–35509. doi: 10.1074/jbc.M402937200. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamontova A., Seguret-Mace S., Esposito B., Chaniale C., Bouly M., Delhaye-Bouchaud N., Luc G., Staels B., Duverger N., Mariani J., Tedgui A. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation. 1998;98:2738–2743. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- Manolopoulos K.N., Karpe F., Frayn K.N. Gluteofemoral body fat as a determinant of metabolic health. Int. J. Obes. 2010;34:949–959. doi: 10.1038/ijo.2009.286. [DOI] [PubMed] [Google Scholar]
- Marciano D.P., Chang M.R., Corzo C.A., Goswami D., Lam V.Q., Pascal B.D., Griffin P.R. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARgamma, RORs, and Rev-erbs. Cell Metab. 2014;19:193–208. doi: 10.1016/j.cmet.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Mariman E.C., Wang P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci. 2010;67:1277–1292. doi: 10.1007/s00018-010-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjosberg J., Bernink J., Peters C., Spits H. Transcriptional control of innate lymphoid cells. Eur. J. Immunol. 2012;42:1916–1923. doi: 10.1002/eji.201242639. [DOI] [PubMed] [Google Scholar]
- Mori H., Prestwich T.C., Reid M.A., Longo K.A., Gerin I., Cawthorn W.P., Susulic V.S., Krishnan V., Greenfield A., Macdougald O.A. Secreted frizzled-related protein 5 suppresses adipocyte mitochondrial metabolism through WNT inhibition. J. Clin. Invest. 2012;122:2405–2416. doi: 10.1172/JCI63604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottillo E.P., Shen X.J., Granneman J.G. Beta3-adrenergic receptor induction of adipocyte inflammation requires lipolytic activation of stress kinases p38 and JNK. Biochim. Biophys. Acta. 2010;1801:1048–1055. doi: 10.1016/j.bbalip.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S.A., Eriksson N., Burow R., Wang S.C., Muscat G.E. Beta-adrenergic signaling regulates NR4A nuclear receptor and metabolic gene expression in multiple tissues. Mol. Cell. Endocrinol. 2009;309:101–108. doi: 10.1016/j.mce.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Sakaue H., Nagare T., Kasuga M. Diet-induced up-regulation of gene expression in adipocytes without changes in DNA methylation. Kobe J. Med. Sci. 2009;54:E241–E249. [PubMed] [Google Scholar]
- Pearen M.A., Ryall J.G., Lynch G.S., Muscat G.E. Expression profiling of skeletal muscle following acute and chronic beta2-adrenergic stimulation: implications for hypertrophy, metabolism and circadian rhythm. BMC Genomics. 2009;10:448. doi: 10.1186/1471-2164-10-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen M.A., Eriksson N.A., Fitzsimmons R.L., Goode J.M., Martel N., Andrikopoulos S., Muscat G.E. The nuclear receptor, Nor-1, markedly increases type II oxidative muscle fibers and resistance to fatigue. Mol. Endocrinol. 2012;26:372–384. doi: 10.1210/me.2011-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen M.A., Goode J.M., Fitzsimmons R.L., Eriksson N.A., Thomas G.P., Cowin G.J., Wang S.-C.M., Tuong Z.K., Muscat G.E. Transgenic muscle-specific Nor-1 expression regulates multiple pathways that effect adiposity, metabolism, and endurance. Mol. Endocrinol. 2013;27:1897–1917. doi: 10.1210/me.2013-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S.A., Massaro J.M., Hoffmann U., Vasan R.S., O'donnel C.J., Fox C.S. Abdominal subcutaneous adipose tissue: a protective fat depot? Diabetes Care. 2009;32:1068–1075. doi: 10.2337/dc08-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido M.R., Diaz-Ruiz A., Jimenez-Gomez Y., Garcia-Navarro S., Gracia-Navarro F., Tinahones F., Lopez-Miranda J., Fruhbeck G., Vazquez-Martinez R., Malagon M.M. Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido M.R., Rabanal-Ruiz Y., Almabouada F., Diaz-Ruiz A., Burrell M.A., Vazquez M.J., Castano J.P., Kineman R.D., Luque R.M., Dieguez C., Vazquez-Martinez R., Malagon M.M. Nutritional, hormonal, and depot-dependent regulation of the expression of the small GTPase Rab18 in rodent adipose tissue. J. Mol. Endocrinol. 2013;50:19–29. doi: 10.1530/JME-12-0140. [DOI] [PubMed] [Google Scholar]
- Puri V., Ranjit S., Konda S., Nicoloro S.M., Straubhaar J., Chawla A., Chouinard M., Lin C., Burkart A., Corvera S., Perugini R.A., Czech M.P. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7833–7838. doi: 10.1073/pnas.0802063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Li B., Dong Y., Xu H., Chen L., Wang H., Li P., Zhao W., Gu Y., Wang C., Guo Z. Deconvolution of the gene expression profiles of valuable banked blood specimens for studying the prognostic values of altered peripheral immune cell proportions in cancer patients. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichur S., Fitzsimmons R.L., Myers S.A., Pearen M.A., Lau P., Eriksson N., Wang S.M., Muscat G.E. Identification and validation of the pathways and functions regulated by the orphan nuclear receptor, ROR alpha1, in skeletal muscle. Nucleic Acids Res. 2010;38:4296–4312. doi: 10.1093/nar/gkq180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonti A.E., Agosti F., De Col A., Silvestri G., Marazzi N., Bini S., Bonomo S., Giunta M., Cella S.G., Sartorio A. Severely obese adolescents and adults exhibit a different association of circulating levels of adipokines and leukocyte expression of the related receptors with insulin resistance. Int. J. Endocrinol. 2013;2013:565967. doi: 10.1155/2013/565967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabio G., Das M., Mora A., Zhang Z., Jun J.Y., Ko H.J., Barrett T., Kim J.K., Davis R.J. A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science. 2008;322:1539–1543. doi: 10.1126/science.1160794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santori F.R. Nuclear hormone receptors put immunity on sterols. Eur. J. Immunol. 2015;45:2730–2741. doi: 10.1002/eji.201545712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro N., Feldstein A.E., Enoksson E., Pierpont B., Kursawe R., Kim G., Caprio S. The association between hepatic fat content and liver injury in obese children and adolescents: effects of ethnicity, insulin resistance, and common gene variants. Diabetes Care. 2013;36:1353–1360. doi: 10.2337/dc12-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D.B., Tan G.D., Acerini C.L., Jebb S.A., Agostini M., Gurnell M., Williams R.L., Umpleby A.M., Thomas E.L., Bell J.D., Dixon A.K., Dunne F., Boiani R., Cinti S., Vidal-Puig A., Karpe F., Chatterjee V.K., O'rahilly S. Human metabolic syndrome resulting from dominant-negative mutations in the nuclear receptor peroxisome proliferator-activated receptor-gamma. Diabetes. 2003;52:910–917. doi: 10.2337/diabetes.52.4.910. [DOI] [PubMed] [Google Scholar]
- Scherer P.E. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes. 2006;55:1537–1545. doi: 10.2337/db06-0263. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Goldberg E.B., Makarova K.S., Lin L., Brown W.J., Jackson C.L. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 2006;7:106–113. doi: 10.1038/sj.embor.7400559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.G., Muscat G.E. Orphan nuclear receptors: therapeutic opportunities in skeletal muscle. Am. J. Physiol. Cell Physiol. 2006;291:C203–C217. doi: 10.1152/ajpcell.00476.2005. [DOI] [PubMed] [Google Scholar]
- Tam C.S., Tordjman J., Divoux A., Baur L.A., Clement K. Adipose tissue remodeling in children: the link between collagen deposition and age-related adipocyte growth. J. Clin. Endocrinol. Metab. 2012;97:1320–1327. doi: 10.1210/jc.2011-2806. [DOI] [PubMed] [Google Scholar]
- Tran T.T., Yamamoto Y., Gesta S., Kahn C.R. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 2008;7:410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuong Z.K., Lau P., Yeo J.C., Pearen M.A., Wall A.A., Stanley A.C., Stow J.L., Muscat G.E. Disruption of Roralpha1 and cholesterol 25-hydroxylase expression attenuates phagocytosis in male Roralphasg/sg mice. Endocrinology. 2013;154:140–149. doi: 10.1210/en.2012-1889. [DOI] [PubMed] [Google Scholar]
- Tuong Z.K., Lau P., Du X., Condon N.D., Goode J.M., Oh T.G., Yeo J.C., Muscat G.E., Stow J.L. RORalpha and 25-hydroxycholesterol crosstalk regulates lipid droplet homeostasis in macrophages. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.V., Deng Y., Wang Q.A., Sun K., Scherer P.E. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology. 2010;151:2933–2939. doi: 10.1210/en.2010-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.A., Scherer P.E., Gupta R.K. Improved methodologies for the study of adipose biology: insights gained and opportunities ahead. J. Lipid Res. 2014;55:605–624. doi: 10.1194/jlr.R046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wree A., Kahraman A., Gerken G., Canbay A. Obesity affects the liver - the link between adipocytes and hepatocytes. Digestion. 2011;83:124–133. doi: 10.1159/000318741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.