Summary
In the current study, we describe a novel biophotonic imaging-based reporter system that is particularly useful for the study of virulence in polymicrobial infections and interspecies interactions within animal models. A suite of luciferase enzymes was compared using three early colonizing species of the human oral flora (Streptococcus mutans, Streptococcus gordonii, and Streptococcus sanguinis) to determine the utility of the different reporters for multiplexed imaging studies in vivo. Using the multiplex approach, we were able to track individual species within a dual species oral infection model in mice with both temporal and spatial resolution. We also demonstrate how biophotonic imaging of multiplexed luciferase reporters could be adapted for real-time quantification of bacterial gene expression in situ. By creating an inducible dual-luciferase expressing reporter strain of S. mutans, we were able to exogenously control and measure expression of nlmAB (encoding the bacteriocin mutacin IV) within mice to assess its importance for the persistence ability of S. mutans in the oral cavity. The imaging system described in the current study circumvents many of the inherent limitations of current animal model systems, which should now make it feasible to test hypotheses that were previously impractical to model.
Introduction
Recent improvements in next generation sequencing technologies have revealed unprecedented insights into the ecology of the human oral microbial flora and have also dramatically impacted upon our understanding of the etiologies of oral diseases (Takahashi and Nyvad, 2011; Zarco et al., 2012; Wade, 2013; Struzycka, 2014; Duran-Pinedo and Frias-Lopez, 2015; Simon-Soro and Mira, 2015). The latest concept of polymicrobial synergy and dysbiosis describes the role of the disease provoking microbiota in the destabilization of oral ecological homeostasis (Hajishengallis and Lamont, 2012; Lamont and Hajishengallis, 2015). For example, while Streptococcus mutans has long been considered the predominant etiological agent of dental caries (Hamada and Slade, 1980), it is now evident that the source of cariogenic dysbiosis is largely determined by the myriad of cooperative and antagonistic interactions among the flora that ultimately favor the overgrowth of cariogenic species like S. mutans (Gross et al., 2012).
Given the polymicrobial nature of complex diseases, such as caries and periodontitis (Peters et al., 2012; Costalonga and Herzberg, 2014; Murray et al., 2014), it still remains a considerable challenge to adequately characterize relevant bacterial virulence mechanisms using animal model systems. Nevertheless, in vivo model systems are still the preferred approach to simulate the variety of forces that shape human oral ecology, such as competitive pressures from the oral flora, similar attachment surfaces, saliva flow, nutrient diversity, physical removal forces, host innate immunity, etc. Rodent model systems have been instrumental for demonstrating the infectious and cariogenic potential of S. mutans in the presence of a high sucrose diet (Hamada et al., 1978; Tanzer, 1979; Loyola and Leyva, 1990). Similarly, recent mouse studies have implicated Porphyromonas gingivalis as a keystone species in periodontitis (Hajishengallis et al., 2011). In both cases, the animal model systems used to study these diseases rely upon end point measurements of the inoculated bacteria to verify their presence at the conclusion of the experiments. Generally, such measurements are made via CFU counts from extracted teeth or simply via PCR (Tanzer, 1979; Daep et al., 2011; Hajishengallis et al., 2011). In our experience, these approaches have been both cumbersome and subject to a high degree of variability. In addition, it is especially challenging to resolve these data spatially, while little, if anything can be concluded about the temporal aspects of the infection, since the animals need to be sacrificed to quantify the bacteria. These factors pose inherent limitations on the types of hypotheses that can be effectively tested using in vivo model systems.
In the present study, our aim was to develop a rodent model system that could circumvent many of these limitations to facilitate studies of oral microbial ecology and bacterial gene expression in vivo. Since we previously demonstrated the utility of firefly luciferase reporter strains for noninvasive in vitro studies of biofilm development and gene expression in oral streptococci (Merritt et al., 2005b; Zheng et al., 2011a; Zheng et al., 2011b), we were interested to further examine the potential utility of luciferase fusions for in vivo studies. Luciferase enzymes allow for genetically encoded bioluminescence (Greer and Szalay, 2002). Similar to the fluorescence emitted by proteins like GFP (Cubitt et al., 1995), the emitted light from luciferase reactions can be quantified (Greer and Szalay, 2002). This has made both fluorescent proteins and luciferases increasingly popular tools for use in biophotonic imaging applications involving live cell cultures and animal model systems (Sato et al., 2004; Kadurugamuwa et al., 2005; Steinhuber et al., 2008; Andreu et al., 2011). Biophotonic imaging has the great advantage of yielding spatially and temporally resolved data, which has revolutionized many animal model systems over the last decade, particularly those used for studies of cancer metastasis and chemotherapeutic delivery (O’Neill et al., 2010; Kocher and Piwnica-Worms, 2013). Bioluminescence reactions are currently among the best options for a variety of biophotonic imaging applications due to their unrivaled signal to noise ratios (Choy et al., 2003). This exceptional sensitivity provided by luciferase reactions has also recently made it feasible to adapt biophotonic imaging for in situ studies of bacterial infections within animal models (Andreu et al., 2011). Here we report the development of a luciferase-based biophotonic imaging platform for animal model studies of the human oral flora. Using three key oral streptococcal species, we created and tested a variety of synthetic codon optimized luciferase enzymes that have never been used in bacteria and demonstrated their utility for noninvasive, multiplexed quantification of multispecies infections and inducible gene expression in vivo.
Results
Construction of Streptococcus mutans, Streptococcus gordonii, and Streptococcus sanguinis firefly and renilla luciferase reporter strains
The excellent signal to noise ratio of luciferases (Choy et al., 2003) motivated us to further explore their performance for biophotonic imaging of oral bacteria. Based upon our previous data (Merritt et al., 2005b), we selected the constitutively expressed lactate dehydrogenase (ldh) gene for fusions with the firefly and renilla luciferases. Lactate dehydrogenase is critical for the metabolic fitness of lactic acid bacteria and its expression in S. mutans is intimately associated with cariogenesis (Johnson and Hillman, 1982). To prevent disruption of normal ldh expression, the firefly and renilla luciferase open reading frames (ORFs) were each inserted immediately downstream of the S. mutans, S. gordonii, and S. sanguinis ldh stop codons to create artificial two-gene operons with ldh. Copies of the ldh Shine-Dalgarno sequences were also added to the luciferase ORFs to ensure their efficient translation. For S. mutans, the luciferase ORFs were inserted using a markerless mutagenesis strategy (Xie et al., 2011), whereas an erythromycin resistance cassette was used in S. gordonii and S. sanguinis (Fig. 1) due to the lack of a markerless mutagenesis system for both species at the time of construction. Since both luciferase enzymes utilize distinct substrates for bioluminescence (D-luciferin for firefly luciferase and coelenterazine for renilla luciferase), we were further interested to test the utility of this combination of reporters for multiplexed dual species experiments.
Fig. 1. Genotype of luciferase reporter strains.
A markerless mutagenesis approach was used to construct the respective luciferase fusions for S. mutans. S. sanguinis and S. gordonii reporters were constructed using an erythromycin resistance cassette (erm) for the selection of mutant clones. The luciferase open reading frames (ORF) were all cloned immediately downstream of the ldh stop codons to create artificial two-gene operons. Each luciferase cassette contained a copy of the ldh Shine-Dalgarno sequence to ensure its efficient translation.
In vitro assessment of bioluminescent S. mutans, S. gordonii, and S. sanguinis reporters
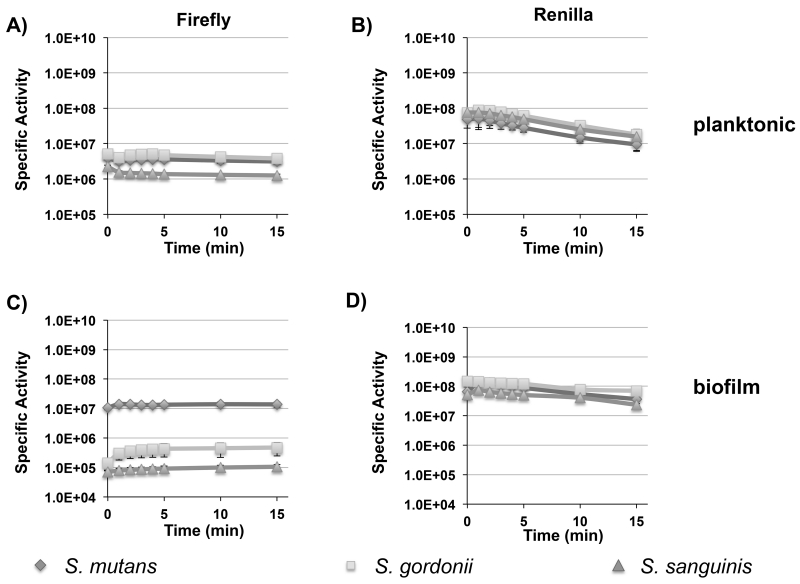
To determine the potential utility of the firefly and renilla luciferase reporters for biophotonic imaging applications, we first assayed the signal stability of planktonic and biofilm cells over a 15 min. time course after luciferase substrate addition (Fig. 2A to D). In planktonic cultures of all three species, the firefly and renilla bioluminescent signal intensities were highly stable for 5 min. However, by 15 min., only the firefly reporters retained all of their original signal intensities (Fig. 2A); the signal intensities of the renilla reporters dropped by approximately 5-fold (Fig. 2B). Despite the faster degradation of the renilla signals, these reporters were >10-fold brighter than their corresponding firefly luciferase reporters. The overall signal intensities varied <2-fold for all of the renilla reporter strains, whereas the firefly luciferase values were similar for S. mutans and S. gordonii, but several fold weaker in S. sanguinis (Fig. 2A and B). In biofilms of all three species, the signal stabilities and intensities of the renilla reporters mirrored the results observed in planktonic cells. However, this was not the case for the firefly luciferase reporters, which exhibited similar signal stabilities among the three species, but also yielded approximately a log reduction in specific activity for S. gordonii and S. sanguinis (Fig. 2C). Overall, the firefly luciferase reporters exhibited superior signal stabilities, while the renilla reporters yielded superior signal intensities (Fig. 2 D). For S. mutans, growth in a planktonic culture or in a biofilm had no impact upon the performance of either the firefly or renilla luciferase reporters, whereas the firefly luciferase reporters exhibited reduced activity in biofilms of S. gordonii and S. sanguinis.
Fig. 2. Temporal signal stabilities of firefly and renilla luciferase reporters in planktonic and biofilm cultures of S. mutans, S. gordonii, and S. sanguinis.
Presented here are the average luminescence values (RLU) and standard deviations normalized to the cell density over a 15 min. time course after luciferase substrate addition (n=3). A) planktonic firefly luciferase activity; B) planktonic renilla luciferase activity; C) biofilm firefly luciferase activity; D) biofilm renilla luciferase activity.
Dual species infection of mice with S. mutans and S. gordonii
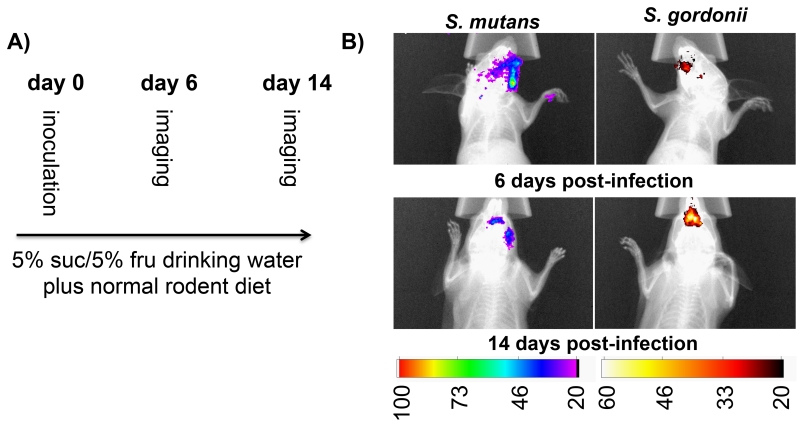
Based upon the favorable signal characteristics of the firefly and renilla reporter strains, we were next interested to test their performance for multiplexed in situ imaging within mouse oral cavities. Our strain comparisons suggested that the optimal dual species combination would be to use the S. mutans firefly luciferase reporter together with the S. gordonii renilla reporter. Thus, both strains were inoculated into the oral cavities of three mice with S. mutans inoculated on the right and S. gordonii on the left of each mouse. This was done to avoid direct competition between both species (Kreth et al., 2005; Kreth et al., 2008) as well as to determine whether luciferase signals would actually localize to the expected sites of inoculation. After inoculation, the mice were fed a normal rodent diet to simulate the natural biofilm removal forces associated with mastication. In addition, the mouse drinking water was supplemented with 5% (wt/vol) sucrose + 5% (wt/vol) fructose to stimulate biofilm formation. On day 6 post-inoculation, the mice were imaged in bioluminescence and X-ray modes with a Carestream In vivo Xtreme imager to localize the bacteria in the oral cavity. The luciferin substrate for firefly luciferase was directly pipetted into the mouse oral cavities immediately prior to imaging. Since we had previously determined that both the firefly and renilla luciferase signals were stable for 5 min. after substrate addition, we performed a 5 min. image acquisition to provide maximal signal sensitivity. Mice were initially imaged for firefly luciferase activity and then again an hour later to confirm that the residual firefly luciferase signal had dissipated. Subsequently, the mice were imaged for renilla luciferase activity following the same protocol. Distinct luciferase signals were detected for each reporter, which were also localized to the expected sides of the mouse oral cavities (i.e. S. mutans on the right and S. gordonii on the left) (Fig. 3B). The imaging procedure was repeated 14 days post-inoculation to confirm that a continuous colonization of our reporter strains was possible using a standard rodent diet together with drinking water supplemented with sugar. The imaging data also confirmed the previous localization results observed on day 6 (Fig 3B).
Fig. 3. Dual species infection of mice with S. mutans and S. gordonii firefly and renilla luciferase reporters.
A) Flow chart illustrating the assay protocol. B) Composite overlay images of the X-ray and bioluminescent images of the dual infections 6 and 14 days post-inoculation. The presented images are representative of the results obtained from 3 mice.
Development of novel luciferases for use in prokaryotes
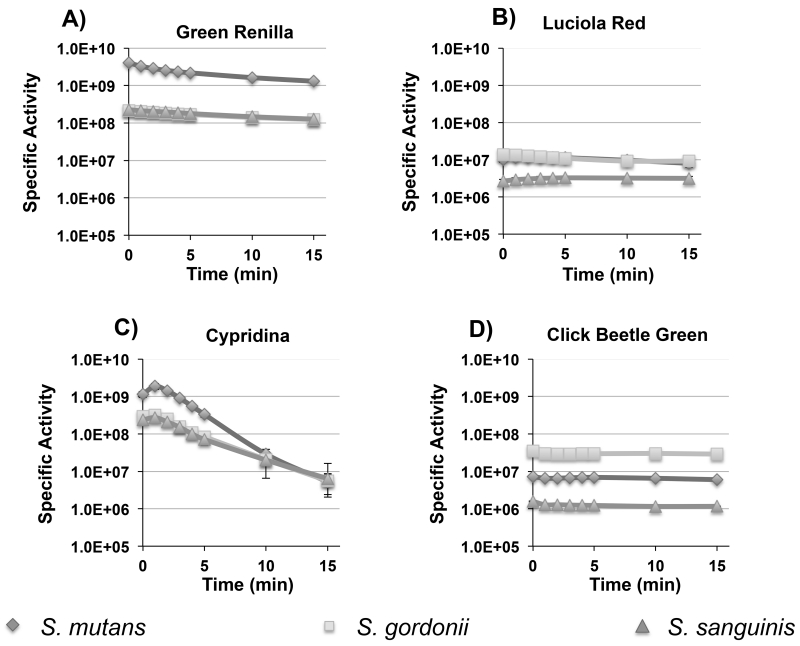
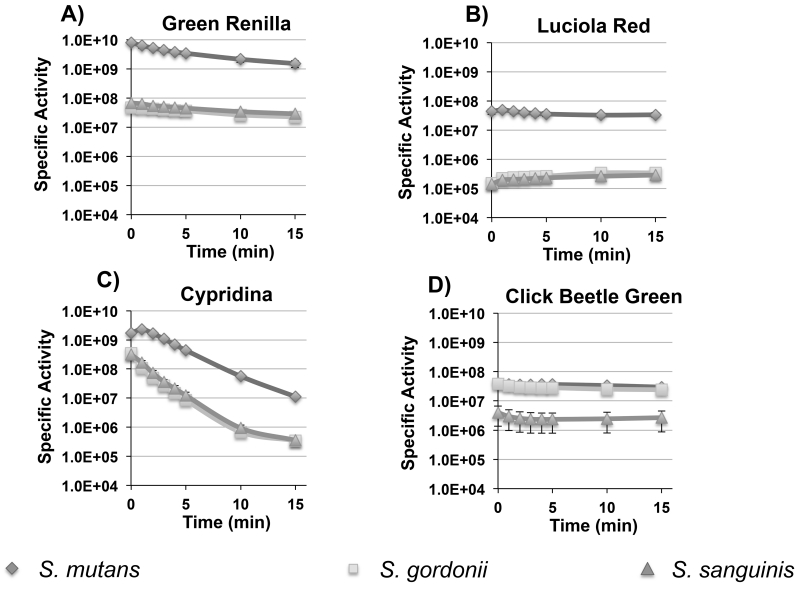
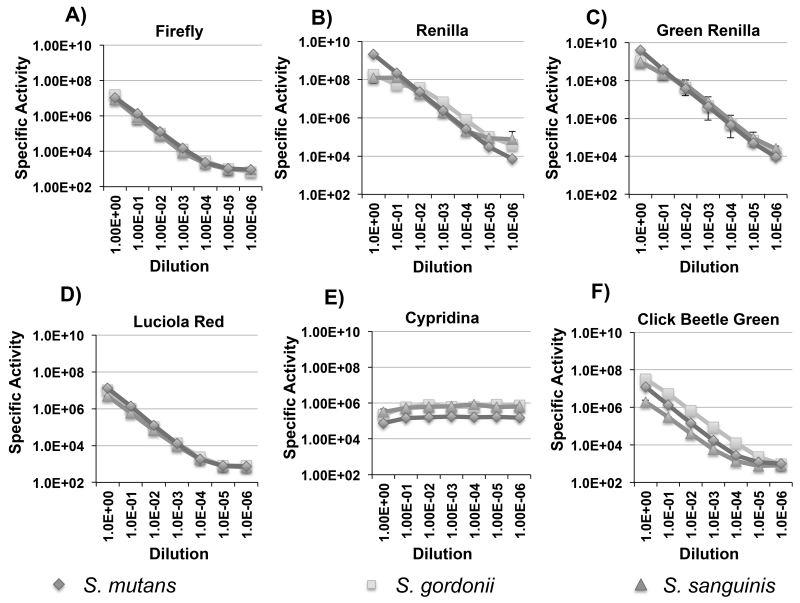
The dual colonization with the S. mutans and S. gordonii luciferase reporters served as a proof of principle for multiplexed in situ imaging of oral bacteria in mice. In an effort to further expand the utility of the system, we were interested to compare the performance of a variety of other luciferase enzymes that had never been tested in bacteria, but still use commercially available substrates. Viable candidates needed to be as bright or brighter than firefly luciferase and have emission spectra suitable for multiplex applications. To this end, we chose click beetle green, green renilla, luciola red, and cypridina luciferases for additional performance testing. Table 2 summarizes the main characteristics of each. Since these enzymes are all derived from eukaryotic organisms, we synthesized codon-optimized versions to ensure their efficient translation in streptococci. As cypridina luciferase is naturally a secreted enzyme (Thompson et al., 1989), the mature form of the enzyme was synthesized without its secretion signal sequence. The respective strains were constructed using the same strategies as the previous firefly and renilla luciferase reporters so their performance could be compared directly (Fig. 1). Similar to the previously tested luciferases, the signal intensities of the new reporters were largely stable for 5 min. in both planktonic and biofilm cells, although the cypridina signal degraded slightly faster. (Fig. 4 A to D and 5 A to D). We also discovered that the cypridina reporters required resuspension in DMSO prior to the addition of the vargulin substrate; otherwise, signal intensity values were extremely low (data not shown). In addition, we were able to increase the signal intensity of the S. mutans renilla and green renilla reporters with the synthetic substrate coelenterazine-h. Curiously, this substrate did not alter the signal values for the S. gordonii and S. sanguinis reporters (data not shown). When comparing the reporter strains, differences in intensities were evident and paralleled those seen with the firefly and renilla luciferases. In biofilms, S. mutans yielded much higher values compared to S. sanguinis and S. gordonii with ~400-fold greater luciferase activity for the luciola red construct, 100-fold higher for green renilla, and >10-fold higher for cypridina (Fig 5A – C). Interestingly, the S. gordonii click beetle green reporter exhibited nearly identical luciferase activity to S. mutans, whereas it was ~10-fold lower in S. sanguinis (Fig. 5D). The specific signal intensities ranged between 1×106 to 4×109 RLU/A600 for planktonic cultures and 6×104 to 7×109 RLU/A600 for biofilm cultures. Overall, the reporter strains all exhibited high signal intensities in planktonic cultures, while biofilm cultures were consistently bright with the green renilla and cypridina reporters demonstrating the versatility of the different reporter genes for in vitro assays. Similar to the previous firefly and renilla luciferase results, no obvious differences in performance were noted in any of the S. mutans reporters when comparing planktonic vs. biofilm cultures.
Table 2.
Luciferases used in this study and their characteristics
| Luciferase | Substrate | ATP | Secreted | Emission (nm) |
ORF (bp) |
adapted from |
Source |
|---|---|---|---|---|---|---|---|
| Renilla | Coelenterazine | No | No | 420 | 936 | Renilla reniformis | pRL-TK Promega (Madison, WI) |
| Cypridina | Vargulin | No | Yes | 460 | 1611 |
Vargula
hilgendorfii |
This study |
| Green renilla |
Coelenterazine | No | No | 527 | 945 | Renilla reniformis | This study |
| Click beetle green |
D-luciferin | Yes | No | 537 | 1629 |
Pyrophorus
plagiophthalamus |
This study |
| Firefly | D-luciferin | Yes | No | 557 | 1653 | Photinus pyralis | (Podbielski et al., 1999) |
| Luciola Red |
D-luciferin | Yes | No | 609 | 1647 | Luciola italica | This study |
Fig. 4. Temporal signal stabilities of planktonic click beetle green, luciola red, green renilla, and cypridina luciferase reporter strains of S. mutans, S. gordonii, and S. sanguinis.
Presented here are the average RLU values and standard deviations normalized to the cell density over a 15 min. time course after substrate addition (n=3). A) green renilla luciferase activity; B) luciola red luciferase activity; C) cypridina luciferase activity; D) click beetle green luciferase activity.
Fig. 5. Temporal signal stabilities of click beetle green, luciola red, green renilla, and cypridina reporter strains of S. mutans, S. gordonii, and S. sanguinis grown in biofilms.
Presented here are the average RLU values and standard deviations normalized to the cell density over a 15 min. time course after substrate addition (n=3). A) green renilla luciferase activity; B) luciola red luciferase activity; C) cypridina luciferase activity; D) click beetle green luciferase activity.
Evaluation of luciferase reporters for quantifying population size
We were next interested to evaluate the correlations between luminescence values and population size to determine whether the luciferase signals from the reporter strains could serve as reliable indices to quantify bacteria in situ. Signal linearity was evaluated using mid-logarithmic planktonic cells serially diluted 10-fold up to a 1×10−6 dilution (Fig. 6 A to F). As shown in Figure 6E, cypridina luciferase was the only reporter that did not exhibit a strong correlation over the dilution series. This was likely due to the fact that it was necessary to perform the serial dilutions and luminescence measurements using growth medium like the other reporters, rather than using DMSO. Unfortunately, DMSO caused the cultures to severely clump together, which did not affect the luminescence measurements, but proved problematic for serial dilution. Both the renilla and green renilla reporters all yielded exceptional linearity over 6 logs of dilution, whereas the firefly, click beetle green, and luciola red reporters could accurately quantify the cell population up to 4 logs of dilution (Fig. 6 A to D and F). Since a mid-logarithmic culture of oral streptococci yields about 2×109 CFU/ml (unpublished results), the renilla and green renilla reporters could accurately quantify just 200 total CFU in our assays. Even with the lower sensitivities of the firefly, click beetle green, and luciola red reporters, we only required ~104 total CFU for an accurate measure of the population. Thus, the combined properties of high signal stability, exceptional sensitivity, and signal linearity over a wide range of cell densities suggested that the reporters would perform well for in vivo mouse colonization studies. Cypridina luciferase was the lone exception. Even though it was nearly as bright as the renilla reporters (Fig. 4A and 5A), the DMSO requirement for activity measurements precluded this enzyme from further consideration for in vivo use (Fig. 6 E).
Fig. 6. Correlation of luciferase signal intensity vs. cell number for S. mutans, S. gordonii, and S. sanguinis.
Presented here are the average RLU values and standard deviations normalized to the cell density over a 10-fold dilution series of planktonic bacteria up to a 10−6 dilution (n=3). A) firefly luciferase activity; B) renilla luciferase activity; C) green renilla luciferase activity; D) luciola red luciferase activity; E) cypridina luciferase activity; F) click beetle green luciferase activity.
Assessment of spatial resolution in situ
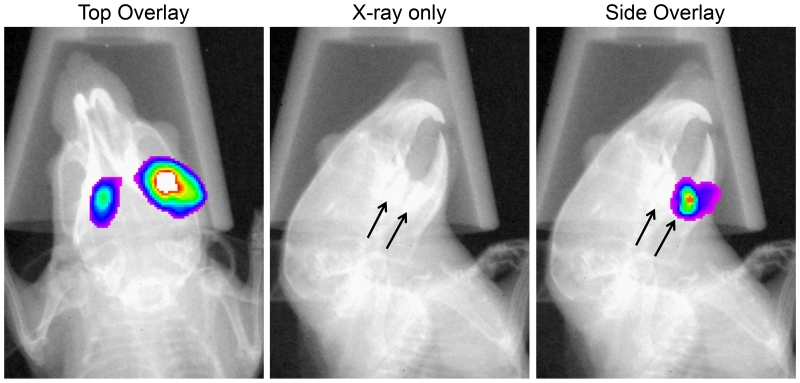
Our previous dual species colonization results indicated that luciferase signals could be localized to the respective sides of inoculation in the mouse. Given the clear signal resolvability in the left/right halves of the oral cavity, we were curious to further examine the spatial resolution of the luciferase signals. To test this, mice (n=3) were inoculated with an S. mutans green renilla luciferase reporter and fed a normal rodent diet together with drinking water supplemented with 5% sucrose and 5% fructose. On day 6 post-inoculation, the mice were imaged using both bioluminescence and X-ray modes to localize the bacteria (Fig. 7). Luciferase signals for all three mice localized to the central part of the alveolar bone, where the molars are located. Similar to our previous results, distinct signals were easily resolvable in the left/right halves of the oral cavities when imaging the mice from the top down. Next, we imaged the mice from the side and we could easily distinguish the molars in the X-ray images, which made it possible to discriminate signals derived from the maxillae and mandibles. By comparing both sets of images, it was possible to determine the particular quadrants where the bacteria were located (Fig. 7). Since mice only posses a single row of molars in each quadrant, signals are resolvable to the specific set of molars where the bacteria have colonized. In this particular experiment, we did not observe signals emanating from the incisors, but there were a couple instances throughout our studies where we observed weak signals on the incisors, such as in Figure 3B (lower left panel). Therefore, stable colonization of the incisors was quite rare overall, possibly due to the rodent habit of constantly gnawing with the incisors. Invariably, most or all of the reporter signals could be localized to one or more of the molars.
Fig. 7. Spatial resolution of orally infected luciferase reporter strains in mice.
Mice (n=3) infected with a green renilla luciferase reporter strain were imaged from the top and side to localize the quadrants of infection. The arrow in the X-ray image depicts the position of the mandibular molars. The presented images are of a single representative mouse in which the sites of colonization can be localized to both of the mandibular molars.
In vitro assessment of a dual luciferase xylose-inducible bacteriocin reporter
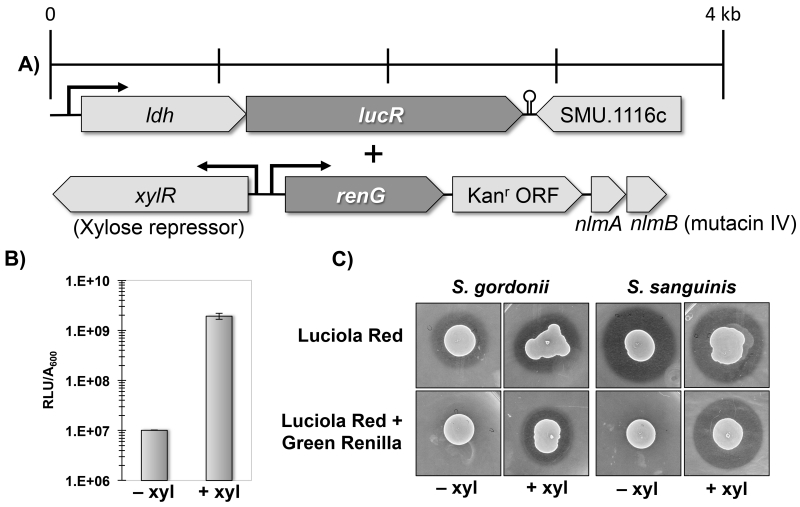
To the best of our knowledge, bacterial gene expression has not been previously quantified in situ within live animals. For quantitative measures of bacterial gene expression, RNA is typically extracted from the site of experimental infection and then further analyzed (Westermann et al., 2012). Such data offer minimal spatial resolution, while temporal measurements are difficult to obtain because sample collection generally requires sacrificing the animals. These limitations were partially circumvented in a recent mouse infection study employing biophotonic imaging of enteropathogenic E. coli to qualitatively examine changes in bacterial gene expression over the course of infection (Nguyen et al., 2015). The principal hindrance for obtaining quantitative measurements of bacterial gene expression in live animals is the requirement for normalization to an internal control to determine the specific activity of the reporter. Since our previous results demonstrated a strong correlation between the signal intensity of the ldh reporters and cell number, we reasoned it should be feasible to create a dual luciferase reporter strain expressing an ldh reporter fusion as an internal control and a separate reporter fusion to a gene of interest. To this end, we opted to combine the constitutively expressed luciola red ldh reporter together with a green renilla luciferase fusion to the nlmAB operon encoding the bacteriocin mutacin IV. Furthermore, the green renilla-nlmAB reporter was placed under the control of the xylose-inducible Xyl-S1 cassette (Xie et al., 2013) to allow for exogenous control of nlmAB expression (Fig. 8 A). With this construct we aimed to address two central questions: i) what role does mutacin IV play in the colonization and persistence of S. mutans in our mouse model, and ii) can we exogenously modulate specific bacterial gene expression within an in vivo system? S. mutans is known to be a copious producer of primarily non-lantibiotic bacteriocins that inhibit the growth of closely related lactic acid bacteria and are hypothesized to play a central role in S. mutans colonization and persistence (Merritt and Qi, 2012). However, this hypothesis had yet to be tested in an animal model system. To first demonstrate the functionality of the inducible mutacin IV reporter strain, we assayed green renilla luciferase activity from mid-log cultures of the dual luciferase strain ± 1% (wt/vol) xylose and observed ~200-fold increase in reporter activity in the presence of xylose (Fig. 8B). In addition, we performed a deferred antagonism assay on BHI plates ± 1% (wt/vol) xylose to test for mutacin IV inhibition of S. gordonii and S. sanguinis, which are both mutacin IV-sensitive competitors of S. mutans in the oral cavity. As expected, growth inhibition halos of both indicator species were present around the single luciferase luciola red ldh reporter strain irrespective of the presence of xylose. In contrast, xylose was essential for mutacin IV production in the dual luciferase strain harboring both the constitutive luciola red construct and the inducible green renilla-nlmAB fusion (Fig. 8C).
Fig. 8. Construction and phenotypic characterization of a xylose-inducible nlmAB reporter strain of S. mutans.
A) Illustration of the reporter constructs for the ldh-luciola red luciferase fusion and the Xyl-S1-green renilla fusion. B) In vitro xylose-dependent induction of renilla green luciferase activity. Presented here are the averages and standard deviations from two independent experiments performed in triplicate. C) Confirmation of the mutacin IV production phenotypes of the dual luciferase-expressing S. mutans. A deferred antagonism assay was performed using both S. sanguinis and S. gordonii as indicator strains for mutacin IV activity. The assay was performed three times with similar results. Shown here are the results from one representative experiment.
In situ measurements of S. mutans bacteriocin gene expression
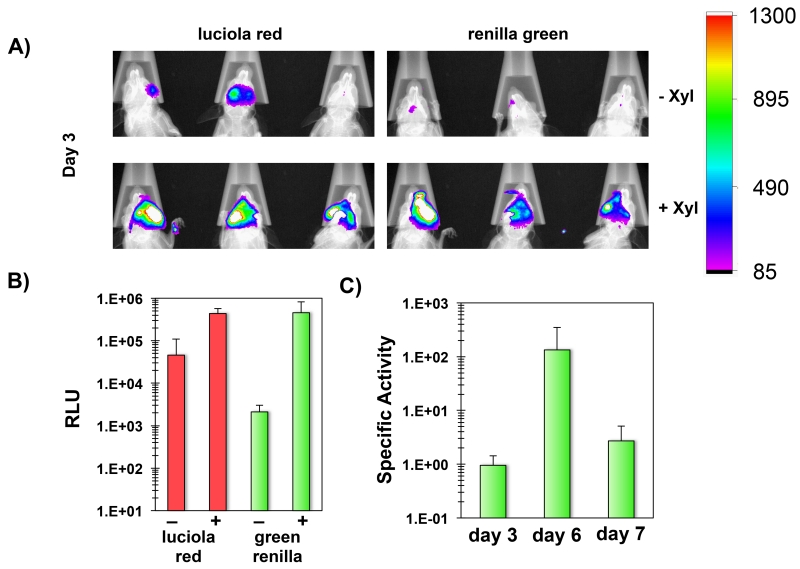
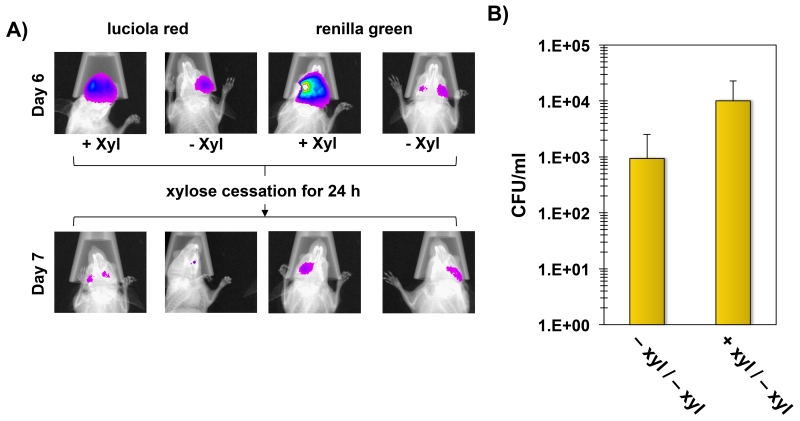
Since the dual luciferase reporter strain yielded the expected mutacin IV phenotypes, this strain was used to colonize six mice to test its utility for temporal studies of gene expression. Mice were separated into two groups receiving drinking water with or without 10% (wt/vol) xylose and imaged on days 3, 6, and 7 post-inoculation. After imaging on day 6, xylose was removed from the drinking water of the mice in the xylose-induced group to further examine the transcriptional response resulting from removal of the inducer. The final imaging performed on day 7 was immediately followed by a CFU determination of S. mutans cells sonicated from the mouse molars. The initial imaging on day 3 revealed a successful colonization of all mice (Fig. 9A). Interestingly, the luciola red reporter values were about 10-fold higher in the xylose group compared to the uninduced control group (Fig. 9B). However, by day 6, the luciola red signals from two of the control mice were too weak to use for normalization. Therefore, the difference in population sizes were likely even greater than on day 3 indicating a distinct survival advantage among the induced group. Similar to the previous in vitro results, the activity of the green renilla-nlmAB reporter was also substantially higher with xylose induction (Fig. 9B). On day 3, the normalized specific activity of the green renilla reporter was 15-fold higher in the xylose group compared to the uninduced control group (calculated using the values presented in Fig. 9B). Due to the weak luciola red signals in the control mice on day 6, we could only calculate the specific activity of one of the mice, which was 60-fold lower than the xylose induced group. After imaging on day 6, xylose was removed from the induced group resulting in a substantial reduction in specific activity of the green renilla reporter on day 7 (Fig. 9C). This further confirmed the role of xylose supplementation for the exogenous control of bacteriocin gene expression. The major reduction in nlmAB expression observed after removal of the inducer also correlated with a ~15-fold reduction in luciola red activity (calculated based on the values in Fig. 10A), which indicated that the S. mutans population had declined rapidly in the absence of bacteriocin gene expression. As a final confirmation of the reporter results, bacteria were extracted from the molars of all mice to compare CFU values. As expected, the xylose-induced group yielded much higher CFU values compared to the control (Fig. 10B). It should also be noted that the bacteria were enumerated on day 7, when xylose had already been withdrawn for 24 hours from the induced group. Presumably, the difference in CFU values between groups would have been even greater on day 6, when the nlmAB operon was still highly expressed in the xylose group (Fig. 9C and 10A).
Fig. 9. In situ quantification of nlmAB gene expression and S. mutans persistence in mice.
A) Detection of luciola red and green renilla activity in situ. A total of 6 mice were infected with the xylose-inducible dual luciferase S. mutans and imaged on day 3 post-inoculation. The presented images are overlays of the individual bioluminescent and X-ray images. −xyl = uninduced control group; +xyl = xylose induced group. B) Day 3 RLU values for the luciola red and green renilla reporters ± xylose. Results are presented as the averages and standard deviations from 3 mice in each group. C) Specific activity of the green renilla reporter for the xylose induced group on days 3, 6, and 7. Results are presented as the averages and standard deviations from 3 mice.
Fig. 10. Effects of xylose cessation on nlmAB expression and bacterial abundance.
A) Effect of xylose cessation. Representative images of one mouse per group are shown on day 6 and again 24 hours. after the cessation of xylose induction in the induced group. B) CFU counts of dual luciferase S. mutans on day 7. The jaws from the mice in both groups were sonicated to remove adherent bacteria. Results are presented as the averages and standard deviations of 3 mice per group. −xyl/−xyl = uninduced control group; +xyl/−xyl = xylose induced group.
Discussion
Biophotonic imaging is routinely employed in cancer studies to track tumor progression and metastasis as well as chemotherapeutic delivery (O’Neill et al., 2010; Kocher and Piwnica-Worms, 2013). With the recent adaptation of this technology for infectious disease research (Andreu et al., 2011), it is now possible to non-invasively quantify both the temporal and spatial aspects of experimental infections by repeatedly imaging the same animals. Our aim was to apply this same technology to develop a model that would facilitate studies of oral bacterial ecology and gene expression in vivo. To this end, we engineered several species of oral streptococci using a variety of novel luciferases that are easily distinguishable by differences in substrate requirements and emission spectra (see Table 2). In this study we focused upon S. mutans as a species commonly isolated from caries lesions as well as the commensal species S. sanguinis and S. gordonii. A variety of clinical and in vitro studies have demonstrated an antagonistic relationship between S. mutans and S. sanguinis/S. gordonii and this antagonism is thought to play a central role in determining caries susceptibility (Caufield et al., 2000; Becker et al., 2002; Kreth et al., 2005; Kreth et al., 2009). While many mechanisms have been proposed to explain the basis for their antagonism (Kreth et al., 2009; Jakubovics et al., 2014), few, if any, of these hypotheses have actually been tested in animal models, largely due to a lack of appropriate tools to easily address these questions in vivo. The current study illustrates how the combination of biophotonic imaging and multiplexed luciferases could be employed for such studies.
The exceptional signal to noise ratio of luciferases makes it possible to accurately quantify a wide range of bacterial cell numbers. The luciferases requiring D-luciferin as a substrate, (firefly, click beetle green, and luciola red luciferases) were capable of quantifying 104 CFU in vitro and yielded a stable signal for at least 15 min. after the addition of substrate. The renilla and green renilla luciferases use coelenterazine as a substrate and were considerably brighter, especially when using the synthetic analog coelenterazine-h. With coelenterazine-h, it was possible to accurately quantify just 102 CFU in vitro. However, the renilla luciferases also yield a less stable signal that begins to diminish beyond 5 min. after substrate addition. Cypridina luciferase, which was the only enzyme using vargulin as substrate, was not suitable for in vivo imaging, since it required the suspension of bacterial cells in DMSO for reliable bioluminescence measurements. In the presence of DMSO, cypridina luciferase did prove to be exceptionally bright however, and may still be useful as a reporter for in vitro assays. Since there were apparently no issues with cypridina luciferase enzymatic activity in oral streptococci, DMSO presumably serves to enhance the diffusion of the vargulin substrate across the cell membrane. Unlike vargulin, both D-luciferin and coelenterazine apparently diffuse easily into the cells and can be directly added to the growth medium for bioluminescence measurements. In addition to the aforementioned luciferases, we also tested red and green shifted variants of firefly luciferase as well as click beetle red luciferase. Unfortunately, these three enzymes all yielded substantially weaker signals relative to native firefly luciferase and therefore they were not pursued further (data not shown). Similarly, bacterial luciferase has been reported to be substantially less sensitive than firefly luciferase for biophotonic imaging in mice (Close et al., 2011), so it was not included in our luciferase comparisons. Though, it should be noted that bacterial luciferase does not require the exogenous addition of luciferase substrate, which may be highly advantageous for certain applications, especially for studies of tissue invasive infections. Based upon our performance tests of the various luciferases, the preferred combination for two signal multiplex imaging is luciola red and green renilla luciferases, due to their distinct substrates and easily resolvable emission spectra. However, using the appropriate filter set, it should be feasible to measure up to four unique signals with the addition of click beetle green and renilla luciferases. Since firefly luciferase emits in the yellow-green range (Table 2), it exhibits a much greater spectral overlap with luciola red compared to click beetle green and is therefore less useful for certain multiplexed applications. Conceivably, the system could be even further expanded to five unique signals if vargulin analogs were developed to circumvent the DMSO requirement for cypridina luciferase measurements.
While multiplex imaging provides a straightforward approach for the simultaneous quantification of multiple species or strains, we also demonstrated how this approach could be adapted for quantitative measurements of gene expression in situ. To the best of our knowledge, the nlmAB expression results are the first quantitative measures of bacterial gene expression in live animals. Using a dual luciferase-expressing strain of S. mutans, we were able to normalize reporter activity to a constitutively expressed internal control, which made it feasible to quantify temporal changes in nlmAB gene expression triggered by xylose induction. Since xylose is nontoxic to both mice and oral streptococci (Zimmer et al., 1993; Johnson et al., 1999; Xie et al., 2013), it serves as an ideal molecule to employ for gene induction studies in mice and can be easily administered simply by supplementing the mouse drinking water. As a test case, we chose to study the effects of exogenous control of mutacin IV production for the persistence ability of S. mutans in mice. Our previous in vitro studies have demonstrated the critical role of mutacin IV in dual species competition assays with commensal streptococci like S. sanguinis and S. gordonii (Kreth et al., 2005). Mutacin IV also has a relatively broad spectrum of activity compared to other nonlantibiotic bacteriocins (Hossain and Biswas, 2011), but its role in promoting S. mutans persistence has never been tested in an animal model system. By placing the nlmAB operon under the control of the Xyl-S1 cassette, it was possible to employ biophotonic imaging to correlate the induction of nlmAB expression with substantial improvements in S. mutans persistence over time. We were also able to remove the inducer from the xylose group to repress nlmAB expression and reverse this effect, as there was a rapid and sharp decline in the S. mutans population just 24 hours. after xylose removal. This approach provided a more direct assessment of nlmAB gene function in vivo because nlmAB gene expression and S. mutans persistence were both monitored simultaneously. One potential limitation to this induction approach is that gene induction becomes dependent upon mouse behavior. Variability may be introduced if some mice drink larger volumes or more often than others. Since the drinking water was also supplemented with sucrose and fructose, it was quite palatable to all of the mice and consumed heavily overall (unpublished observations). This may have helped to mitigate some potential variability introduced by differences in individual mouse behavior. Furthermore, our success with the induction system serves as a proof principle for the dual luciferase approach as a reliable method to monitor in situ gene expression patterns of genes controlled by their native promoters. This could be an important tool to determine whether hypothesized virulence genes are actually expressed in the animals and if so, how their temporal expression patterns are influenced by growth in the oral cavity. Likewise, the spatial resolution afforded by biophotonic imaging could provide much greater insight into the pathologies triggered by experimental infections in the oral cavity. For example, in caries studies, the highest levels of tooth demineralization would be expected to occur on the teeth harboring the greatest abundance of reporter bacteria, whereas those teeth exhibiting little or no colonization would be expected to exhibit proportionally lower caries scores. Based upon our imaging results, colonization seems to be highly heterogeneous within the oral cavity. Thus, measures of pathology would be much more conclusive if one were able to use biophotonic imaging to correlate the site(s) of infection with the areas of tissue destruction.
Experimental procedures
Bacterial species and culture conditions
All bacterial strains used in this study are listed in Table 1 and were either grown in ambient air with 5% CO2 in a CO2 adjustable incubator (S. gordonii and S. sanguinis) or in an anaerobic chamber with 85% N2, 10% CO2, and 5% H2 (S. mutans) at 37°C in Brain Heart Infusion medium (BHI; Difco, Sparks, MD) unless otherwise stated. For antibiotic selection, cultures were supplemented with the following antibiotics: 5 μg ml−1 erythromycin (S. gordonii and S. sanguinis) and 800 μg ml−1 kanamycin (S. mutans). For counterselection of markerless mutants, BHI plates were supplemented 0.02 M p-chlorophenylalanine (p-Cl-Phe).
Table 1.
Strains used in this study
| Strain | Relevant characteristics | Reference |
|---|---|---|
| DL1 | wild type S. gordonii | (Pakula and Walczak, 1963) |
| SK36 | wild type S. sanguinis | Genome reference strain (Xu et al., 2007) |
| UA159 | wild type S. mutans | Genome reference strain (Ajdic et al., 2002) |
| JK-G-1 | DL1∷Φ(ldhrbs – firefly luc); ErmR | This study |
| JK-G-2 | DL1∷Φ(ldhrbs – click beetle green luc); ErmR | This study |
| JK-G-3 | DL1∷Φ(ldhrbs – renilla luc); ErmR | This study |
| JK-G-4 | DL1∷Φ(ldhrbs – click beetle green luc); ErmR | This study |
| JK-G-5 | DL1∷Φ(ldhrbs – green renilla luc); ErmR | This study |
| JK-G-6 | DL1∷Φ(ldhrbs – cypridina luc); ErmR | This study |
| JK-G-7 | DL1∷Φ(ldhrbs – luciola red luc); ErmR | This study |
| JK-S-1 | SK36∷Φ(ldhrbs – firefly luc); ErmR | This study |
| JK-S-2 | SK36∷Φ(ldhrbs – click beetle green luc); ErmR | This study |
| JK-S-3 | SK36∷Φ(ldhrbs – renilla luc); ErmR | This study |
| JK-S-4 | SK36∷Φ(ldhrbs – click beetle green luc); ErmR | This study |
| JK-S-5 | SK36∷Φ(ldhrbs – green renilla luc); ErmR | This study |
| JK-S-6 | SK36∷Φ(ldhrbs – cypridina luc); ErmR | This study |
| JK-S-7 | SK36∷Φ(ldhrbs – luciola red luc); ErmR | This study |
| ldhIFDC2 | UA159∷Φ(IFDC2 cassette); ErmR, p-Cl-PheS | This study |
| IdhFlySm | UA159∷Φ(ldhrbs – firefIy luc); p-CI-PheR | This study |
| IdhRenSm | UA159∷Φ(ldhrbs – renilla luc); p-CI-PheR | This study |
| IdhCBGSm | UA159∷Φ(ldhrbs – click beetle green luc); p-CI- PheR |
This study |
| IdhRenGSm | UA159∷Φ(ldhrbs – green renilla luc); p-CI-PheR | This study |
| IdhLucRSm | UA159∷Φ(ldhrbs – luciola red luc); p-CI-PheR | This study |
| IdhCypSm | UA159∷Φ(ldhrbs – cypridina luc); p-CI-PheR | This study |
| iRenGIV | UA159∷Φ(ldhrbs – luciola red luc; Xyl-S1 – green renilla upstream of nlmAB); p-CI-PheR, KanR |
This study |
DNA manipulations
PCR was performed with a G-Storm GS1 thermocycler (GeneTechnologies; Essex, UK) according to the manufacturer’s protocol. Phusion DNA polymerase was purchased from New England Biolabs (Ipswich, MA) and Accuprime DNA polymerase from Life Technologies (Grand Island, NY). Phusion and Accuprime DNA polymerases were used for overlap extension PCR-ligation. Oligonucleotides (Table S1) were designed using sequence data obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/genome/) and synthesized by Integrated DNA Technologies (Coralville, IA).
Bioluminescent reporter genes used in the study
The following reporter genes were used to construct the various bioluminescent reporter strains. The firefly luciferase gene optimized as described in (Bonin et al., 1994) was PCR-amplified from pFW5-luc (Podbielski et al., 1999). The renilla luciferase gene was PCR-amplified from plasmid pRL-TK (Promega, Madison, WI; kindly provided by Dr. Ralf Janknecht, University of Oklahoma Health Sciences Center). Synthetic codon optimized versions of click beetle green, green renilla, luciola red, and cypridina luciferases were all synthesized by Integrated DNA Technologies (Coralville, IA) and provided in the plasmid pIDTBlue. Codon optimization was performed using software provided by IDT and the genome sequence of S. mutans UA159 (Ajdic et al., 2002). The sequences for these genes can be found in the supplemental materials.
Construction of constitutive bioluminescent reporter strains
All primers used for strain construction are listed in Table S1. The bioluminescent reporter strains for S. sanguinis and S. gordonii were constructed via a four-piece overlap extension PCR ligation strategy (Fig. 1). The respective reporter gene was placed under the control of the constitutive ldh (lactate dehydrogenase) promoter from S. gordonii or S. sanguinis such that the luciferase open reading frame (ORF) was inserted downstream of the ldh stop codon to leave the ldh gene intact. Briefly, PCR was used to amplify about 1 kb immediately upstream and downstream of the S. gordonii/S. sanguinis ldh stop codons. The respective reporter genes were amplified from the sources mentioned above. Each luciferase forward primer introduced a copy of the ldh Shine-Dalgarno sequence onto the 5’ end of the ORF to promote efficient translation. Constructs also included the ermAM gene cassette (Martin et al., 1987) to facilitate the selection of mutants on erythromycin. All PCR amplicons contained complementary sequences that facilitated PCR ligation. PCR amplicons of each fragment were purified with the QIAGEN PCR purification kit and mixed in a 1:1:1:1 ratio. The mixture served as a template for a second round PCR reaction containing the appropriate flanking ldh upstream forward and downstream reverse oligonucleotides. The resulting PCR amplicons were directly transformed into S. gordonii or S. sanguinis using previously published protocols (Wang et al., 2002; Zhu et al., 2011). Successful transformation was confirmed by testing several transformants for reporter gene activity. The bioluminescent reporter strains for S. mutans were constructed using a previously described markerless mutagenesis strategy (Xie et al., 2011). A similar overlap extension PCR ligation strategy as described above was used to insert each luciferase ORF between two 1 kb fragments homologous to the upstream and downstream region surrounding the ldh stop codon. Each constitutive reporter construct used a similar assembly scheme. The ldh upstream homologous fragments were amplified with the primers ldh upF and each of the corresponding luciferase upR primers. The luciferase open reading frames were amplified with the corresponding luciferase F/R primers and the downstream ldh homologous fragments were all amplified with ldh dnF/R. Overlap extension PCR of the resulting PCR amplicons was performed with the primers ldh upF/ldh dnR. The assembled PCR amplicons were transformed into the recipient strain ldhIFDC2 and selected on plates containing p-Cl-Phe to obtain the following strains: ldhFlySm (firefly luciferase), ldhRenSm (renilla luciferase), ldhCBGSm (click beetle green luciferase), ldhRenGSm (green renilla luciferase), ldhLucRSm (luciola red luciferase), and ldhCypSm (cyridina luciferase). The ldhIFDC2 recipient strain was constructed as follows. The ldh upstream homologous fragment was amplified with the primers ldh upF/ldh-IFDC2 upR2, the counterselectable IFDC2 cassette was amplified with the primers IFDC2 F2/IFDC2-ldh R, and the downstream ldh homologous fragment was amplified with the primers ldh dnF/R. Overlap extension PCR of the resulting PCR amplicons was performed with the primers ldh upF/ldh dnR and transformed into the wild-type strain UA159. Transformants were selected on plates containing erythromycin.
Construction of a xylose-inducible mutacin IV (nlmAB)-green renilla luciferase fusion
A four piece overlap extension PCR reaction was used to assemble a xylose-inducible mutacin IV operon containing the green renilla luciferase ORF, a terminatorless kanamycin resistance ORF (Merritt et al., 2005a), and the nlmAB ORFs encoding mutacin IV (Fig. 1). The construct was integrated onto the chromosome at the native nlmAB locus to replace the endogenous nlmAB operon promoter. The primers used to amplify the upstream and downstream nlmAB homologous fragments (nlmA upF/R and nlmA dnF/R), the Xyl-S1 cassette (XylR F/R), and the green renilla + kanamycin resistance cassette (RenG ORF F/Kan ORF R) are all listed in Table S1. The Xyl-S1 cassette was amplified from plasmid pZX9 (Xie et al., 2013), the green renilla + kanamycin resistance cassette was amplified from strain ldhRenGkan (unpublished strain), and the nlmAB upstream and downstream homologous fragments were amplified from the wild-type strain UA159. Complementary sequences in the primers facilitated the final construct assembly in a second round PCR reaction containing all four PCR amplicons as templates and the primers nlmA upF/nlmA dnR (Table S1). The resulting PCR amplicon was directly transformed into the constitutive luciola red luciferase expression strain ldhLucRSm and selected on plates containing 1% (wt/vol) xylose and 800 μg ml−1 kanamycin to produce the final strain iRenGIV.
Deferred antagonism assay
To confirm the expected mutacin IV production phenotype from strain iRenGIV, the parent strain ldhLucRSm and the inducible mutacin IV strain iRenGIV were spotted onto both BHI plates and BHI plates supplemented with 1% (wt/vol) xylose. After 24 hours of growth, the plates were overlaid with a soft agar suspension of either S. gordonii DL1 or S. sanguinis SK36. The presence of growth inhibition halos is indicative of mutacin IV production.
In vitro bioluminescent reporter assays of planktonic and biofilm cells
To assay for firefly, click beetle green, and luciola red luciferases, D-luciferin (2 mM suspended in 0.1 M citrate buffer; pH 6.0) was added similarly as previously described (Merritt et al., 2005b). To assay for native renilla and green renilla luciferases, 100 μl of culture was mixed with 1.5 μl of coelenterazine or the synthetic version coelenterazine-h (0.75 mg ml−1 diluted in ethanol). Coelenterazine substrates were purchased from NanoLight Technologies. To assay for cypridina luciferase activity, 100 μl of culture was centrifuged, resuspended in 100% DMSO, and then mixed with 10 μL cypridina luciferin (vargulin) substrate solution (1.4 mg ml−1 diluted in ethanol). Cypridina luciferin (vargulin) was purchased from NanoLight Technologies. Bioluminescence was determined with a Modulus Luminometer (Turner BioSystems). Planktonic cells were grown as described above. To assay the luciferase activity of S. sanguinis and S. gordonii biofilms, cells were inoculated in 2 ml BHI plus 1% (wt/vol) sucrose in a 24-well plate and grown as static cultures in 5% CO2 at 37°C. After overnight incubation, the medium was removed and replaced with 1 ml pre-warmed BHI plus 1% (wt/vol) sucrose and further incubated for 1 hour to reenergize cells depleted of ATP. Subsequently, the cells were resuspended by vigorous pipetting and luciferase activity determined. To assay luciferase activity from S. mutans biofilms cells, biofilms were grown statically in microfuge tubes and measured directly as previously described (Merritt et al., 2005b). Microfuge tubes containing 100 μl medium supplemented with 1% (wt/vol) sucrose were inoculated with a 500× dilution of a stationary phase culture and incubated anaerobically overnight. The following day, the spent medium was aspirated and replaced with an equal volume of fresh medium and further incubated for an additional hour at 37 °C to reenergize cells depleted of ATP. After the incubation period, luciferin substrate solutions were added directly into the microfuge tubes for luciferase measurements.
In vivo bioluminescent imaging
All animal care and procedures were previously reviewed and approved by the University of Oklahoma Health Sciences Center Institutional Animal Care and Use Committee (IACUC approval # 11-059).
i) Preparation of mice
Three-week-old female weanling BALB/CyJ mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). Mice were fed pellets of a standard Laboratory Rodent Diet 5001 throughout the assay period for all experiments. 24 hours after arrival, the mouse drinking water was supplemented with ampicillin (100 μg ml−1) for 24 hours. The next day, mice were anesthetized with 3% (vol/vol) isoflurane before cleaning their teeth with cotton swabs (Puritan, head width 2mm) soaked in 4% (vol/vol) H2O2. The cleaning procedure was performed twice with a brief recovery period given between cleanings. After the second round of cleaning, the mice were left in the cage for 1 hour prior to inoculation with bacteria.
ii) Preparation of bacteria
Overnight cultures were grown in semi-defined medium (SDM) (Merritt et al., 2009) and then diluted 1:40 in 10 ml SDM supplemented with 0.5% fructose. Cultures were grown to early log phase (A600nm ≈ 0.3) before adding 1% (wt/vol) sucrose and further incubated for another hour. Cells were concentrated by centrifugation and resuspended in 1 ml SDM medium.
iii) Mouse inoculation
Mice were first anesthetized with 3% (vol/vol) isoflurane for inoculation procedures. After anesthetization, mice were held horizontally with either the left or the right side facing down. 30 μl of concentrated bacterial solution was pipetted directly into the mice oral cavities and then they were immediately returned to the isoflurane anesthesia chamber and left for 2 min before finally returning the mice to their cages for recovery. After a brief recovery period, mice were inoculated on their opposite sides in the same manner. After inoculation, mice were kept without food and water for 60 min. For the remainder of the experiments, mice were fed a standard Laboratory Rodent Diet 5001, except that the drinking water was supplemented with 5% sucrose and 5% fructose. For xylose inducible gene expression studies, 10% (wt/vol) xylose was added to the water for the indicated time frames.
iv) Imaging
All animals were initially anesthetized using 3% (vol/vol) isoflurane and 20 μl 20% glucose was pipetted into the oral cavities to reenergize reporter bacteria before imaging. The mice were placed back into their cages for 15 min and subsequently anesthetized using 3% (vol/vol) isoflurane throughout the imaging procedure. For biophotonic imaging of mice carrying luciola red and firefly luciferase reporters, 25 μl of 2 mM D-luciferin solution suspended in 0.1 M citrate buffer (pH 6.0) was directly pipetted into the oral cavities. For measurements of native renilla or green renilla luciferases, 1.5 μl of coelenterazine-h solution (0.75 mg ml−1 diluted in ethanol) was added to 25 ul of phosphate buffered saline (PBS) and pipetted directly into the oral cavities. After substrate addition, the mice were immediately placed into a Carestream In Vivo Xtreme Imaging System (Carestream Health, Inc., Rochester, NY, USA) for biophotonic data collection. Colocalization images of X-ray and bioluminescence data were analyzed and exported using Molecular Imaging (MI) Software (Bruker Corporation).
Supplementary Material
Acknowledgements
This work was supported by an NIH-NIDCR grant DE021726 to J.K. and NIH-NIDCR grants DE018893 and DE022083 to JM. The authors wish to acknowledge Dr. Rajagopal Ramesh and the University of Oklahoma Health Sciences Center Molecular Imaging Core Facility for continuous support and Yifan Xu for technical assistance.
Footnotes
Table S1: Oligonucleotides used in the study.
Open reading frames of the codon optimized luciferases
The authors declare no conflict of interest.
References
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu N, Zelmer A, Wiles S. Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol Rev. 2011;35:360–394. doi: 10.1111/j.1574-6976.2010.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin AL, Gossen M, Bujard H. Photinus pyralis luciferase: vectors that contain a modified luc coding sequence allowing convenient transfer into other systems. Gene. 1994;141:75–77. doi: 10.1016/0378-1119(94)90130-9. [DOI] [PubMed] [Google Scholar]
- Caufield PW, Dasanayake AP, Li Y, Pan Y, Hsu J, Hardin JM. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun. 2000;68:4018–4023. doi: 10.1128/iai.68.7.4018-4023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy G, O’Connor S, Diehn FE, Costouros N, Alexander HR, Choyke P, Libutti SK. Comparison of noninvasive fluorescent and bioluminescent small animal optical imaging. Biotechniques. 2003;35:1022–1026. 1028–1030. doi: 10.2144/03355rr02. [DOI] [PubMed] [Google Scholar]
- Close DM, Hahn RE, Patterson SS, Baek SJ, Ripp SA, Sayler GS. Comparison of human optimized bacterial luciferase, firefly luciferase, and green fluorescent protein for continuous imaging of cell culture and animal models. J Biomed Opt. 2011;16:047003. doi: 10.1117/1.3564910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162:22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo AE, Frias-Lopez J. Beyond microbial community composition: Functional activities of the oral microbiome in health and disease. Microbes Infect. 2015 doi: 10.1016/j.micinf.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer LF, 3rd, Szalay AA. Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence. 2002;17:43–74. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One. 2012;7:e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Slade HD. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ooshima T, Torii M, Imanishi H, Masuda N, Sobue S, Kotani S. Dental caries induction in experimental animals by clinical strains of Streptococcus mutans isolated from Japanese children. Microbiol Immunol. 1978;22:301–314. doi: 10.1111/j.1348-0421.1978.tb00375.x. [DOI] [PubMed] [Google Scholar]
- Hossain MS, Biswas I. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl Environ Microbiol. 2011;77:2428–2434. doi: 10.1128/AEM.02320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics NS, Yassin SA, Rickard AH. Community interactions of oral streptococci. Adv Appl Microbiol. 2014;87:43–110. doi: 10.1016/B978-0-12-800261-2.00002-5. [DOI] [PubMed] [Google Scholar]
- Johnson KP, Hillman JD. Competitive properties of lactate dehydrogenase mutants of the oral bacterium Streptococcus mutans in the rat. Arch Oral Biol. 1982;27:513–516. doi: 10.1016/0003-9969(82)90093-0. [DOI] [PubMed] [Google Scholar]
- Johnson SA, van Tets IG, Nicolson SW. Sugar preferences and xylose metabolism of a mammal pollinator, the Namaqua rock mouse (Aethomys namaquensis) Physiol Biochem Zool. 1999;72:438–444. doi: 10.1086/316685. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Modi K, Yu J, Francis KP, Orihuela C, Tuomanen E, et al. Noninvasive monitoring of pneumococcal meningitis and evaluation of treatment efficacy in an experimental mouse model. Mol Imaging. 2005;4:137–142. doi: 10.1177/13505068211024891. [DOI] [PubMed] [Google Scholar]
- Kocher B, Piwnica-Worms D. Illuminating cancer systems with genetically engineered mouse models and coupled luciferase reporters in vivo. Cancer Discov. 2013;3:616–629. doi: 10.1158/2159-8290.CD-12-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009;28:397–403. doi: 10.1089/dna.2009.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola JP, Leyva P. Dental caries in an animal model. Rev ADM. 1990;47:190–194. [PubMed] [Google Scholar]
- Martin B, Alloing G, Mejean V, Claverys JP. Constitutive expression of erythromycin resistance mediated by the ermAM determinant of plasmid pAM beta 1 results from deletion of 5′ leader peptide sequences. Plasmid. 1987;18:250–253. doi: 10.1016/0147-619x(87)90068-0. [DOI] [PubMed] [Google Scholar]
- Merritt J, Qi F. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol. 2012;27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Qi F, Shi W. A unique nine-gene comY operon in Streptococcus mutans. Microbiology. 2005a;151:157–166. doi: 10.1099/mic.0.27554-0. [DOI] [PubMed] [Google Scholar]
- Merritt J, Niu G, Okinaga T, Qi F. Autoaggregation response of Fusobacterium nucleatum. Appl Environ Microbiol. 2009;75:7725–7733. doi: 10.1128/AEM.00916-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Kreth J, Qi F, Sullivan R, Shi W. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. J Microbiol Methods. 2005b;61:161–170. doi: 10.1016/j.mimet.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J Microbiol. 2014;52:188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Rizvi J, Hecht G. Expression of enteropathogenic Escherichia coli map is significantly different than that of other type III secreted effectors in vivo. Infect Immun. 2015;83:130–137. doi: 10.1128/IAI.02467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill K, Lyons SK, Gallagher WM, Curran KM, Byrne AT. Bioluminescent imaging: a critical tool in pre-clinical oncology research. J Pathol. 2010;220:317–327. doi: 10.1002/path.2656. [DOI] [PubMed] [Google Scholar]
- Pakula R, Walczak W. On the nature of competence of transformable streptococci. J Gen Microbiol. 1963;31:125–133. doi: 10.1099/00221287-31-1-125. [DOI] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielski A, Woischnik M, Leonard BA, Schmidt KH. Characterization of nra, a global negative regulator gene in group A streptococci. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- Sato A, Klaunberg B, Tolwani R. In vivo bioluminescence imaging. Comp Med. 2004;54:631–634. [PubMed] [Google Scholar]
- Simon-Soro A, Mira A. Solving the etiology of dental caries. Trends Microbiol. 2015;23:76–82. doi: 10.1016/j.tim.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Steinhuber A, Landmann R, Goerke C, Wolz C, Fluckiger U. Bioluminescence imaging to study the promoter activity of hla of Staphylococcus aureus in vitro and in vivo. Int J Med Microbiol. 2008;298:599–605. doi: 10.1016/j.ijmm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Struzycka I. The oral microbiome in dental caries. Pol J Microbiol. 2014;63:127–135. [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- Tanzer JM. Essential dependence of smooth surface caries on, and augmentation of fissure caries by, sucrose and Streptococcus mutans infection. Infect Immun. 1979;25:526–531. doi: 10.1128/iai.25.2.526-531.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EM, Nagata S, Tsuji FI. Cloning and expression of cDNA for the luciferase from the marine ostracod Vargula hilgendorfii. Proc Natl Acad Sci U S A. 1989;86:6567–6571. doi: 10.1073/pnas.86.17.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Wang BY, Chi B, Kuramitsu HK. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol Immunol. 2002;17:108–112. doi: 10.1046/j.0902-0055.2001.00001.x. [DOI] [PubMed] [Google Scholar]
- Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of pathogen and host. Nat Rev Microbiol. 2012;10:618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- Xie Z, Qi F, Merritt J. Development of a tunable wide-range gene induction system useful for the study of streptococcal toxin-antitoxin systems. Appl Environ Microbiol. 2013;79:6375–6384. doi: 10.1128/AEM.02320-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Okinaga T, Qi F, Zhang Z, Merritt J. Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl Environ Microbiol. 2011;77:8025–8033. doi: 10.1128/AEM.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Alves JM, Kitten T, Brown A, Chen Z, Ozaki LS, et al. Genome of the opportunistic pathogen Streptococcus sanguinis. J Bacteriol. 2007;189:3166–3175. doi: 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18:109–120. doi: 10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]
- Zheng L, Itzek A, Chen Z, Kreth J. Environmental influences on competitive hydrogen peroxide production in Streptococcus gordonii. Appl Environ Microbiol. 2011a;77:4318–4328. doi: 10.1128/AEM.00309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng LY, Itzek A, Chen ZY, Kreth J. Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. Int J Oral Sci. 2011b;3:82–89. doi: 10.4248/IJOS11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Fan J, Herzberg MC, Kreth J. Characterization of competence and biofilm development of a Streptococcus sanguinis endocarditis isolate. Mol Oral Microbiol. 2011;26:117–126. doi: 10.1111/j.2041-1014.2010.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer JP, Lewis SM, Moyer JL. Comparison of gavage, water bottle, and a high-moisture diet bolus as dosing methods for quantitative D-xylose administration to B6D2F1 (Mus musculus) mice. Lab Anim. 1993;27:164–170. doi: 10.1258/002367793780810423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.