INTRODUCTION
Recent molecular genetics studies have made remarkable progress by identifying several candidate susceptibility genes for schizophrenia [1, 2]. Some of these genes have now been extensively examined with respect to their neurobiological roles, phenotypic expression of risk genotypes, and their regulations in patients’ brain tissues [3–5]. As the evidence accumulates for the pathophysiologic roles of these susceptibility genes, it has also become clear that schizophrenia involves multiple genes and pathways as well as their interactions. Particularly relevant to this proposition are the outcomes of recent genome-wide association studies (GWAS) [6, 7] and rare variants investigations [8, 9], which highlight the involvement of multiple genes, genetic mechanisms, and their interactions in schizophrenia.
Schizophrenia is a complex trait disorder, characterized by multiple dimensions of neuro-cognitive symptoms. For susceptibility genes and other etiologic factors to conspire to form such specific sets of clinical manifestations, there are likely to be interactive dynamics through which genes and pathways integrate. If so, the pathophysiologic roles of a susceptibility gene will be determined not by its own neurobiological functions by the totality of its interactions with other factors rather than solely. To date, most pathophysiologic studies of candidate susceptibility genes have focused on a single candidate gene or pathway per se. It will be important to evaluate the impact of susceptibility genes in the context of their interactions with other factors.
Such perspectives have recently emerged for a few candidate genes [10–14]. These studies have proposed biological substrates, upon which candidate genes or their pathways can converge, called an “interactome”. The purpose of this article is to apply this perspective to the NRG1 –erbB4 signaling pathway, to consider its interactions with other pathways and to propose a possible interactome for the schizophrenia candidate pathway, schizophrenia interactome.
Neuregulin 1 (NRG1) is a schizophrenia susceptibility gene, for which risk genotypes or haplotypes have been identified [15–17] and its associations with the illness have been replicated in many populations [15, 18, 19] [20–24]. The potential pathophysiologic role of NRG1 is further supported by its diverse neurobiological functions [25, 26], some of which have been implicated in the pathophysiology of the illness [27]. These functions include neuronal migration [28], neuro- glial trophic effects, myelination [29] and modulation of glutamatergic [30, 31] or GABAergic neurotransmitter systems [32–34]. Genetic evidence also supports erbB4 as a candidate susceptibility gene and suggests positive epistatic interactions between NRG1 and erbB4 in schizophrenia [35, 36]. The tyrosine kinase receptor, erbB4, along with erbB2 and erbB3, transduces neuregulin1 and influences glutamatergic and GABAergic transmission as well as neurotrophic effects, which have also been implicated in schizophrenia. Together, these studies support the NRG1 –erbB4 signaling pathway as a whole for its role in schizophrenia.
While genetic, neurobiological and endophenotypic investigations support the role of the NRG1 –erbB4 pathway in schizophrenia, they also provide clues that this pathway might not act in isolation. In genetic studies, positive association of NRG1 with schizophrenia has been replicated in various populations; still, no single risk genotype has proved to be solely responsible for the illness [27, 37]. Genotype – phenotype association studies have demonstrated links between risk genotypes of NRG1 and some of the disease manifestations of schizophrenia. However, most disease endophenotypes were not associated with the NRG1 or erbB4 risk haplotypes as a whole, but rather with a single nucleotide polymorphism (SNP) within the hapblocks. Together, these studies suggest that the NRG1 – erbB4 pathway might play a role in schizophrenia, but in conjunction with other genes and factors.
Several investigators have provided excellent reviews on genetic, neurobiological, and pathophysiologic studies of neuregulin 1 and erbB4 in schizophrenia [38–41]. In this article, we will evaluate the pathophysiologic evidence for NRG1 –erbB4 signaling with respect to its interactions with other factors and will consider a putative interactome with which NRG1 – erbB4 signaling is associated.
Genetic evidence for NRG1 - erbB4 as a susceptibility pathway in schizophrenia
While NRG1 – erbB signaling has been extensively studied over decades [25, 29, 40, 42, 43], it was genetic findings that first implicated NRG1 in the pathophysiology of schizophrenia [16, 17]. Steffanson et al provided the first evidence for the association of NRG1 with schizophrenia in an Icelandic population [16]. Linkage analyses and subsequent association studies pointed to NRG1 as a susceptibility gene for schizophrenia and further identified an at risk haplotype, consisting of 5 SNPs and two microsatellites in the 5’ regulatory region of the gene, called Hap(Ice).
Subsequently, many groups have tested the association of NRG1 with schizophrenia and have reported positive results in many populations [15, 18, 19] with a few exceptions [44, 45]. Perhaps, as should be expected for a complex trait disorder, no single causative allele has yet been identified [46, 47]. Instead, at risk SNPs and haplotypes have varied between populations: a few studies reported positive associations for the seven marker HapIce (or three marker Haplotype) [15, 19] and others reported associations for other variants either at the 5’ end of the gene or other parts of the gene [18, 22].
Prompted by positive association studies for NRG1, several groups have examined erbB4, one of the receptors for NRG1, for its association with schizophrenia. In an Ashkenazi population, Silberberg et al. reported positive associations of alleles and haplotypes of erbB4 in an Ashkenazi population [48], which was followed by the reports of positive associations of erbB4 in two independent family samples [36].
Evidence for positive gene-gene interactions between NRG1, a ligand, and its receptor, erbB4, can lend further support for the pathophysiologic influence of the pathway. In populations from the UK [35, 49], a significant interaction was found between IVS12-15C>T in ErbB4 and HAPICE in NRG1 [35] or suggestive evidence for epistasis between ErbB4 with NRG1, NRG2 or NRG3 [49]. In a Japanese population, rs2919381 of NRG1 and rs7560730 of ErbB4, when combined, increased the risk for schizophrenia, while NRG1 alone was not associated with schizophrenia [50]. While more studies are needed to fully elucidate these gene gene interactions, these studies may provide further support NRG 1 – erbB4 signaling as a candidate susceptibility pathway for schizophrenia.
Our understanding of the genetic predisposition for complex trait disorders is guided by two perspectives; namely common alleles of small effects vs. rare alleles of large effects [51, 52]. The advent of new technologies, the microchip and its deep sequencing methods, have recently enabled genome-wide association studies (GWAS), the most systematic approach for common alleles, and extensive rare variants studies for schizophrenia. The results for these studies together have revealed the complexity to the schizophrenia genetics, in which the heritability of schizophrenia reflects a combination of common alleles and rare variants [37].
Several GWAS studies on large schizophrenia patient populations have so far identified several genes with the p value of less than 10−7. These studies, however, have failed to report positive associations for any of the widely recognized susceptibility candidate genes, including NRG1 and erbB4. Recent studies have shown that rare variants are more often associated with schizophrenia than with controls. It is also interesting that one of the rare variant studies has reported deletions in the erbB4 gene in a patient [9].
It remains unclear how the negative results of GWAS studies for candidate susceptibility genes are to be interpreted. Nor is it presently evident what relative roles the common vs. rare alleles contribute to schizophrenia. It is apparent, however, that no single causative allele or haplotype of NRG1 or erbB4 precipitates the illness [46, 47]. Consistent with this notion is the fact that risk genotypes have small odds ratios (mostly less than 2.0) and that the results of several meta-analyses have shown associations for NRG1 and/or erbB4 that were positive but nevertheless weak [46, 53–55]. At present, it will be reasonable to consider that NRG1 and erbB4 confer genetic susceptibility, via common or rare risk variants. It is also worth considering whether the resultant small effects of genetic variants do combine with other factors in order to precipitate biological substrates for schizophrenia.
Phenotypic expression of risk sequence variations of NRG1
A direct way to assess the pathophysiologic effects of genetic variants would be to examine human subjects with risk variants for symptoms of schizophrenia. Along these lines, many groups have tested the association of NRG1 risk variants with electrophysiological and structural endophenotypes of schizophrenia in human subjects. These measures have included prepulse inhibition (PPI) as a measure of sensory motor gating [56], P300 waveform during tasks of stimuli discrimination [57] and structural assessment of the brain as endophenotypes of schizophrenia.
These studies have reported positive associations of individual SNPs of NRG1 with some of previously described components of these endophenotypes, although the results have varied between populations. In patients with schizophrenic illnesses, [58], a SNP within the HapIce, SNP 221533, was associated with altered P300 latency, although the association was negative for HapIce per se. Association of NRG1 variants with altered PPI has varied, with the association of a missense genotype of NRG1, Rs3924999 being either positive [59] or negative [60].
The core haplotype was also examined for its association with volumetric abnormalities in the brain, but the results have varied. In a study of schizophrenia patients and controls, HapIce was found to be associated with smaller hippocampal volumes in both groups [61]. On the other hand, in a much larger sample, consisting of 120 families with schizophrenia probands, the core haplotype was not associated either with volumetric changes nor with age of onset of the illness [62].
Interestingly, a series of studies have shown rather consistent association of SNP8NRG243177, a SNP within HapIce, with various changes in the brain. The T allele of the SNP was associated with altered fronto-temporal brain function as well as the development of psychotic symptoms in subjects with high risk for schizophrenia [63]. There was also an association with increased lateral ventricle volume in patients with first episode schizophrenia [64]. The same allele was also associated with changes in spatial working memory capacity in healthy control male subjects [65]. However, in another cohort of patients with chronic schizophrenia, SNP8NRG243177 was not associated with either schizophrenia, age of onset or neurocognition [66].
Accumulating evidence suggests that a few SNPs within the core haplotype, SNP8NRG243177 in particular, could impact on at least some components of schizophrenia endophenotypes. However, associations of endophenotypes with the core haplotype as a whole or other SNPs within the hapblock have varied. Thus, phenotypic expression of these SNPs can not be fully explained by the transmission of the haplotype, which might suggest the involvement of other factors. The presence of variability between populations is also noteworthy. Phenotypic effects of these SNPs appear to be more pronounced in certain populations; suggesting that the observed positive associations are linked to certain biological contexts characteristic of certain populations.
In sum, genotype-phenotype association studies suggest the roles of NRG1 – erbB signaling in schizophrenia but also support the notion that phenotypic expression of the candidate pathway might involve other factors.
Behavioral manifestations of dysregulated NRG1 or erbB4
Transgenic approaches are a powerful tool to evaluate the behavioral effects of susceptibility genes [67]. Several transgenic lines have been developed for NRG1 and erbB4 by targeting specific domains of the molecules [68, 69]. These include NRG1 (+/−, EGF), a transgenic line with a mutation in the EGF like domain, [70, 71], NRG1 (+/−, TM), with a deletion in the transmembrane domain [16], NRG1 (+/−) Ig, with an alteration in the immunoglobulin domain, [72], and NRG1 (+/−, type III) with a mutation in type III of NRG1 [73]. Another transgenic line manifesting dysregulated NRG1 is the BACE1 null, in which the beta-Site APP-cleaving enzyme 1 (BACE1), responsible for proteolytic processing of NRG1, was targeted [74].
These transgenic mouse models have been examined for behavioral phenotypes utilizing comprehensive testing batteries. Overall, the studies show that the mutations of various domains are associated with several endophenotypic behavioral characteristics of schizophrenia. Such characteristics include altered locomotor activity, which was suppressed by antipsychotic treatment [16], deficits in PPI, mismatch negativity [75], contextual fear conditioning [75], cognitive impairment [74], and social behaviors [75–77]. Furthermore, NRG1 (+/−, type III) showed enlarged lateral ventricles and decreased dendritic spine density, structural characteristics of brains of schizophrenia patients [73]. These together suggest that NRG1 signaling is linked to components of schizophrenia endophenotypes and therefore support the pathophysiologic role of NRG1 in schizophrenia.
It was also noted, however, that the transgenic lines differed in their manifestation of behavioral endophenotypes. PPI deficit was robust in NRG1 (+/−, type III) [73] and BACE 1 mutants [74]; less noticeable in NRG1 (+/−, TM); and either slight or unobserved in NRG1 (+/−, EGF)[69, 75]. Locomotor hyperactivity was pronounced in NRG1 (+/−, TM) and NRG1 (+/−, EGF), but it was not observed in NRG1 (+/−, Ig) at baseline [78]. Impaired habituation was shown in NRG1 (+/−, TM) but not in NRG1 (+/−, EGF) [79].
Mice mutated for erbB2 or erbB3, two other receptors for NRG1, demonstrated none of the behavioral changes seen in NRG1 mutants [79]. Thus, behavioral effects of NRG1 mutations are more likely to be mediated by erbB4. Mutants for erbB4 showed hyperactivity similar to NRG1 mutants, yet exhibited no deficits in PPI [16]. However, when erbB4 was perturbed in a conditional knockout paradigm specific for the CNS, the animals showed an overall decreased level of activity, the opposite of other mutants’ behavioral trait [80, 81].
When viewed together, these studies in transgenic animals support the notion that perturbations in NRG1 - erbB4 signaling contribute to the behavioral phenotypes of schizophrenia. A closer look at individual mutants, however, reveals additional subtleties to the relationship between this gene and patient behavioral outcomes. It appears that alterations in NRG1 – erbB4 signaling can lead to different behavioral manifestations, depending on the affected domains, time, and biological context of the dysregulation.
Transgenic studies help us to evaluate the pathophysiologic roles of candidate genes. It is a challenge, however, to relate behavioral outcomes of mutants to endophenotypic expression of genetic variants in patients. Variations in patients’ risk genotypes are subtle, while gene alterations in transgenic mice are more pronounced. Thus, behavioral manifestations of risk genotypes in patients could be much less robust than those seen in transgenic mice. Furthermore, it becomes increasingly clear that risk variants of candidate genes can differ between populations and vary among individuals, leading to diverse behavioral effects.
Transgenic studies provide clarity for the function of candidate genes. Risk genotypes of candidate genes, however, could have subtle and diverse effects on the endophenotypes of schizophrenia. These heterogeneous effects must be integrated in some fashion to form disease specific endophenotypes. If so, it will be reasonable to consider that such integration occurs via interactions with other candidate genes or pathways in specific interactive dynamics.
Dysregulations of NRG1 and erbB4 in postmortem brains of patients
Despite various confounds inherent to the study paradigm, postmortem studies can offer direct evidence for dysregulated candidate genes and pathways in patients’ brains. Several groups have assessed gene expression and proteins of NRG1 and erbB4 in postmortem brains and reported differences between schizophrenia and healthy control groups. In the dorsolateral PFC (DLPFC), Hashimoto et al examined mRNAs for types I, II and III of NRG1 and found that that there was a significant increase in type I mRNAs, while type II and type III mRNAs remained unaltered [82]. Type I mRNAs were also found to be increased in the DLPFC of schizophrenia patients in another cohort [83]. NRG1 protein expression has also been examined, although the results were somewhat varied. Chong et al showed significant increases in NRG1 protein expression, particularly for smaller MW bands in western blotting analyses [84] whereas in another cohort of much older subjects, no discernable changes were found in NRG1 expression among patients [85].
Examination of ErbB4 protein expression in postmortem brains of schizophrenia patients has also yielded mixed results. In one study, western blot analyses of the PFC showed the full length erbB4, 180 kDa, was increased by 30% in schizophrenia [84]. In addition, this study showed that lower molecular weight bands; 21, 55 and 60 kDa bands were also altered and that the ratios 21 kDa/180 kDa and 55 kDa/180 kDa became significantly reduced [84].
In a very similar study design, we have conducted immunoblot analyses of erbB4 proteins in synaptosomal, synaptic membranes and post-synaptic density fractions of 14 matched pairs of patients and controls [85]. Our study failed to detect differences in erbB4 protein expression between the schizophrenia and control groups. This apparent discrepancy between the two studies could be attributable to the differences in demographics of subjects, specifically age of subjects (48 vs. 79) and differences in the PMI (30 vs. 10) [84].
Interestingly, significant increases were reported in mRNAs for ErbB4 JMa/CYTI in postmortem brain studies of two separate cohorts of patients with schizophrenia. ErbB4 JMa/CYTI. consists of a metalloprotease cleavable extracellular domain (JM-a) and a cytoplasmic domain (CYT-1) that contains a phosphotidylinositol-3 kinase (PI3K) binding site [86]. This study also reported positive association of intronic SNPs with increased mRNAs for JMa/CYTI, suggesting that splice-variant specific expression of ErbB4 is the basis for the association of this gene with schizophrenia [86].
Alternatively, dysregulations in NRG1 signaling may be a reflection of altered function of the receptors. To test this, we employed an experimental paradigm in which postmortem brain tissues were incubated with NRG1, and the erbB4 activation was biochemically monitored [85]. We found that ligand-induced erbB4 activation, measured by tyrosine phosphorylation and the activation of downstream signaling, was significantly enhanced in the PFC of schizophrenia subjects. Interestingly, this finding was accompanied by the increased association of erbB4 with PSD-95, which plays a critical role in the activation of erbB4; suggesting that enhanced erbB4 activity might be due to altered protein associations of erbB4 [85].
In sum, these studies demonstrate that NRG1- erbB4 signaling is dysregulated at multiple levels from gene expression to protein-protein interactions in the brains of schizophrenia patients.
Interactions with other pathways for NRG1-erbB4 dysregulations in schizophrenia
The results of postmortem brain studies provide a few clues that dysregulations in NRG1 – erbB4 signaling might be partly mediated by interactions with other pathways. Genetic variants of NRG1 or erbB4 are thought to alter the expression or the function of NRG1 or erbB4. The observed dysregulations, however, can not be explained solely by the risk haplotypes because risk genotypes of NRG1 or erbB4 affect only subgroups of patients [16, 35, 48], while altered NRG1 – erbB4 signaling appears to be more prevalent among patients [84–86].
It is also of note that the observed dysregulations in NRG1 and erbB4 are unidirectional: with increases in mRNAs and proteins of NRG1 and erbB4 and in the activity of erbB4. It is unlikely that a single abnormality, either in NRG1 or in erbB4, would be able to cause consistent changes in the levels of the ligand, receptors and their functions. If the primary dysregulation is an increase in NRG1, for instance, erbB4 signaling is likely to decrease as a compensatory mechanism. A relevant example is the observation that increased NRG1 was accompanied by decreased association of erbB4 with PSD-95 in BACE1 (−/−) mice [74]. The dysregulations in patients are therefore more likely to be the cumulative results of alterations not only from within the NRG1 – erbB4 pathway but also from other pathways associated with NRG1 – erbB4.
There could be several mechanisms by which other genes or pathways impact on NRG1 – erbB4. NRG1 – erbB4 dysregulations could be compensatory responses to changes in other pathways that transduce signals via erbB4. For instance, down-regulations of NRG2 or NRG3 that also transduce signals via erbB4 could enhance NRG1- erbB4 signaling at all levels. Relevant to this is the recent report that NRG3 was associated with schizophrenia [87]. In addition, dysregulations in the NRG1 – erbB4 pathway as a whole could be associated with alterations in the downstream pathways of erbB4. In this regard, it is interesting that AKT signaling, a downstream pathway of erbB4, has been reported to be attenuated in the postmortem brains of SCZ patients [88, 89].
Another mechanism could be one in which NRG1 - erbB4 signaling is part of a larger molecular complex that encompasses multiple pathways. These pathways may interact with each other in the network and their interactions are coordinated to serve specific cellular functions. In this scheme, erbB4 can associate with other molecules, through which it can modulate other pathways and/or can be affected by those that are critical for schizophrenia. In this regard, the relationship between erbB4 and glutamatergic signaling pathway is of particular import.
ErbB4 and glutamatergic receptors are highly concentrated in the postsynaptic density (PSD) and are physically associated, albeit indirectly. ErbB4 can impact the establishment and activity of glutamatergic receptors [31, 90–92] and alter glutamatergic receptor function [86], NMDAR in particular, can also impact on erbB4 [93]. The relationship between erbB4 and NMDAR is of particular import, considering the increasing evidence supporting NMDAR hypofunction as a pathophysiologic mechanism for schizophrenia. Of the various possible mechanisms by which NRG1 –erbB4 can interact with other pathways, therefore, we will focus on its relationship with glutamatergic signaling and examine these receptor complexes as a possible interactome of schizophrenia.
NRG1 – erbB signaling modulates glutamatergic signaling
Several groups have examined the acute effects of NRG1 on glutamatergic signaling and shown important interactions between the two pathways. Huang et al employed electrophysiological recordings in rodent hippocampal slices and demonstrated that NRG1 stimulation suppressed long term potentiation (LTP) [31] via non-NMDA mediated mechanisms. In this study, NRG1 did not alter NMDA mediated synaptic responses, which suggested that NRG1 modulates LTP via AMPA receptors. Kwon et al [90]investigated the effects of the NRG1 in the hippocampal CA1 region in more detail. It was shown that NRG1β reverses LTP in vivo by promoting the internalization of GluR-1 containing AMPARs.
In another preparation, Gu et al [91]showed that EGF domain of NRGβ-1 produced a significant reduction of the NMDA receptor mediated ionic currents and synaptic currents in PFC pyramidal neurons in primary cultures derived from rat PFC [91]. The effects of NRG1 on NMDAR signaling were also examined in human postmortem brain tissues. Our group showed that NRG1 attenuates ligand induced phosphorylation of NMDA receptors and its association with signaling partners in human postmortem brain tissues [85]. These observations together, demonstrate that NRG1 overall attenuates the activation of glutamatergic pathways impacting on NMDA receptor signaling.
It is largely unknown how exactly the NRG1- erbB4 signaling modulates NMDA receptor activation. Such modulation, however, could occur via molecular events that govern NMDA receptor activity such as tyrosine phosphorylation of NR2 subunits. Of various kinases influencing NMDA receptor activity, Src/Fyn kinases are particularly interesting, because they are modulated by multiple PKC, Pyk2 and PKA thus can serve as a hub of various pathways influencing NMDA receptor activation [94, 95]).
NRG1 stimulation can enhance Fyn kinase activity and increase phosphorylation of Y1472 of NR2B [96]. Interestingly, when NRG1 was applied to brain tissues in addition to NMDA and glycine, tyrosine phosphorylations of NR2 subunits was lower than those in the tissues that were incubated solely with NMDA and glycine. This modulation, shown in human postmortem [85] and mouse brains (unpublished observation), was accompanied by decreases in phosphorylation of src and Pyk2. This suggests NRG1 - erbB4 mediated modulation of NMDA receptor signaling via src/Fyn and its upstream kinases.
Evidence has accumulated that the NRG1 – erbB4 signaling is crucial for the development and maintenance of excitatory synapses [28, 92, 97]. Over-expression of erbB4 can enhances AMPA synaptic currents and increase dendritic spine size, while reduced NRG1/ErbB4 activity can destabilize synaptic AMPA receptors leading to the loss of synaptic NMDA currents and dendritic spines [98]. Synaptic activity is largely dependent on the size and morphology of dendritic spines, which in turn are modulated by synaptic activity. NRG1/erbB4 activity can mediate the interactions between the synaptic activity and dendritic spines [98]. These observations together suggest that NRG1 – erbB4 signaling provides trophic support for glutamatergic pathway for development, maturation and stabilization.
The results of these studies reveal seemingly divergent effects of NRG1 – erbB signaling on the glutamatergic pathway. In acute applications, NRG1, partially mediated by erbB4, attenuates glutamatergic transmission. Long term, erbB4 may provide trophic support to synaptic structure and function during pre- and post-natal development and thus enhance AMPA and NMDA currents. It is presently unclear which of these is more relevant to the pathophysiology of schizophrenia.
The results of postmortem brain studies showed overall increases in NRG1 – erbB4 signaling in schizophrenia [85]. This may lend more support to the acute effects of NRG1 on the glutamatergic pathway than to the long term trophic effects. In the scheme of the acute effects, the increased NRG1 – erbB4 signaling will lead to NMDA receptor hypofunction as postulated for schizophrenia. In contrast, the increased expression or signaling of the NRG1 –erbB4, based on the long term effects, might lead to increases in dendritic spines and other features of synpatic strengths, which would be the opposite of previous postmortem findings [99].
It will be also of note, however, that postmortem brains may not exhibit erbB4’s long term trophic or developmental effects, because postmortem subjects are well past the stages of brain development in which the disease first manifests. Highly relevant to this are recent observations that spine densities of postsynaptic neurons in postmortem brain tissues are decreased in schizophrenia patients [100, 101]. Together, these studies illustrate various ways in which NRG1 –erbB can impact on glutamatergic function and further confirms robust interactions between the two pathways.
NMDA receptor complexes as a schizophrenia interactome
ErbB4 and NMDA receptors are part of the macromolecular complexes consisting of hundreds of proteins linked to diverse signaling pathways. In these complexes, the proteins are highly organized around scaffolding proteins, particularly those containing PDZ domains. Such intermolecular organization determines the recruitment of proteins into the complexes as well as the proximity between signaling partners.
The physical relationships between the molecules are crucial to their interactions and could therefore determine the activity of the signaling proteins. Moreover, the overall constellation of the proteins could influence the receptor complex as a whole in terms of how it responds to other entities, such as ligand stimulation or downstream transcriptional changes.
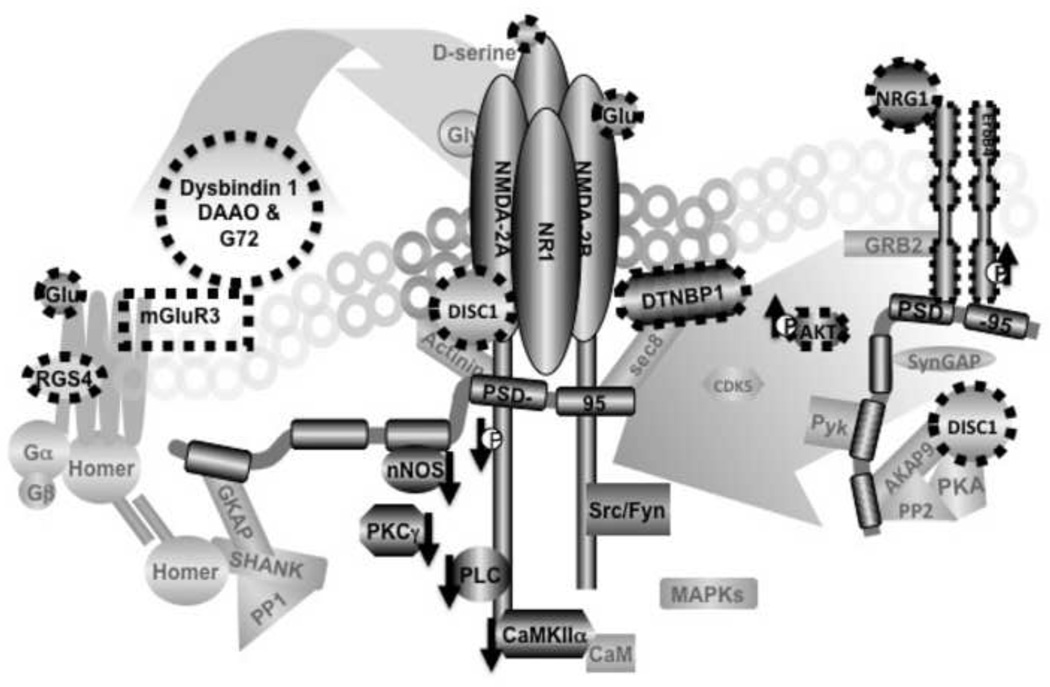
Therefore, these macromolecular complexes of erbB4, NMDAR and other glutamatergic receptors could serve to integrate the dysregulations of various molecular processes linked to the receptor complexes, as might happen with schizophrenia (illustrated in Figure 1).
Figure 1. Protein interactions of the NMDA receptor (NMDAR) complex as an interactome for the pathophysiology of schizophrenia.
Several candidate susceptibility genes converge on NMDAR complexes either in ligand mediated mechanisms (pre-receptor) or in post-receptor protein interactions. Dysbindin 1 modulates glutamate release, DAAO and G72 regulate D-serine and thus alter NMDAR activation. These dysregulations originating from ligand mediated processes will lead to altered associations between NMDARs and their signaling partners; PLCγ nNOS and PKC and others. Note that associations of NMDAR1 with PLCγ, nNOS and CAMKII were found to be decreased in the postmortem brains of schizophrenia patients (see downward arrows). ErbB4, DISC1 and dysbindin 1 could also directly impact on protein associations in the NMDAR complexes; ErbB4 via PSD-95, DISC1 via AKAP9 or Actinin and dysbindin 1 via sec8. Thus, alterations in the susceptibility genes and their pathways, via ligand mediated or post-receptor mechanisms, can converge on protein associations within the NMDAR complexes. It is of note that there are numerous potential protein-protein interactions in NMDAR complexes that might be associated with various psychiatric illnesses [112]. This diagram represents a permutation of numerous possible arrangements.
It has been proposed that NMDAR signaling could be a point of convergence for various candidate susceptibility genes for schizophrenia [2, 102]. This proposition was based on the observation that several candidate susceptibility genes, in addition to erbB4, such as DAAO, G72 and dysbindin-1 can affect glutamatergic signaling (Fig.1). DAAO metabolizes D-serine, an endogenous modulator of the NMDA receptor. G72, another candidate gene can activate DAAO, thereby also influencing glutamatergic signaling [103–105]. Dysbindin-1 can modulate glutamate release via its interactions with the vesicular transport machinery [106].
Protein-protein interactions of NMDAR complexes could be molecular substrates upon which the effects of dysregulated candidate genes converge. The influence of DAAO, G-72 and dysbindin-1 on the activation of NMDAR, could lead to changes in the modulation of NMDAR with its signaling partners, such as PLCγ or nNOS (Fig.1). Interestingly, our postmortem brain studies have shown decreases in the association of NMDAR1 with PLCγ and nNOS in response to NMDA stimulation in postmortem brains [85] as well as in their basal states (unpublished observations).
Some of the susceptibility genes can more directly impact protein associations of NMDAR complexes (Fig.1). DISC-1 associates with AKAP-9 and alpha-actinin-1 [13], binding partners of NMDAR. Altered DISC-1 expression can therefore modulate the relationships of AKAP-9 and alpha-actinin-1 with other signaling proteins of NMDAR complexes [107]. Dysbindin-1, which was also found in the PSD [108], can bind the exocyst protein sec8 [107], which is critical to NMDAR trafficking. Thus, the decreased level of dysbindin observed in schizophrenia [106, 109, 110] can impact on both the protein association and trafficking of NMDAR. In this regard, study of subcellular distribution of NMDAR subunits and their associated proteins in postmortem brains of patients will be an interesting avenue of research as the methodologies for such studies are now available [111].
The impact of erbB4 on protein interactions of NMDAR complexes could be mediated by its relationship with PSD-95. The association with PSD-95 is crucial for the activity of erbB4 as well as for NMDAR and it has been shown that the PSD-95 protein complexes also contain a number of proteins that have been implicated for schizophrenia [112]. The increased erbB4 -PSD-95 association observed in schizophrenia was accompanied by enhanced association of erbB4 with NMDAR. Also relevant to this is the observation that NRG1 stimulation induced further decreases in the activation of NMDAR in patients’ brain tissues, in which erbB4 - PSD-95 association was enhanced.
The interactions, between susceptibility candidate genes and protein interactions of NMDAR, might also be reciprocal. Altered protein composition in the NMDAR complexes or in NMDAR function can influence the activity of the candidate pathway. As an example, decreased expression of NMDAR’s signaling partners, such as PLCγ or PKC, or their recruitment into the receptor complexes, could reduce the activation of downstream pathways following receptor activation. Thus, the effects of dysbindin-1, DISC-1, or other candidate genes that influence NMDAR activation would be further attenuated.
Altered NMDAR function could also impact on protein associations of candidate genes within NMDAR complexes and thereby can modulate their functions. For example, glutamate induced down-regulation of cdk-5 enhances phosphorylation of PSD-95 at the N-terminal, which facilitates the association of PSD-95 with erbB4 [113]. Decreased NMDAR activation therefore can enhance cdk-5 activity [114], which in turn could increase the association of erbB4 with PSD-95 as observed in schizophrenia. Interestingly, we recently observed that erbB4: PSD-95 association was also strikingly enhanced in NMDAR hypomorphs expressing significantly decreased levels of NMDAR (unpublished observation). Thus, the enhancement in erbB4 activity and/or in erbB4 - PSD-95 association can not only precipitate NMDAR hypofunction, as noted above, but also can result from it.
Reciprocal interactions between NMDAR complexes and the susceptibility genes could be critical for establishing the pathophysiology of schizophrenia. A unilateral dysregulation of a pathway by another could conceivably be compensated for by another biological process thus dampening its phenotypic expression. Mutually facilitating bidirectional modulations, however, could defy compensatory processes, thus maintaining abnormal signaling as a newly established homeostasis. Protein – protein interactions in the NMDAR complexes can mediate such reciprocal interactions between pathways and therefore are equipped to respond to, integrate and reciprocally modulate dysregulations in the candidate susceptibility genes. Thus, the protein stoichiometry of NR complexes can reflect dysregulations of many molecular pathways in postsynaptic neurons, thus can represent molecular pathology of various neuropsychiatric illnesses. Of these, the pathophysiologic role of NMDA receptor complexes is particularly interesting as we find that NRG1-erbB4 signaling and dysregulations of other candidate genes and pathways can ultimately converge on NMDAR complexes. Thus we propose the protein stoichiometry of NMDAR complexes as a schizophrenia interactome.
Concluding Remarks
NRG1 – erbB4 signaling has been extensively studied as a candidate pathway of schizophrenia. While these studies endorse possible roles of NRG1 – erbB4 signaling in schizophrenia, they also indicate that this pathway does not act alone but in conjunction with other candidate pathways. Most pathophysiologic studies of candidate genes, including NRG1 and erbB4, have thus far focused on the candidate gene or pathway per se. Pathophysiologic investigation of candidate genes should be considered in the context of interactions with other pathways. Such approaches will require identification of schizophrenia interactomes and re-evaluation of candidate genes in the context of pathophysiologic roles of the interactomes.
We propose the protein-protein interactions in NMDAR complexes as a possible schizophrenia interactome in which dysregulations of NRG1 –erbB4 and other susceptibility genes can integrate. This proposition is prompted by several observations. First, evidence accumulates to support NMDAR hypofunction in the pathophysiology of schizophrenia. Second, the protein stoichiometry of NMDAR complexes is well poised to integrate changes in neighboring candidate pathways. Third, various candidate genes and pathways can impact on protein composition of NMDAR complexes (Fig. 1).
Pathophysiologic investigation of the interactome will require research paradigms that can capture the interactions between the genes and pathways. Generating mutations in multiple candidate genes either using double knock-outs or recreating the sequence variations of patients employing knock-in approaches may be a rewarding research avenue. It will also be important to fine tune the expression of candidate genes in transgenic mice in a brain region specific manner.
Also critical will be the development of experimental techniques that can capture protein complexes and decipher both protein composition and the interactions among the proteins. In this regard, recent technological advancements in the biochemical fractionation of brain tissues and mass-spectrometry analyses of protein complexes are timely. Until recently, examination of these protein-protein interactions has been hindered by the complexity of the PSD and NMDAR complexes. Recent technical advancements, however, have enabled biochemical enrichment of the PSD and proteomic analysis of protein complexes of brain tissues [115]. More recently, these methods have been adapted human postmortem brain tissues [111], aided by advances in proteomic analyses of both the PSD [111, 116, 117] and the protein complexes located therein [112, 117].
Combined with transgenic strategies and with in vivo and ex vivo pharmacologic manipulations, this approach could elucidate the role of NRG1 –erbB4 or other candidate pathways in the context of the disease interactome. Such investigations could lead us not only to a better pathophysiologic understanding but also to the identification of certain protein associations as potential therapeutic targets.
Acknowledgement
This work was supported by a grant from Stanley Medical Research Institute (CGH) and a grant from the National Institute of Mental Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Owen MJ, Craddock N, O'Donovan MC. Schizophrenia: genes at last? Trends Genet. 2005;21(9):518–525. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Harrison PJ, Owen MJ. Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet. 2003;361(9355):417–419. doi: 10.1016/S0140-6736(03)12379-3. [DOI] [PubMed] [Google Scholar]
- 3.Sawamura N, Sawa A. Disrupted-in-schizophrenia-1 (DISC1): a key susceptibility factor for major mental illnesses. Ann N Y Acad Sci. 2006;1086:126–133. doi: 10.1196/annals.1377.018. [DOI] [PubMed] [Google Scholar]
- 4.Arnold SE, Talbot K, Hahn CG. Neurodevelopment, neuroplasticity, and new genes for schizophrenia. Prog Brain Res. 2005;147:319–345. doi: 10.1016/S0079-6123(04)47023-X. [DOI] [PubMed] [Google Scholar]
- 5.Harrison PJ. Schizophrenia susceptibility genes and their neurodevelopmental implications: focus on neuregulin 1. Novartis Found Symp. 2007;288:246–255. discussion 255-9, 276-81. [PubMed] [Google Scholar]
- 6.Shi J, Badner JA, Liu C. PDLIM5 and susceptibility to bipolar disorder: a family-based association study and meta-analysis. Psychiatr Genet. 2008;18(3):116–121. doi: 10.1097/YPG.0b013e3282fa184b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefansson H, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460(7256):744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 10.Lee SA, et al. POINeT: protein interactome with sub-network analysis and hub prioritization. BMC Bioinformatics. 2009;10:114. doi: 10.1186/1471-2105-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang CY, et al. PhosphoPOINT: a comprehensive human kinase interactome and phospho-protein database. Bioinformatics. 2008;24(16):i14–i20. doi: 10.1093/bioinformatics/btn297. [DOI] [PubMed] [Google Scholar]
- 12.Camargo LM, Wang Q, Brandon NJ. What can we learn from the disrupted in schizophrenia 1 interactome: lessons for target identification and disease biology? Novartis Found Symp. 2008;289:208–216. doi: 10.1002/9780470751251.ch17. discussion 216-21, 238-40. [DOI] [PubMed] [Google Scholar]
- 13.Brandon NJ. Dissecting DISC1 function through protein-protein interactions. Biochem Soc Trans. 2007;35(Pt 5):1283–1286. doi: 10.1042/BST0351283. [DOI] [PubMed] [Google Scholar]
- 14.Guo AY, et al. The dystrobrevin-binding protein 1 gene: features and networks. Mol Psychiatry. 2009;14(1):18–29. doi: 10.1038/mp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stefansson H, et al. Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet. 2003;72(1):83–87. doi: 10.1086/345442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefansson H, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71(4):877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stefansson H, et al. Neuregulin 1 and schizophrenia. Ann Med. 2004;36(1):62–71. doi: 10.1080/07853890310017585. [DOI] [PubMed] [Google Scholar]
- 18.Corvin AP, et al. Confirmation and refinement of an 'at-risk' haplotype for schizophrenia suggests the EST cluster, Hs.97362, as a potential susceptibility gene at the Neuregulin-1 locus. Mol Psychiatry. 2004;9(2):208–213. doi: 10.1038/sj.mp.4001412. [DOI] [PubMed] [Google Scholar]
- 19.Williams NM, et al. Support for genetic variation in neuregulin 1 and susceptibility to schizophrenia. Mol Psychiatry. 2003;8(5):485–487. doi: 10.1038/sj.mp.4001348. [DOI] [PubMed] [Google Scholar]
- 20.Fukui N, et al. Supportive evidence for neuregulin 1 as a susceptibility gene for schizophrenia in a Japanese population. Neurosci Lett. 2006;396(2):117–120. doi: 10.1016/j.neulet.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Li CH, Liao HM, Chen CH. Identification of molecular variants at the promoter region of the human alpha 7 neuronal nicotinic acetylcholine receptor subunit gene but lack of association with schizophrenia. Neurosci Lett. 2004;372(1–2):1–5. doi: 10.1016/j.neulet.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Petryshen TL, et al. Support for involvement of neuregulin 1 in schizophrenia pathophysiology. Mol Psychiatry. 2005;10(4):366–374. doi: 10.1038/sj.mp.4001608. 328. [DOI] [PubMed] [Google Scholar]
- 23.Yang JZ, et al. Association study of neuregulin 1 gene with schizophrenia. Mol Psychiatry. 2003;8(7):706–709. doi: 10.1038/sj.mp.4001377. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, et al. A case control and family based association study of the neuregulin1 gene and schizophrenia. J Med Genet. 2004;41(1):31–34. doi: 10.1136/jmg.2003.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buonanno A, Fischbach GD. Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol. 2001;11(3):287–296. doi: 10.1016/s0959-4388(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 26.Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284(1):14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 27.Harrison PJ, Law AJ. Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biol Psychiatry. 2006;60(2):132–140. doi: 10.1016/j.biopsych.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Bendito G, et al. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell. 2006;125(1):127–142. doi: 10.1016/j.cell.2006.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birchmeier C, Nave KA. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56(14):1491–1497. doi: 10.1002/glia.20753. [DOI] [PubMed] [Google Scholar]
- 30.Garcia RA, Vasudevan K, Buonanno A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proc Natl Acad Sci U S A. 2000;97(7):3596–3601. doi: 10.1073/pnas.070042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang YZ, et al. Regulation of neuregulin signaling by PSD-95 interacting with ErbB4 at CNS synapses. Neuron. 2000;26(2):443–455. doi: 10.1016/s0896-6273(00)81176-9. [DOI] [PubMed] [Google Scholar]
- 32.Longart M, Buonanno A. [Neuregulins: A family of factors with critical functions during nervous system development and in the cellular transformation and differentiation] Rev Neurol. 2002;34(1):91–97. [PubMed] [Google Scholar]
- 33.Xie F, Raetzman LT, Siegel RE. Neuregulin induces GABAA receptor beta2 subunit expression in cultured rat cerebellar granule neurons by activating multiple signaling pathways. J Neurochem. 2004;90(6):1521–1529. doi: 10.1111/j.1471-4159.2004.02685.x. [DOI] [PubMed] [Google Scholar]
- 34.Woo RS, et al. Neuregulin-1 enhances depolarization-induced GABA release. Neuron. 2007;54(4):599–610. doi: 10.1016/j.neuron.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Norton N, et al. Evidence that interaction between neuregulin 1 and its receptor erbB4 increases susceptibility to schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(1):96–101. doi: 10.1002/ajmg.b.30236. [DOI] [PubMed] [Google Scholar]
- 36.Nicodemus KK, et al. Further evidence for association between ErbB4 and schizophrenia and influence on cognitive intermediate phenotypes in healthy controls. Mol Psychiatry. 2006;11(12):1062–1065. doi: 10.1038/sj.mp.4001878. [DOI] [PubMed] [Google Scholar]
- 37.O'Donovan MC, Craddock NJ, Owen MJ. Genetics of psychosis; insights from views across the genome. Hum Genet. 2009;126(1):3–12. doi: 10.1007/s00439-009-0703-0. [DOI] [PubMed] [Google Scholar]
- 38.Buonanno A, et al. Neuregulins and neuronal plasticity: possible relevance in schizophrenia. Novartis Found Symp. 2008;289:165–177. doi: 10.1002/9780470751251.ch13. discussion 177-9, 193-5. [DOI] [PubMed] [Google Scholar]
- 39.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9(6):437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talmage DA. Mechanisms of neuregulin action. Novartis Found Symp. 2008;289:74–84. doi: 10.1002/9780470751251.ch6. discussion 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang UE, et al. Molecular mechanisms of schizophrenia. Cell Physiol Biochem. 2007;20(6):687–702. doi: 10.1159/000110430. [DOI] [PubMed] [Google Scholar]
- 42.Fischbach GD. NRG1 and synaptic function in the CNS. Neuron. 2007;54(4):495–497. doi: 10.1016/j.neuron.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Birchmeier C. ErbB receptors and the development of the nervous system. Exp Cell Res. 2009;315(4):611–618. doi: 10.1016/j.yexcr.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 44.Thiselton DL, et al. No evidence for linkage or association of neuregulin-1 (NRG1) with disease in the Irish study of high-density schizophrenia families (ISHDSF) Mol Psychiatry. 2004;9(8):777–783. doi: 10.1038/sj.mp.4001530. image 729. [DOI] [PubMed] [Google Scholar]
- 45.Iwata N, et al. No association with the neuregulin 1 haplotype to Japanese schizophrenia. Mol Psychiatry. 2004;9(2):126–127. doi: 10.1038/sj.mp.4001456. [DOI] [PubMed] [Google Scholar]
- 46.Gardner M, et al. Extreme population differences across Neuregulin 1 gene, with implications for association studies. Mol Psychiatry. 2006;11(1):66–75. doi: 10.1038/sj.mp.4001749. [DOI] [PubMed] [Google Scholar]
- 47.Munafo MR, et al. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11(6):539–546. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- 48.Silberberg G, et al. The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(2):142–148. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- 49.Benzel I, et al. Interactions among genes in the ErbB-Neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav Brain Funct. 2007;3:31. doi: 10.1186/1744-9081-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shiota S, et al. Association and interaction analyses of NRG1 and ERBB4 genes with schizophrenia in a Japanese population. J Hum Genet. 2008;53(10):929–935. doi: 10.1007/s10038-008-0332-9. [DOI] [PubMed] [Google Scholar]
- 51.Craddock N, O'Donovan MC, Owen MJ. Phenotypic and genetic complexity of psychosis. Invited commentary on … Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:200–203. doi: 10.1192/bjp.bp.106.033761. [DOI] [PubMed] [Google Scholar]
- 52.McClellan JM, Susser E, King MC. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- 53.Li D, Collier DA, He L. Meta-analysis shows strong positive association of the neuregulin 1 (NRG1) gene with schizophrenia. Hum Mol Genet. 2006;15(12):1995–2002. doi: 10.1093/hmg/ddl122. [DOI] [PubMed] [Google Scholar]
- 54.Munafo MR, Attwood AS, Flint J. Neuregulin 1 genotype and schizophrenia. Schizophr Bull. 2008;34(1):9–12. doi: 10.1093/schbul/sbm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tosato S, Dazzan P, Collier D. Association between the neuregulin 1 gene and schizophrenia: a systematic review. Schizophr Bull. 2005;31(3):613–617. doi: 10.1093/schbul/sbi043. [DOI] [PubMed] [Google Scholar]
- 56.Braff DL, Geyer MA. Sensorimotor gating and schizophrenia. Human and animal model studies. Arch Gen Psychiatry. 1990;47(2):181–188. doi: 10.1001/archpsyc.1990.01810140081011. [DOI] [PubMed] [Google Scholar]
- 57.Thaker GK. Neurophysiological endophenotypes across bipolar and schizophrenia psychosis. Schizophr Bull. 2008;34(4):760–773. doi: 10.1093/schbul/sbn049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bramon E, et al. Neuregulin-1 and the P300 waveform--a preliminary association study using a psychosis endophenotype. Schizophr Res. 2008;103(1–3):178–185. doi: 10.1016/j.schres.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 59.Hong LE, et al. Evidence of missense mutations on the neuregulin 1 gene affecting function of prepulse inhibition. Biol Psychiatry. 2008;63(1):17–23. doi: 10.1016/j.biopsych.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quednow BB, et al. Sensorimotor Gating Depends on Polymorphisms of the Serotonin-2A Receptor and Catechol-O-Methyltransferase, but Not on Neuregulin-1 Arg38Gln Genotype: A Replication Study. Biol Psychiatry. 2009 doi: 10.1016/j.biopsych.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gruber O, et al. Neuregulin-1 haplotype HAP(ICE) is associated with lower hippocampal volumes in schizophrenic patients and in non-affected family members. J Psychiatr Res. 2008;43(1):1–6. doi: 10.1016/j.jpsychires.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 62.Voineskos D, et al. Neuregulin 1 and age of onset in the major psychoses. J Neural Transm. 2009 doi: 10.1007/s00702-008-0182-9. [DOI] [PubMed] [Google Scholar]
- 63.Hall J, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9(12):1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 64.Mata I, et al. A Neuregulin 1 Variant Is Associated with Increased Lateral Ventricle Volume in Patients with First-Episode Schizophrenia. Biol Psychiatry. 2008 doi: 10.1016/j.biopsych.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 65.Stefanis NC, et al. Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biol Psychiatry. 2007;62(7):784–792. doi: 10.1016/j.biopsych.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 66.Crowley JJ, et al. The neuregulin 1 promoter polymorphism rs6994992 is not associated with chronic schizophrenia or neurocognition. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1298–1300. doi: 10.1002/ajmg.b.30727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Tuathaigh CM, et al. Susceptibility genes for schizophrenia: characterisation of mutant mouse models at the level of phenotypic behaviour. Neurosci Biobehav Rev. 2007;31(1):60–78. doi: 10.1016/j.neubiorev.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 68.O'Tuathaigh CM, Desbonnet L, Waddington JL. Neuregulin-1 signaling in schizophrenia: 'Jack of all trades' or master of some? Expert Rev Neurother. 2009;9(1):1–3. doi: 10.1586/14737175.9.1.1. [DOI] [PubMed] [Google Scholar]
- 69.Duffy L, et al. Behavioral profile of a heterozygous mutant mouse model for EGF-like domain neuregulin 1. Behav Neurosci. 2008;122(4):748–759. doi: 10.1037/0735-7044.122.4.748. [DOI] [PubMed] [Google Scholar]
- 70.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378(6555):386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 71.Erickson SL, et al. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development. 1997;124(24):4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- 72.Kramer R, et al. Neuregulins with an Ig-like domain are essential for mouse myocardial and neuronal development. Proc Natl Acad Sci U S A. 1996;93(10):4833–4838. doi: 10.1073/pnas.93.10.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen YJ, et al. Type III neuregulin-1 is required for normal sensorimotor gating, memory-related behaviors, and corticostriatal circuit components. J Neurosci. 2008;28(27):6872–6883. doi: 10.1523/JNEUROSCI.1815-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Savonenko AV, et al. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc Natl Acad Sci U S A. 2008;105(14):5585–5590. doi: 10.1073/pnas.0710373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ehrlichman RS, et al. Neuregulin 1 transgenic mice display reduced mismatch negativity, contextual fear conditioning and social interactions. Brain Res. 2009 doi: 10.1016/j.brainres.2009.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O'Tuathaigh CM, et al. Phenotypic characterization of spatial cognition and social behavior in mice with 'knockout' of the schizophrenia risk gene neuregulin 1. Neuroscience. 2007;147(1):18–27. doi: 10.1016/j.neuroscience.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 77.O'Tuathaigh CM, et al. Disruption to social dyadic interactions but not emotional/anxiety-related behaviour in mice with heterozygous 'knockout' of the schizophrenia risk gene neuregulin-1. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(2):462–466. doi: 10.1016/j.pnpbp.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 78.Rimer M, et al. Neuregulin-1 immunoglobulin-like domain mutant mice: clozapine sensitivity and impaired latent inhibition. Neuroreport. 2005;16(3):271–275. doi: 10.1097/00001756-200502280-00014. [DOI] [PubMed] [Google Scholar]
- 79.Gerlai R, Pisacane P, Erickson S. Heregulin, but not ErbB2 or ErbB3, heterozygous mutant mice exhibit hyperactivity in multiple behavioral tasks. Behav Brain Res. 2000;109(2):219–227. doi: 10.1016/s0166-4328(99)00175-8. [DOI] [PubMed] [Google Scholar]
- 80.Golub MS, Germann SL, Lloyd KC. Behavioral characteristics of a nervous system-specific erbB4 knock-out mouse. Behav Brain Res. 2004;153(1):159–170. doi: 10.1016/j.bbr.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 81.Thuret S, et al. The neuregulin receptor, ErbB4, is not required for normal development and adult maintenance of the substantia nigra pars compacta. J Neurochem. 2004;91(6):1302–1311. doi: 10.1111/j.1471-4159.2004.02809.x. [DOI] [PubMed] [Google Scholar]
- 82.Hashimoto R, et al. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9(3):299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- 83.Law AJ, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5' SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103(17):6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chong VZ, et al. Elevated neuregulin-1 and ErbB4 protein in the prefrontal cortex of schizophrenic patients. Schizophr Res. 2008;100(1–3):270–280. doi: 10.1016/j.schres.2007.12.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hahn CG, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12(7):824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- 86.Law AJ, et al. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16(2):129–141. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 87.Chen PL, et al. Fine mapping on chromosome 10q22-q23 implicates Neuregulin 3 in schizophrenia. Am J Hum Genet. 2009;84(1):21–34. doi: 10.1016/j.ajhg.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emamian ES, et al. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36(2):131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 89.Hallmayer J. Getting our AKT together in schizophrenia? Nat Genet. 2004;36(2):115–116. doi: 10.1038/ng0204-115. [DOI] [PubMed] [Google Scholar]
- 90.Kwon OB, et al. Neuregulin-1 reverses long-term potentiation at CA1 hippocampal synapses. J Neurosci. 2005;25(41):9378–9383. doi: 10.1523/JNEUROSCI.2100-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gu Z, et al. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25(20):4974–4984. doi: 10.1523/JNEUROSCI.1086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li B, et al. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54(4):583–597. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Braun I, et al. Alterations of hippocampal and prefrontal GABAergic interneurons in an animal model of psychosis induced by NMDA receptor antagonism. Schizophr Res. 2007;97(1–3):254–263. doi: 10.1016/j.schres.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 94.Salter MW, Kalia LV. Src kinases: a hub for NMDA receptor regulation. Nat Rev Neurosci. 2004;5(4):317–328. doi: 10.1038/nrn1368. [DOI] [PubMed] [Google Scholar]
- 95.Bennett M. Positive and negative symptoms in schizophrenia: the NMDA receptor hypofunction hypothesis, neuregulin/ErbB4 and synapse regression. Aust N Z J Psychiatry. 2009;43(8):711–721. doi: 10.1080/00048670903001943. [DOI] [PubMed] [Google Scholar]
- 96.Bjarnadottir M, et al. Neuregulin1 (NRG1) signaling through Fyn modulates NMDA receptor phosphorylation: differential synaptic function in NRG1+/− knock-outs compared with wild-type mice. J Neurosci. 2007;27(17):4519–4529. doi: 10.1523/JNEUROSCI.4314-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anton ES, et al. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124(18):3501–3510. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- 98.Krivosheya D, et al. ErbB4-neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J Biol Chem. 2008;283(47):32944–32956. doi: 10.1074/jbc.M800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57(1):65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 100.Sweet RA, et al. Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology. 2009;34(2):374–389. doi: 10.1038/npp.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bennett AOM. Dual constraints on synapse formation and regression in schizophrenia: neuregulin, neuroligin, dysbindin, DISC1, MuSK and agrin. Aust N Z J Psychiatry. 2008;42(8):662–677. doi: 10.1080/00048670802203467. [DOI] [PubMed] [Google Scholar]
- 102.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10(1):40–68. doi: 10.1038/sj.mp.4001558. image 5. [DOI] [PubMed] [Google Scholar]
- 103.Madeira C, et al. Increased brain D-amino acid oxidase (DAAO) activity in schizophrenia. Schizophr Res. 2008;101(1–3):76–83. doi: 10.1016/j.schres.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 104.Schumacher J, et al. Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol Psychiatry. 2004;9(2):203–207. doi: 10.1038/sj.mp.4001421. [DOI] [PubMed] [Google Scholar]
- 105.Chumakov I, et al. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci U S A. 2002;99(21):13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Talbot K, et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113(9):1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Camargo LM, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12(1):74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 108.Talbot K, et al. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum Mol Genet. 2006;15(20):3041–3054. doi: 10.1093/hmg/ddl246. [DOI] [PubMed] [Google Scholar]
- 109.Weickert CS, et al. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr Res. 2008;98(1–3):105–110. doi: 10.1016/j.schres.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weickert CS, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004;61(6):544–555. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 111.Hahn CG, et al. The post-synaptic density of human postmortem brain tissues: an experimental study paradigm for neuropsychiatric illnesses. PLoS ONE. 2009;4(4):e5251. doi: 10.1371/journal.pone.0005251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fernandez E, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morabito MA, Sheng M, Tsai LH. Cyclin-dependent kinase 5 phosphorylates the N-terminal domain of the postsynaptic density protein PSD-95 in neurons. J Neurosci. 2004;24(4):865–876. doi: 10.1523/JNEUROSCI.4582-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xie F, Padival M, Siegel RE. Association of PSD-95 with ErbB4 facilitates neuregulin signaling in cerebellar granule neurons in culture. J Neurochem. 2007;100(1):62–72. doi: 10.1111/j.1471-4159.2006.04182.x. [DOI] [PubMed] [Google Scholar]
- 115.Phillips GR, et al. Proteomic comparison of two fractions derived from the transsynaptic scaffold. J Neurosci Res. 2005;81(6):762–775. doi: 10.1002/jnr.20614. [DOI] [PubMed] [Google Scholar]
- 116.Cheng D, et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5(6):1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 117.Dosemeci A, et al. Preparation of postsynaptic density fraction from hippocampal slices and proteomic analysis. Biochem Biophys Res Commun. 2006;339(2):687–694. doi: 10.1016/j.bbrc.2005.11.069. [DOI] [PubMed] [Google Scholar]