Abstract
Auxin responses have been arbitrarily divided into two categories: genomic and non-genomic effects. Genomic effects are largely mediated by SCFTIR1/AFB-Aux/IAA auxin receptor complexes whereas it has been postulated that AUXIN BINDING PROTEIN 1 (ABP1) controls the non-genomic effects. However, the roles of ABP1 in auxin signaling and plant development were recently called into question. In this paper, we present recent progress in understanding the SCFTIR1/AFB-Aux/IAA pathway. In more detail, we discuss the current understanding of ABP1 research and provide an updated view of ABP1-related genetic materials. Further, we propose a model in which auxin efflux carriers may play a role in auxin perception and we briefly describe recent insight on processes downstream of auxin perception.
Introduction
Auxin plays essential roles in many developmental processes. Much effort has been directed toward understanding the precise molecular mechanisms by which auxin regulates diverse aspects of plant growth and development. A key aspect of the effort is to understand auxin perception. There are two well-studied auxin perception systems: SCFTIR1/AFB-Aux/IAA auxin receptor complexes (SCF [(SKP, CULLIN, F-BOX), TIR1/AFB (TRANSPORT INHIBITOR RESPONSE 1/AUXIN-RELATED F-BOX PROTEINS), AUX/IAA (AUXIN/INDOLE-3-ACETIC ACID)] and the AUXIN BINDING PROTEIN 1 (ABP1) system, which were proposed to control auxin-mediated transcription and non-genomic effects, respectively [1,2]. In this paper, we review the recent progress in our understanding of the regulation of the stability and assembly of the SCFTIR1/AFB-Aux/IAA auxin receptor complexes. We will also address the controversies surrounding ABP1, which was accepted as a plasma-membrane-associated auxin receptor that forms an auxin receptor complex with a small family of receptor-like kinases (TMKs) to regulate many developmental processes including pavement cell development, cytoskeleton re-organization, and polar auxin transport [1–5]. However, a recent study showed that ABP1 is not required for auxin signaling and plant development [6], prompting re-analyses of the previous abp1 genetic materials that formed the basis for assigning ABP1 functions. We detail the experimental results that have adequately resolved the underlying causes of the conflicting results regarding the roles of ABP1 in auxin signaling and plant development. Finally, we present several recent studies of auxin signaling events downstream of the SCFTIR1/AFB-Aux/IAA auxin receptor complexes.
AUXIN BINDING PROTEIN 1 (ABP1)
Since the discovery of ABP1 in 1972, research into this proposed auxin receptor has been surrounded with controversy [7–9]. ABP1 is a 22 kDa protein with a C-terminal KDEL ER-retention signal [10,11]; over 90% of ABP1 is ER-localized [12]. Yet nearly all ABP1 functional studies focused on the small fraction of plasma-membrane-bound ABP1 [1]. ABP1 has been suggested to have essential roles in almost every aspect of plant growth and development including embryogenesis [13], root growth and maintenance of the root meristem [14], hypocotyl elongation [15], pavement cell development [4], vascular patterning [16], cytoskeleton re-organization and polar auxin transport [3,5], control of flowering time [17], and flower development [16].
Many resources have been dedicated to study ABP1 and more than 100 ABP1 papers have been published. During the era of Arabidopsis genetics, researchers generated various knockdown lines and mutants for determining ABP1 functions and the action mechanisms: 1) T-DNA insertion mutants abp1-1 and abp1-1s [13,18,19]; 2) abp1-5, carrying a point mutation in the auxin-binding pocket [4]; 3) Three knockdown lines (abp1-AS, SS12K, SS12S) [16]; 4) ABP1 overexpression lines [3,4,20,21]. All of these lines were reported to have obvious phenotypes and were the basis for all functional studies of ABP1 since 2000. Therefore, it was shocking when ABP1 was reported not to be required for either auxin signaling or Arabidopsis development [6]. Making use of the CRISPR/Cas9 (Clustered regularly-interspaced short palindromic repeats/CRISPR associated protein 9) technology [22], Gao et al. generated a true abp1 null allele (abp1-c1), consisting of a 5-bp deletion in the first exon of the ABP1 gene [6]. This abp1-c1 allele did not produce detectable full length or truncated ABP1 protein [6]. Both the abp1-c1 mutant and the abp1-TD1 mutant, carrying a T-DNA insertion in the first exon of ABP1, are indistinguishable from WT in overall morphology, development, and auxin responses [6]. The results presented in the Gao et al. paper were unequivocal whereas previous results appeared solid as well. It has been a difficult task for the plant biology community to reconcile the apparent contradictory findings regarding the roles of ABP1 in auxin signaling and plant development [9,23,24]. Fortunately, the discrepancies between the Gao et al. paper [6] and previous ABP1 papers appeared to have been largely resolved.
The first genetic evidence revealing ABP1 importance in plant development was the embryo lethality of the abp1-1 T-DNA insertion mutant [13]. Indeed, abp1-1 was a cornerstone of ABP1 research. Prior to the abp1-1 description, skepticism about ABP1 as an auxin receptor was pervasive, and ABP1 was even called a red herring [7,8]. The abp1-1 mutant contains a T-DNA insertion 51 bp downstream of the ATG start codon and a 35S:ABP1 cDNA construct was reported to have complemented the abp1-1 embryo lethal phenotypes [13]. Further, the abp1-1s allele harbors a T-DNA in the promoter region of ABP1 [18] and was endorsed as a second abp1 null allele. A cross between abp1-1 and abp1-1s failed to complement the embryo lethal phenotype, suggesting that these two mutants were disrupted in the same gene, although there are no reports on complementation of abp1-1s using WT ABP1 genomic DNA or cDNA. Although both abp1-1s and abp1-1 were cited as null mutants, analyses of ABP1 mRNA and ABP1 protein levels were precluded by the homozygous lethality of the two mutations [9,19,25]. Recent re-analyses of abp1-1 and abp1-1s have uncovered the underlying basis of their embryo lethality [25,26], which is not from disruption of ABP1. The T-DNA insertion in abp1-1 not only disrupted the ABP1 gene, but also deleted the entire neighboring BSM gene [26], which is an essential gene [27]. In fact, abp1-1 embryo phenotypes closely resembled to those observed in bsm mutants [27]. Introduction of the BSM gene under the control of the CaMV 35S promoter into the abp1-1 mutant plants completely rescued the embryo lethal phenotype [26]. Furthermore, abp1-1 failed to accumulated detectable ABP1 protein, but the abp1-1−/− CaMV 35S:BSM plants did not display obvious developmental defects [26]. These results unequivocally demonstrated that the abp1-1 embryo lethal phenotype is not caused by the ABP1 disruption, but rather by the deletion of the neighboring BSM gene [26]. Analyses of reciprocal crosses between abp1-1, abp1-1s, abp1-c1, abp1-TD1, and bsm-1 [25] further support the conclusion that the embryo lethality observed in abp1-1 and abp1-1s is caused by disruption of the BSM gene. A caveat to the latter study is the possibility that ABP1 function may have been compromised in the bsm-1 mutant, which was not examined. This point is significant because T-DNA insertions can profoundly affect expression levels of adjacent genes [28]. Nevertheless, the data in Gao et al. paper [6], combined with results from the re-analyses of the abp1-1 and abp1-1s [25,26], allow for the confident conclusion that Arabidopsis plants do not require ABP1 for normal growth and development under laboratory conditions.
The abp1-5 mutant also played a major role in a series of ABP1 studies. It was reported that the development of pavement cell and polar auxin transport were affected in abp1-5 [1,3–5]. Moreover, exogenous auxin failed to rescue abp1-5 developmental defects [1,3–5]. However, recent whole genome sequencing of the abp1-5 mutant revealed that there are more than 8000 polymorphisms between abp1-5 and the parental line Arabidopsis Col-0 [29], suggesting that the mutant has not been properly backcrossed as previously reported [4]. Some of the phenotypes associated with abp1-5 are likely not caused by the abp1 lesion. For example, the longer hypocotyl phenotype observed in light-grown abp1-5 seedlings [15] is linked to a secondary mutation in the PHYTOCHROME B gene present in the abp1-5 background, and is not linked to the abp1-5 mutation itself [29]. Because abp1-5 contains numerous background mutations, conclusions derived from this allele should be treated with caution.
Three abp1 knockdown lines (SS12K, SS12S, abp1-AS) played instrumental roles in connecting ABP1 to many developmental processes [16]. The SS12K and SS12S lines, often referred as ABP1 antibody lines, were generated by expressing the single chain fragment variable regions (scFv12) of ABP1 monoclonal antibody under the control of an ethanol-inducible promoter [16]. The SS12K contains the KDEL ER retention signal whereas SS12S lacks the KDEL motif [16]. The SS12K and SS12S lines behaved very similarly, even though one was targeted to the ER and the other was targeted to the apoplast region [16]. Another knockdown line is abp1-AS, which expresses an ABP1 antisense RNA under the control of the aforementioned ethanol inducible promoter [16]. Although the initial study using the ABP1 knockdown lines reported that multiple lines from each construct produced similar phenotypes [16], subsequent studies almost exclusively used SS12K6, SS12S9, and abp1-AS [1,3–5,14,30,31]. Recently, the three knockdown lines were crossed to abp1-c1 and abp1-TD1 null alleles [32]. Analysis of the segregation patterns resulting from the crosses clearly demonstrated that the phenotypes observed in these lines are not caused by affecting ABP1 and are likely caused by off-target effects [32].
Results from overexpressing ABP1 in various systems including tobacco leaves, BY2 cells, and Arabidopsis have been used to support that ABP1 plays roles in cell expansion and other developmental processes [3,33]. Results from ABP1 overexpression should be treated with caution as the different lines did not always produce consistent results. For example, overexpression of full length ABP1 in Arabidopsis did not result in obvious developmental phenotypes, but overexpression of ABP1 lacking the KDEL ER-retention signal led to auxin-related phenotypes including three cotyledons, shorter roots, reduced apical dominance, sterility, and seedling lethality [3]. Interestingly, the ABP1ΔKDEL overexpression phenotypes are consistent with a decrease in auxin signaling in plants. The shorter roots and three cotyledon phenotypes were also observed in abp1-5 and the antibody lines [1,3,16].
A key argument supporting the importance and requirement of ABP1 is the presence of homologues in all plant species [32,34]. This argument was based on available genomic sequences at that time, and recently has been refuted by the absence of ABP1 in Marchantia polymorpha, a liverwort species [35], indicating that ABP1 is dispensable, at least in some plant species.
The contradictory results between Gao et al paper and the previous studies have been reasonably resolved; however, the physiological functions of ABP1 remain unknown because all of the available abp1 knockouts display no aberrant phenotypes under normal laboratory conditions. The simplest explanation is that ABP1 is not required for normal plant development. However, two competing hypotheses addressing the lack of phenotypes in the known abp1 null mutants are worth discussing: 1) The abp1-c1, abp1-TD1, and abp1-1 may not be null [9]; 2) Proteins with functions overlapping/redundant with ABP1 compensate for the loss of ABP1 [32,36,37]. All of the available abp1 null alleles (abp1-1, abp1-c1, and abp1-TD1) have the mutations in the first exon of ABP1 [6,26]. Although ABP1 protein was undetectable by Western analysis in these mutants, it remains possible that the ABP1 antibody used in these experiments might be specific to the ABP1 N-terminal region and thus might have failed to detect potentially functional truncated ABP1 proteins. No full-length or partial ABP1 mRNAs could be identified in abp1-TD1 [6]. However, transcript coverage by the primer pairs used for the RT-PCR reactions might have been insufficient or mRNA quantities may have been below the detection limit in these assays [9]. Therefore, it is still possible that the reported abp1 null mutants make functional truncated ABP1 proteins. The only unequivocal means of guaranteeing an abp1 null mutant is to delete the entire ABP1 coding region from the genome. Advancements in CRISPR/Cas9 gene editing technology have made it feasible to obtain such mutants [22]. Indeed, we successfully generated two new abp1 alleles, which contained 1141 bp and 711 bp deletions in the ABP1 gene, respectively (Zhao, unpublished results). These two new abp1 mutants conclusively demonstrated that ABP1 is not required for normal Arabidopsis development.
The hypothesis that proteins with functions overlapping/redundant with ABP1 compensate for the loss of ABP1 has been raised repeatedly [32,36,37]. ABP1 is a single copy gene in Arabidopsis, and there are no cryptic copies of ABP1 in the Arabidopsis genome [25]. Using ABP1 protein sequence as a query to BLASTP the Arabidopsis proteins failed to yield any significant hits other than ABP1 itself (using an E-value ≤ 1 as the cutoff). Therefore, there are no detectable ABP1 homologs in the Arabidopsis genome. ABP1 is a member of the Cupin superfamily, which is a functionally diverse protein superfamily with little homology amongst family members [38,39]. Further, GLP4, a Golgi-located Cupin family protein, has been reported to bind auxin [40]. Thus, it was proposed that a lack of phenotypes in abp1 null mutants might be due to redundant functions of Cupin proteins such as GLP4 [32,36,37]. However, the lack of homology between GLP4 and ABP1 beyond the two short Cupin motifs and their distinct sub-cellular locations suggest that it is very unlikely that ABP1 and Cupin family proteins have overlapping or redundant functions.
In conclusion, it appears that ABP1 research field has to be reset back to the 20th century. This is based on the Gao et al. findings showing no observable phenotypes in the abp1 null mutants, the recent re-analysis showing problems of the previously used genetic materials, and that ABP1 is dispensable in some plant species.
Auxin efflux carriers as auxin receptors
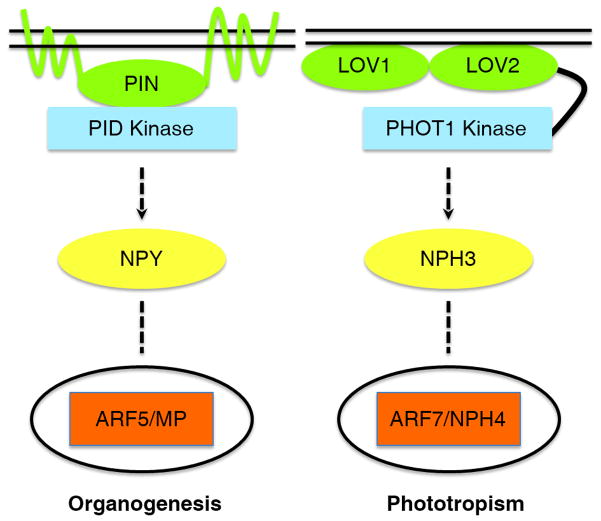
Because ABP1 is likely not the auxin receptor for auxin-mediated non-genomic effects, it is worth revisiting the hypothesis that auxin efflux carriers may serve as auxin receptors. Such a hypothesis was first proposed in the 1960s, modified in the 1980s, and re-proposed in the 1990s, but it failed to gain traction [7]. PIN (PIN-FORMED) proteins are well-characterized auxin efflux carriers, playing essential roles in many developmental processes [41,42]. Can the PIN proteins also serve as auxin receptors? PIN proteins binds auxins and therefore meet one of the key criteria as auxin receptors. More importantly, genetic studies suggest that members of several gene families such as PID (PINOID), NPY (NAKED PINS IN YUC MUTANTS), and ARF (AUXIN RESPONSE FACTOR) can potentially function downstream of PIN1. Disruption of PIN1 in Arabidopsis leads to the development of pin-like inflorescences [41]. The same pin-like phenotypes have been observed in pid [43], npy1 npy3 npy5 triple mutants [44,45], and arf5/mp (auxin response factor 5/monopteros) [46]. Genetic studies clearly place PID and NPY genes in the same pathway regulating auxin-mediated organogenesis [44,45]. Furthermore, the PID/NPY-mediated signaling pathway is strikingly analogous to the blue-light mediated phototropic pathway (Figure 1). In phototropic response, the phototropins have the N-terminal light-perception LOV domains and the C-terminal Ser/Thr protein kinase domain [47]. Upon receiving light signals by the LOV domains, the phototropin kinase is activated, which leads to a change in phosphorylation status of NPH3 (NONPHOTOTROPIC HYPOCOTYL 3) and phototropic responses [47,48]. The phototropin kinase domain is similar to the protein kinase PID. Further, NPH3 is homologous to the NPYs whereas ARF5/MP is very similar to ARF7/NPH4 (Figure 1). These two pathways differ in that phototropins contain both light-perceiving LOV domains and a kinase domain while PID only has a kinase domain (Figure 1). PID may have a partner that perceives a signal such that PID combined with its partner is analogous to the phototropins.
Figure 1. A schematic presentation of the hypothesis that PIN and PID may form a plasma-membrane localized auxin receptor complex important for auxin-mediated Arabidopsis organogenesis.
The organogenesis pathway is strikingly analogous to the blue light-mediated phototropic response. Arabidopsis plants with compromised PIN1 or PID or NPY or ARF5/MP fail to produce flowers whereas mutations in PHOT1 or NPH3 or ARF7/NPH4 cause defects in normal phototropic responses to blue light. PID is homologous to PHOT1 and NPYs are homologous to NPH3. Moreover, ARF5/MP is very similar to ARF7/NPH4. The starting point for phototropic response is the perception of light by the LOV domains in PHOT1. Light activates the kinase activity in PHOT1 and consequently phosphorylation status of NPH3 is altered. The role of ARF7/NPH4 in phototropism is not understood because phototropic response is generally believed to be non-genomic. PID lacks a receptor domain and we hypothesize that PIN proteins may function as an auxin receptor based on genetic evidences. The function of PIN/PID complex in the auxin-mediated organogenesis pathway is equivalent to PHOT1 in the phototropism pathway.
PIN proteins serve as good candidates as PID partners. First, both pid and pin1 develop the same pin-like inflorescences, suggesting that they participate in the same pathway [41,43]. Second, PID interacts and phosphorylates PIN1 [49]. Third, the genetic interactions among pin1, pid, and npy suggest that they participate in the same pathway [44,45,50,51]. The hypothesis that PIN proteins partner with PID to form auxin perception complexes can be tested experimentally. For example, determining whether NPYs are phosphorylated in a PIN1 or PID-dependent manner will strengthen or weaken this hypothesis. Previous studies focused on the capacity and directionality of PIN-mediated auxin transport. This new model suggests that auxin transport may be coupled with a signal transduction pathway, which can reasonably account for the observed pin-like phenotypes in various Arabidopsis mutants.
SCFTIR1/AFB-Aux/IAA auxin receptor complexes
Auxin perception by SCFTIR1/AFB-Aux/IAA complexes and their essential roles in auxin-mediated transcription regulation and plant development have been well established [2]. The SCFTIR1/AFB-mediated auxin signaling pathway requires a TIR1/AFB F-box protein, an Aux/IAA transcriptional repressor, and an ARF transcription factor. Auxin-binding brings Aux/IAA proteins to the SCFTIR1/AFB ubiquitin E3-ligase complex and results in degradation of the Aux/IAA repressors and activation of the ARFs. Recent progress in auxin perception and transcription regulation by SCFTIR1/AFB have been reviewed elsewhere [2]. Here we focus on several recent studies on mechanisms by which SCFTIR1/AFB-Aux/IAA auxin receptor complexes may be regulated.
TIR1 protein abundance appeared to be controlled by an autocatalytic mechanism. Yu et al. uncovered a series of mutations in the first helix of TIR1 that disrupt TIR1 association with CULLIN 1 [52]. Consequently, the mutated proteins were stabilized, indicating that un-tethering TIR1 from the SCF complex affects the TIR1 stability and that TIR1 itself is likely a substrate for the SCFTIR1 ubiquitin E3 ligase.
Aux/IAA recruitment to the SCFTIR1/AFB ubiquitin E3 ligase is a key step in auxin signaling and is dependent on the presence of auxin and the domain II degron [2]. Recently, it was shown that the configuration of a conserved proline residue in the degron dictates Aux/IAA recruitment to the SCFTIR1/AFB complex, adding another layer of complexity to auxin signaling [53]. Mutations in rice LATERAL ROOTLESS2 (LRT2), which encodes a cyclophilin-type peptidyl-prolyl cis/trans isomerase, result in auxin resistance and lateral root development defects [53]. LRT2 catalyzes the cis/trans isomerization of the Trp104–Pro105 peptide bond of OsIAA11, promoting the formation of the Pro105-cis configuration, which is preferred during the assembly of a SCFTIR1/AFBAux/IAA auxin receptor complex [53]. These new insights into SCFTIR1/AFB regulation may provide additional mechanisms for auxin response regulation.
Auxin signaling downstream of auxin perception
ARF proteins activate or repress gene expression by binding cis-regulatory auxin response elements (AuxREs) in the promoters of auxin-responsive genes and serve as key mediators of auxin transcriptional outputs [54,55]. Recent advances in understanding ARF function and regulation include insights provided by ARF recruitment of chromatin remodeling factors and roles for tasiRNA regulation of ARFs to make the auxin-regulated gene expression network robust.
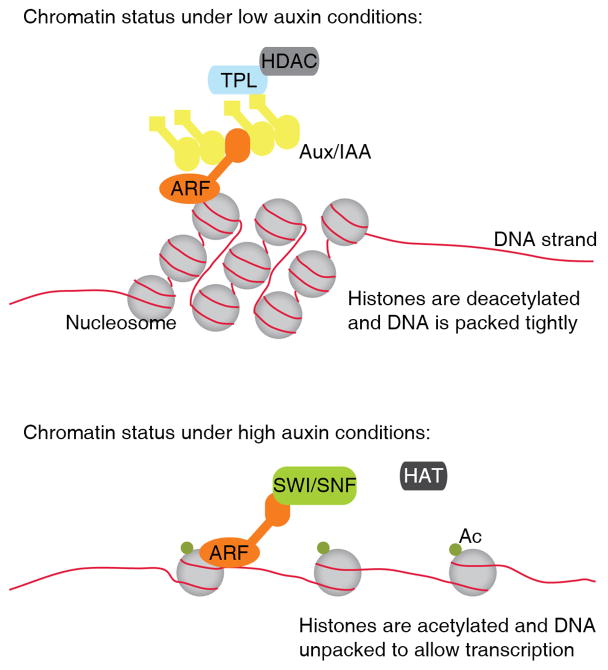
Chromatin remodeling as a mechanism of auxin-responsive gene regulation is only beginning to be understood [56]. Aux/IAA repressors are recruited to specific chromosome regions by interaction with ARF proteins, and in turn recruit TOPLESS and TOPLESS-related co-repressors (Figure 2) [57,58]. These corepressors recruit histone deacetylases that act to condense DNA to prevent gene transcription. A long-standing model of auxin signaling involves de-repression of ARF proteins by Aux/IAA proteolysis. After Aux/IAA disappearance, how then, is transcription to occur if the chromatin is still in a condensed state? An answer to this question may come from a recent study that identified roles for the SWI/SNF nucleosome-remodeling complex in promoting transcription of ARF targets (Figure 2) [59]. The BRAHMA and SPLAYED subunits of the SWI/SNF complex directly interact with ARF5 in a manner dependent on Aux/IAA removal [59]; because SWI/SNF complexes act to de-condense DNA, its recruitment likely acts to reverse the gene repression previously imposed by Aux/IAAs (Figure 2). Although this has yet to be demonstrated as a widespread mechanism of promoting auxin-regulated transcriptional responses, it provides an attractive mechanism to counteract the effects of Aux/IAA proteins and to allow for recruitment of the transcriptional machinery.
Figure 2. Roles for chromatin packing in auxin-mediated transcriptional response.
Under low auxin conditions, Aux/IAA proteins are abundant and interact with ARF proteins. Aux/IAAs recruit the TOPLESS (TPL) family of corepressors, which in turn recruit Histone deacetylases (HDACs), resulting in tightly packed chromatin. Furthermore, Aux/IAA proteins likely block the interaction between ARF and SWI/SNF chromatin remodeling complexes. When auxin levels are elevated, Aux/IAA proteins are targeted for proteasomal degradation, allowing for SWI/SNF interaction with ARF proteins. SWI/SNF-enabled chromatin remodeling, through histone acetyltransferase (HAT) activity, results in chromatin unpacking to allow gene target access by additional transcription factors.
In addition to protein regulation by interaction with repressors and chromatin remodeling factors, ARFs can be regulated posttranscriptionally. Several repressing ARFs have been shown to be regulated by miRNAs [60]. In addition, tasiRNAs have been demonstrated as important regulators of repressing ARFs that serve to increase the robustness of auxin response. Working with the bryophyte model plant Physcomitrella patens, Plavskin et al. [61] showed that loss of the tasiRNA system in Physcomitrella resulted not only in a decreased auxin response, caused by an increased accumulation of repressor ARF, but also resulted in greater stochasticity in auxin responses. With the use of an elegant combination of experimental data and modeling [61], the authors created a model in which tasiRNAs act to buffer the noise in the auxin gene response network; this role may explain why tasiRNAs have been co-opted frequently into various gene response networks.
Highlights.
Genetic evidence supporting the proposed roles for ABP1 in auxin signaling has been invalidated
The PIN auxin efflux carriers may be the missing auxin receptors for non-genomic auxin responses
Several mechanisms control SCFTIR1/AFB-Aux/IAA auxin receptor complex assembly and stability
Chromatin-remodeling and small RNAs participate in regulating auxin-mediated transcription
Acknowledgments
We would like to thank Drs. Brian Crawford and Juan-José Ripoll for discussions and comments. This work was supported by the National Institutes of Health (R01GM114660 to Y. Zhao and R01GM112898 to L. Strader).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusova H, et al. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science. 2014;343:1025–1028. doi: 10.1126/science.1245125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell. 2015;27:9–19. doi: 10.1105/tpc.114.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Covanova M, et al. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell. 2010;143:111–121. doi: 10.1016/j.cell.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell. 2010;143:99–110. doi: 10.1016/j.cell.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Grandont L, Li H, Hauschild R, Paque S, Abuzeineh A, Rakusova H, Benkova E, Perrot-Rechenmann C, Friml J. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature. 2014;516:90–93. doi: 10.1038/nature13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6••.Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci U S A. 2015;112:2275–2280. doi: 10.1073/pnas.1500365112. Taking advantage of CRISPR/Cas9 technology to generate new abp1 null alleles, this study provided important evidence that loss of ABP1 results in no obvious defects in auxin response and Arabidopsis morphology under standard laboratory conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertel R. Auxin binding protein 1 is a red herring. Journal of Experimental Botany. 1995;46:461–462. [Google Scholar]
- 8.Timpte C. Auxin binding protein: curiouser and curiouser. Trends Plant Sci. 2001;6:586–590. doi: 10.1016/s1360-1385(01)02150-1. [DOI] [PubMed] [Google Scholar]
- 9.Habets ME, Offringa R. Auxin Binding Protein 1: A Red Herring After All? Mol Plant. 2015;8:1131–1134. doi: 10.1016/j.molp.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Lobler M, Klambt D. Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). I. Purification by immunological methods and characterization. J Biol Chem. 1985;260:9848–9853. [PubMed] [Google Scholar]
- 11.Jones AM, Venis MA. Photoaffinity labeling of indole-3-acetic acid-binding proteins in maize. Proc Natl Acad Sci U S A. 1989;86:6153–6156. doi: 10.1073/pnas.86.16.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones AM, Herman EM. KDEL-Containing Auxin-Binding Protein Is Secreted to the Plasma Membrane and Cell Wall. Plant Physiol. 1993;101:595–606. doi: 10.1104/pp.101.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 2001;15:902–911. doi: 10.1101/gad.866201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tromas A, Braun N, Muller P, Khodus T, Paponov IA, Palme K, Ljung K, Lee JY, Benfey P, Murray JA, et al. The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS One. 2009;4:e6648. doi: 10.1371/journal.pone.0006648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Effendi Y, Jones AM, Scherer GF. AUXIN-BINDING-PROTEIN1 (ABP1) in phytochrome-B-controlled responses. J Exp Bot. 2013;64:5065–5074. doi: 10.1093/jxb/ert294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Rechenmann C, Fleming AJ. Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell. 2008;20:2746–2762. doi: 10.1105/tpc.108.059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Effendi Y, Rietz S, Fischer U, Scherer GF. The heterozygous abp1/ABP1 insertional mutant has defects in functions requiring polar auxin transport and in regulation of early auxin-regulated genes. Plant J. 2011;65:282–294. doi: 10.1111/j.1365-313X.2010.04420.x. [DOI] [PubMed] [Google Scholar]
- 18.Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassi M, Ali O, Boudon F, Cloarec G, Abad U, Cellier C, Chen X, Gilles B, Milani P, Friml J, et al. An auxin-mediated shift toward growth isotropy promotes organ formation at the shoot meristem in Arabidopsis. Curr Biol. 2014;24:2335–2342. doi: 10.1016/j.cub.2014.08.036. [DOI] [PubMed] [Google Scholar]
- 20.Effendi Y, Ferro N, Labusch C, Geisler M, Scherer GF. Complementation of the embryo-lethal T-DNA insertion mutant of AUXIN-BINDING-PROTEIN 1 (ABP1) with abp1 point mutated versions reveals crosstalk of ABP1 and phytochromes. J Exp Bot. 2014;66:403–418. doi: 10.1093/jxb/eru433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grones P, Chen X, Simon S, Kaufmann WA, De Rycke R, Nodzynski T, Zazimalova E, Friml J. Auxin-binding pocket of ABP1 is crucial for its gain-of-function cellular and developmental roles. J Exp Bot. 2015;66:5055–5065. doi: 10.1093/jxb/erv177. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 23.Liu CM. Auxin Binding Protein 1 (ABP1): a matter of fact. J Integr Plant Biol. 2015;57:234–235. doi: 10.1111/jipb.12339. [DOI] [PubMed] [Google Scholar]
- 24.Tena G. Auxin signalling: ABP1 springs a surprise. Nature Plants. 2015;1:15028. doi: 10.1038/nplants.2015.28. [DOI] [PubMed] [Google Scholar]
- 25•.Michalko J, Dravecka M, Bollenbach T, Friml J. Embryo-lethal phenotypes in early abp1 mutants are due to disruption of the neighboring BSM gene. F1000Res. 2015;4:1104. doi: 10.12688/f1000research.7143.1. This study demonstrated that the molecular basis for the embryo lethality observed in the abp1-1 allele is likely caused by disruption of the neighboring BSM gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Dai XH, Zhang Y, Zhang D, Chen JL, Gao XH, Estelle M, Zhao YD. Embryonic lethality of Arabidopsis abp1-1 is caused by deletion of the adjacent BSM gene. Nature Plants. 2015:1. doi: 10.1038/nplants.2015.183. This study rescued the embryo lethal phenotype of abp1-1 by expressing the neighboring BSM gene. Furthermore, this study showed that the T-DNA insertion in abp1-1 simultaneously deleted the BSM gene and inactivated ABP1, but the abp1-1 plants with the BSM transgene had no obvious phenotypes, demonstrating unambiguously that disruption of ABP1 does not cause embryo lethality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubes J, Beeckman T, Jansch L, Frentzen M, et al. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci U S A. 2011;108:6674–6679. doi: 10.1073/pnas.1103442108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Kawabayashi T, Shikanai T, Hara-Nishimura I. Decreased Expression of a Gene Caused by a T-DNA Insertion in an Adjacent Gene in Arabidopsis. PLoS One. 2016;11:e0147911. doi: 10.1371/journal.pone.0147911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Enders TA, Oh S, Yang Z, Montgomery BL, Strader LC. Genome Sequencing of Arabidopsis abp1-5 Reveals Second-Site Mutations That May Affect Phenotypes. Plant Cell. 2015;27:1820–1826. doi: 10.1105/tpc.15.00214. Whole genome sequencing revealed over 8000 background SNPs in abp1-5 coding regions. This study revealed that at least some of these background mutations are likely responsible for phenotypes attributed to the mutation in ABP1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Naramoto S, Robert S, Tejos R, Lofke C, Lin D, Yang Z, Friml J. ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr Biol. 2012;22:1326–1332. doi: 10.1016/j.cub.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 31.Tromas A, Paque S, Stierle V, Quettier AL, Muller P, Lechner E, Genschik P, Perrot-Rechenmann C. Auxin-binding protein 1 is a negative regulator of the SCF(TIR1/AFB) pathway. Nat Commun. 2013;4:2496. doi: 10.1038/ncomms3496. [DOI] [PubMed] [Google Scholar]
- 32••.Michalko J, Glanc M, Perrot-Rechenmann C, Friml J. Strong morphological defects in conditional Arabidopsis abp1 knock-down mutants generated in absence of functional ABP1 protein. F1000Res. 2016;5:86. doi: 10.12688/f1000research.7654.1. Using a segregating population from crosses between an abp1 null allele and conditional knock-down mutants, the authors determined that the morphological phenotypes previously associated with conditional down-regulation of ABP1 can be recapitulated in the absence of a functional ABP1 gene, suggesting that those phenotypes are likely caused by off-target effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones AM, Im KH, Savka MA, Wu MJ, DeWitt NG, Shillito R, Binns AN. Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science. 1998;282:1114–1117. doi: 10.1126/science.282.5391.1114. [DOI] [PubMed] [Google Scholar]
- 34.Tromas A, Paponov I, Perrot-Rechenmann C. AUXIN BINDING PROTEIN 1: functional and evolutionary aspects. Trends Plant Sci. 2010;15:436–446. doi: 10.1016/j.tplants.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 35••.Kato H, Ishizaki K, Kouno M, Shirakawa M, Bowman JL, Nishihama R, Kohchi T. Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in Marchantia polymorpha. PLoS Genet. 2015;11:e1005084. doi: 10.1371/journal.pgen.1005084. The authors of this study functionally characterized the auxin transcriptional response machinery in the liverwort Marchantia polymorpha. Intriguingly, many components of the pathway except ABP1 were present as single orthologs and phylogeny of these genes supports the possibility that this was the ancestral state of the pathway for land plants and that ABP1 is dispensable. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Wang F, Zheng S, Xu T, Yang Z. Pavement cells: a model system for non-transcriptional auxin signalling and crosstalks. J Exp Bot. 2015;66:4957–4970. doi: 10.1093/jxb/erv266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan X, Chen J, Yang Z. Auxin regulation of cell polarity in plants. Curr Opin Plant Biol. 2015;28:144–153. doi: 10.1016/j.pbi.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. Evolution of functional diversity in the cupin superfamily. Trends Biochem Sci. 2001;26:740–746. doi: 10.1016/s0968-0004(01)01981-8. [DOI] [PubMed] [Google Scholar]
- 39.Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Yin K, Han X, Xu Z, Xue H. Arabidopsis GLP4 is localized to the Golgi and binds auxin in vitro. Acta Biochim Biophys Sin (Shanghai) 2009;41:478–487. doi: 10.1093/abbs/gmp036. [DOI] [PubMed] [Google Scholar]
- 41.Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 42.Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- 43.Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Y, Qin G, Dai X, Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200:229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- 47.Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 48.Motchoulski A, Liscum E. Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science. 1999;286:961–964. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- 49.Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 50.Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development. 2004;131:5021–5030. doi: 10.1242/dev.01388. [DOI] [PubMed] [Google Scholar]
- 51.Furutani M, Kajiwara T, Kato T, Treml BS, Stockum C, Torres-Ruiz RA, Tasaka M. The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development. 2007;134:3849–3859. doi: 10.1242/dev.009654. [DOI] [PubMed] [Google Scholar]
- 52.Yu H, Zhang Y, Moss BL, Bargmann BO, Wang R, Prigge M, Nemhauser JL, Estelle M. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat Plants. 2015:1. doi: 10.1038/nplants.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Jing H, Yang X, Zhang J, Liu X, Zheng H, Dong G, Nian J, Feng J, Xia B, Qian Q, et al. Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat Commun. 2015;6:7395. doi: 10.1038/ncomms8395. This report describes roles for the LATERAL ROOTLESS2 (LRT2), a cyclophilin-type PPIase, in regulating auxin response in rice. In particular, LRT2 was found to catalyze isomerization of the peptidyl-prolyl bond of OsIAA11 to regulate its incorporation into the SCFTIR1 coreceptor, thus affecting stability of this repressor. Further, this study underscores the importance of using systems other than Arabidopsis for discovery of auxin regulatory mechanisms. [DOI] [PubMed] [Google Scholar]
- 54.Li SB, Xie ZZ, Hu CG, Zhang JZ. A Review of Auxin Response Factors (ARFs) in Plants. Front Plant Sci. 2016;7:47. doi: 10.3389/fpls.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandler JW. Auxin response factors. Plant Cell Environ. 2015 doi: 10.1111/pce.12662. [DOI] [PubMed] [Google Scholar]
- 56.Weijers D, Wagner D. Transcriptional Responses to the Auxin Hormone. Annu Rev Plant Biol. 2016 doi: 10.1146/annurev-arplant-043015-112122. [DOI] [PubMed] [Google Scholar]
- 57.Kagale S, Rozwadowski K. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics. 2011;6:141–146. doi: 10.4161/epi.6.2.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 59••.Wu MF, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. Elife. 2015;4:e09269. doi: 10.7554/eLife.09269. This study demonstrated that SWI/SNF chromatin remodelers interact with ARF5 to unlock the chromatin surrounding ARF5 and to allow for gene activation. This finding has led to fundamental insights on chromatin-level control of auxin responses and has uncovered an additional mechanism of regulating auxin transcriptional responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamuro C, Zhu JK, Yang Z. Epigenetic Modifications and Plant Hormone Action. Mol Plant. 2016;9:57–70. doi: 10.1016/j.molp.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Plavskin Y, Nagashima A, Perroud PF, Hasebe M, Quatrano RS, Atwal GS, Timmermans MC. Ancient trans-Acting siRNAs Confer Robustness and Sensitivity onto the Auxin Response. Dev Cell. 2016;36:276–289. doi: 10.1016/j.devcel.2016.01.010. In this study, tasiRNA regulation of repressor ARF accumulation is discovered as a mechanism to make the auxin signaling pathway more robust. An elegant combination of experimental approaches and modeling suggests that tasiRNAs promote sensitivity and robustness in Physcomitrella protonema. [DOI] [PMC free article] [PubMed] [Google Scholar]