Abstract
Alteration of the gut microbial community structure and function through antibiotic use increases susceptibility to colonization by Clostridium difficile and other enteric pathogens. However, the mechanisms that mediate colonization resistance remain elusive. As the leading definable cause of infectious diarrhea, toxigenic C. difficile represents a burden for patients and health care systems, underscoring the need for better diagnostics and treatment strategies. Next-generation sequence data has increased our understanding of how the gut microbiota is influenced by many factors including diet, disease, aging and drugs. However, a microbial-based biomarker differentiating C. difficile infection from antibiotic-associated diarrhea remains elusive. Metabolomics profiling, which is highly responsive to changes in physiological conditions, have shown promise in differentiating subtle disease phenotypes that exhibit a nearly identical microbiome community structure, suggesting metabolite-based biomarkers may be an ideal diagnostic for identifying patients with CDI. This review focuses on the current understanding of structural and functional changes to the gut microbiota during C. difficile infection obtained from studies assessing the microbiome and metabolome of samples from patients and murine models.
Keywords: Clostridium difficile, microbiome, metabolome, CDI
Introduction
Clostridium difficile is the major cause of infectious diarrhea in the United States, causing 12.1% of health care-associated infection (1, 2). The mortality rate for health-care associated C. difficile infection (CDI) is estimated at 9.3%, contributing to 29,000 deaths in the U.S. (3). C. difficile pathogenicity is attributed to the production of two enterotoxins, TcdA and TcdB that inactivate GTP binding proteins, triggering a cascade of events culminating in diarrhea and colitis that can range from mild to life-threatening illness (4–7). The host immune response to C. difficile toxins is reported to modulate CDI susceptibility (8–10) and an inadequate humoral immune response to C. difficile toxins and/or a lack of anti-toxin antibodies influences development of CDI (9–12).
Disruption of the gut microbiota is another underlying component of Clostridium difficile pathogenesis. Therefore, it is not surprising that both established and proposed risk factors for developing CDI are associated with an altered intestinal ecosystem (Table 1, Fig. 1). Two major risk factors for CDI; antibiotic exposure and advanced age are both known to impact the gut microbiome and metabolome (13–16). Compared to healthy adults, the gut microbiota of the elderly and individuals receiving antibiotics display decreased microbial diversity, evenness and richness. Moreover, taxonomic changes within the microbiomes share similar characteristics including, enrichment of Proteobacteria and decreased abundances of Firmicutes and Bifidobacteria (Table 1) (13–15, 17). Decreases in microbial diversity and comparable taxonomic alterations are reported for other CDI risk factors, including inflammatory bowel disease, chemotherapy and use of proton pump inhibitors (Table 1). Microbiome dysbiosis likely increases CDI susceptibility by altering several aspects of colonization resistance (18–20), including but not limited to antimicrobial production (21), competition for nutrients (22–25) and bile acid metabolism (26–30). Moreover, altered communication between the immune system and the microbiome may also contribute to increased susceptibility to CDI following antibiotic exposure (31–35).
Table 1.
Taxonomic alterations associated with CDI risk factors as compared to a healthy gut microbiome
| CDI Risk Factor | Decreased Taxa | Increased Taxa | References |
|---|---|---|---|
| Advanced age | Ruminococcaceae, Bifidobacterium, Lactobacillus, Faecalibacterium | Bacteroidetes, Proteobacteria, | (125–128) |
| Antibiotic exposure | Bacteroidiaceae, Clostridiales, Ruminococcaceae, Lachnospiraceae, Bifidobacteria | Enterobacteriaceae, Enterococcaceae, Lactobaciliaceae, Streptococcaceae | (14–16, 63, 64) |
| IBD | Firmicutes, Lachnospiraceae, Rumonicoccaceae, and Clostridiales; Bifidobacteria | Enterobacteriaceae, including E. coli; Fusobacterium, Mycobacterium | (129–140) |
| Proton Pump Inhibitors | Ruminococcacae, Clostridiales | Lactobacillales, Enterobacteriaceae, Streptococcaceae, Enterococcaceae | (141–143) |
| Chemotherapy | Clostridiales, Lachnospiraceae, Ruminococcaceae, Bifidobacteriaceae | Bacteroidetes, Enterococcaceae Enterobacteriaceae | (144, 145) |
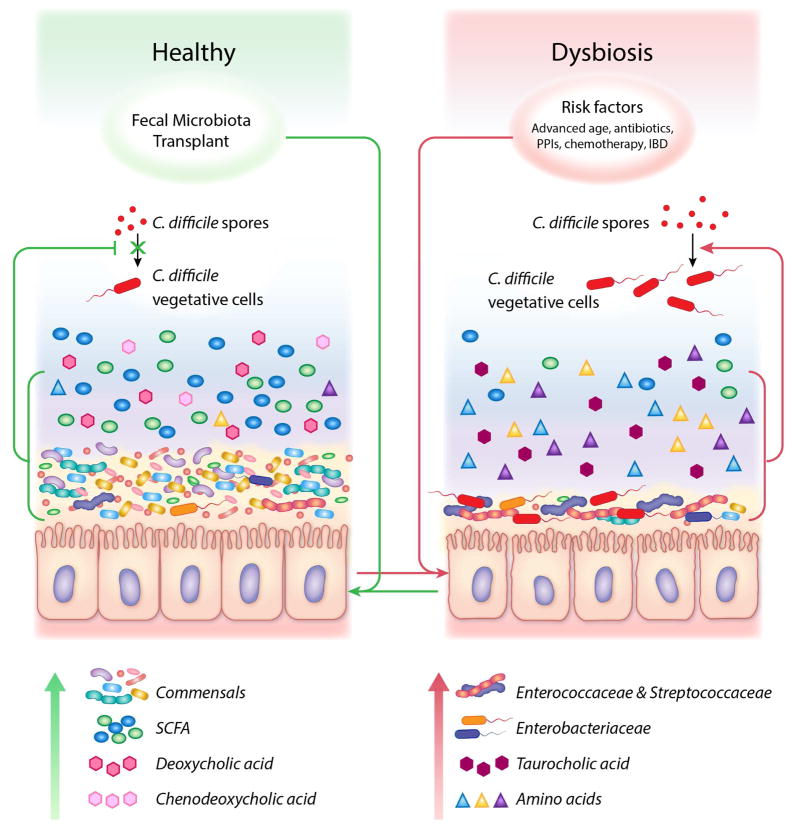
Figure 1. Microbial and metabolite status during health and disease.
A healthy large intestine characterized by commensal bacteria and short chain fatty acids, chenodeoxycholate and deoxycholate; a metabolic environment that inhibits germination of C. difficile spores, expansion of vegetative cells and subsequent colonization (Left). Following exposure to CDI risk factors, the microbiome is altered, exhibiting increased abundances of Enterobacteriaceae, Enterococcaceae and Streptococcaceae, and a metabolic state enriched in amino acids and primary bile acids that favor C. difficile germination, colonization and toxin production (Right). Following Fecal Microbiota Transplantation (FMT), the structure and function of the intestinal ecosystem is restored to a disease-resistant state.
Understanding how alterations to the gut microbiota contribute to CDI susceptibility is expected to identify novel therapeutic strategies and biomarkers that may predict treatment outcome and improve diagnostics. The currently preferred diagnostic platform, nucleic acid amplification of the C. difficile toxin-encoding genes, cannot distinguish between colonization and disease (36). Not only have the rates of CDI steadily increased since adoption of this method (37–40), a recent study found that up to 25% of patients were misdiagnosed for CDI (41). Although, inclusion of the toxin immunoassay (42, 43) and/or glutamate dehydrogenase (44) assay with nucleic acid amplification provides results that correlate better with clinical disease (40), there is currently no clinical diagnostic available that quickly and reliably identifies patients at risk for CDI recurrence. Several studies suggest that 15–35% of patients who initially respond to therapy will experience a recurrent episode following cessation of antibiotics (45–47). Subsequently, up to 50% of these patients will experience further relapse resulting in substantially higher morbidity and mortality (48). Frequent readmission to a primary care facility are common, contributing to an economic burden that is up to three times higher for recurrent CDI than what is estimated for primary CDI (3, 49).
In addition, therapeutics for primary and recurrent CDI are lacking. First line therapy typically includes vancomycin or off-label use of metronidazole while fidaxomicin is currently considered the best practice for treating a recurrent infection. However, efficacy of fidaxomicin treatment decreases significantly with each recurrent episode underscoring the need to identify patients at risk for recurrence as indiscriminant treatment of all primary cases with fidaxomicin may be cost prohibitive (50–53).
The clinical success of Fecal Microbiota Transplantation (FMT), which involves the transfer of fecal bacteria from a healthy donor to a CDI patient, presents some of the strongest support for the modulation of C. difficile susceptibility by the microbiome (54–61). Most recurrent CDI patients receiving FMT have repeatedly failed antibiotic-therapy and recover clinically with disease eradication after one treatment. A defined cocktail of well-studied organisms is expected to be safer and have fewer consequences. Moreover, if at-risk patients can be identified, it is likely that a prebiotic could prevent an initial infection. Here we discuss what is currently known about the structure and function of the microbiome during C. difficile infection.
C. difficile and the gut microbiome
Several recent studies utilized next generation sequencing to compare the microbiome profiles of individuals without diarrhea (NDC) to patients with CDI and patients with C. difficile negative diarrhea (CDN) (62–64). Although these studies identify clear differences between NDC and active diarrhea, they do not readily distinguish CDI from CDN (63, 64). Compared with controls, both CDI and CDN samples exhibit lower diversity and decreased species richness, as well as a high degree of heterogeneity among individual samples (62–64). In addition, the diarrheal samples exhibit a low abundance of Bacteroidiaceae, Lachnospiraceae and Ruminococcaceae, which dominate the microbiomes of NDC samples. Specimens from patients with active diarrhea have increased abundances of Lactobacilliaceae, Enterococcaceae, Streptococcaceae and Proteobacteria compared to controls (Fig. 1) (62–64). Notably, decreases in Clostridiales have also been reported in patients with diarrhea (62, 63) and patients at risk for developing CDI (65). The nonspecific disruption of the gut microbiota, regardless of C. difficile status, may suggest that many patients with active diarrhea are susceptible to CDI and that the presence of C. difficile does not alter the fecal microbiome structure.
Inclusion of asymptomatic carriers, toxigenic C. difficile positive patients receiving antibiotics without signs of diarrhea, is likely to provide important epigenetic information regarding CDI susceptibility. A recent study by Zhang et al. found that samples from asymptomatic carriers exhibited decreased diversity, similar to samples obtained from CDI patients, but were structurally more similar to healthy control samples (66). The asymptomatic carrier specimens contained fewer Proteobacteria than CDI samples and a greater abundance of Bifidobacteria, which were completely lacking in the CDI specimens (66). These data suggest that the presence or absence of certain microbial taxa is more important than microbial diversity when considering CDI susceptibility. Another study found microbial taxa belonging to the Clostridium XIVa group correlated with C. difficile carriage, but not development of CDI, in a population of patients with similar levels of alpha diversity and who received chemotherapy and antibiotics (67). It is also possible that a protective toxin immune response contributes to adult asymptomatic C. difficile carriage in the absence of microbial diversity.
Low microbial diversity and asymptomatic carriage of toxigenic C. difficile is also prevalent during the first year of life (68–71) when the gut microbiome is dominated by Bifidobacterium and Lactobacillus (72–75). Although passive transfer of maternal antibodies (76, 77) and a lack of toxin receptors have been proposed (78), the mechanism for asymptomatic C. difficile carriage in this population remains unknown. A single study characterized the intestinal microbiota of children with CDI aged 28–48 months (79) and reported changes in the microbiota that were similar to changes in adult specimens. They observed that fecal microbiota diversity and richness in children with CDI was significantly reduced and exhibited greater heterogeneity compared to samples from healthy children (79). The pediatric CDI samples also displayed similar taxonomic alterations; reduced Bacteroidiaceae, Ruminococcaceaa, and Lachnospiraceae and increased Enterococcaceae, Enterobacteriaceae, Streptococcaceae and Lactobacillaceae (79). However, asymptomatic carriage of C. difficile is highest in infants (<12 mo), which were not included in this study (68, 70).
Few studies have investigated structural changes within the gut microbiome of recurrent patients. A recent study comparing samples from healthy, primary and recurrent CDI patients reported that recurrent CDI samples were significantly less diverse that primary CDI samples. Similar findings were reported by a 2008 study comparing 16S clone libraries (80, 81). Allegretti et al. also found that both primary and recurrent samples contained significantly fewer Clostridiales and members of the Collinsella genus than the healthy control specimens (81). However, the majority of next generation sequence data regarding the gut microbiome of recurrent CDI patients has largely been provided by studies analyzing changes to the gut microbiota following FMT. Similar to primary CDI and antibiotic associated diarrhea, pre-FMT samples show decreased diversity, richness and evenness compared to healthy donors and post-FMT samples (54, 56, 57, 61, 82, 83). In addition, pre-FMT samples exhibit reduced levels of Bacteroidiaceae, Lachnospiraceae and Ruminococcaceae and are enriched for Enterococcaceae, Streptococcaceae and Veillonellaceae compared to donors and samples collected following FMT (56, 57, 61, 82, 83). While these studies clearly demonstrate the importance of the microbiome in modulating CDI and provide unique insight into recovery of the gut microbiota following this therapy, they do not provide insight into disease recurrence as these studies lack longitudinal samples. Moreover, the majority of pre-FMT samples are collected during vancomycin administration which will further alter the gut microbiota.
Disease susceptibility and the gut metabolome
The inter-individual variability of the human gut microbiota (84) is a major hindrance to identifying species that can either function as biomarkers of CDI or provide colonization resistance. Because different microbial communities can provide similar functions, examining the metabolic status of health and disease may be more useful in terms of identifying biomarkers of disease susceptibility than microbial community structure. In support of this, a recent CDI study was able to differentiate three similar patient groups irrespective of age, gender, antibiotic use, disease duration or medical history through global metabolic profiling (85). A comparison of patients with diarrhea that were: (1) positive for C. difficile and toxin production, (2) positive for C. difficile but negative for toxin production, and (3) C. difficle negative, identified metabolites that were C. difficile specific, such as N-palmitoyl glutamic acid, phlorizin, ceramide and Leonuriside A. They also found that toxin production was associated with deficiencies in choline and acetyl-putrescine. Notably, these differences were only observed in stool samples that were pre-treated with sonication and multiple centrifugation steps to obtain a sample consisting mostly of microbial cells. Analysis from non-pretreated stool samples did not differentiate these three groups (85).
The majority of metabolite data has been obtained from murine models and has identified pathways that may be important for CDI susceptibility including bile acid metabolism, amino acid metabolism and carbohydrate fermentation (Fig. 1).
Bile acid metabolism
The contribution of altered bile acid metabolism due to administration of antibiotics is one mechanism C. difficile may exploit during infection. It is proposed that, in a healthy microbiota, the primary bile acid, chenodeoxycholate and secondary bile acids, deoxycholate and lithocholate, inhibit spore germination and growth of C. difficile in the large bowel. However, administration of antibiotics alters microbial structure and bile acid metabolism. As a result, chenodeoxycholate concentration and transformation of cholate to deoxycholate is reduced, creating a colonic environment that favors spore germination and bacterial expansion (Fig. 1). Several studies report increased tauro-conjugated primary bile acids and decreased secondary bile acids, including deoxycholate, following antibiotic administration in animal models (16, 28–30) and patient samples (83). These results are supported by in vitro (86) (87–90) and in vivo studies (26, 91) assessing the effects of primary and secondary bile acids on C. difficile growth and germination. Strong evidence for modulation of CDI susceptibility by fecal bile acid composition was recently described by Weingarden. They reported that secondary bile acids, lithocholate, deoxycholate and isodeoxycholate, were absent in pre-FMT samples while primary bile acids, cholate and chenodeoxycholate, were significantly decreased in post-FMT and donor samples (83). Similarly, Allegretti et al. found that specimens from primary and recurrent CDI patients contained significantly higher levels of primary bile acids and lower levels of secondary bile acids compared to samples obtained from healthy controls. They also noted that primary bile acids were significantly elevated in samples from recurrent cases compared to samples obtained from patients experiencing the first episode of CDI (81).
Amino acids
Like bile salts, amino acids play an important role in the life cycle of C. difficle. Glycine, in combination with certain bile acids, promotes C. difficile germination (88, 92). In addition, histidine and to a lesser extent, arginine, aspartic acid and valine, can further enhance germination in the presence of both glycine and conjugated bile acids (93). An increased abundance of histidine was associated with patient samples that were C. difficle positive but not those that were C. difficle negative (85) while glycine and valine, among other amino acids, are associated with the cecal contents of C. difficile susceptible animals (29). In addition, N-acetylated forms of methionine, leucine and isoleucine were increased in the cecal contents of antibiotic-treated susceptible mice while n-acetylated aspartate decreased (30). Global metabolic profiling also suggests that metabolism of another amino acid, tryptophan, may play a role in colonization resistance. The intestinal microbiota synthesize several compounds from tryptophan (30, 94, 95), including indol-containing metabolites, such as inole-3-propionic acid and kynurenate, which were decreased following antibiotic treatment during a time period where mice were susceptible to C. difficile colonization (30).
During this time period, other tryptophan metabolites, indole lactate and N-acetyltryptophan, which presumably result from host enzyme activity, concurrently increased, suggesting that in the absence of microbial activity, tryptophan becomes available for utilization by host proteins (30). Unlike amino acids, microbial-derived tryptophan products increased when colonization resistance was restored suggesting tryptophan metabolism may serve as a biomarker for colonization resistance. Other investigators have observed increased levels of tryptophan in the cecal contents of CDI-susceptible mice (29, 30) and in the feces of antibiotic treated rats (96).
Carbohydrate fermentation
Microbial fermentation of diet and host-derived carbohydrates is the major source of short chain fatty acids (SCFAs) in the gut. SCFAs are reduced following antibiotic treatment in both humans and animal models (96–100) and have been linked to C. difficile colonization resistance (18, 29, 30, 101, 102) (Fig. 1). They also inhibit growth of C. difficile in vitro (102). One possible mechanism for regulation of C. difficile susceptibility by SCFA is modulation of luminal pH. When concentrations of SCFAs decline, pH increases, resulting in an environment that is favorable for growth of Enterobacteriaceae and Clostridia, including C. difficile (103–105).
Another possible mechanism is through production of the SCFA, butyrate. Butyric acid has anti-inflammatory effects, decreases permeability through modulation of tight junction protein production and increases antimicrobial peptide levels and mucin production (Fig. 1) (106, 107). Butyrate-producing bacteria are found within the Lachnospiraceae and Ruminococcaceae families; taxa that are greatly reduced in stool specimens from hospitalized patients at risk for developing CDI and patients with diarrhea, including those diagnosed with CDI (62–65). However, data assessing the role for SCFAs in C. difficile infection using animal models have yielded mixed results (102, 108, 109).
Succinate, an organic acid resulting from microbial carbohydrate fermentation is an important intermediate metabolite in the gut (110) that promotes infection by C. difficle in vivo (111). Studies utilizing a B. thetaiotamicron mono-colonized mouse on a polysaccharide-rich diet exhibited increased levels of succinate and upregulated transcript levels of genes involved in conversion of succinate to butyrate by C. difficile. Furthermore, a C. difficile succinate transporter mutant exhibited decreased proliferation in the B. thetaiotamicron mono-colonized mouse and mice treated with streptomycin or polyethylene glycol, compounds that increase cecal succinate levels, suggesting that the inability to utilize succinate negatively affects proliferation of C. difficile in the gut (111).
Discussion
There is a strong association between perturbation of the gut microbiota and susceptibility to C. difficile infection. These important, early studies using patient samples and animal models have characterized the structural, and to a lesser degree, functional changes within the microbiota during CDI (56–58, 62–66, 79). However, much remains to be done. To date, the studies assessing the gut community structure are limiting and cannot address the role of the microbiome in CDI risk and prevention and offer little insight into recurrent CDI. The increased morbidity, mortality and lack of treatment options associated with recurrent CDI underscore the importance of characterizing the intestinal ecosystem of this population. Studies including longitudinal samples encompassing primary and recurrent episodes may identify microbial or metabolic markers that are predictive of disease relapse. Moreover, inclusion of samples prior to and following treatment failure may identify biomarkers that are predictive of treatment outcome allowing earlier and more precise utilization of the limited treatment options available. Not only would this be particularly instructive when considering treatments like fidaxomicin, it would provide much-needed insight into the basis of disease relapse and treatment failure.
Currently, there is a paucity of data examining the microbiome structure and function of pediatric CDI patients despite the continued rise of CDI among children (112–114). Furthermore, molecular diagnostics remain problematic due to concerns about detection of colonization rather than true disease. A study by Leibowitz et al. found that hospitalized children aged 1–18 years (19% with diarrhea and 24% without diarrhea), tested positive for C. difficile by tcdB-specific PCR (115). Furthermore, high rates of C. difficile colonization have been reported in pediatric populations with additional co-morbidities, such as cancer and IBD (112–114). The ability to identify symptomatic CDI in a population with high rates of asymptomatic C. difficile carriage would improve diagnostics and treatment for at-risk children. Furthermore, a better understanding of disease resistance in the infant population represents a unique opportunity to identify key host and microbial metabolic pathways, and microbial species that may protect young children from developing clinical disease despite a lack of microbial diversity.
Pyrosequencing has advanced our understanding of biodiversity and microbial community structures considerably. However, there are multiple potential sources of bias in 16S rDNA sequencing and analysis, including DNA extraction technique (116, 117), PCR parameters (118, 119), 16S variable region primers (120, 121), 16S rDNA copy number (122) and clustering algorithm (123, 124). Moreover, the short read length limits resolution of some bacterial species. Moving beyond 16S rDNA sequencing will provide important information regarding the roles of microbes in CDI. Unlike 16S rDNA sequencing, whole genome sequencing (WGS) provides functional information derived from assessing gene content and allow taxonomic classification at the species level. The utility of determining changes to gene content was highlighted recently by Buffie et al. who found increased abundance of the bile acid inducible operon (bai), but not genes predicted to encode bile salt hydrolases, in specimens obtained from asymptomatic C. difficile carriers compared CDI specimens (67). Sorg and Sonnenshein demonstrated that bile acid transformation by the bai-containing Clostridium scindens negatively affected C. difficile germination (86). This strain provided moderate protection from disease in a murine model of CDI (67) suggesting that bai+ bacteria may be useful in probiotic mixtures used as therapy or for prevention of CDI. Data obtained from metabolomics will also assign functional information to microbiome studies and because metabolites are highly responsive to changes in physiological conditions, they are ideally suited to distinguish subtle disease phenotypes. Ultimately, a systems biology approach combining metabolomics and metagenomics is expected to elucidate the epigenetic influences of microbiome-mediated CDI susceptibility and resistance.
The currently published data describing the microbiome changes associated with CDI using murine models and patient samples has provided the basis for future studies that will offer insight into disease susceptibility through identification of species-level community changes, alterations in genetic pathways and differences in metabolic by-products associated with gut metabolism. Not only will these studies broaden our understanding of how the microbiome contributes to health and disease but should identify therapeutic and diagnostic targets for CDI and other diseases modulated by the intestinal ecosystem.
Acknowledgments
Karen Prince’s assistance in preparing figures is appreciated. This work was supported by Grants RO1AI100914, DK096323 and P30DK56338 from the National Institute of Allergy and Infectious Diseases and National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health (NIH) as well as the Pediatric GI Training Grant T32-DK07664.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Magill SS, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly CP, LaMont JT. Clostridium difficile--more difficult than ever. N Engl J Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 3.Lessa FC, Winston LG, McDonald LC E. I. P. C. d. S. Team. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:2369–2370. doi: 10.1056/NEJMc1505190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly CP, LaMont JT. Clostridium difficile infection. Annu Rev Med. 1998;49:375–390. doi: 10.1146/annurev.med.49.1.375. [DOI] [PubMed] [Google Scholar]
- 5.Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Just I, et al. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 7.Just I, et al. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem. 1995;270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 8.Madan R, WA Immune responses to Clostridium difficile infection. Trends Mol Med. 2012;18:658–666. doi: 10.1016/j.molmed.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyne L, Warny M, Qamar A, Kelly CP. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med. 2000;342:390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 10.Kelly CP, Kyne L. The host immune response to Clostridium difficile. J Med Microbiol. 2011;60:1070–1079. doi: 10.1099/jmm.0.030015-0. [DOI] [PubMed] [Google Scholar]
- 11.Wullt M, Noren T, Ljungh A, Akerlund T. IgG antibody response to toxins A and B in patients with Clostridium difficile infection. Clin Vaccine Immunol. 2012;19:1552–1554. doi: 10.1128/CVI.00210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leav BA, et al. Serum anti-toxin B antibody correlates with protection from recurrent Clostridium difficile infection (CDI) Vaccine. 2010;28:965–969. doi: 10.1016/j.vaccine.2009.10.144. [DOI] [PubMed] [Google Scholar]
- 13.Henrich TJ, Krakower D, Bitton A, Yokoe DS. Clinical risk factors for severe Clostridium difficile-associated disease. Emerg Infect Dis. 2009;15:415–422. doi: 10.3201/eid1503.080312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011; 108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antunes LC, et al. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salazar N, et al. Microbial targets for the development of functional foods accordingly with nutritional and immune parameters altered in the elderly. J Am Coll Nutr. 2013;32:399–406. doi: 10.1080/07315724.2013.827047. [DOI] [PubMed] [Google Scholar]
- 18.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Britton RA, Young VB. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol. 2012;20:313–319. doi: 10.1016/j.tim.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caballero S, Pamer EG. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol. 2015;33:227–256. doi: 10.1146/annurev-immunol-032713-120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzman NH. Paneth cell defensins and the regulation of the microbiome: detente at mucosal surfaces. Gut Microbes. 2010;1:401–406. doi: 10.4161/gmic.1.6.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. 1988;56:2610–2614. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambol SP, Merrigan MM, Tang JK, Johnson S, Gerding DN. Colonization for the prevention of Clostridium difficile disease in hamsters. J Infect Dis. 2002;186:1781–1789. doi: 10.1086/345676. [DOI] [PubMed] [Google Scholar]
- 26.Giel JL, Sorg JA, Sonenshein AL, Zhu J. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One. 2010;5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Theriot CM, Bowman AA, Young VB. Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere. 2016 doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theriot CM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jump RL, et al. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice. PLoS One. 2014;9:e101267. doi: 10.1371/journal.pone.0101267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hasegawa M, et al. Protective role of commensals against Clostridium difficile infection via an IL-1β-mediated positive-feedback loop. J Immunol. 2012;189:3085–3091. doi: 10.4049/jimmunol.1200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67:3504–3511. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bibbo S, et al. Role of microbiota and innate immunity in recurrent Clostridium difficile infection. J Immunol Res. 2014;2014:462740. doi: 10.1155/2014/462740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koo HL, et al. Real-time polymerase chain reaction detection of asymptomatic Clostridium difficile colonization and rising C. difficile-associated disease rates. Infect Control Hosp Epidemiol. 2014;35:667–673. doi: 10.1086/676433. [DOI] [PubMed] [Google Scholar]
- 37.Planche TD, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis. 2013;13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murad YM, et al. Impact of polymerase chain reaction testing on Clostridium difficile infection rates in an acute health care facility. Am J Infect Control. 2015;43:383–386. doi: 10.1016/j.ajic.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Dubberke ER, Burnham CA. Diagnosis of Clostridium difficile Infection: Treat the Patient, Not the Test. JAMA Intern Med. 2015;175:1801–1802. doi: 10.1001/jamainternmed.2015.4607. [DOI] [PubMed] [Google Scholar]
- 40.Polage CR, et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern Med. 2015;175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson M, Olefson S, Machan JT, Kelly CR. A High Rate of Alternative Diagnoses in Patients Referred for Presumed Clostridium difficile Infection. J Clin Gastroenterol. 2015 doi: 10.1097/MCG.0000000000000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whittier S, et al. Evaluation of four commercially available enzyme immunoassays for laboratory diagnosis of Clostridium difficile-associated diseases. J Clin Microbiol. 1993;31:2861–2865. doi: 10.1128/jcm.31.11.2861-2865.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilligan PH. Optimizing the Laboratory Diagnosis of Clostridium difficile Infection. Clin Lab Med. 2015;35:299–312. doi: 10.1016/j.cll.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Fenner L, Widmer AF, Goy G, Rudin S, Frei R. Rapid and reliable diagnostic algorithm for detection of Clostridium difficile. J Clin Microbiol. 2008;46:328–330. doi: 10.1128/JCM.01503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McFarland LV, et al. Recurrent Clostridium difficile disease: epidemiology and clinical characteristics. Infect Control Hosp Epidemiol. 1999;20:43–50. doi: 10.1086/501553. [DOI] [PubMed] [Google Scholar]
- 46.Kelly CP. Can we identify patients at high risk of recurrent Clostridium difficile infection? Clin Microbiol Infect. 2012;18(Suppl 6):21–27. doi: 10.1111/1469-0691.12046. [DOI] [PubMed] [Google Scholar]
- 47.Garey KW, Sethi S, Yadav Y, DuPont HL. Meta-analysis to assess risk factors for recurrent Clostridium difficile infection. J Hosp Infect. 2008;70:298–304. doi: 10.1016/j.jhin.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 48.Garey KW, et al. A randomized, double-blind, placebo-controlled pilot study to assess the ability of rifaximin to prevent recurrent diarrhoea in patients with Clostridium difficile infection. J Antimicrob Chemother. 2011;66:2850–2855. doi: 10.1093/jac/dkr377. [DOI] [PubMed] [Google Scholar]
- 49.Ghantoji SS, Sail K, Lairson DR, DuPont HL, Garey KW. Economic healthcare costs of Clostridium difficile infection: a systematic review. J Hosp Infect. 2010;74:309–318. doi: 10.1016/j.jhin.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 50.Figueroa I, et al. Relapse versus reinfection: recurrent Clostridium difficile infection following treatment with fidaxomicin or vancomycin. Clin Infect Dis. 2012;55(Suppl 2):S104–109. doi: 10.1093/cid/cis357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cornely OA, et al. Clinical efficacy of fidaxomicin compared with vancomycin and metronidazole in Clostridium difficile infections: a meta-analysis and indirect treatment comparison. J Antimicrob Chemother. 2014;69:2892–2900. doi: 10.1093/jac/dku261. [DOI] [PubMed] [Google Scholar]
- 52.Cornely OA, et al. Fidaxomicin versus vancomycin for infection with Clostridium difficile in Europe, Canada, and the USA: a double-blind, non-inferiority, randomised controlled trial. Lancet Infect Dis. 2012;12:281–289. doi: 10.1016/S1473-3099(11)70374-7. [DOI] [PubMed] [Google Scholar]
- 53.Cornely OA, Miller MA, Louie TJ, Crook DW, Gorbach SL. Treatment of first recurrence of Clostridium difficile infection: fidaxomicin versus vancomycin. Clin Infect Dis. 2012;55(Suppl 2):S154–161. doi: 10.1093/cid/cis462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youngster I, et al. Fecal microbiota transplant for relapsing Clostridium difficile infection using a frozen inoculum from unrelated donors: a randomized, open-label, controlled pilot study. Clin Infect Dis. 2014;58:1515–1522. doi: 10.1093/cid/ciu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Youngster I, et al. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312:1772–1778. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- 56.Song Y, et al. Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent Clostridium difficile infection. PLoS One. 2013;8:e81330. doi: 10.1371/journal.pone.0081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dutta SK, et al. Efficacy of combined jejunal and colonic fecal microbiota transplantation for recurrent Clostridium difficile Infection. Clin Gastroenterol Hepatol. 2014;12:1572–1576. doi: 10.1016/j.cgh.2013.12.032. [DOI] [PubMed] [Google Scholar]
- 58.Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004;42:1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walia R, et al. Efficacy of fecal microbiota transplantation in 2 children with recurrent Clostridium difficile infection and its impact on their growth and gut microbiome. J Pediatr Gastroenterol Nutr. 2014;59:565–570. doi: 10.1097/MPG.0000000000000495. [DOI] [PubMed] [Google Scholar]
- 60.Walia R, Kunde S, Mahajan L. Fecal microbiota transplantation in the treatment of refractory Clostridium difficile infection in children: an update. Curr Opin Pediatr. 2014;26:573–578. doi: 10.1097/MOP.0000000000000127. [DOI] [PubMed] [Google Scholar]
- 61.Shankar V, et al. Species and genus level resolution analysis of gut microbiota in Clostridium difficile patients following fecal microbiota transplantation. Microbiome. 2014;2:13. doi: 10.1186/2049-2618-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu S, et al. Identification of key taxa that favor intestinal colonization of Clostridium difficile in an adult Chinese population. Microbes Infect. 2016;18:30–38. doi: 10.1016/j.micinf.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Antharam VC, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol. 2013;51:2884–2892. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schubert AM, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014;5:e01021–01014. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vincent C, et al. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome. 2013;1:18. doi: 10.1186/2049-2618-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L, et al. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe. 2015;34:1–7. doi: 10.1016/j.anaerobe.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rousseau C, et al. Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis. 2012;55:1209–1215. doi: 10.1093/cid/cis637. [DOI] [PubMed] [Google Scholar]
- 69.Rexach CE, Tang-Feldman YJ, Cantrell MC, Cohen SH. Epidemiologic surveillance of Clostridium difficile diarrhea in a freestanding pediatric hospital and a pediatric hospital at a university medical center. Diagn Microbiol Infect Dis. 2006;56:109–114. doi: 10.1016/j.diagmicrobio.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Larson HE, Barclay FE, Honour P, Hill ID. Epidemiology of Clostridium difficile in infants. J Infect Dis. 1982;146:727–733. doi: 10.1093/infdis/146.6.727. [DOI] [PubMed] [Google Scholar]
- 71.Collignon A, et al. Heterogeneity of Clostridium difficile isolates from infants. Eur J Pediatr. 1993;152:319–322. doi: 10.1007/BF01956743. [DOI] [PubMed] [Google Scholar]
- 72.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Jumaili IJ, Shibley M, Lishman AH, Record CO. Incidence and origin of Clostridium difficile in neonates. J Clin Microbiol. 1984;19:77–78. doi: 10.1128/jcm.19.1.77-78.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sjogren YM, et al. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy. 2009;39:1842–1851. doi: 10.1111/j.1365-2222.2009.03326.x. [DOI] [PubMed] [Google Scholar]
- 76.Jaureguy F, et al. Effects of intrapartum penicillin prophylaxis on intestinal bacterial colonization in infants. J Clin Microbiol. 2004;42:5184–5188. doi: 10.1128/JCM.42.11.5184-5188.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Penders J, Stobberingh EE, van den Brandt PA, van Ree R, Thijs C. Toxigenic and non479 toxigenic Clostridium difficile: determinants of intestinal colonisation and role in childhood atopic manifestations. Gut. 2008;57:1025–1026. doi: 10.1136/gut.2007.143214. [DOI] [PubMed] [Google Scholar]
- 78.Eglow R, et al. Diminished Clostridium difficile toxin A sensitivity in newborn rabbit ileum is associated with decreased toxin A receptor. J Clin Invest. 1992;90:822–829. doi: 10.1172/JCI115957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ling Z, et al. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci Rep. 2014;4:7485. doi: 10.1038/srep07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang JY, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 81.Allegretti JR, et al. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther. 2016 doi: 10.1111/apt.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shahinas D, et al. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. MBio. 2012;3 doi: 10.1128/mBio.00338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weingarden AR, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306:G310–319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rojo D, et al. Clostridium difficile heterogeneously impacts intestinal community architecture but drives stable metabolome responses. ISME J. 2015;9:2206–2220. doi: 10.1038/ismej.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 2010;192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol. 1982;15:443–446. doi: 10.1128/jcm.15.3.443-446.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wilson KH. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol. 1983;18:1017–1019. doi: 10.1128/jcm.18.4.1017-1019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Howerton A, Patra M, Abel-Santos E. A new strategy for the prevention of Clostridium difficile infection. J Infect Dis. 2013;207:1498–1504. doi: 10.1093/infdis/jit068. [DOI] [PubMed] [Google Scholar]
- 92.Wheeldon LJ, Worthington T, Hilton AC, Elliott TS, Lambert PA. Physical and chemical factors influencing the germination of Clostridium difficile spores. J Appl Microbiol. 2008;105:2223–2230. doi: 10.1111/j.1365-2672.2008.03965.x. [DOI] [PubMed] [Google Scholar]
- 93.Wheeldon LJ, Worthington T, Lambert PA. Histidine acts as a co-germinant with glycine and taurocholate for Clostridium difficile spores. J Appl Microbiol. 2011;110:987–994. doi: 10.1111/j.1365-2672.2011.04953.x. [DOI] [PubMed] [Google Scholar]
- 94.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chung KT, Anderson GM, Fulk GE. Formation of indoleacetic acid by intestinal anaerobes. J Bacteriol. 1975;124:573–575. doi: 10.1128/jb.124.1.573-575.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Swann JR, et al. Variation in antibiotic-induced microbial recolonization impacts on the host metabolic phenotypes of rats. J Proteome Res. 2011;10:3590–3603. doi: 10.1021/pr200243t. [DOI] [PubMed] [Google Scholar]
- 97.Hove H, Tvede M, Mortensen PB. Antibiotic-associated diarrhoea, Clostridium difficile, and short-chain fatty acids. Scand J Gastroenterol. 1996;31:688–693. doi: 10.3109/00365529609009151. [DOI] [PubMed] [Google Scholar]
- 98.Hoverstad T, et al. Influence of oral intake of seven different antibiotics on faecal short-chain fatty acid excretion in healthy subjects. Scand J Gastroenterol. 1986;21:997–1003. doi: 10.3109/00365528608996411. [DOI] [PubMed] [Google Scholar]
- 99.Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr. 1986;116:1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 100.Romick-Rosendale LE, et al. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril) Magn Reson Chem. 2009;47(Suppl 1):S36–46. doi: 10.1002/mrc.2511. [DOI] [PubMed] [Google Scholar]
- 101.May T, Mackie RI, Fahey GC, Cremin JC, Garleb KA. Effect of fiber source on short-chain fatty acid production and on the growth and toxin production by Clostridium difficile. Scand J Gastroenterol. 1994;29:916–922. doi: 10.3109/00365529409094863. [DOI] [PubMed] [Google Scholar]
- 102.Rolfe RD. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect Immun. 1984;45:185–191. doi: 10.1128/iai.45.1.185-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cherrington CA, Hinton M, Pearson GR, Chopra I. Short-chain organic acids at ph 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J Appl Bacteriol. 1991;70:161–165. doi: 10.1111/j.1365-2672.1991.tb04442.x. [DOI] [PubMed] [Google Scholar]
- 104.Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol. 2009;11:2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- 105.Gupta P, et al. Does Alkaline Colonic pH Predispose to Clostridium difficile Infection? South Med J. 2016;109:91–96. doi: 10.14423/SMJ.0000000000000414. [DOI] [PubMed] [Google Scholar]
- 106.Cook SI, Sellin JH. Review article: short chain fatty acids in health and disease. Aliment Pharmacol Ther. 1998;12:499–507. doi: 10.1046/j.1365-2036.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 107.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 108.Borriello SP, Barclay FE. An in-vitro model of colonisation resistance to Clostridium difficile infection. J Med Microbiol. 1986;21:299–309. doi: 10.1099/00222615-21-4-299. [DOI] [PubMed] [Google Scholar]
- 109.Lawley TD, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reichardt N, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ferreyra JA, et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sandora TJ, et al. Epidemiology and risk factors for Clostridium difficile infection in children. Pediatr Infect Dis J. 2011;30:580–584. doi: 10.1097/INF.0b013e31820bfb29. [DOI] [PubMed] [Google Scholar]
- 113.Sammons JS, Toltzis P, Zaoutis TE. Clostridium difficile Infection in children. JAMA Pediatr. 2013;167:567–573. doi: 10.1001/jamapediatrics.2013.441. [DOI] [PubMed] [Google Scholar]
- 114.Sammons JS, et al. Diagnosis and Management of Clostridium difficile Infection by Pediatric Infectious Diseases Physicians. J Pediatric Infect Dis Soc. 2014;3:43–48. doi: 10.1093/jpids/pit065. [DOI] [PubMed] [Google Scholar]
- 115.Leibowitz J, Soma VL, Rosen L, Ginocchio CC, Rubin LG. Similar proportions of stool specimens from hospitalized children with and without diarrhea test positive for Clostridium difficile. Pediatr Infect Dis J. 2015;34:261–266. doi: 10.1097/INF.0000000000000556. [DOI] [PubMed] [Google Scholar]
- 116.Scupham AJ, Jones JA, Wesley IV. Comparison of DNA extraction methods for analysis of turkey cecal microbiota. J Appl Microbiol. 2007;102:401–409. doi: 10.1111/j.1365-2672.2006.03094.x. [DOI] [PubMed] [Google Scholar]
- 117.Salonen A, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 118.Wu JY, et al. Effects of polymerase, template dilution and cycle number on PCR based 16 S rRNA diversity analysis using the deep sequencing method. BMC Microbiol. 2010;10:255. doi: 10.1186/1471-2180-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pinto AJ, Raskin L. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One. 2012;7:e43093. doi: 10.1371/journal.pone.0043093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tremblay J, et al. Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol. 2015;6:771. doi: 10.3389/fmicb.2015.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim M, Morrison M, Yu Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods. 2011;84:81–87. doi: 10.1016/j.mimet.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 122.Větrovsky T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS One. 2014;9:e94249. doi: 10.1371/journal.pone.0094249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Westcott S, Schloss P. De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ. 2015;3 doi: 10.7717/peerj.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.O’Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 126.Biagi E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Tongeren SP, Slaets JP, Harmsen HJ, Welling GW. Fecal microbiota composition and frailty. Appl Environ Microbiol. 2005;71:6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zwielehner J, et al. Combined PCR-DGGE fingerprinting and quantitative-PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Exp Gerontol. 2009;44:440–446. doi: 10.1016/j.exger.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 129.Walker AW, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lepage P, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 132.Baumgart M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 133.Darfeuille-Michaud A, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 134.Martinez-Medina M, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm Bowel Dis. 2009;15:872–882. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 135.Martin HM, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 136.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sasaki M, et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest. 2007;87:1042–1054. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- 138.Ohkusa T, et al. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol. 2002;17:849–853. doi: 10.1046/j.1440-1746.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 139.Andoh A, et al. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:955–962. doi: 10.1002/ibd.20151. [DOI] [PubMed] [Google Scholar]
- 140.Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- 141.Jackson MA, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2015 doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Imhann F, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2015 doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Freedberg DE, et al. Proton Pump Inhibitors Alter Specific Taxa in the Human Gastrointestinal Microbiome: A Crossover Trial. Gastroenterology. 2015;149:883–885.e889. doi: 10.1053/j.gastro.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Montassier E, et al. 16S rRNA gene pyrosequencing reveals shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb Ecol. 2014;67:690–699. doi: 10.1007/s00248-013-0355-4. [DOI] [PubMed] [Google Scholar]
- 145.Montassier E, et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment Pharmacol Ther. 2015;42:515–528. doi: 10.1111/apt.13302. [DOI] [PubMed] [Google Scholar]