Abstract
Clostridium difficile is an anaerobic, Gram positive, spore-forming bacillus that is the leading cause of nosocomial gastroenteritis. Clostridium difficile infection (CDI) is associated with increasing morbidity and mortality, consequently posing an urgent threat to public health. Recurrence of CDI after successful treatment with antibiotics is high, thus necessitating discovery of novel therapeutics against this pathogen. Susceptibility to CDI is associated with alterations in the gut microbiota composition and bile acid metabolome, specifically a loss of microbial derived secondary bile acids. This review aims to summarize in vitro, ex vivo, and in vivo studies done by our group and others that demonstrate how secondary bile acids affect the different stages of the C. difficile life cycle. Understanding the dynamic interplay of C. difficile and microbial derived secondary bile acids within the gastrointestinal tract will shed light on how bile acids play a role in colonization resistance against C. difficile. Rational manipulation of secondary bile acids may prove beneficial as a treatment for patients with CDI.
Keywords: Clostridium difficile, secondary bile acids, colonization resistance, gut microbiota, antibiotics
Graphical Abstract
Introduction
Clostridium difficile is an anaerobic, Gram positive, spore forming bacillus that was first isolated from newborn infants by Hall and O’Toole in 1935[1]. Currently, C. difficile is a leading nosocomial enteric pathogen that causes significant human morbidity, mortality, and results in over $4.8 billion per year in healthcare costs[2–5]. In 2013, the Centers for Disease Control and Prevention (CDC) categorized C. difficile as an urgent antibiotic resistance threat negatively impacting public health[5]. A major risk factor for infection with C. difficile is the use of antibiotics[6, 7]. Antibiotics lead to significant and long lasting shifts in the gastrointestinal (GI) microbiota and metabolome[8–10] resulting in a loss of colonization resistance against C. difficile[11–14]. Colonization resistance is the ability of the indigenous gut microbiota to protect against invasion by enteric pathogens[15]. Although the exact mechanisms of colonization resistance against C. difficile are unknown, there is increasing evidence that gut microbiota derived secondary bile acids play an important role[11, 12, 16, 17].
Antibiotic treatment with vancomycin and metronidazole is considered standard of care for C. difficile infection (CDI)[18]. Unfortunately this treatment further disrupts the gut microbiota composition and recurrence of CDI after cessation of antibiotics is high, occurring in 20–30% of patients[2, 18–21]. Consequently, antibiotic treatment is insufficient for some patients with CDI thus necessitating the discovery of novel therapeutics against C. difficile. In this review we aim to highlight the dynamic interplay between C. difficile and the secondary bile acids within the GI tract. In particular, we will review in vitro, ex vivo and in vivo studies done by our group and others that focus on how bile acids affect the different stages of the C. difficile life cycle. Rational manipulation of secondary bile acids in the GI tract may prove beneficial as a therapeutic strategy against C. difficile [12, 22].
Formation of Microbial Derived Secondary Bile Acids
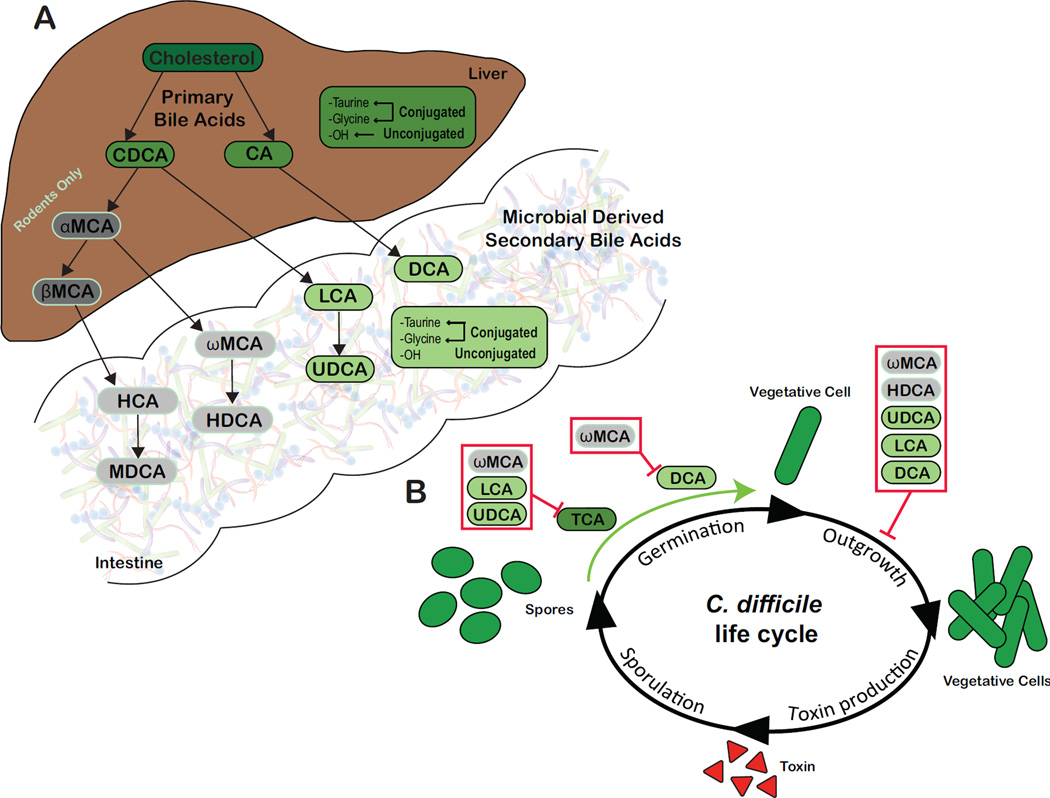
Bile acids are water-soluble, cholesterol derived amphipathic molecules synthesized by hepatocytes[23]. Cholate (CA) and chenodeoxycholate (CDCA) are the primary bile acids synthesized in humans and rodents (Figure 1A)[23–25]. In rodents, a significant amount of CDCA is converted by 6-β-hydroxylation to muricholate (MCA) [26]. The host further metabolizes primary bile acids via N-acyl amination to glycine or taurine forming conjugated bile acids, such as glycocholate (GCA) or taurocholate (TCA). Primary bile acids enter the small intestine where they aid in fat emulsification and absorption[23, 27]. Bile acids are also biological detergents and induce expression of antimicrobial peptides, thus contributing to the host defense system against both commensal microbes and some enteric pathogens[28, 29]. Once host derived primary bile acids enter into the GI tract, members of the gut microbiota transform them into over 50 chemically diverse secondary bile acids[23, 30]. Secondary bile acids are formed by two main bacterial reactions: deconjugation predominantly within the small intestine and epimerization/dehydroxylation within the large intestine[23].
Fig 1.
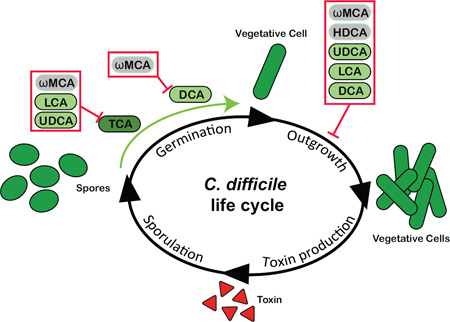
A: Production of microbial derived secondary bile acids. Primary bile acids, chenodeoxycholate (CDCA) and cholate (CA) are synthesized from cholesterol by hepatocytes in humans and rodents. In rodents, a portion of CDCA is further converted into α-muricholate (αMCA) and β-muricholate (βMCA), which are not recognized in humans (represented in gray). Primary bile acids can be unconjugated or further modified via conjugation to taurine or glycine within the liver. Once synthesized, host derived primary bile acids (represented in darker shades) enter into bile. Bile is stored in the gallbladder until release in the duodenum following ingestion of a meal. Once within the GI tract, the gut microbiota can convert host derived primary bile acids into secondary bile acids (represented in lighter shades). Microbial derived secondary bile acids can also be unconjugated or conjugated to taurine or glycine. B: Effects of various secondary bile acids on the life cycle of C. difficile in vitro. TCA is essential for germination of C. difficile spores (green arrow). TCA-mediated spore germination can be blocked by specific secondary bile acids (red box). DCA can also stimulate germination of C. difficile spores, a process that is inhibited by ωMCA in mice. Outgrowth of C. difficile vegetative cells is inhibited by multiple secondary bile acids (red box).
Abbreviations: CA, cholate; CDCA, chenodeoxycholate; DCA, deoxycholate; HCA, hyocholate; HDCA, hyodeoxycholate; LCA, lithocholate; MDCA, murideoxycholate; UDCA, ursodeoxycholate; αMCA, α-muricholate; βMCA, β-muricholate; ωMCA, ω-muricholate
Deconjugation of conjugated primary bile acids occurs rapidly by extracellular bile salt hydrolases (BSH), which are widespread in the gut microbiota[23, 28, 31]. Based on metagenomic screening, 3 major phyla in the gut microbiota possess BSHs: Firmicutes (30%), Bacteroidetes (14.4%), and Actinobacteria (8.9%)[31]. Within these phyla, BSHs from the following genera are heavily studied: Clostridium, Bacteroides, Lactobacillus, Bifidobacterium, and Enterococcus [28]. BSHs appear to enhance bacterial colonization within the lower GI tract potentially by detoxification of bile acids[23]. Thus the presence of BSHs are included in probiotic selection criteria to improve strain competitiveness within the gut[32]. However, the host and microbial physiologic function of BSHs is still being investigated[23, 28].
The second bile acid modifying reaction results in oxidization and epimerization of specific hydroxyl groups by three distinct bacterial hydroxysteroid dehydrogenases (HSHD), 3-α, 7-α, and 12-α[23]. HSHDs can produce 27 unique metabolites from the primary bile acid CA alone[23, 33]. In the colon nearly 100% of bile acids are bacterial derived and a broad spectrum of bacteria can dehydrogenate unconjugated bile acids[23, 34]. In contrast, 7α-dehydroxylation is performed by only a few anaerobic species, representing less than 0.025% of the total gut microbiota and 0.0001% of total colonic microbiota[23, 35, 36]. These are largely represented by Clostridium spp. (C. hiranonis, C. hylemonae, C. sordelli, C. absonum, and C. scindens) and Eubacterium spp., which are all members of the Firmicutes phylum[12, 23, 33, 37–43]. Removal of the 7-α hydroxyl group of primary bile acids requires multiple intracellular enzymatic steps, which are encoded in the bai (bile acid inducible) operon[12, 23, 33, 44–46]. Ultimately these reactions lead to the formation of secondary bile acids, deoxycholate (DCA) from CA and lithocholate (LCA) from CDCA (Figure 1A)[34, 47]. DCA and LCA can be modified further by gut microbes into additional secondary bile acids, such as ursodeoxycholate (UDCA) from LCA (Figure 1A)[47].
The diverse chemical structures of bile acids are a collaborative effort by the host (production of primary bile acids) and the gut microbiota (production of secondary bile acids). The lack of secondary bile acids in a germfree mouse GI tract illustrates the bacterial contribution to bile acid modification[48]. Alterations in bile acid profiles (primary vs. secondary) are also observed in the antibiotic treated mouse gut, which have an altered gut microbial composition[11, 49, 50]. The host and microbial physiologic functions of secondary bile acids remain complex. In the host, secondary bile acids DCA and LCA can be cytotoxic leading to oxidative stress, membrane damage, and colonic carcinogenesis[51]. However, the secondary bile acid UDCA can protect colonic cells against apoptosis and oxidative damage[51]. For gut microbes, secondary bile acids are postulated to have three potential roles: serve as terminal electron acceptors for production of energy, form less hydrophobic membrane damaging bile acids pools, and alter the virulence of enteric pathogens[23, 28]. Secondary bile acids modulate the virulence of the enteric pathogen C. difficile, by inhibiting different stages of its life cycle[11, 23, 28, 52–55]. These examples highlight the diverse and potentially divergent roles of bile acids in relation to host and microbial physiology.
Impact of Secondary Bile Acids on the C. difficile Life Cycle
Bile acids play a dynamic yet critical role in the life cycle of C. difficile. The impact of bile acids on C. difficile dates back to 1982 when Wilson et al. demonstrated that bile acids TCA, desoxycholate or deoxycholate (DCA), and CA stimulated germination of C. difficile spores in vitro[56]. It is well accepted that the primary bile acid TCA triggers C. difficile spore germination and permits outgrowth of vegetative cells, which can culminate in toxin production at high cell densities[53] (Figure 1B). Variations in the efficiency of primary bile acids to stimulate spore germination in vitro are also documented in clinical isolates of C. difficile[57–59].
In Vitro Studies
Despite the ability of CA derived primary bile acids to initiate spore germination, many other bacterial derived secondary bile acids are able to inhibit spore germination (ωMCA, LCA, UDCA) and growth (ωMCA, HDCA, UDCA, LCA, and DCA) of C. difficile in vitro (Figure 1B and Table 1)[11, 12, 50, 53–55, 60–62]. In particular, the secondary bile acids ωMCA, LCA, and UDCA inhibit TCA-mediated spore germination and [50, 54, 55] ωMCA interferes with DCA-mediated spore germination [50]. Growth of C. difficile is altered by most secondary bile acids including ωMCA, HDCA, UDCA, LCA, and DCA [12, 22, 50, 53, 54, 63].
Table 1.
The impact of secondary bile acids on the life cycle of C. difficile using in vitro approaches.
| Strain (ribotype) | Methods to measure spore germination and growth |
Secondary bile acid concentrations |
Main findings of the study | Ref. |
|---|---|---|---|---|
| CD196 UK14 (027) |
|
0.1% DCA |
|
53 |
| 1% DCA |
|
|||
| UK1 (027) |
Relative spore germination determined by drop in OD600 assay in BHIS media |
2 mM UDCA 0.2 mM LCA |
UDCA and LCA can inhibit TCA-mediated spore germination | 55 |
| VPI 10463 (003) 630 (012) |
Relative spore germination determined by drop in OD580 assay in BHIS media |
6 mM 7-keto-LCA | 7-keto-LCA did not induce nor inhibit spore germination | 62 |
| UK1(027) M68 (017) |
|
0.29, 0.2 mM ωMCA |
|
54 |
| 2 mM ωMCA | ||||
| VPI 10463 (003) |
|
0.001, 0.01, 0.1% DCA 0.001, 0.01% LCA |
DCA and LCA inhibited growth in a dose dependent manner | 12 |
| VPI 10463 (003) |
|
*0.001, 0.004% ωMCA *0.001, 0.01% LCA *0.0001, 0.001% HDCA |
|
50 |
|
*0.001, 0.01% HDCA *0.01% UDCA *0.001% LCA *0.1% DCA |
|||
| 10 strains (NAP1/027) |
|
*0.5, 1, 2 mM DCA *0.5, 1, 2 mM LCA 0.5, 1, 2 mM UDCA |
|
22, 63 |
Concentrations based on in vivo targeted bile acid LC-MS assay (see Table 2)
Abbreviations: OD, Optical density; BHIS, Brain heart infusion-supplemented
Bile acid concentrations in the murine gut determined by targeted bile acid liquid chromatography-mass spectrometry (LC-MS) revealed that at physiologic concentrations ωMCA and LCA inhibited TCA-mediated C. difficile spore germination, while HDCA, UDCA, LCA, and DCA decreased C. difficile growth in a dose dependent manner[50]. Additionally, bile acid concentrations in CDI patients’ feces before and after fecal microbiota transplantation (FMT) were tested against ten clinical isolates of C. difficile in vitro[17, 63]. Primary bile acids TCA (0.55 +/− 0.25 mM), CA (1.45 +/−0.29 mM), and CDCA (0.37 +/− 0.09 mM) were detected in the feces prior to FMT[17, 63]. At physiological concentrations, TCA, CA, and CDCA induced germination of C. difficile spores from all clinical isolates in vitro[63]. Following FMT, only secondary bile acids DCA (1.24 +/− 0.24 mM) and LCA (0.95 +/−0.15 mM) were detected in feces[17, 63]. At physiological concentrations, DCA and LCA abated spore germination and growth of C. difficile in 9 out of 10 clinical isolates in vitro[63].
Collectively, these studies emphasize the major impact that secondary bile acids have on the life cycle of C. difficile in vitro. Direct comparison of these studies is challenging since different strains of C. difficile were used (Table 1). Evaluation of additional strains of C. difficile exposed to physiologically relevant concentrations of secondary bile acids is warranted.
Ex vivo Studies
In order to evaluate the impact of secondary bile acids on the life cycle of C. difficile in the presence of the gut microbiota outside of the host, ex vivo models are utilized. Ex vivo studies remove intestinal content from mice at necropsy for use in C. difficile spore germination and outgrowth assays in vitro (Table 2). Multiple studies have shown prior to antibiotic treatment murine ileal content supports C. difficile spore germination ex vivo whereas cecal content inhibits spore germination and outgrowth [11, 50, 64, 65]. After disruption of the gut microbiota with specific antibiotics, cecal content allows for spore germination and outgrowth of C. difficile [11, 50, 64, 65]. Since microbial derived secondary bile acids are predominantly produced in the large intestine, we will focus on this section of the GI tract.
Table 2.
The impact of secondary bile acids on the life cycle of C. difficile using ex vivo and in vivo approaches.
| Host | Antibiotic treatment |
Bile acid analysis | Main findings of the study | Strain (ribotype) |
Ref. |
|---|---|---|---|---|---|
| Ex vivo studies | |||||
| CD-1 female mice |
Clindamycin | Measure NADH during oxidation of hydroxyl groups of bile salts by HSDHs |
Able to stimulate a high level of colony formation from spores in antibiotic treated mouse cecal contents, made up of primary bile acids and a reduction in secondary bile acids. |
CD196 (027) |
64 |
| 5–14 wk C57BL/6 WT male and female mice (colony established from Jax) |
Cefoperazone | Untargeted and targeted bile acid LC- MS assay (limited bile acids library) |
Cecal content of mice after antibiotics had a decrease in secondary bile acid DCA and increased primary bile acids, TCA and CA, and allowed for spore germination and outgrowth and growth of vegetative cells. No spore germination and outgrowth was seen in mouse cecal content prior to antibiotics. |
VPI 10463 (003) BI-9 (027) |
11 |
| See above | Cefoperazone | Targeted bile acid LC-MS assay (expanded bile acid library) |
Cecal content of mice prior to antibiotic treatment contained higher concentrations of secondary bile acids, including DCA, UDCA, LCA and ωMCA. Inhibition of spore germination and outgrowth was seen in cecal content. |
VPI 10463 (003) |
65 |
| See above |
|
Targeted bile acid LC-MS assay (expanded bile acid library) |
Cecal content that provided resistance against spore germination and outgrowth had an average concentration of secondary bile acids: ωMCA 0.004%, HDCA 0.002%, UDCA 0.004%, LCA 0.001%, and DCA 0.023%. Cecal content that allowed for susceptibility to spore germination and outgrowth showed a significant loss in the secondary bile acids listed above and increased TCA. |
VPI 10463 (003) |
50 |
| In vivo studies | |||||
| 5–14 wk C57BL/6 WT male and female mice (colony established from Jax) |
Cefoperazone | Targeted bile acid LC-MS assay (expanded bile acid library) |
Susceptibility to C. difficile colonization in mice was associated with significant changes to the gut metabolome, specifically a decrease in secondary bile acid DCA and an increase in primary bile acid TCA. |
VPI 10463 (003) |
11 |
| Fecal transplant patients and donors |
LC-MS assay | Increased fecal DCA and LCA were associated with recovery from C. difficile infection in post-FMT patients. |
NAP1 (027) |
17 | |
| 6–8 wk C57BL/6J female mice from Jax |
Combination of kanamycin, gentamycin, colistin, metronidazole, vancomycin in followed by single dose of clindamycin |
LC-MS assay |
C. scindens alone and in concert with three other bacteria restored partial colonization resistance against C. difficile in mice. This was associated with restored relative abundance of secondary bile acids DCA and LCA in the cecum and no changes in primary bile acid relative abundance. |
VPI 10463 (003) |
12 |
| CDI Relapse patient |
LC-MS assay | Oral therapy of UDCA prevents relapse of C. difficile infection in a patient with ileal pouchitis (n=1). |
NAP1 (027) |
22 | |
Abbreviations: HSDH: Hydroxysteroid dehydrogenases; LC-MS: Liquid chromatography–mass spectrometry
Giel et al. determined that cecal content from clindamycin treated mice could stimulate some germination and outgrowth of C. difficile CD196 spores ex vivo [64]. Based on an enzymatic assay, they found that the cecal content was dominated by primary bile acids (100 µM)[64]. Cecal content from cefoperazone treated C57BL/6 mice also allowed for spore germination and growth of C. difficile VPI 10463 vegetative cells [11]. Targeted bile acid metabolomics revealed the cecal content had decreased secondary bile acid DCA and increased primary bile acids TCA and CA[11]. Koenigsknecht et al. examined cecal content of C57BL/6 mice using a targeted bile acid LC-MS assay with an extended bile acid library that included 30 unique bile acids[65]. Prior to antibiotic treatment the cecal content of mice was made up of many secondary bile acids including DCA, UDCA, LCA, and ωMCA[65]. Multiple studies suggest that cecal content from mice prior to antibiotic treatment does not support spore germination or outgrowth of C. difficile [11, 65].
Taking it a step further, Theriot et al. 2016 treated groups of mice with a variety of different antibiotics (cefoperazone plus 1–6 weeks recovery off of antibiotic, clindamycin, vancomycin, metronidazole, and kanamycin) to create distinct microbial and metabolic (bile acids) environments. Only specific antibiotic treatments (cefoperazone, clindamycin and vancomycin) allowed for spore germination and outgrowth of C. difficile VPI 10463 in mouse cecal content ex vivo. Cecal contents were associated with significantly more primary bile acid TCA and a loss of all secondary bile acids[50]. Cecal content that did not support C. difficile spore germination and outgrowth was associated with secondary bile acids, such as ωMCA (average concentration 0.004%), HDCA (0.002%), UDCA (0.004%), LCA (0.001%), and DCA (0.023%).
In summary, the ex vivo studies reveal that non-antibiotic treated cecal content with secondary bile acids, specifically ωMCA, HDCA, UDCA, LCA, and DCA conferred resistance to spore germination and outgrowth of C. difficile[11, 50, 64, 65]. Whereas, after specific antibiotic treatment cecal content with low secondary bile acids and high primary bile acids TCA and CA were able to support some stages of the C. difficile life cycle[11, 50]. The alteration of microbial derived secondary bile acids in the GI tract impacts C. difficile spore germination and outgrowth.
In vivo Studies
The impact of secondary bile acids on the life cycle of C. difficile is also evident in vivo (Table 2). Susceptibility of mice to C. difficile colonization after antibiotics is associated with alterations in gut bile acids, specifically a decrease in secondary bile acids and an increase in primary bile acids[11]. The same trend is being seen in patients with recurrent CDI, where high levels of primary bile acids and reduced secondary bile acids were observed in feces when compared to healthy individuals[17, 66]. After successful treatment of CDI with FMT, patients restored the level of fecal secondary bile acids, specifically, DCA and LCA [17]. Weingarden et al. 2014 suggested that FMT restores the gut microbiota, specifically bacteria that are important for conversion of primary bile acids into secondary bile acids[17].
More recently, comparison of the human and murine intestinal microbiota in CDI susceptible and resistant states revealed that the loss of several bacterial taxa was associated with infection[12]. Buffie et al. 2015 used mathematical modeling to demonstrate that C. scindens, a 7α-dehydroxylating gut microbe capable of transforming primary bile acids into secondary bile acids, was associated with resistance to CDI[12]. C57BL/6 female mice treated with an antibiotic cocktail were deemed susceptible to CDI (Table 2). Administration of C. scindens alone or with a consortium of three other bacteria (Barnesiella intestihominis, Pseudoflavonifractor capillosus, Blautia hansenii) in antibiotic treated mice resulted in partial protection against CDI[12]. The observed colonization resistance against C. difficile was associated with restoration of the relative abundance of secondary bile acids DCA and LCA in the cecum[12].
The current literature collectively suggests that bile acids play an important role in the C. difficile life cycle in vitro, ex vivo, and in vivo. Bile acids directly impact C. difficile physiology and thus the pathogenesis. Further studies exploring the dynamics between the gut microbiota and the bile acid metabolome are essential for identifying novel therapeutics against this enteric pathogen.
Antibiotic Mediated Alterations in the Gut Microbiota Alters the Bile Acid Metabolome Contributing to a Loss of Colonization Resistance Against C. difficile
Antibiotics cause collateral damage to the indigenous gut microbiota and loss of colonization resistance against pathogens, such as C. difficile[8, 9, 12, 67]. Susceptibility to CDI after antibiotic treatment in mouse models is associated with a decrease in gut bacterial diversity, an increase in the relative abundance of members from the Proteobacteria phylum and a decrease in the Bacteroidetes phylum [11, 13, 14, 68]. However, it is important to acknowledge that no single gut microbial community permits susceptibility to CDI[67]. Based on the current literature, it is postulated that depletion of specific gut microbes responsible for converting primary bile acids into secondary bile acids reduces colonization resistance against C. difficile.
In 2010, Sorg and Sonenshein first demonstrated inhibition of C. difficile using secondary bile acid producing bacterium C. scindens in vitro [55]. As mentioned previously, Buffie et al. 2015 demonstrated that the presence of C. scindens significantly correlated with resistance to CDI in vivo[12]. C. scindens encodes the bai operon responsible for formation of microbial derived secondary bile acids[69]. Using metagenomic analysis in the antibiotic treated mouse gut, the abundance of the bai operon genes correlated strongly with resistance to CDI, however BSH encoding genes did not. Furthermore, using PCR for baiCD, the gene that specifically encodes the 7-α dehydroxylating enzyme, they established that mice with restored colonization resistance against C. difficile following antibiotic treatment were baiCD+ compared to susceptible mice which lacked this gene[12].
More recently, C57BL/6 mice treated with various antibiotics (detailed in the Ex vivo section), resulted in distinct gut microbial compositions and thus impacted bile acid profiles[50]. Following antibiotic treatment, gut microbial composition analysis revealed that a significant loss of secondary bile acids correlated with a loss of members from the Lachnospiraceae and Ruminococcaceae families. Interestingly, several of these family members are known to be involved in the formation of secondary bile acids[34]. Overall, this study demonstrated that antibiotics induced changes in the gut microbial composition and subsequently modified bile acid profiles. Such alterations had a direct impact on the C. difficile life cycle ex vivo and are regionally specific within the mouse GI tract[50].
The current literature supports the hypothesis that following antibiotics, alterations in gut microbial composition and a subsequent alteration in the bile acid metabolome result in a loss of colonization resistance against C. difficile[11, 12, 50]. Restoration of gut microbes that possess the ability to modulate intestinal bile acid profiles, specifically via production of secondary bile acids, may prove beneficial in the treatment of CDI.
Manipulation of Microbial Derived Secondary Bile Acids to Restore Colonization Resistance Against C. difficile
Studies showing the contribution of secondary bile acids to colonization resistance against C. difficile are increasing. However, evidence of administering bile acids or bile acid modifying bacteria to manipulate bile acid profiles against this enteric pathogen is limited[12, 22, 70]. In a single case report, daily UDCA administration successfully eliminated and prevented recurrence of C. difficile ileal pouchitis [22]. The C. difficile strain from the patient was isolated and UDCA was able to inhibit spore germination and vegetative growth in vitro (Table 1)[22]. Others have administered bacteria to restore colonization against C. difficile in humans and in antibiotic treated and germfree mice, however the impact of these microbes on bile acids was not investigated[71–76].
Conclusions
The necessity of novel therapeutics against C. difficile is evident. The dynamic and pivotal role bile acids play in the C. difficile life cycle creates a potential target for such therapeutics. Although the exact mechanisms of colonization resistance are unknown, current literature suggests that microbial derived secondary bile acids could play an important role. Studies evaluating the rational manipulation of bile acid pools by either administration of bile acids directly or by administering bile acid modifying bacteria are needed. Such orchestration of collaborative bile acid metabolism may provide an innovative therapeutic strategy against C. difficile infection. Additional studies investigating the interplay between C. difficile, bile acids, the gut microbiota, and the host are essential for understanding the complexity of colonization resistance. Such information may also prove beneficial in other disease processes displaying bile acid dysmetabolism such as metabolic disease, obesity, and inflammatory bowel disease[77].
Highlights.
The chemical diversification of bile acids is a collaborative effort by the host (production of primary bile acids) and the gut microbiota (production of secondary bile acids).
Bile acids play an important and dynamic role in the C. difficile life cycle and this can be seen with in vitro, ex vivo, and in vivo approaches.
Alterations in the gut microbiota that result in a loss of secondary bile acids are associated with a loss of colonization resistance against C. difficile.
Acknowledgments
JAW is funded by Ruth L. Kirschstein National Research Service Award Research Training grant T32OD011130 by NIH. CMT is funded by a career development award in metabolomics grant K01GM109236 by the NIGMS of the NIH. The manuscript content is solely the responsibility of the authors and is not necessarily a reflection of the NIH.
Abbreviations
- GI
Gastrointestinal
- CDI
Clostridium difficile infection
- LC-MS
Liquid chromatography mass spectrometry
- CA
cholate
- CDCA
chenodeoxycholate
- DCA
deoxycholate
- HCA
hyocholate
- HDCA
hyodeoxycholate
- LCA
lithocholate
- MDCA
murideoxycholate
- UDCA
ursodeoxycholate
- αMCA
α-muricholate
- βMCA
β-muricholate
- ωMCA
ω-muricholate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hall IC, O’Toole E. Intestinal flora in new-born infants: With a description of a new pathogenic anaerobe, bacillus difficilis. American Journal of Diseases of Children. 1935;49:390–402. [Google Scholar]
- 2.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile Infection in the United States. New England Journal of Medicine. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerding DN, Lessa FC. The epidemiology of Clostridium difficile infection inside and outside health care institutions. Infect Dis Clin North Am. 2015;29:37–50. doi: 10.1016/j.idc.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubberke ER, Olsen MA. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis 55 Suppl. 2012;2:S88–S92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.U.S.D.o.H.a.H. Services. Antibiotic Resistance Threats in the United States. Centers for Disease Control and Prevention; 2013. [online] [Google Scholar]
- 6.Tartof SY, Rieg GK, Wei R, Tseng HF, Jacobsen SJ, Yu KC. A Comprehensive Assessment Across the Healthcare Continuum: Risk of Hospital-Associated Clostridium difficile Infection Due to Outpatient and Inpatient Antibiotic Exposure. Infect Control Hosp Epidemiol. 2015;36:1409–1416. doi: 10.1017/ice.2015.220. [DOI] [PubMed] [Google Scholar]
- 7.Abou Chakra CN, Pepin J, Sirard S, Valiquette L. Risk factors for recurrence, complications and mortality in Clostridium difficile infection: a systematic review. PLoS One. 2014;9:e98400. doi: 10.1371/journal.pone.0098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antunes LC, Han J, Ferreira RB, Lolic P, Borchers CH, Finlay BB. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes. 2014;2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–414. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorg JA. Microbial bile acid metabolic clusters: the bouncers at the bar. Cell Host Microbe. 2014;16:551–552. doi: 10.1016/j.chom.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. American journal of physiology Gastrointestinal and liver physiology. 2014;306:G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kociolek LK, Gerding DN. Breakthroughs in the treatment and prevention of Clostridium difficile infection. Nat Rev Gastroenterol Hepatol. 2016 doi: 10.1038/nrgastro.2015.220. [DOI] [PubMed] [Google Scholar]
- 19.Kelly CP, LaMont JT. Clostridium difficile--more difficult than ever. N Engl J Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 20.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, et al. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med. 2011;364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 21.Theriot CM, Schumacher CA, Bassis CM, Seekatz AM, Young VB. Effects of tigecycline and vancomycin administration on established Clostridium difficile infection. Antimicrob Agents Chemother. 2015;59:1596–1604. doi: 10.1128/AAC.04296-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weingarden AR, Chen C, Zhang N, Graiziger CT, Dosa PI, Steer CJ, et al. Ursodeoxycholic Acid Inhibits Clostridium difficile Spore Germination and Vegetative Growth, and Prevents the Recurrence of Ileal Pouchitis Associated With the Infection. J Clin Gastroenterol. 2015 doi: 10.1097/MCG.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res. 2010;51:226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annual review of biochemistry. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 26.Alnouti Y, Csanaky IL, Klaassen CD. Quantitative-profiling of bile acids and their conjugates in mouse liver, bile, plasma, and urine using LC-MS/MS. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2008;873:209–217. doi: 10.1016/j.jchromb.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann AF. The enterohepatic circulation of bile acids in mammals: form and functions. Front Biosci (Landmark Ed) 2009;14:2584–2598. doi: 10.2741/3399. [DOI] [PubMed] [Google Scholar]
- 28.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS microbiology reviews. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Setchell KD, Lawson AM, Tanida N, Sjovall J. General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. J Lipid Res. 1983;24:1085–1100. [PubMed] [Google Scholar]
- 31.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72:1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 2013;4:382–387. doi: 10.4161/gmic.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol. 2015;11:685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridlon JM, Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharmaceutica Sinica B. 2015;5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Gut microbiota, cirrhosis, and alcohol regulate bile acid metabolism in the gut. Dig Dis. 2015;33:338–345. doi: 10.1159/000371678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7alpha-dehydroxylating bacteria in human feces. Clinica chimica acta; international journal of clinical chemistry. 2003;331:127–134. doi: 10.1016/s0009-8981(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 38.Kitahara M, Takamine F, Imamura T, Benno Y. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. International journal of systematic and evolutionary microbiology. 2001;51:39–44. doi: 10.1099/00207713-51-1-39. [DOI] [PubMed] [Google Scholar]
- 39.Kitahara M, Takamine F, Imamura T, Benno Y. Assignment of Eubacterium sp. VPI 12708 related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens, proposal of Clostridium hylemonae sp. nov., isolated from human faeces. International journal of systematic and evolutionary microbiology 50 Pt. 2000;3:971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- 40.Ridlon JM, Kang DJ, Hylemon PB. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe. 2010;16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macdonald IA, White BA, Hylemon PB. Separation of 7 alpha- and 7 beta-hydroxysteroid dehydrogenase activities from Clostridium absonum ATCC# 27555 and cellular response of this organism to bile acid inducers. J Lipid Res. 1983;24:1119–1126. [PubMed] [Google Scholar]
- 42.Dawson JA, Mallonee DH, Bjorkhem I, Hylemon PB. Expression characterization of a C24 bile acid 7 alpha-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J Lipid Res. 1996;37:1258–1267. [PubMed] [Google Scholar]
- 43.Mallonee DH, Hylemon PB. Sequencing expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J Bacteriol. 1996;178:7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fiorucci S, Distrutti E. Bile Acid-Activated Receptors, Intestinal Microbiota, and the Treatment of Metabolic Disorders. Trends Mol Med. 2015 doi: 10.1016/j.molmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doerner KC, Takamine F, LaVoie CP, Mallonee DH, Hylemon PB. Assessment of fecal bacteria with bile acid 7 alpha-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol. 1997;63:1185–1188. doi: 10.1128/aem.63.3.1185-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap--bile acids in metabolic control. Nat Rev Endocrinol. 2014;10:488–498. doi: 10.1038/nrendo.2014.60. [DOI] [PubMed] [Google Scholar]
- 48.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Limaye PB, Renaud HJ, Klaassen CD. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol. 2014;277:138–145. doi: 10.1016/j.taap.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theriot C, Bowman A, Young V. Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere. 2016;1:00045–00015. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrasa JI, Olmo N, Lizarbe MA, Turnay J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol In Vitro. 2013;27:964–977. doi: 10.1016/j.tiv.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 52.Shen A. A Gut Odyssey: The Impact of the Microbiota on Clostridium difficile Spore Formation and Germination. PLoS Pathog. 2015;11:e1005157. doi: 10.1371/journal.ppat.1005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Francis MB, Allen CA, Sorg JA. Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One. 2013;8:e73653. doi: 10.1371/journal.pone.0073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 2010;192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on on a medium selective for Clostridium difficile. J Clin Microbiol. 1982;15:443–446. doi: 10.1128/jcm.15.3.443-446.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carlson PE, Jr, Kaiser AM, McColm SA, Bauer JM, Young VB, Aronoff DM, et al. Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe. 2015;33:64–70. doi: 10.1016/j.anaerobe.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heeg D, Burns DA, Cartman ST, Minton NP. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One. 2012;7:e32381. doi: 10.1371/journal.pone.0032381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharjee D, Francis MB, Ding X, McAllister KN, Shrestha R, Sorg JA. Reexamining the Germination Phenotypes of Several Clostridium difficile Strains Suggests Another Role for the CspC Germinant Receptor. J Bacteriol. 2015;198:777–786. doi: 10.1128/JB.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Howerton A, Ramirez N, Abel-Santos E. Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol. 2011;193:274–282. doi: 10.1128/JB.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramirez N, Liggins M, Abel-Santos E. Kinetic evidence for the presence of putative germination receptors in Clostridium difficile spores. J Bacteriol. 2010;192:4215–4222. doi: 10.1128/JB.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weingarden AR, Dosa PI, DeWinter E, Steer CJ, Shaughnessy MK, Johnson JR, et al. Changes in Colonic Bile Acid Composition following Fecal Microbiota Transplantation Are Sufficient to Control Clostridium difficile Germination and Growth. PLoS One. 2016;11:e0147210. doi: 10.1371/journal.pone.0147210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giel JL, Sorg JA, Sonenshein AL, Zhu J. Metabolism of Bile Salts in Mice Influences Spore Germination in Clostridium difficile . PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun. 2015;83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, et al. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther. 2016 doi: 10.1111/apt.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Theriot CM, Young VB. Interactions Between the Gastrointestinal Microbiome and Clostridium difficile . Annu Rev Microbiol. 2015;69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bassis CM, Theriot CM, Young VB. Alteration of the murine gastrointestinal microbiota by tigecycline leads to increased susceptibility to Clostridium difficile infection. Antimicrob Agents Chemother. 2014;58:2767–2774. doi: 10.1128/AAC.02262-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang DJ, Ridlon JM, Moore DR, 2nd, Barnes S, Hylemon PB. Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochimica et biophysica acta. 2008;1781:16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howerton A, Patra M, Abel-Santos E. A new strategy for the prevention of Clostridium difficile infection. The Journal of infectious diseases. 2013;207:1498–1504. doi: 10.1093/infdis/jit068. [DOI] [PubMed] [Google Scholar]
- 71.Nagaro KJ, Phillips ST, Cheknis AK, Sambol SP, Zukowski WE, Johnson S, et al. Nontoxigenic Clostridium difficile protects hamsters against challenge with historic epidemic strains of toxigenic BI/NAP1/027 C. difficile . Antimicrob Agents Chemother. 2013;57:5266–5270. doi: 10.1128/AAC.00580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corthier G, Dubos F, Raibaud P. Modulation of cytotoxin production by Clostridium difficile in the intestinal tracts of gnotobiotic mice inoculated with various human intestinal bacteria. Appl Environ Microbiol. 1985;49:250–252. doi: 10.1128/aem.49.1.250-252.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun. 2012;80:3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet (London, England) 1989;1:1156–1160. doi: 10.1016/s0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 77.Joyce SA, Gahan CG. Bile Acid Modifications at the Microbe-Host Interface: Potential for Nutraceutical and Pharmaceutical Interventions in Host Health. Annual review of food science and technology. 2016 doi: 10.1146/annurev-food-041715-033159. [DOI] [PubMed] [Google Scholar]