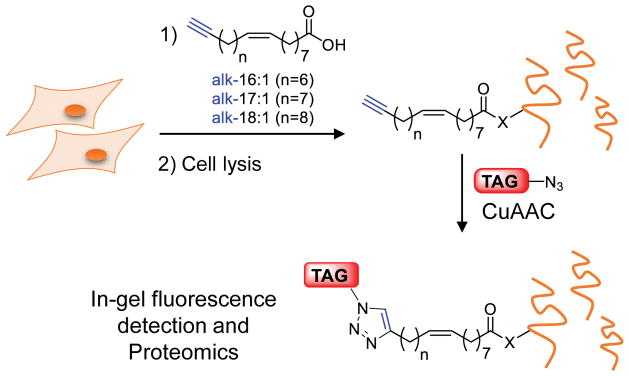
Abstract
Dietary unsaturated fatty acids, such as oleic acid, have been shown to be covalently incorporated into a small subset of proteins but the generality and diversity of this protein modification has not been studied. We synthesized unsaturated fatty acid chemical reporters and determined their protein targets in mammalian cells. The unsaturated fatty acid chemical reporters can induce the formation of lipid droplets and be incorporated site-specifically onto known fatty-acylated proteins and label many proteins in mammalian cells. Quantitative proteomics analysis revealed that unsaturated fatty acids modify similar protein targets to saturated fatty acids, including several immunity-associated proteins, which demonstrates unsaturated fatty acids may directly modify many proteins to exert their unique and often beneficial physiological effects in vivo.
Keywords: Fatty-acylation, unsaturated fatty acids, chemical reporter, quantitative proteomics
Graphical abstract
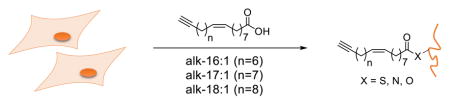
Unsaturated fatty acid chemical reporters are incorporated site-specifically onto known fatty-acylated proteins and label many proteins in mammalian cells. Quantitative proteomics analysis revealed that unsaturated fatty acids modify similar protein targets to saturated fatty acids
Protein fatty-acylation, such as S-palmitoylation, N-myristoylation, O-fatty acylation and lysine (Lys) side-chain fatty-acylation, are important posttranslational modifications that regulate the trafficking and function of membrane-associated proteins in eukaryotes.[1,2] Recent proteomic methods have revealed a greater diversity and number of fatty-acylated proteins, and highlighted new functions and regulatory mechanisms for these lipid-modified proteins.[2–4] Several methods have been developed to visualize and capture fatty-acylated proteins. One approach builds upon traditional radioactive fatty acids, and employs alkyne-tagged fatty acids for metabolic feeding followed by bioorthogonal labeling of cell lysates with azide-fluorophores or affinity tags for detection or enrichment, respectively. This method has been widely used to characterize N-myristoylated,[5–8] S-palmitoylated,[5,9–11] O-palmitoylated,[12] Lys-fatty-acylated proteins,[13,14] as well as GPI-anchored proteins[6] in a wide range of cell lines and organisms depending on the fatty acid reporter chain length.[4] For example, alkyne-fatty acid reporters alk-12 (C12:0) preferentially labels N-myristoylated proteins,[5–7] while alk-16 (C16:0) labels S-palmitoylated proteins as well as N/O-fatty-acylated proteins.[5,9–11]
Despite these advances in chemical proteomics, the covalent addition of unsaturated fatty acids to proteins has not been systematically explored. The composition of fatty acids, saturated versus unsaturated, can significantly affect the behavior of cells and yield drastically different physiological responses in vivo.[15] For example, high levels of saturated fatty acids are associated with inflammatory diseases and metabolic syndrome,[15] while unsaturated fatty acids often yield protective effects in vivo.[16] Since unsaturated fatty acids have also been reported to covalently modify a few proteins in mammalian cells such as Fyn, Gα-protein, GAP43, Ras,[17–19] Wnt[20] and others,[19,21–23] dietary unsaturated fatty acids may also be broadly incorporated into many proteins. To investigate the covalent modification of proteins with unsaturated fatty acids, we synthesized chemical reporters of monounsaturated fatty acids that function similarly to oleic acid in mammalian cells and demonstrate these monounsaturated fatty acid reporters can label diverse proteins in mammalian cells, which may directly affect the function of fatty-acylated proteins.
The synthesis of cis-unsaturated alkynyl-fatty acids can be achieved by Wittig coupling of the phosphoylide and alkynyl-aldehydes of varying chain length (SI Figure S1). We envisioned that the exact chain length (C18, alk-16:1) would be a reasonable mimic of oleic acid in cells. Nevertheless, we also synthesized a previously reported analog of oleic acid, alk-17:1,[24] as well as an analog bearing two additional carbons (alk-18:1) (Scheme 1). Within cells, oleic acid was shown to induce lipid droplets.[25] Analysis of the formation of lipid droplets with Nile Red (lipid droplet-selective dye) staining in LPS/IFN-γ stimulated Raw264.7 macrophages demonstrates that the unsaturated fatty acid reporters, but not alk-16, can generate lipid droplets (Figure 1, SI Figure S2). These observations are consistent with previous studies that demonstrate that alk-17:1 is primarily incorporated into triacylglycerides (TAGs), which are the main components of lipid droplets in mammalian cells.[24] Interestingly, only alk-16:1 could induce comparable levels of lipids droplets at similar concentrations to oleic acid, suggesting that this unsaturated fatty acid reporter is a better mimic of the natural lipid substrate. Induction of lipid droplets in NIH/3T3 fibroblast gave comparable results (SI Figure S3).
Scheme 1.
Chemical reporter strategy for functional analysis of protein modified by unsaturated fatty acids. Alkyne-tagged reporters of unsaturated fatty acids, alk-16:1, alk-17:1 and alk-18:1 can metabolically label proteins in live cells. Following cell lysis, labelled proteins can be captured by an azido-functionalized reagent via CuAAC for fluorescence detection or enrichment for proteomics analysis.
Figure 1.
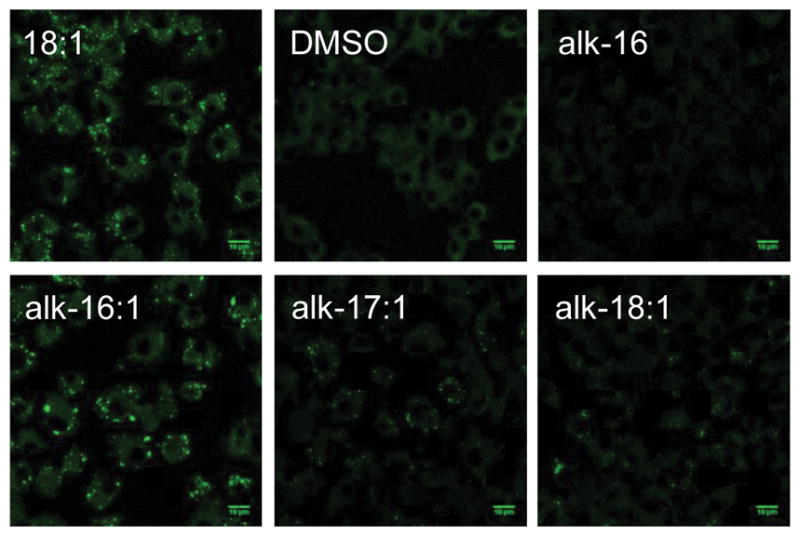
(A) Alkyne-tagged reporters of unsaturated fatty acids, alk-16:1, alk-17:1 and alk-18:1 (B) Analysis of lipid droplet induction by Nile red staining. RAW264.7, stimulated for 8 h with LPS/IFN-γ, were treated with DMSO, oleic acid (18:1), alk-16:1, alk-17:1 and alk-18:1 (200 μM) before being fixed with formaldehyde, stained with Nile Red and analyzed by confocal fluorescence microscopy. Cells are present in alk-16 samples, as judged by bright field images, but no Nile Red staining was observed.
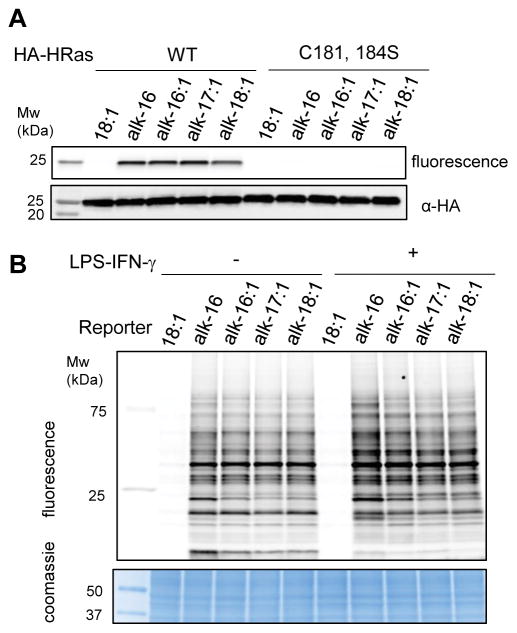
Next, we evaluated if the unsaturated fatty acid reporters could label HRas, a S-palmitoylated protein[26] which has been shown to be modified with oleic acid.[19] HeLa cells were transfected with wild-type (wt) HA-HRas, labeled with the fatty acid reporters, immunoprecipitated, reacted with azido-rhodamine, analyzed by in-gel fluorescence and western blot (Figure 2A). Unsaturated fatty acid reporters can specifically label HRas at comparable level to alk-16 (Figure 3A). Since oleic acid had been shown to modify S-palmitoylated Cys residues of HRas,[19] we also evaluated Cys181,184Ser mutants of HRas. No fluorescence labeling was observed with either fatty acid reporters, indicating that the unsaturated fatty acid reporters are specifically incorporated into key S-fatty acylation sites of HRas.
Figure 2.
(A) HeLa were transfected with HA-tagged HRas (wt and C181,184S mutant) for 24h. 4h before lysis, cells were treated with alkyne-tagged fatty acid, immunoprecipitated, and reacted with az-rhodamine. Chemical reporters (alk-16:1, alk-17:1, alk-18:1) label HA-HRas on key Cys residues. Anti-HA western blot is included as a loading control. (B) In-gel fluorescence analysis of −/+ LPS/IFN-γ stimulated Raw264.7 treated with oleic acid (18:1), alk-16, alk-16:1, alk-17:1, alk-18:1 (50 μM, 4 h). Coomassie staining was included as a loading control.
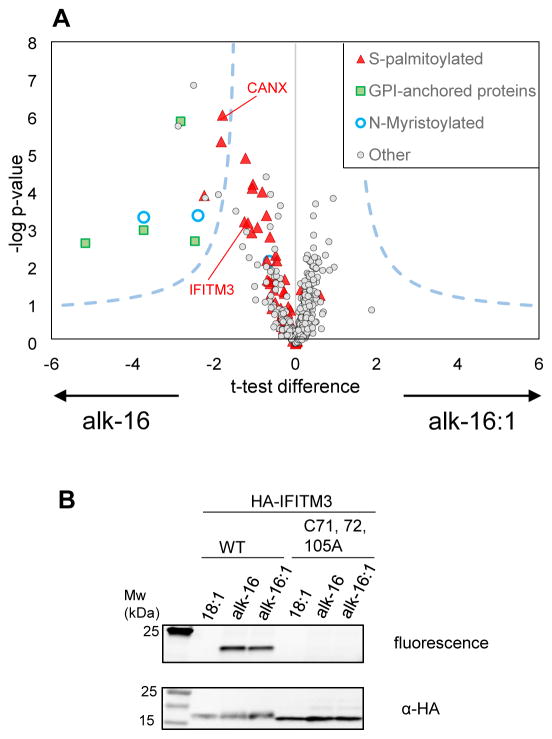
Figure 3.
(A) Volcano plot representing results of the label-free quantitative proteomics analysis of proteins labeled by alk-16 and alk-16:1 in LPS/IFN-γ stimulated Raw264.7 (n=4 biological replicates). Dashed lines represent the t-test significance cut-off (Benjamini–Hochberg FDR 0.05, s0 of 1). The majority of proteins (light grey dots) are not differentially enriched by one of the reporter. 13 proteins were significantly enriched in the alk-16 sample, including proteins known to be N-myristoylated (blue), S-palmitoylated (red) or have a GPI-anchor (green). S-palmitoylated only refers to proteins reported as S-palmitoylated in Uniprot or have been shown to be S-palmitoylated using targeted studies.[27] Only proteins identified with medium or high confidence (significantly enriched over background (oleic acid treated cells)) are shown. Proteins more enriched in the alk-16 or alk-16:1 treated samples have a negative (left) or positive (right) t-test difference respectively. (B) Validation of IFITM3 labeling by the alkynyl-reporters of unsaturated fatty acid alk-16:1. HeLa cells were transfected with HA-IFITM3, labeled with alk-16 or alk-16:1, lysed, immunoprecipitated and reacted with azido-rhodamine.
We then evaluated the scope of protein labeling with the unsaturated fatty acid reporters. Protein labeling with alk-16:1 in Raw264.7 macrophages occurred in a time- and dose-dependent manner (SI Figure S4A). Importantly, fluorescence labeling of proteins with alk-16:1, alk17:1 and alk18:1 decreases with increasing amounts of oleic acid, suggesting that the reporters mimic the natural lipid substrate (SI Figure S4B). Interestingly, alk16:1 labeling can also be competed with palmitic acid, but alk-16 labeling can only be competed with palmitic acid and not with oleic acid (SI Figure S4C and 4D). Labeling with alk-16:1, alk-17:1 and alk-18:1 appeared to be partially NH2OH-sensitive, suggesting that, similarly to alk-16, these unsaturated fatty acid reporters can be attached to Cys residues (SI Figure S4E). In-gel fluorescence analysis of proteins labeled with the unsaturated (alk-16:1, alk-17:1, alk-18:1) and saturated (alk-16, a palmitic acid chemical reporter) fatty acid reporters demonstrates that the unsaturated fatty acid chemical reporters covalently label many proteins (Figure 2B). The in-gel fluorescence protein labeling patterns are highly similar between the fatty acid reporters in different cell types (SI Figure S5), suggesting that these saturated and unsaturated fatty acids share many protein targets.
To identify the proteins modified by unsaturated fatty acids in mammalian cells, we performed LFQ proteomics (n= 4 biological replicates) in activated Raw264.7 macrophages (SI Table S1). Cells were stimulated with LPS/IFN-γ for 12 h and treated with oleic acid, alk-16, alk-16:1, alk17:1 or alk18:1 for 8h, similarly to the conditions used for the induction of lipid droplets. The data was highly reproducible between replicates (SI Figure S6). Following statistical analysis to identify only proteins labeled over background (oleic acid) (two sample test, permutation-based, 250 permutations, FDR = 0.001 and s0 value = 2), the data was further classified in high, medium and low confidence based on peptide LFQ and spectra counts (SI methods and SI Table S4). Out of the 347 proteins identified with high/medium confidence in the alk-16 labeled sample, 270 proteins had been identified in previous proteomics datasets.[27] Interestingly, alk-16, alk-16:1, alk17:1 and alk18:1 shared many proteins substrates (SI Figure S7).
To identify if proteins were differentially labeled by alk-16 or alk-16:1, we performed a permutation-based t-test (250 permutations, FDR 0.05, s0 of 1) (Figure 3A). Out of the >1000 proteins identified in both samples, 13 proteins were significantly more enriched in the alk-16 sample, including CANX[27] and proteins known to be N-myristoylated,[7] or GPI-anchored. The analysis of this proteomic dataset for known S-palmitoylated proteins, suggests that S-palmitoylated proteins are preferentially labeled by saturated fatty acids (Figure 3A). Gene ontology annotation of proteins identified by proteomics (SI Table 1) showed that several proteins were involved in the immune response, including IFITM3, a known S-palmitoylated proteins key in the host response to virus infection,[28,29] which antiviral activity may be modulated by oleic acid.[30] To confirm that IFITM3 could be modified by oleic acid, cells were transfected with wt and palmitoylation deficient mutant (C71,72,105A) HA-IFITM3. HA-IFITM3 can be labelled by both alk-16 and alk16:1 specifically on Cys residues (Figure 3B). Similar results were obtained with alk-17:1 and alk-18:1 (SI Figure S8). Taken together, these results demonstrate that many proteins can be modified with unsaturated fatty acids in mammalian cells treated with high levels of unsaturated fatty acid.
Conclusions
Protein fatty-acylation is prominent and can occur in different forms to regulate cellular functions. We have described an improved oleic acid reporter (alk-16:1) and demonstrate unsaturated fatty acid reporters can site-specifically label known S-fatty-acylated proteins in mammalian cells. Quantitative analysis of the fatty acid reporter-modified proteins showed that S-fatty-acylated proteins are preferentially labeled by saturated fatty acids, suggesting the enzymes that regulate these dynamically lipid-modified proteins prefer saturated fatty acid substrates in cells. Nonetheless, our studies demonstrate unsaturated fatty acids can label many proteins in mammalian cells, suggesting these dietary fatty acids may directly regulate protein function by covalent modification when incorporated at high levels.
Acknowledgments
We thank Hang lab members for helpful discussions and Milica Tesic Mark, Brian Dill, Joseph Fernandez and Henrik Molina at the Proteomics Resource Center of The Rockefeller University for mass spectrometry analysis. This work was supported by a Marie Skłodowska-Curie Individual Fellowship to E.T and National Science Foundation Graduate fellowship to A.P. H.C.H. acknowledges grant support from NIH-NIGMS R01GM087544 and Starr Cancer Consortium I7-A717.
Footnotes
Supporting information for this article is given via a link at the end of the document. Proteomics mass spectrometry datasets have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository, with the dataset identifier PXD003545.
References
- 1.Chamberlain LH, Shipston MJ. Physiol Rev. 2015;95:341–76. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thinon E, Hang HC. Biochem Soc Trans. 2015;43:253–61. doi: 10.1042/BST20150004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernandez JL, Majmudar JD, Martin BR. Curr Opin Chem Biol. 2013;17:20–6. doi: 10.1016/j.cbpa.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng T, Thinon E, Hang HC. Curr Opin Chem Biol. 2016;30:77–86. doi: 10.1016/j.cbpa.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 6.Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, Moss DK, Bottrill AR, Heal WP, Broncel M, et al. Nat Chem. 2014;6:112–21. doi: 10.1038/nchem.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thinon E, Serwa RA, Broncel M, Brannigan JA, Brassat U, Wright MH, Heal WP, Wilkinson AJ, Mann DJ, Tate EW. Nat Commun. 2014;5:4919. doi: 10.1038/ncomms5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin DD, Vilas GL, Prescher JA, Rajaiah G, Falck JR, Bertozzi CR, Berthiaume LG. FASEB J. 2008;22:797–806. doi: 10.1096/fj.07-9198com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin BR, Cravatt BF. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannoush RN, Arenas-Ramirez N. ACS Chem Biol. 2009;4:581–587. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 12.Gao X, Hannoush RN. Nat Chem Biol. 2014;10:61–8. doi: 10.1038/nchembio.1392. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Yang T, Li X, Peng T, Hang HC, Li XD. Angew Chem Int Ed. 2014 doi: 10.1002/anie.201408763R1. in press. [DOI] [Google Scholar]
- 15.Stienstra R, Tack CJ, Kanneganti TD, Joosten LAB, Netea MG. Cell Metab. 2012;15:10–8. doi: 10.1016/j.cmet.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 16.Delmastro-Greenwood M, Freeman BA, Wendell SG. Annu Rev Physiol. 2014;76:79–105. doi: 10.1146/annurev-physiol-021113-170341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang X, Lu Y, Wilkes M, Neubert TA, Resh MD. J Biol Chem. 2004;279:8133–9. doi: 10.1074/jbc.M311180200. [DOI] [PubMed] [Google Scholar]
- 18.Liang X, Lu Y, Neubert TA, Resh MD. J Biol Chem. 2002;277:33032–40. doi: 10.1074/jbc.M204607200. [DOI] [PubMed] [Google Scholar]
- 19.Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. J Biol Chem. 2001;276:30987–94. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- 20.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, Takao T, Takada S. Dev Cell. 2006;11:791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Montigny C, Decottignies P, Le Maréchal P, Capy P, Bublitz M, Olesen C, Møller JV, Nissen P, le Maire M. J Biol Chem. 2014;289:33850–61. doi: 10.1074/jbc.M114.590307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schey KL, Gutierrez DB, Wang Z, Wei J, Grey AC. Biochemistry. 2010;49:9858–65. doi: 10.1021/bi101415w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bereta G, Palczewski K. Biochemistry. 2011;50:3764–76. doi: 10.1021/bi200245t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiele C, Papan C, Hoelper D, Kusserow K, Gaebler A, Schoene M, Piotrowitz K, Lohmann D, Spandl J, Stevanovic A, et al. ACS Chem Biol. 2012;7:2004–2011. doi: 10.1021/cb300414v. [DOI] [PubMed] [Google Scholar]
- 25.Pol A, Gross SP, Parton RG. J Cell Biol. 2014;204:635–46. doi: 10.1083/jcb.201311051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS, Hang HC. Proc Natl Acad Sci. 2016;113:4302–4307. doi: 10.1073/pnas.1602244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanc M, David F, Abrami L, Migliozzi D, Armand F, Bürgi J, van der Goot FG. F1000Research. 2015;4:261. doi: 10.12688/f1000research.6464.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, López CB, Hang HC. Nat Chem Biol. 2010;6:610–4. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yount JS, Karssemeijer RA, Hang HC. J Biol Chem. 2012;287:19631–41. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. PLoS Pathog. 2014;10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]