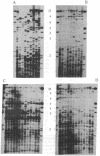
Abstract
The Puerto Rican parrot was reduced to approximately 13 animals in 1975 and as a conservation measure, a captive population was established from a few founders taken from the wild between 1973 and 1983. The number of successful breeding pairs in captivity has been low, and the captive breeding program has not been as productive as that of the closely related Hispaniolan parrot. Therefore, a genetic study was initiated to examine the relative levels of relatedness of the captive founders using levels of bandsharing in DNA fingerprints. Unrelated captive founder Puerto Rican parrots had the same average level of bandsharing (0.41) as second-degree relatives of the Hispaniolan parrot (0.38, P > 0.05), with an inbreeding coefficient of 0.04. High levels of bandsharing (> 40%) between pairs of males and females correlated with reproductive failure, suggesting that inbreeding depression is partly responsible for the low number of breeding pairs. Consequently, DNA profiling can be used to guide the captive breeding program for the Puerto Rican parrot, and other endangered species, by identifying pairs of males and females with low levels of bandsharing.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Flint J., Boyce A. J., Martinson J. J., Clegg J. B. Population bottlenecks in Polynesia revealed by minisatellites. Hum Genet. 1989 Oct;83(3):257–263. doi: 10.1007/BF00285167. [DOI] [PubMed] [Google Scholar]
- Galbraith D. A., Boag P. T., Gibbs H. L., White B. N. Sizing bands on autoradiograms: a study of precision for scoring DNA fingerprints. Electrophoresis. 1991 Feb-Mar;12(2-3):210–220. doi: 10.1002/elps.1150120218. [DOI] [PubMed] [Google Scholar]
- Gilbert D. A., Lehman N., O'Brien S. J., Wayne R. K. Genetic fingerprinting reflects population differentiation in the California Channel Island fox. Nature. 1990 Apr 19;344(6268):764–767. doi: 10.1038/344764a0. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Jakobsson S., Temrin H. No evidence for illegitimate young in monogamous and polygynous warblers. Nature. 1990 Jan 11;343(6254):168–170. doi: 10.1038/343168a0. [DOI] [PubMed] [Google Scholar]
- Hillel J., Plotzy Y., Haberfeld A., Lavi U., Cahaner A., Jeffreys A. J. DNA fingerprints of poultry. Anim Genet. 1989;20(2):145–155. doi: 10.1111/j.1365-2052.1989.tb00852.x. [DOI] [PubMed] [Google Scholar]
- Honma M., Ishiyama I. Application of DNA fingerprinting to parentage and extended family relationship testing. Hum Hered. 1990;40(6):356–362. doi: 10.1159/000153959. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Morton D. B. DNA fingerprints of dogs and cats. Anim Genet. 1987;18(1):1–15. doi: 10.1111/j.1365-2052.1987.tb00739.x. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Turner M., Debenham P. The efficiency of multilocus DNA fingerprint probes for individualization and establishment of family relationships, determined from extensive casework. Am J Hum Genet. 1991 May;48(5):824–840. [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Individual-specific 'fingerprints' of human DNA. Nature. 1985 Jul 4;316(6023):76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L., Weatherall D. J., Ponder B. A. DNA "fingerprints" and segregation analysis of multiple markers in human pedigrees. Am J Hum Genet. 1986 Jul;39(1):11–24. [PMC free article] [PubMed] [Google Scholar]
- Kuhnlein U., Zadworny D., Dawe Y., Fairfull R. W., Gavora J. S. Assessment of inbreeding by DNA fingerprinting: development of a calibration curve using defined strains of chickens. Genetics. 1990 May;125(1):161–165. doi: 10.1093/genetics/125.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Estimation of relatedness by DNA fingerprinting. Mol Biol Evol. 1988 Sep;5(5):584–599. doi: 10.1093/oxfordjournals.molbev.a040518. [DOI] [PubMed] [Google Scholar]
- Lynch M. The similarity index and DNA fingerprinting. Mol Biol Evol. 1990 Sep;7(5):478–484. doi: 10.1093/oxfordjournals.molbev.a040620. [DOI] [PubMed] [Google Scholar]
- Reeve H. K., Westneat D. F., Noon W. A., Sherman P. W., Aquadro C. F. DNA "fingerprinting" reveals high levels of inbreeding in colonies of the eusocial naked mole-rat. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2496–2500. doi: 10.1073/pnas.87.7.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. S., Bargiello T. A., Clark B. T., Jackson F. R., Young M. W. An unusual coding sequence from a Drosophila clock gene is conserved in vertebrates. Nature. 1985 Oct 3;317(6036):445–448. doi: 10.1038/317445a0. [DOI] [PubMed] [Google Scholar]
- Wetton J. H., Carter R. E., Parkin D. T., Walters D. Demographic study of a wild house sparrow population by DNA fingerprinting. Nature. 1987 May 14;327(6118):147–149. doi: 10.1038/327147a0. [DOI] [PubMed] [Google Scholar]