Abstract
Sugars fuel life and exert numerous regulatory actions that are fundamental to all life forms. There are two principal mechanisms underlie sugar “perception and signal transduction” in biological systems. Direct sensing and signaling is triggered via sugar-binding sensors with a broad range of affinity and specificity, whereas sugar-derived bioenergetic molecules and metabolites modulate signaling proteins and indirectly relay sugar signals. This review discusses the emerging sugar signals and potential sugar sensors discovered in plant systems. The findings leading to informative understanding of physiological regulation by sugars are considered and assessed. Comparative transcriptome analyses highlight the primary and dynamic sugar responses and reveal the convergent and specific regulators of key biological processes in the sugar-signaling network.
Introduction
Sugars produced from plant photosynthesis play a central role to support and integrate the functions and actions of internal and external regulatory signals in driving diverse biological processes from embryogenesis to senescence. Although the knowledge on how plants produce, transport, metabolize, store and sense diverse sugar signals has been significantly advanced [1–9], the spectrum of sugar signals, sensors and molecular mechanisms mediating primary signaling remained to be fully explored. Many informative review articles presented recent progress on broad aspects of sugar-related research in plant biology, encompassing source-sink communication [9,10], sugar-hormone interactions [11], new sugar transporters and their functions [8], sugar regulation of plant development [9,12–15], chloroplast-nuclear signaling [16], sucrose, starch and trehalose metabolism and signaling [2,3,5–7,10,13,15,17], clock-sugar connections [18], as well as sugar and stress [19]. New discoveries on key regulators of sugar and energy signaling have also been thoroughly reviewed [4,5,20–27]. Extensive efforts of past research on sugar regulation have mainly focused on long-term phenotypic characterization in mutants and transgenic plants. The accumulated knowledge will provide an excellent and comprehensive platform for future research, especially on elucidating the molecular, cellular and biochemical basis of sugar sensing and signaling underlying the plasticity and potential in plant growth and development. Emphasis in this review is placed on the emerging understanding of the dynamic, primary and integrated sugar signaling mechanisms and transcriptional networks triggered by direct and indirect sugar signals via sugar, energy and metabolite sensors.
Sugar signals and intracellular sensors
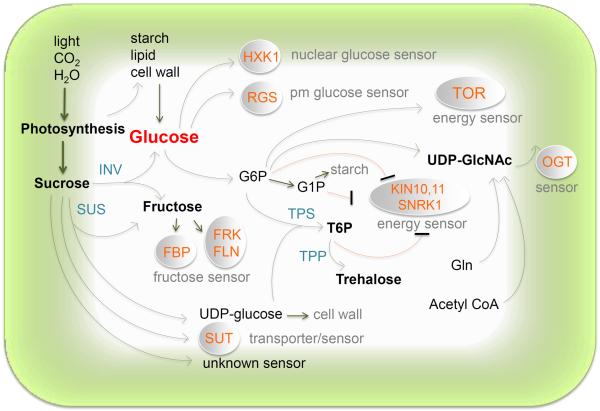
The complex and intertwined plant metabolic and regulatory pathways provide plastic capacity to generate and regulate a wide range of sugar signals originated from different sources, including active photosynthetic cells, dynamic storage reservoir, and organs for nutrient remobilization (Figure 1) [2,4,6,7,9,19,28–30,31*,32*]. Understanding the physiological status and cellular/subcellular actions of each sugar signal relies on the recognition that sugar providing and perceiving cells, as well as sugar metabolic pathways and transport systems in different organs, tissues and cells, are subject to diverse modulations by other nutrient supplies, developmental stages, environmental cues, hormonal regulation, and interactions with microbes and animals [2,7–9,19,23,33,34*]. For instance, high sugar signals can either promote leaf development and photosynthesis with abundant nitrogen supplies or lead to photosynthesis gene repression and developmental arrest at low nitrate levels [35–37]. Plant sugar responses are also significantly influenced by phosphate levels [33]. Although sucrose is the main sugar for systemic transport from source to sink in plants [38], many of the sugar responses observed in plants are channeled through invertases or sucrose synthases [7,39] to generate glucose and other signaling sugars to trigger signal transduction via direct perception by diverse sensors or indirect signaling by energy and metabolite sensors. However, compelling evidence also supports multiple sucrose signaling pathways (Figure 1) [3,5].
Figure 1.
Sugar signals and sensors. Distinct sugar signals are generated locally or systemically via diverse sources. FBP, fructose-1,6-bisphosphtase; FLN, fructokinase-like protein; FRK, fructokinase; G6P, glucose 6-phosphate; G1P, glucose 1-phosphate; Gln, glutamine; HXK1, hexokinase1; INV, invertase; KIN10,11, Arabiodpsis protein kinase 10,11; OGT, O-linked N-acetylglucosamine (O-GlcNAc) transferase; pm, plasma membrane; RGS, regulator of G-protein signaling; SnRK1, sucrose-non-fermentation-related protein kinase1; SUS, sucrose synthase; SUT, sucrose transporter; T6P, trehalose 6-phosphate; TOR, target of rapamycin; TPS, T6P synthase.
Hexokinases (HXKs) are the first demonstrated intracellular glucose sensors in plants [4,23,36,37,40–45]. Plant genomes encode multiple hexokinases (HXKs) and HXK-like (HKL) proteins that appear to serve overlapping and distinct functions in signaling and metabolism [4,36,37,40–45,46]. In Arabidopsis, HXK1 plays dual roles in signaling and metabolism, which can be uncoupled by the S177A mutation that abolishes the glucose phosphorylation activity but possesses full glucose sensor function based on diverse sugar responses [4,23,36,40,41]. The various functions of the Arabidopsis HXK1 glucose sensor are likely evolutionarily conserved and shared by specific HXKs in moss, maize, rice, tomato, poplar, Selaginella moellendorffi and tobacco [36,37, 41–45]. A recent structural study showed that both HXK1 and HXK1(S177A) formed co-crystals with glucose in the single glucose binding pocket and induced similar conformational changes. The findings support the full sensor function of HXK1(S177A) without glucose phosphorylation [36,47]. However, the relatively low Kd in the range of 15–89 μM glucose was measured by the isothermal titration calorimetry (ITC) assay based on transient glucose binding in vitro. It seems inconsistent with the physiological requirement of glucose concentrations for biological responses in cells and plants, which would need further investigation [31*,36,40,48,49]. The determination of the physiological Kd of HXK1 as a glucose sensor by in vitro and in vivo analyses will facilitate the molecular dissection of the direct and indirect glucose signaling responses and regulatory networks.
Interestingly, recent research has uncovered new physiological functions of sugar phosphorylation and metabolism mediated by HXK1. For instance, a rare sugar D-allose requires HXK1-mediated sugar phosphorylation but not the sensor function to trigger a long-term activation of abscisic acid biosynthesis and signaling in Arabidopsis and rice leading to growth inhibition [50]. Another critical role of HXK1-based metabolic activity was revealed in the imps1 mutant, which is deficient for the enzyme, myo-inositol 1-phosphate synthase (MIPS) catalyzing the limiting step of myo-inositol synthesis, and exhibits light-dependent formation of lesions on leaves due to salicylic acid-dependent programmed cell death (PCD) [51]. The somi1 (suppressor of mips1) mutant suppressed cell death and defense responses of mips1 and was mapped to the T231I mutation critical for glucose phosphorylation. Consistently, mips1 did not affect the HXK1 glucose sensor function. HXK1 (S177A) with reduced glucose metabolism alleviates PCD in mips1 [51]. The new findings suggest that HXK1 appears to gate multiple glucose metabolic pathways in plant cells. The channeling and regulation of glucose to myo-inositol metabolism represents an emerging regulatory network, which may explain previously unknown connections between sugar, stress and immune responses in plants [19].
In addition to glucose, ample evidence has indicated that sucrose is perceived as a distinct sugar signal, which cannot be substituted by glucose or fructose, in controlling flowering, seed and storage organ development, branching, and pigmentation [3,5–7]. Compelling examples are the sucrose-specific repression of the beet leaf BvSUT1 gene encoding the sugar proto-sucrose sympoter [52] and the bZIP11 protein translation via the 5'UTR upstream open reading frame (uORF) [53]. Sucrose also specifically stabilizes DELLA proteins to activate MYB75 expression and anthocyanin biosynthesis, but inhibits cell expansion [54*], whereas both sucrose and glucose activate auxin biosynthesis and cell expansion involving HXK1 and complex actions of PIF transcription factors [11,55]. In contrast to the long-standing theory that auxin controls apical dominance, artificially increasing sucrose levels repress the expression of BRANCHED1 transcription factor and promote axillary bud outgrowth [34*]. It is interesting to note that sucrose activates calcium signaling and calcium-dependent protein kinases [3] and plants possess a large number of proteins and enzymes with conserved sugar binding domains that play critical roles in sucrose transport and metabolism [7,8]. Future investigation may explore the roles of SUT proton-symporters, voltage-activated calcium channels, and membrane-associated sucrose synthases (SUS) as potential sucrose sensors and signaling effectors distinct from glucose sensors and signaling mechanisms (Figure 1) [56–58].
Recent research has implicated a tight link between endogenous sucrose and trehalose 6-phosphate (T6P) levels, and it was proposed that T6P is a signal of sucrose availability and influences the relative amounts of sucrose and starch [6,59]. Extensive genetic and transgenic manipulations of trehalose metabolic enzymes have provided fascinating findings that many metabolic and developmental phenotypes are associated with altered levels of T6P, trehalose synthase (TPS) or T6P phosphatase (TPP) (Figure 1), including gene expression, metabolism, seed development, shoot expansion and flowering in Arabidopsis, tobacco and maize plants [5,6,10,12,13,15,59–62,63*,64,65]. New development based on genetic, biochemical and genomic studies has led to important findings that T6P inhibits the activity of the evolutionarily conserved energy sensor complex SNRK1 (Sucrose NonFermenting1-Related Kinase1) via unknown protein regulators [21,62,66*, 67]. It remains a possibility that the surprisingly large plant gene family encoding TPS and TPP proteins lacking prominent enzymatic activities may act as T6P regulators or sensors and modulate SNRK1 activity (Figure 1) [67–69].
Novel sugar sensors
Besides glucose, sucrose and T6P, other sugar signals and putative sensors implicated in regulating plant gene expression, metabolism and development are also emerging [32*,70–72,73*]. An important example is the discovery of new transcription factors containing β–amylase (BAM)-like domain characteristic of starch degradation enzymes in higher plants but no in algae and mosses. The BAM domain of BAM8 appears to mediate DNA-binding and transcriptional activation based on a synthetic reporter driven by BZR1-BAM responsive cis-elements in plant cells and transgenic plants, whereas BAM7 interacts with BAM8 but may act as a transcription repressor [73*]. Future studies could lead to new advances in supporting their potential role as sensors of starch metabolism.
Intrigued by the observation that gin2 is insensitive to glucose but still sensitive to fructose, an integrated study using cell-based functional screen and genetic mutations has identified the nuclear localized fructose1-6-bisphosphatase (FBP/FIS1, FRUCTOSE-INSENSITIVE1) as a putative fructose sensor uncoupled from its catalytic activity [71]. It will be interesting to determine whether FBP is connected to the fructose-specific signaling suppressor, the Arabidopsis NAC89 transcription factor, in the nucleus sharing downstream interactions with abscisic acid and ethylene signaling pathways [74]. Currently, there is no evidence for the involvement of fructokinases (FRKs), catalyzing irreversible fructose phosphorylation and playing a key role in vascular development, in sugar sensing [4]. However, proteomic and genetic studies have identified FRK-like proteins (FLN1 and FLN2) in the plastid-encoded RNA polymerase complexes and regulate plastic gene transcription and chloroplast development [75].
Sensors of extracellular sugars
Besides intracellular sugar sensing, regulator of G-protein signaling (RGS1) as a seven-transmembrane domain protein on the plasma membrane, has been proposed to play a critical role as external glucose sensor in plants. An important advance is the recent determination of RGS1 phosphorylation by WNK8 (WITH NO LYSINE8), which leads to RGS1 endocytosis and G-protein-mediated sugar signaling and cell proliferation [76]. By combining thorough dose-duration experiments with mathematical modeling, it has been shown that 6% glucose stimulates rapid RGS1 endocytosis through WNK8 and WNK10, whereas 2% glucose slowly activates the pathway through WNK1, allowing the cells to respond similarly to transient, high-intensity signals and sustained, low-intensity signals [77*]. The RGS1 signaling pathway appears to be unique in the requirement of extremely high glucose and the rgs1 mutant diminishes the regulation of a few glucose responsive genes in genome-wide analyses. As the RGS1 specific marker gene At4g01080 is strongly activated by 100–300 mM D-glucose, D-fructose and sucrose, RGS1 may be a plasma membrane sensor or partner responding to changes of multiple extracellular sugars [78]. It will be a promising investigation to determine whether cell-wall invertases or sucrose transporters are required to generate high local sugar signals to stimulate RGS1 signaling [7]. A significant recent study has implicated a role of RGS1 in soybean nodulation [79*]. Distinct from RGS1 phosphorylation by WNKs to trigger endocytosis in Arabidopsis sugar signaling [77*], Nod factor receptor1 (NFR1) phosphorylates RGS1 to accelerate GTPase activity and maintains Gα proteins in inactive trimeric conformation. It will be interesting to determine whether RGS1 also functions as a sensor of extracellular sugars in the soybean nodulation process and how RGS1 senses and transduces sugar signals.
Indirect sugar sensing via energy sensors
Manipulations of key enzymes involved in sugar and starch metabolism have started to provide new insights into how the physiological sugar levels modulated by light, CO2 and photoperiod alter gene expression and plant developmental processes [7, 9,36,38,45, 60,61,63*, 80–85]. Many key questions remain to be answered regarding the signaling actions of physiological levels of sugar signals in extracellular spaces and in different subcellular compartments [7,38]. Besides direct sensing by sugar sensors such as HXK1, intracellular sugar levels can be perceived as metabolic input by energy sensing regulators to coordinate energy status and plant metabolism and growth. Recent research on the evolutionarily conserved energy sensor TOR (target of rapamycin) protein kinase is especially informative in uncovering new aspects of glucose signaling in plants [20,22,23,25–27,31*]. Analyses with chemical inhibitors demonstrate that glucose metabolism through glycolysis and the electron transport chain in the mitochondria is required to activate TOR signaling and control global gene transcription. The use of a thymidine analogue, 5-ethynyl-2'-deoxyuridine (EdU) also enables in situ visualization of photosynthesis- or glucose-stimulated cell-cycle S-phase entry in the primary root meristem from quiescence after the depletion of maternal sugar supplies (Figure 2) [31*]. Glucose-TOR signaling is activated below 1 mM glucose via metabolic and energy signaling relay, which is coordinated by shoot-root sugar communication [31*]. Future investigation will expand our understanding on the connections between physiological sugar levels and putative sugar regulators or particular energy sensors and specific signaling pathways in different biological contexts and processes.
Figure 2.
Post-germination seedling development relies on photoautotrophic transition and photosynthesis. After seed sugar depletion at 3 DAG, exogenously supplied Glc (1–15 mM glucose) promotes similar growth based on endogenous sugars derived from photosynthesis in Arabidopsis seedlings. C, CO2; DAG, days after germination; D, DCMU, a photosynthesis inhibitor; EdU, 5-ethynyl-2'-deoxyuridine; L, light.
Under sugar deprivation conditions, the evolutionarily conserved energy sensor complex SNRK1 plays central regulatory functions in metabolism, stress signaling and plant development [9,10,15,21,23,24]. In Arabidopsis, KIN10/11 protein kinases provide catalytic activities in the SNRK1 complex and orchestrate global gene expression changes to activate catabolism but repress anabolism [21,23, 24,70]. Recent exciting progresses have identified new transcription factors as Arabidopsis KIN10 phosphorylation targets, including bZIP63, MYC2, NAC2/ATAF1, FUS3 and IDD transcription factors involved in low energy responses in darkness, submergence, starvation, and flowering [15,84,86,87*, 88*,89]. Direct phosphorylation by KIN10 protein kinase promotes bZIP63-bZIPS dimerization and the transcriptional activation of bZIPs, NAC2 and FUS3 [84,87*, 88*]. Phosphorylation by KIN10 reduces MYC2 protein stability and transcriptional activity of IDD8 [86]. Surprisingly, only KIN10 but not KIN11 directly phosphorylates bZIP63 and IDD, even though the single kin10 or kin11 mutants do not show overt phenotypes [70,87*, 89]. How KIN10 phosphorylation contributes to the opposite leaf senescence phenotypes of bzip63 and bZIP63 overexpression requires further molecular, biochemical and physiological insights [70,87*]. Besides transcriptional controls, new mechanisms of regulation by mRNA stability and miRNAs provide additional layers of molecular controls in dynamic sugar responses [9, 80,81, 90,91,92*].
Another novel finding in the indirect sugar signaling mechanism came from the functional characterization of Arabidopsis SEC (SECRET AGENT) encoding a specific O-linked N-acetylgluocosamine (O-GlcNAc) transferase (OGT). SEC promotes gibberellin signaling by O-GlcNAcylating DELLA transcription repressors and prevents the interaction and suppression of multiple transcription factors, BZR1, PIF4, PIF5 and JAZ1, involved in brassinosteroid, light and jasmonate signaling, respectively [32*]. Further research advances will resolve the remaining puzzles regarding the physiological and biochemical functions of another Arabidopsis OGT paralog SPY, carrying out an opposite repressor role in gibberellin signaling but related functions with SEC in embryogenesis. As O-GlcNAc is synthesized from glucose, lipid and glutamine (Figure 1), and DELLAs are the convergent regulators in hormonal, sugar and stress signaling crosstalk [11,93,94], OGT likely act as a pivotal sensor in modulating and integrating nutrient, hormonal and stress signaling pathways central to plant growth and development.
Primary and dynamic sugar signaling network
Comprehensive transcriptome analyses provide a powerful approach to explore dynamic and primary sugar responses and to discover new regulators in sugar-mediated processes. Over the past decade, many microarray studies have been performed under different conditions. Arabidopsis seedlings and adult leaves were analyzed with different concentrations of exogenous sugars, as well as cell-based transient expression systems by manipulating sensors and signaling components [31*,49,70,90,95–98]. An integrated analysis of four representative genome-wide expression profiling data reveals a convergent energy-signaling network modulating nearly 1,300 genes in rapid sugar responses (2–4 hr) (Figure 3). Importantly, TOR and KIN10 protein kinases are central regulators in sugar-mediated energy signaling, but act antagonistically in the regulation of convergent primary sugar responsive genes [31*,49,70,90,91,97,98]. Among the key functional classes regulated by the KIN10-TOR and sucrose/glucose (KTSG) convergent network, genes involved in protein synthesis, cell cycle, signaling, transcription, glycolysis, TCA cycle, mitochondria electron transport chain, as well as secondary carbon metabolism are activated by both glucose and sucrose. On the other hand, genes participating in transcription, diverse transporter functions, as well as the degradation of protein, amino acid, lipid and cell wall are repressed by sugars (Figure 3).
Figure 3.
Dynamic Transcriptional control by sugars. Arabidopsis ATH1 transcriptome data are clustered based on key functional gene sets regulated by glucose, sucrose, TOR and KIN10 in Arabidopsis thaliana. Genes representing KIN10 targets [70], TOR targets [31*], E2Fa targets [31*], sucrose [49] and glucose (4h data)[96] regulation, cell cycle, protein synthesis, primary metabolism and secondary metabolism are included using hierarchical analyses. Gene lists are chosen from supplemental data from representative studies (Log2 ≥ 1 or ≤ −1; q-value ≤ 0.05). Totally 3240 up regulated genes and 2560 down regulated genes are shown in the heatmap, with 428 convergent up-regulated genes and 863 convergent down-regulated genes as indicated by red lines.
Besides the convergent sugar signaling program, it is important to note that the differences in gene expression profiles may reflect the existence of truly distinct regulatory programs controlled by either sucrose or glucose, or may represent specific features of each experimental system and approach for data generation. For instance, some genes activated by glucose-TOR signaling are missing from seedlings stimulated by both sucrose and glucose. It is likely that the primary root meristem cells expressing cell cycle genes and the TOR targeted transcription factor E2FA target genes are more significantly represented in 3-day seedlings [31*] vs. 8–9 day seedlings [49,96]. In future research, it will be crucial to uncover new biological regulation of specific TOR and KIN10 target genes in not only seedlings but also adult plants, apical meristems and diverse cell types that may act in multiple metabolic, stress and developmental pathways [5,12,15,60,61,63*,70,99]. Notably, sucrose treated seedlings appear to modulate many more uniquely regulated genes that may provide important information for future dissection of the sucrose-specific pathways [3,5–7].
Despite the overt convergence between the TOR and KIN10 target genes in rapid sugar responses, how Arabidopsis TOR and KIN10 protein kinases regulate the vast primary transcriptional programs of diverse genes in an opposite manner represents a major challenge. In mesophyll protoplasts, KIN10 overexpression inhibits TOR-mediated phosphorylation of S6K1 (Xiong and Sheen, unpublished) [70,100]. However, it remains unclear whether KIN10 directly phosphorylates and inactivates TOR kinase through the phosphorylation of RAPTOR as a regulatory subunit in the TOR sensor complex [26,101]. Although prior studies have emphasized TOR functions in ribosome biogenesis, protein stability and translational control [22,25–27,102–104], the identification of E2FA as a direct TOR kinase substrate [31*] opens up new mechanisms of direct and rapid phosphorylation of transcription factors by sugars in central metabolic and growth pathways. Importantly, this type of regulation is independently controlled or co-regulated by the SNRK1 energy sensor and the HXK1 glucose sensor (Figure 4). It is most likely that the modulation of related transcription factors on distinct phosphorylation sites by TOR and SNRK1 to mediate contrast regulation in response to sugar availability and energy status. Sensitive and quantitative phosphoproteomics will further facilitate the integration of SNRK1-TOR signaling networks [105].
Figure 4.
Convergent regulation in the sugar signaling network. The trifurcated model focuses on integrating glucose signaling mediated by glucose and energy sensors. Glucose is produced by photosynthesis or from storage source and transported as sucrose or glucose to the sink tissues and other organs to promote growth and to maintain energy and metabolic homeostasis. The regulatory mechanisms and functions of three master regulators, HXK1, KIN10/11 and TOR, modulated by glucose signals are shown. The glucose signaling networks are tightly intertwined with environmental light, nutrients, stresses and microbes, as well as internal hormones, peptides and clock.
Glc, glucose; HXK, hexokinase; HKLs, hexokinaselike; KIN, Arabidopsis protein kinase; QC, quiescent center; TOR, target of rapamycin.
Future challenges
The biological functions of plant sugar signals and sensors in embryogenesis, seedling establishment, growth, metabolism, juvenile-adult transition, flowering and senescence have emerged. The molecular regulatory mechanisms of the plant sugar-signaling network are starting to be elucidated in the meristem, expanding and differentiated cells (Figure 4). The application of versatile and integrated molecular, cellular, genetic, genomic, phospho-proteomic and systems analyses will facilitate the discoveries of new regulators and molecular links in diverse mechanisms mediating sugar signaling. Major puzzles await to be resolved include how the different sugar sensors distinguish regulatory ligands with high specificity in different physiological concentration ranges, where these sensors act at the subcellular, cellular and organismal levels [40,77*,102,106–108], what the components are in these sensor complexes [26,40,76,77*,109*,110,111], how they mediate the first steps of signal transduction, what the mechanisms are in the convergent or specific regulations by TOR, SNRK1 and HXK1 (Figure 4), as well as how parallel or integrative signaling by other novel sugar sensors and signaling components modulate a large array of downstream effectors and responses (Figure 1). Finally, development of sensitive and quantitative technologies for single-cell based genetic and chemical perturbations and for transcriptome, epigenome and metabolite profiling, as well as application of genetic encoded biosensors for dynamic imaging of sugar, energy or metabolite signaling will likely lead to new discoveries. Much information will be gained in understanding the plant energy-stress signaling network by elucidating the antagonistic functions of TOR and KIN10 as key energy sensors and central regulators of transcriptional, translational and metabolic programs in response to other nutrients, hormones, clock, microbes and diverse environmental cues (Figure 4).
Highlights.
Emerging sugar signals are discussed.
Direct sugar sensing and signaling is triggered via sugar-binding sensors.
Sugar-derived metabolites modulate energy sensors and indirectly relay signalling.
Comparative transcriptome profiles highlight the dynamic & diverse sugar responses.
Acknowledgements
We thank Matthew McCormack for guidance in transcriptom analyses. Funding by the NIH and NSF grants and WJC Special Project RDA-Korea to J.S. supports the research projects on the sugar signaling networks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as *
- 1.Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 2.Stitt M, Zeeman SC. Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol. 2012;15:282–292. doi: 10.1016/j.pbi.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Tognetti JA, Pontis HG, Martinez-Noel GM. Sucrose signaling in plants: A world yet to be explored. Plant Signal Behav. 2013;8:e23316. doi: 10.4161/psb.23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Granot D, Kelly G, Stein O, David-Schwartz R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J Exp Bot. 2014;65:809–819. doi: 10.1093/jxb/ert400. [DOI] [PubMed] [Google Scholar]
- 5.Lastdrager J, Hanson J, Smeekens S. Sugar signals and the control of plant growth and development. J Exp Bot. 2014;65:799–807. doi: 10.1093/jxb/ert474. [DOI] [PubMed] [Google Scholar]
- 6.Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M. Trehalose metabolism in plants. Plant J. 2014;79:544–567. doi: 10.1111/tpj.12509. [DOI] [PubMed] [Google Scholar]
- 7.Ruan Y-L. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol. 2014;65:33–67. doi: 10.1146/annurev-arplant-050213-040251. [DOI] [PubMed] [Google Scholar]
- 8.Chen L-Q, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of sugars. Annu Rev Biochem. 2015;84:865–894. doi: 10.1146/annurev-biochem-060614-033904. [DOI] [PubMed] [Google Scholar]
- 9.Yu S-M, Lo S-F, Ho T-H. Source–Sink Communication: Regulated by Hormone, Nutrient, and Stress Cross-Signaling. Trends Plant Sci. 2015;20:844–857. doi: 10.1016/j.tplants.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Lawlor DW, Paul MJ. Source/sink interactions underpin crop yield: the case for trehalose 6-phosphate/SnRK1 in improvement of wheat. Front Plant Sci. 2014;5:418. doi: 10.3389/fpls.2014.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljung K, Nemhauser JL, Perata P. New mechanistic links between sugar and hormone signalling networks. Curr Opin Plant Biol. 2015;25:130–137. doi: 10.1016/j.pbi.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Eveland AL, Jackson DP. Sugars, signalling, and plant development. J Exp Bot. 2012;63:3367–3377. doi: 10.1093/jxb/err379. [DOI] [PubMed] [Google Scholar]
- 13.O'Hara LE, Paul MJ, Wingler A. How do sugars regulate plant growth and development? New insight into the role of trehalose-6-phosphate. Mol Plant. 2013;6:261–274. doi: 10.1093/mp/sss120. [DOI] [PubMed] [Google Scholar]
- 14.Sablowski R, Dornelas MC. Interplay between cell growth and cell cycle in plants. J Exp Bot. 2014;65:2703–2714. doi: 10.1093/jxb/ert354. [DOI] [PubMed] [Google Scholar]
- 15.Tsai AY, Gazzarrini S. Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front Plant Sci. 2014;5:119. doi: 10.3389/fpls.2014.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Häusler RE, Heinrichs L, Schmitz J, Flügge U-I. How sugars might coordinate chloroplast and nuclear gene expression during acclimation to high light intensities. Mol Plant. 2014;7:1121–1137. doi: 10.1093/mp/ssu064. [DOI] [PubMed] [Google Scholar]
- 17.Tiessen A, Padilla-Chacon D. Subcellular compartmentation of sugar signalling: Links among carbon cellular status, route of sucrolysis, sink-source allocation, and metabolic partitioning. Front Plant Sci. 2013;3:306. doi: 10.3389/fpls.2012.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haydon MJ, Hearn TJ, Bell LJ, Hannah MA, Webb AA. Metabolic regulation of circadian clocks. Semin Cell Dev Biol. 2013;24:414–421. doi: 10.1016/j.semcdb.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Valluru R, Van den Ende W. Myo-inositol and beyond–emerging networks under stress. Plant Sci. 2011;181:387–400. doi: 10.1016/j.plantsci.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Dobrenel T, Marchive C, Azzopardi M, Clément G, Moreau M, Sormani R, Robaglia C, Meyer C. Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR. Front Plant Sci. 2013;4:93. doi: 10.3389/fpls.2013.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, Elias CA, Baena-González E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci. 2014;5:190. doi: 10.3389/fpls.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henriques R, Bögre L, Horváth B, Magyar Z. Balancing act: matching growth with environment by the TOR signalling pathway. J Exp Bot. 2014;65:2691–2701. doi: 10.1093/jxb/eru049. [DOI] [PubMed] [Google Scholar]
- 23.Sheen J. Master regulators in plant glucose signaling networks. J Plant Biol. 2014;57:67–79. doi: 10.1007/s12374-014-0902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomé F, Nägele T, Adamo M, Garg A, Marco-llorca C, Nukarinen E, Pedrotti L, Peviani A, Simeunovic A, Tatkiewicz A, Tomar M, Gamm M. The low energy signaling network. Front Plant Sci. 2014;5:353. doi: 10.3389/fpls.2014.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Y, Sheen J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014;164:499–512. doi: 10.1104/pp.113.229948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong Y, Sheen J. Novel links in the plant TOR kinase signaling network. CurrOpin Plant Biol. 2015;28:83–91. doi: 10.1016/j.pbi.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrada A, Montané M-H, Robaglia C, Menand B. Spatial Regulation of Root Growth: Placing the Plant TOR Pathway in a Developmental Perspective. Int JMol Sci. 2015;16:19671–19697. doi: 10.3390/ijms160819671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee E-J, Matsumura Y, Soga K, Hoson T, Koizumi N. Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 2007;48:405–413. doi: 10.1093/pcp/pcm009. [DOI] [PubMed] [Google Scholar]
- 29.Graham IA. Seed storage oil mobilization. Annu Rev Plant Biol. 2008;59:115–142. doi: 10.1146/annurev.arplant.59.032607.092938. [DOI] [PubMed] [Google Scholar]
- 30.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *31.Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors discovered that plant TOR signaling is regulated by shoot photosynthesis-derived glucose signals to rapidly control global transcriptome reprogramming and root meristem activation. Furthermore, TOR directly phosphorylates and activates transcription factor E2Fa to induce S-phase gene expression and DNA replication. This finding represents an unconventional mode of TOR regulation beyond stimulating translation to promote cell cycle progression through S6K1 and 4E-BP in mammals.
- *32.Zentella R, Hu J, Hsieh W-P, Matsumoto PA, Dawdy A, Barnhill B, Oldenhof H, Hartweck LM, Maitra S, Thomas SG, et al. O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 2016;30:164–176. doi: 10.1101/gad.270587.115. [DOI] [PMC free article] [PubMed] [Google Scholar]; RGA, one of the DELLA family transcription repressors in the GA signaling pathway, undergoes O-GlcNAcylation modification mediated by O-GlcNAc transferase (OGT) SECRET AGENT (SEC). O-GlcNAcylation of RGA impairs its interacting with other master transcription regulators in various signaling pathways including light, JA and BR. These findings provided the first example of protein O-GlcNAcylation modification in Arabidopsis, identified the corresponding OGT, and uncovered the role of O-GlcNAcylation in modulating plant growth.
- 33.Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 2007;143:156–171. doi: 10.1104/pp.106.090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *34.Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc Natl Acad Sci USA. 2014;111:6092–6097. doi: 10.1073/pnas.1322045111. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors made an unexpected finding that apical dominance is independent of the level of auxin, and instead appears to be determined by sugar availability. Experiments showed that shoot tip's demand for sugars limits sugar availability to axillary buds, therefore predominantly maintains the apical dominance.
- 35.Martin T, Oswald O, Graham IA. Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon: nitrogen availability. Plant Physiol. 2002;128:472–481. doi: 10.1104/pp.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-X, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- 37.Cho Y-H, Sheen J, Yoo S-D. Low glucose uncouples hexokinase1-dependent sugar signaling from stress and defense hormone abscisic acid and C2H4 responses in Arabidopsis. Plant Physiol. 2010;152:1180–1182. doi: 10.1104/pp.109.148957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Han L, Slewinski TL, Sun J, Zhang J, Wang Z-Y, Turgeon R. Symplastic phloem loading in poplar. Plant Physiol. 2014;166:306–313. doi: 10.1104/pp.114.245845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barratt DH, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA. 2009;106:13124–13129. doi: 10.1073/pnas.0900689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho Y-H, Yoo S-D, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 41.Cho J-I, Ryoo N, Eom J-S, Lee D-W, Kim H-B, Jeong S-W, Lee Y-H, Kwon Y-K, Cho M-H, Bhoo SH, et al. Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 2009;149:745–759. doi: 10.1104/pp.108.131227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karve R, Lauria M, Virnig A, Xia X, Rauh BL, Moore Bd. Evolutionary lineages and functional diversification of plant hexokinases. Mol Plant. 2010;3:334–346. doi: 10.1093/mp/ssq003. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson A, Olsson T, Ulfstedt M, Thelander M, Ronne H. Two novel types of hexokinases in the moss Physcomitrella patens. BMC Plant Biol. 2011;11:32. doi: 10.1186/1471-2229-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karve A, Xia X, Moore Bd. Arabidopsis Hexokinase-Like1 and Hexokinase1 form a critical node in mediating plant glucose and ethylene responses. PlantPhysiol. 2012;158:1965–1975. doi: 10.1104/pp.112.195636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YM, Heinzel N, Giese JO, Koeber J, Melzer M, Rutten T, Von Wiren N, Sonnewald U, Hajirezaei MR. A dual role of tobacco hexokinase 1 in primary metabolism and sugar sensing. Plant Cell Environ. 2013;36:1311–1327. doi: 10.1111/pce.12060. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z-W, Yuan S, Xu F, Yang H, Zhang N-H, Cheng J, Lin H-H. The plastid hexokinase pHXK: a node of convergence for sugar and plastid signals in Arabidopsis. FEBS Lett. 2010;584:3573–3579. doi: 10.1016/j.febslet.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 47.Feng J, Zhao S, Chen X, Wang W, Dong W, Chen J, Shen J-R, Liu L, Kuang T. Biochemical and structural study of Arabidopsis hexokinase 1. ActaCrystallogr D Biol Crystallogr. 2015;71:367–375. doi: 10.1107/S1399004714026091. [DOI] [PubMed] [Google Scholar]
- 48.Jang J-C, Sheen J. Sugar sensing in higher plants. Plant Cell. 1994;6:1665–1679. doi: 10.1105/tpc.6.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, Stitt M. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 2007;49:463–491. doi: 10.1111/j.1365-313X.2006.02979.x. [DOI] [PubMed] [Google Scholar]
- 50.Fukumoto T, Kano A, Ohtani K, Inoue M, Yoshihara A, Izumori K, Tajima S, Shigematsu Y, Tanaka K, Ohkouchi T, et al. Phosphorylation of d-allose by hexokinase involved in regulation of OsABF1 expression for growth inhibition in Oryza sativa L. Planta. 2013;237:1379–1391. doi: 10.1007/s00425-013-1853-9. [DOI] [PubMed] [Google Scholar]
- 51.Bruggeman Q, Prunier F, Mazubert C, de Bont L, Garmier M, Lugan R, Benhamed M, Bergounioux C, Raynaud C, Delarue M. Involvement of Arabidopsis hexokinase1 in cell death mediated by myo-inositol accumulation. Plant Cell. 2015;27:1801–1814. doi: 10.1105/tpc.15.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaughn MW, Harrington GN, Bush DR. Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc Natl Acad Sci USA. 2002;99:10876–10880. doi: 10.1073/pnas.172198599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009;150:1356–1367. doi: 10.1104/pp.109.136036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *54.Li Y, Van den Ende W, Rolland F. Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Mol Plant. 2014;7:570–572. doi: 10.1093/mp/sst161. [DOI] [PubMed] [Google Scholar]; The authors reported a novel finding that protein stability of the DELLA family transcription factor RGA appears to be specific for sucrose, as glucose only has limited effect. High sucrose can repress plant growth and promote anthocyanin formation in the cells through RGA-mediated transcriptional regulation on its downstream targets, including MYB75, a key transcription factor required for anthocyanin biosynthesis.
- 55.Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell. 2012;24:4907–4916. doi: 10.1105/tpc.112.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB. SUT2, a putative sucrose sensor in sieve elements. Plant Cell. 2000;12:1153–1164. doi: 10.1105/tpc.12.7.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furuichi T, Cunningham KW, Muto S. A putative two pore channel AtTPC1 mediates Ca2+ flux in Arabidopsis leaf cells. Plant Cell Physiol. 2001;42:900–905. doi: 10.1093/pcp/pce145. [DOI] [PubMed] [Google Scholar]
- 58.Duncan KA, Huber SC. Sucrose synthase oligomerization and F-actin association are regulated by sucrose concentration and phosphorylation. Plant Cell Physiol. 2007;48:1612–1623. doi: 10.1093/pcp/pcm133. [DOI] [PubMed] [Google Scholar]
- 59.Yadav UP, Ivakov A, Feil R, Duan GY, Walther D, Giavalisco P, Piques M, Carillo P, Hubberten H-M, Stitt M, Lunn JE. The sucrose–trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J Exp Bot. 2014;65:1051–1068. doi: 10.1093/jxb/ert457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gómez LD, Baud S, Gilday A, Li Y, Graham IA. Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J. 2006;46:69–84. doi: 10.1111/j.1365-313X.2006.02662.x. [DOI] [PubMed] [Google Scholar]
- 61.Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature. 2006;441:227–230. doi: 10.1038/nature04725. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 2009;149:1860–1871. doi: 10.1104/pp.108.133934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *63.Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339:704–707. doi: 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]; The authors provided convincing data that TREHALOSE-6-PHOSPHATE SYNTHASE 1 (TPS1) is required for the timely initiation of flowering in Arabidopsis. The genetic evidence showed that loss of TPS1 leads to extremely late flowering phenotype. It was demonstrated that the T6P pathway affects flowering both in the leaves and at the shoot meristem, by regulates the expression of flowering genes, such as FT.
- 64.Nuccio ML, Wu J, Mowers R, Zhou H-P, Meghji M, Primavesi LF, Paul MJ, Chen X, Gao Y, Haque E, et al. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat Biotechnol. 2015;33:862–869. doi: 10.1038/nbt.3277. [DOI] [PubMed] [Google Scholar]
- 65.Figueroa CM, Feil R, Ishihara H, Watanabe M, Kölling K, Krause U, Höhne M, Encke B, Plaxton WC, Zeeman SC, et al. Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 2015;85:410–423. doi: 10.1111/tpj.13114. [DOI] [PubMed] [Google Scholar]
- *66.Nunes C, Primavesi LF, Patel MK, Martinez-Barajas E, Powers SJ, Sagar R, Fevereiro PS, Davis BG, Paul MJ. Inhibition of SnRK1 by metabolites: tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiol Biochem. 2013;63:89–98. doi: 10.1016/j.plaphy.2012.11.011. [DOI] [PubMed] [Google Scholar]; This study developed biochemical assays to comprehensively investigate the inhibition of SnRK1 activity by various sugar metabolites. It was shown that SnRK1 was inhibited by low concentrations of trehalose 6-phosphate (T6P), glucose 6-phosphate (G6P) and glucose 1-phosphate (G1P), each with distinct kinetics. Furthermore, the inhibition of SnRK1 by Ribose 5-phosphate (R5P) is ATP concentration dependent, providing a potential important caveat for kinase assay interpretation.
- 67.Jeong E-Y, Seo PJ, Woo JC, Park C-M. AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Biol. 2015;15:110. doi: 10.1186/s12870-015-0503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duarte GT, Matiolli CC, Pant BD, Schlereth A, Scheible W-R, Stitt M, Vicentini R, Vincentz M. Involvement of microRNA-related regulatory pathways in the glucose-mediated control of Arabidopsis early seedling development. J Exp Bot. 2013;64:4301–4312. doi: 10.1093/jxb/ert239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cookson SJ, Yadav UP, Klie S, Morcuende R, Usadel B, Lunn JE, Stitt M. Temporal kinetics of the transcriptional response to carbon depletion and sucrose readdition in Arabidopsis seedlings. Plant Cell Environ. 2016;39:768–786. doi: 10.1111/pce.12642. [DOI] [PubMed] [Google Scholar]
- 70.Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 71.Cho Y-H, Yoo S-D. Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLoS Genet. 2011;7:e1001263. doi: 10.1371/journal.pgen.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reinhold H, Soyk S, Šimková K, Hostettler C, Marafino J, Mainiero S, Vaughan CK, Monroe JD, Zeeman SC. β-Amylase–like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell. 2011;23:1391–1403. doi: 10.1105/tpc.110.081950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *73.Soyk S, Šimková K, Zürcher E, Luginbühl L, Brand LH, Vaughan CK, Wanke D, Zeeman SC. The enzyme-like domain of Arabidopsis nuclear β-amylases is critical for DNA sequence recognition and transcriptional activation. Plant Cell. 2014;26:1746–1763. doi: 10.1105/tpc.114.123703. [DOI] [PMC free article] [PubMed] [Google Scholar]; This comprehensive study showed that two Arabidopsis BZR1-BAM family transcription factors BAM7 and BAM8 contain the b-amylase-like (BAM) domain but opposite functions in transcriptional regulation. The BAM domain has no apparent catalytic activity, but is critical for DNA recognition and transcription modulation. As BAM8's function as a transcriptional activator requires an intact substrate binding site, BZR1-BAMs could serve as novel transcription regulators with metabolite sensing functions.
- 74.Li P, Wind JJ, Shi X, Zhang H, Hanson J, Smeekens SC, Teng S. Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proc Natl Acad Sci USA. 2011;108:3436–3441. doi: 10.1073/pnas.1018665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gilkerson J, Perez-Ruiz JM, Chory J, Callis J. The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biol. 2012;12:102. doi: 10.1186/1471-2229-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, Taylor JP, Jones AM. Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol. 2012;14:1079–1088. doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *77.Fu Y, Lim S, Urano D, Tunc-Ozdemir M, Phan NG, Elston TC, Jones AM. Reciprocal encoding of signal intensity and duration in a glucose-sensing circuit. Cell. 2014;156:1084–1095. doi: 10.1016/j.cell.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; By combining dose-duration experiments with mathematical modeling, the authors showed that upon high exogenous glucose treatment, the regulator of G-protein signaling (RGS1) is phosphorylated by WNKs (WITH NO LYSINE) in a reciprocal dose and duration dependent manner. The phosphorylation results in RGS1 endocytosis and trigger G-protein-mediated glucose signaling and cell proliferation.
- 78.Grigston JC, Osuna D, Scheible W-R, Liu C, Stitt M, Jones AM. D-Glucose sensing by a plasma membrane regulator of G signaling protein, AtRGS1. FEBS Lett. 2008;582:3577–3584. doi: 10.1016/j.febslet.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *79.Choudhury SR, Pandey S. Phosphorylation-Dependent Regulation of G-Protein Cycle during Nodule Formation in Soybean. Plant Cell. 2015;27:3260–3276. doi: 10.1105/tpc.15.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study showed that a soybean heterotrimeric G-protein complex and the regulator RGS protein are involved in regulation of nodulation. Phosphorylation of RGS by Nod factor receptor 1 (NFR1) keeps the Gα proteins in their inactive, trimeric conformation, which subsequently leads to nodule development.
- 80.Yu S, Cao L, Zhou C-M, Zhang T-Q, Lian H, Sun Y, Wu J, Huang J, Wang G, Wang J-W. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. Elife. 2013;2:e00269. doi: 10.7554/eLife.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang L, Xu M, Koo Y, He J, Poethig RS. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife. 2013;2:e00260. doi: 10.7554/eLife.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible W-R, Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kelly G, David-Schwartz R, Sade N, Moshelion M, Levi A, Alchanatis V, Granot D. The pitfalls of transgenic selection and new roles of AtHXK1: a high level of AtHXK1 expression uncouples hexokinase1-dependent sugar signaling from exogenous sugar. Plant Physiol. 2012;159:47–51. doi: 10.1104/pp.112.196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsai AY, Gazzarrini S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012;69:809–821. doi: 10.1111/j.1365-313X.2011.04832.x. [DOI] [PubMed] [Google Scholar]
- 85.Matsoukas IG, Massiah AJ, Thomas B. Starch metabolism and antiflorigenic signals modulate the juvenile-to-adult phase transition in Arabidopsis. Plant Cell Environ. 2013;36:1802–1811. doi: 10.1111/pce.12088. [DOI] [PubMed] [Google Scholar]
- 86.Im JH, CHO YH, KIM GD, KANG GH, HONG JW, YOO SD. Inverse modulation of the energy sensor aSnf1-related protein kinase 1 on hypoxia adaptation and salt stress tolerance in Arabidopsis thaliana. Plant Cell Environ. 2014;37:2303–2312. doi: 10.1111/pce.12375. [DOI] [PubMed] [Google Scholar]
- *87.Mair A, Pedrotti L, Wurzinger B, Anrather D, Simeunovic A, Weiste C, Valerio C, Dietrich K, Kirchler T, Nägele T, et al. SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. Elife. 2015;4:e05828. doi: 10.7554/eLife.05828. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided comprehensive genetic, biochemistry and genomic evidence to show that Arabidopsis transcription factor bZIP63 is a key regulator of low-energy response. The direct phosphorylation of bZIP63 by SnRK1 reshapes its dimerization preference with other transcription factors, and subsequently alters downstream target gene expression and primary metabolism.
- *88.Garapati P, Feil R, John EL, Van Dijck P, Balazadeh S, Mueller-Roeber B. Transcription factor ATAF1 integrates carbon starvation responses with trehalose metabolism. Plant Physiol. 2015;169:379–390. doi: 10.1104/pp.15.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors revealed that the carbon starvation induced protein ATAF1 (Arabidopsis Transcription Activation Factor1) directly activates TREHALASE1 expression, and induce transcriptome reprograming featuring energy and carbon starvation responses. Up-regulation of ATAF1 results in decreased trehalose-6-phosphate levels and reduced sugar starvation metabolome, as well as global transcriptome reprograming featuring energy and carbon starvation responses. The study demonstrated that ATAF1 is a key regulator of carbon starvation responses and trehalose metabolism.
- 89.Jeong E-Y, Seo PJ, Woo JC, Park C-M. AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Biol. 2015;15:110. doi: 10.1186/s12870-015-0503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duarte GT, Matiolli CC, Pant BD, Schlereth A, Scheible W-R, Stitt M, Vicentini R, Vincentz M. Involvement of microRNA-related regulatory pathways in the glucose-mediated control of Arabidopsis early seedling development. J Exp Bot. 2013;64:4301–4312. doi: 10.1093/jxb/ert239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cookson SJ, Yadav UP, Klie S, Morcuende R, Usadel B, Lunn JE, Stitt M. Temporal kinetics of the transcriptional response to carbon depletion and sucrose readdition in Arabidopsis seedlings. Plant Cell Environ. 2016;39:768–786. doi: 10.1111/pce.12642. [DOI] [PubMed] [Google Scholar]
- *92.Confraria A, Martinho C, Elias A, Rubio-Somoza I, Baena-González E. miRNAs mediate SnRK1-dependent energy signaling in Arabidopsis. Front Plant Sci. 2013;4:197. doi: 10.3389/fpls.2013.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]; By comparing transcriptional reprogramming in response to energy deprivation in WT and the microRNA biogenesis mutant dcl1-9, the authors made a novel link of microRNA regulation with SnRK1 signaling, and identified 155 putative microRNA targeted genes. Analyses revealed these microRNAs targeted genes involved in diverse translational and organelle functions, including miR319 targeted transcription factors TCPs.
- 93.Claeys H, De Bodt S, Inzé D. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 2014;19:231–239. doi: 10.1016/j.tplants.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 94.Xu H, Liu Q, Yao T, Fu X. Shedding light on integrative GA signaling. Curr Opin Plant Biol. 2014;21:89–95. doi: 10.1016/j.pbi.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 95.Price J, Laxmi A, St Martin SK, Jang J-C. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell. 2004;16:2128–2150. doi: 10.1105/tpc.104.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. Establishing glucose-and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res. 2006;16:414–427. doi: 10.1101/gr.4237406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kunz S, Pesquet E, Kleczkowski LA. Functional dissection of sugar signals affecting gene expression in Arabidopsis thaliana. PloS One. 2014;9:e100312. doi: 10.1371/journal.pone.0100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kunz S, Gardeström P, Pesquet E, Kleczkowski LA. Hexokinase 1 is required for glucose-induced repression of bZIP63, At5g22920, and BT2 in Arabidopsis. Front Plant Sci. 2015;6:525. doi: 10.3389/fpls.2015.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2003;100:6849–6854. doi: 10.1073/pnas.1132018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem. 2012;287:2836–2842. doi: 10.1074/jbc.M111.300749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ren M, Qiu S, Venglat P, Xiang D, Feng L, Selvaraj G, Datla R. Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 2011;155:1367–1382. doi: 10.1104/pp.110.169045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013;32:1087–1102. doi: 10.1038/emboj.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim Y-K, Kim S, Shin Y-j, Hur Y-S, Kim W-Y, Lee M-S, Cheon C-I, Verma DPS. Ribosomal protein S6, a target of rapamycin, is involved in the regulation of rRNA genes by possible epigenetic changes in Arabidopsis. J Biol Chem. 2014;289:3901–3912. doi: 10.1074/jbc.M113.515015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cho H-Y, Wen T-N, Wang Y-T, Shih M-C. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J Exp Bot. 2016;67(9):2745–2760. doi: 10.1093/jxb/erw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bitrián M, Roodbarkelari F, Horváth M, Koncz C. BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 2011;65:829–842. doi: 10.1111/j.1365-313X.2010.04462.x. [DOI] [PubMed] [Google Scholar]
- 107.Cho Y-H, Hong J-W, Kim E-C, Yoo S-D. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 2012;158:1955–1964. doi: 10.1104/pp.111.189829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams SP, Rangarajan P, Donahue JL, Hess JE, Gillaspy GE. Regulation of sucrose non-fermenting related kinase 1 genes in Arabidopsis thaliana. Front Plant Sci. 2014;5:324. doi: 10.3389/fpls.2014.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *109.Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F. The hybrid Four-CBS-Domain KINβγ subunit functions as the canonical γ subunit of the plant energy sensor SnRK1. Plant J. 2013;75:11–25. doi: 10.1111/tpj.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]; Through comprehensive analyses, the authors convincingly showed that the Arabidopsis AMPK/SNF1/SnRK1 protein kinase complexes contain KIN βγ subunit for the heterotrimeric complex formation. Using integrated analyses including SnRK1 complex reconstitution, mutant complementation, phylogenetic reconstruction, and a seedling starvation assay, it was shown that only the hybrid βγ subunit is required for SnRK1 signaling, but not the canonical γ subunit.
- 110.Lin C-R, Lee K-W, Chen C-Y, Hong Y-F, Chen J-L, Lu C-A, Chen K-T, Ho T-HD, Yu S-M. SnRK1A-interacting negative regulators modulate the nutrient starvation signaling sensor SnRK1 in source-sink communication in cereal seedlings under abiotic stress. Plant Cell. 2014;26:808–827. doi: 10.1105/tpc.113.121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Emanuelle S, Hossain MI, Moller IE, Pedersen HL, Meene AM, Doblin MS, Koay A, Oakhill JS, Scott JW, Willats WG, et al. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015;82:183–192. doi: 10.1111/tpj.12813. [DOI] [PubMed] [Google Scholar]