Abstract
Advanced, anaplastic lymphoma kinase (ALK)-positive lung cancer is currently treated with the first-generation ALK inhibitor crizotinib followed by more potent, second-generation ALK inhibitors (e.g., ceritinib, alectinib) upon progression. Second-generation inhibitors are generally effective even in the absence of crizotinib-resistant ALK mutations, likely reflecting incomplete inhibition of ALK by crizotinib in many cases. Herein, we analyzed 103 repeat biopsies from ALK-positive patients progressing on various ALK inhibitors. We find that each ALK inhibitor is associated with a distinct spectrum of ALK resistance mutations and that the frequency of one mutation - ALK G1202R - increases significantly after treatment with second-generation agents. To investigate strategies to overcome resistance to second-generation ALK inhibitors, we examine the activity of the third-generation ALK inhibitor lorlatinib in a series of ceritinib-resistant, patient-derived cell lines, and observe that the presence of ALK resistance mutations is highly predictive for sensitivity to lorlatinib, whereas those cell lines without ALK mutations are resistant.
Keywords: Anaplastic lymphoma kinase, ALK, acquired resistance, crizotinib, ceritinib, alectinib, brigatinib, lorlatinib, NSCLC
INTRODUCTION
Anaplastic lymphoma kinase (ALK) rearrangements define a distinct molecular subtype of non-small cell lung cancer (NSCLC; ref 1, 2). Recently, the therapeutic landscape for advanced ALK-positive NSCLC has been transformed by the development of increasingly potent and selective ALK inhibitors. Crizotinib was the first ALK inhibitor to enter clinical development (3). In randomized phase III trials, crizotinib produced significant improvements in objective response rates (ORRs) and progression-free survival (PFS) compared to cytotoxic chemotherapy, establishing crizotinib as a standard treatment for advanced ALK-positive NSCLC (4, 5).
While most patients respond to crizotinib, patients ultimately relapse on therapy, generally within one to two years (4, 5). Analysis of post-progression biopsy specimens has proven extremely valuable, facilitating a greater understanding of molecular mechanisms of crizotinib resistance (6, 7). Broadly speaking, such mechanisms have been classified as involving either on-target genetic alterations (e.g., ALK resistance mutations, ALK gene amplification) or off-target mechanisms of resistance (e.g., up-regulation of bypass signaling pathways, such as EGFR, KIT, IGF-1R, SRC, MEK/ERK and others; ref 6, 8–11). In published series to date, on-target resistance mechanisms have been found in approximately one-third of patients progressing on crizotinib (6, 7).
Recently, several second-generation ALK inhibitors have demonstrated impressive activity in ALK-positive NSCLC (12–16). Two of these agents, ceritinib and alectinib, recently received approval by the U.S. Food and Drug Administration (FDA) for the treatment of crizotinib-refractory, ALK-rearranged NSCLC. A third agent, brigatinib, has received breakthrough-therapy designation by the FDA. In preclinical models, second-generation ALK inhibitors overcome several crizotinib-resistant ALK mutations (17, 18). Furthermore, in phase I–II studies, these agents have demonstrated high ORRs (48–71%) in crizotinib-resistant patients (12–16). Importantly, second-generation ALK inhibitors have also been active in patients without ALK resistance mutations or fusion gene amplification (12), suggesting that many cancers become resistant to crizotinib due to inadequate suppression of ALK. However, despite the efficacy of second-generation ALK inhibitors, patients almost invariably relapse. Thus far, descriptions of molecular mechanisms of resistance to second-generation ALK inhibitors have been limited to in vitro studies, case reports and small clinical series, making it difficult to determine the scope of such alterations (17, 19–23).
Herein, we present the largest series of repeat biopsies from ALK-positive NSCLC patients with resistance to ALK inhibitors, a majority of whom had acquired resistance. Using a combination of genetic sequencing, histological analyses and functional drug screens, we find that the frequency and spectrum of ALK resistance mutations evolve as patients relapse on different ALK inhibitors. Moreover, in a series of ceritinib-resistant, patient-derived cell lines, we demonstrate that the presence of ALK resistance mutations is associated with sensitivity to the novel, third-generation ALK inhibitor lorlatinib. In contrast, cell lines without ALK resistance mutations are resistant to lorlatinib. Together, these findings suggest a role for tailoring ALK inhibitor therapy based upon the underlying mechanisms of resistance.
RESULTS
Baseline Clinical Characteristics
Between January 2009 and June 2016, 83 ALK-positive patients underwent repeat biopsies following disease progression on first- or second-generation ALK inhibitors (Table S1). All biopsies were procured from progressing lesions. Baseline clinical characteristics of these patients are summarized in Table S2. A total of 103 biopsies were performed. Six (7%) patients underwent two separate biopsies while on the same ALK inhibitor (crizotinib N=4, ceritinib N=1, brigatinib N=1). Fourteen patients (18%) had paired repeat biopsies after disease progression on crizotinib and a second-generation ALK inhibitor (ceritinib N=9, alectinib N=3, brigatinib N=2; Table S3).
ALK Resistance Mutations in Crizotinib-Resistant Specimens
We first investigated the frequency of ALK resistance mutations in 51 ALK-positive patients progressing on crizotinib. Twenty-one patients received crizotinib as part of a clinical trial, with 18 (86%) experiencing an objective response by Response Evaluation Criteria in Solid Tumors (RECIST) version 1.0 (24). Among the remaining 30 patients, the median duration of crizotinib treatment was 7.6 months (range 1.5 to 21.4 months). Two of these patients (6.7%) experienced disease progression on the first repeat tumor assessment, indicative of potential intrinsic resistance to therapy. Most biopsies (85%) were performed while patients were still receiving crizotinib or within one month of discontinuation. Biopsy sites included pleural fluid (31%), liver (22%) and nodal tissue (18%; Table S2).
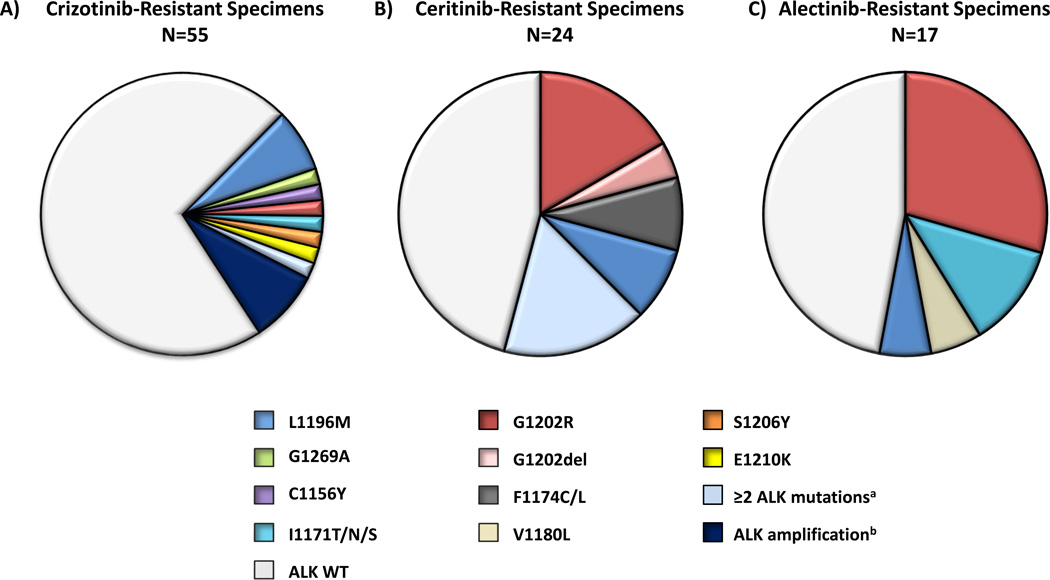
ALK resistance mutations were identified in 11 (20%) specimens (Figure 1A) among 10 (20%) patients. Consistent with prior reports (6, 7), the most common ALK resistance mutations were L1196M and G1269A, but these were present in only 7% and 4% of all of the crizotinib-resistant specimens, respectively. The remaining ALK resistance mutations included: C1156Y (2%), G1202R (2%), I1171T (2%), S1206Y (2%), and E1210K (2%). Four patients underwent two separate biopsies on crizotinib. In three patients, both samples were negative for ALK mutations; one patient harbored ALK L1196M in two separate pleural fluid specimens obtained approximately one month apart. No ALK resistance mutations were found among the two patients with intrinsic resistance.
Figure 1.
Overview of on-target mechanisms of resistance among ALK-positive specimens obtained from patients progressing on: A) crizotinib, B) ceritinib, and C) alectinib. Pie charts depict the frequency and distribution of ALK resistance mutations and ALK fusion gene amplification in each cohort. Four patients underwent two separate biopsies while on crizotinib; one patient underwent two separate biopsies while on ceritinib. Note: If a specimen is listed as having ≥2 ALK resistance mutations, the individual mutations are not listed separately. aOne post-crizotinib specimen harbored ALK G1269A and 1151Tins mutations. Four post-ceritinib samples contained ≥2 ALK resistance mutations. These included: I1171N+C1156Y, D1203N+F1174C, F1174L+G1202R, C1156Y+G1202del+V1180L mutations. bALK fluorescence in situ hybridization (FISH) to assess for fusion gene amplification was performed in only crizotinib-resistant specimens (N=36), of which 8% had amplification. Ceritinib- and alectinib-resistant specimens were not assessed for ALK amplification by FISH. ALK, anaplastic lymphoma kinase; WT, wild-type.
Thirty-six crizotinib-resistant specimens underwent repeat ALK fluorescence in situ hybridization (FISH). All were positive for ALK rearrangements. Three specimens (8.3%) demonstrated ALK gene amplification (defined as ALK/centromere 2 ratio > 2.0). No ALK-amplified tumor harbored a concomitant ALK resistance mutation. Thus, among 36 crizotinib-resistant specimens tested for both ALK resistance mutations and ALK gene amplification, 31% harbored on-target genetic alterations contributing to resistance. As ORRs to second-generation ALK inhibitors are reported to be 48–71% following progression on crizotinib (12–16), this low rate of on-target resistance mechanisms is consistent with previous studies demonstrating that crizotinib-resistant tumors without ALK point mutations also respond to second-generation ALK inhibitors (12).
ALK Resistance Mutations are More Common after Treatment with Second-Generation ALK Inhibitors
To evaluate whether the scope of ALK resistance mutations changes following treatment with second-generation ALK inhibitors, we assembled cohorts of ALK-positive patients who underwent post-progression biopsies on ceritinib (N=23), alectinib (N=17), or brigatinib (N=6). Baseline clinical characteristics for each cohort are summarized in Table S2. Repeat biopsies were performed either on drug or within one month of discontinuation in 86%, 94% and 100% of patients receiving ceritinib, alectinib and brigatinib, respectively.
Among 23 patients undergoing ceritinib-resistant biopsies, 21 (91%) had received prior crizotinib. Details regarding prior crizotinib exposure and intervening therapies are provided in Table S4. In nine cases, pre-ceritinib/post-crizotinib biopsies were also available. Notably, only two (22%) of these crizotinib-resistant specimens harbored on-target mechanisms of resistance. MGH011 was found to have an ALK S1206Y mutation, while MGH034 harbored ALK fusion gene amplification. Three (13%) patients had previously received both crizotinb and alectinib, but none underwent biopsies following progression on either of those agents. Among the entire cohort of patients with ceritinib-resistant biopsies (N=23), 13 were treated with ceritinib as part of a clinical trial, and 8 (62%) of these patients experienced an objective response (RECIST v1.0). The median duration of ceritinib treatment among the remaining 10 patients was 8.0 months (range 1.1–9.2 months), suggesting that the majority of patients had acquired resistance to therapy. Two patients progressed on ceritinib on the first tumor assessment, indicative of intrinsic resistance.
In total, 24 separate post-ceritinib biopsies were performed. Over one-half of ceritinib-resistant specimens (13/24; 54%) harbored ALK resistance mutations (Figure 1B), and 17% contained ≥2 ALK resistance mutations. The most common ALK mutations were G1202R (21%) and F1174C/L (16.7%), both of which have been previously described in ALK-positive NSCLC (6, 17). G1202R maps to the solvent-exposed region of the ALK kinase, where the bulkier, charged side chain is thought to lead to steric hindrance of most ALK inhibitors (6, 17, 25). ALK F1174 mutations map adjacent to the C-terminus of the αC helix and may stabilize an active conformation that increases the ATP-binding affinity of ALK (22, 26). In addition to G1202R and F1174C/L, we observed ALK C1156Y mutations in two (8%) specimens. In preclinical models, we previously demonstrated that ceritinib has less activity against ALK C1156Y (17). It should be noted, however, that both C1156Y-containing specimens in this series also harbored additional ALK mutations (MGH084-1: C1156Y and I1171N; MGH932-1: C1156Y, V1180L, and G1202del) that may have contributed to resistance.
In this analysis, we also identified a novel ALK G1202del mutation in two (8%) specimens. To directly evaluate whether ALK G1202del confers resistance to ceritinib, we engineered Ba/F3 cells to express EML4-ALK (E13;A20) harboring G1202del and examined ALK phosphorylation after treatment with various ALK inhibitors. We observed that G1202del confers moderate levels of resistance to ceritinib, alectinib and brigatinib (Figures S1A–D and S2A–C). By contrast, crizotinib potency was less impacted by G1202del. Structural data on the G1202del mutation is not available. Based upon structural modeling studies, however, we speculate that deletion of the glycine at position 1202 could shift the aspartic acid at position 1203 into the 1202 position, leading to disruption of TKI binding (T Johnson, personal communication). Importantly, while our functional models suggest that ALK G1202del may be a novel resistance mutation, G1202del appears to be quite distinct from ALK G1202R. Indeed, in Ba/F3 models, ALK G1202R results in much higher degrees of resistance to all currently available first- and second-generation ALK inhibitors (Figure S1D).
We next evaluated a cohort of 17 ALK-positive patients who underwent repeat biopsies following progression on alectinib. All 17 patients (100%) had previously received crizotinib; three patients (18%) had received both crizotinib and ceritinib. Only two patients (MGH087 and MGH090) underwent pre-alectinib/post-crizotinib biopsies, both of which were negative for ALK resistance mutations. Details regarding prior ALK inhibitor exposure and intervening therapies are available in Table S5 and Figure S3. Within this cohort, the ORR with alectinib was 40% (RECIST v1.1; ref 27). Among 17 alectinib-resistant biopsies, ALK resistance mutations were found in 9 (53%) specimens (Figure 1C). Moreover, the most common ALK resistance mutation was G1202R, which was found in 29% of cases. Interestingly, among three patients with RECIST-defined progressive disease on a first repeat tumor assessment (i.e., intrinsic resistance), two harbored G1202R. Other ALK resistance mutations identified within the complete alectinib-resistant cohort included I1171T/S (12%), V1180L (6%), and L1196M (6%). Of note, several different ALK I1171 mutations (e.g., I1171T/N/S) have been reported previously among ALK-positive patients progressing on alectinib (23, 28). ALK V1180L has also been previously shown to confer resistance to alectinib in vitro (29), but this represents the first clinical observation of an ALK V1180L mutation in an alectinib-resistant patient. Interestingly, alectinib has demonstrated significant activity against the ALK gatekeeper mutation L1196M in preclinical models (18), yet this was observed in one (6%) post-alectinib biopsy. Of note, this patient (MGH988) had previously received crizotinib for 12.1 months, but no post-crizotinib/pre-alectinib biopsy was available. Therefore, it is possible that the L1196M mutation existed prior to treatment with alectinib in this case.
We next evaluated a third cohort (N=6) of ALK-positive patients who underwent biopsies following progression on brigatinib. Five of six patients had previously received crizotinib, and none had received another second-generation ALK inhibitor. Treatment histories for each patient are summarized in Table S6. Median duration of treatment was 20.2 months (range 12.1 to 44.4 months). No patients had intrinsic resistance to therapy. One patient (MGH086) underwent two separate brigatinib-resistant biopsies at the same anatomic site (see Compound ALK Resistance Mutations). Overall, ALK resistance mutations were seen in 5/7 (71%) brigatinib-resistant specimens. Like patients progressing on ceritinib and alectinib, the most common ALK resistance mutation was G1202R, which was observed in three specimens.
Given the retrospective nature of this analysis, there is limited clinical data on the therapies patients received after disease progression on ceritinib, alectinib and/or brigatinib. Immediate post-progression therapies are summarized in Tables S4–6. Patients generally received systemic chemotherapy (25%) or they were enrolled onto clinical trials (31%). Three patients (6%) received commercially available ALK inhibitors, while the remaining 38% of patients did not receive subsequent therapy and/or were lost to follow-up. Thus, we are unable to draw meaningful associations between our findings in post-progression biopsies and clinical outcomes to subsequent therapies.
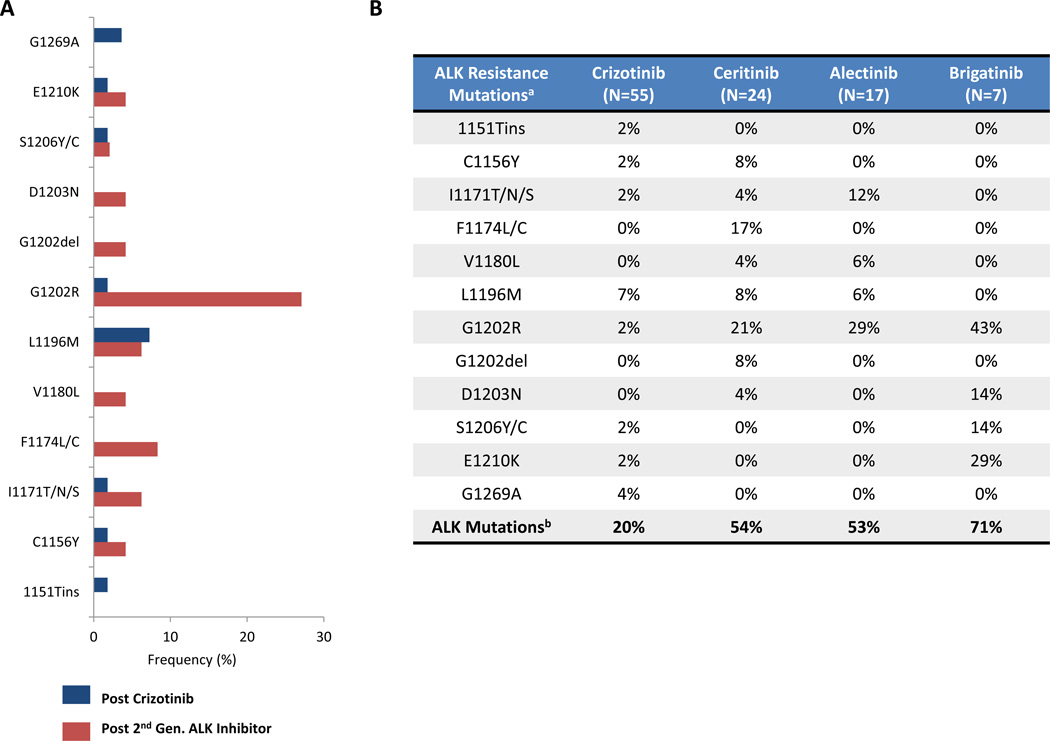
Collectively, across all three biopsy cohorts (N=48), ALK resistance mutations were present in 56% of ALK-positive patients progressing on second-generation ALK inhibitors (ceritinib 54%, alectinib 53%, and brigatinib 71%). Thus, while ALK resistance mutations are observed in only 20% of ALK-positive patients progressing on crizotinib, treatment with more potent second-generation ALK inhibitors is associated with a significantly higher frequency of ALK resistance mutations (P = 0.0002) and a different spectrum of such mutations (Figure 2A). Indeed, consistent with preclinical data, ALK G1202R emerged as the most common ALK resistance mutation among patients receiving second-generation ALK inhibitors (Figure 2A–B).
Figure 2.
ALK resistance mutations are more common after treatment with second-generation ALK inhibitors compared to crizotinib. A) Comparison of the frequency and distribution of ALK resistance mutations in biopsy specimens obtained after disease progression on crizotinib (blue) or second-generation ALK inhibitors (red). Frequencies are expressed based upon the total numbers of biopsies in each cohort. B) Breakdown of specific ALK resistance mutations in ALK-positive patients progressing on crizotinib, ceritinib, alectinib or brigatinib. aFor patients with ≥2 ALK resistance mutations in a biopsy, each individual mutation is incorporated into the frequencies above. bEach specimen with ≥2 ALK resistance mutations is considered only once in determining the total number of specimens with ALK resistance mutations. WT, wild-type.
Compound ALK Resistance Mutations Following Sequential ALK Inhibitor Therapy
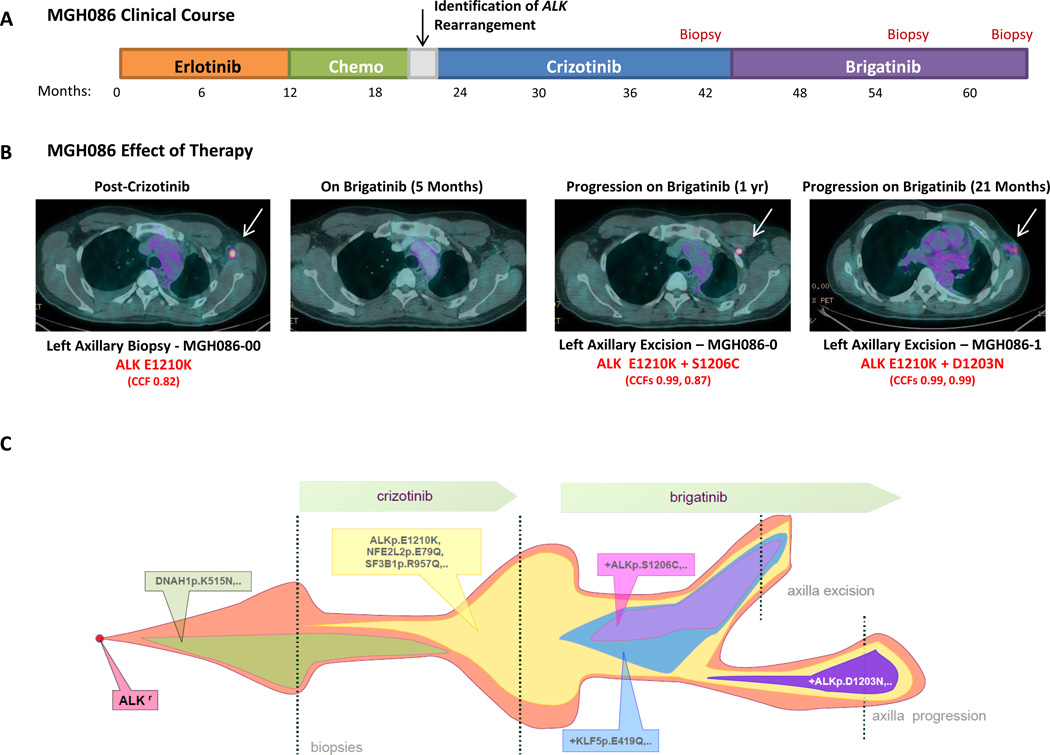
Based upon experience from other targeted therapy settings (e.g., chronic myeloid leukemia [CML] and EGFR-mutant lung cancer; ref 30–32), we hypothesized that sequential ALK inhibitor therapy may predispose patients to develop compound mutations. Indeed, we recently described the development of a compound ALK resistance mutation in a patient treated with crizotinib, ceritinib and lorlatinib (33). To investigate the frequency of such dual alterations, we examined biopsies from patients progressing on second-generation ALK inhibitors, identifying 6/48 (12.5%) specimens harboring ≥2 ALK resistance mutations (Table S7). In each case, patients had received crizotinib and a second-generation ALK inhibitor. In three specimens (MGH905-1, MGH086-0, and MGH086-1), ALK resistance mutations were in close enough proximity to confirm that they were present on the same allele or whole-exome sequencing (WES) demonstrated similar cancer cell fractions of each resistance mutation, suggesting that they were present on the same allele of the ALK fusion gene. The remaining cases did not have sufficient tissue available to make this determination.
In one of the patients above (MGH086), we were also able to investigate clonal evolution of compound ALK resistance mutations over time within the same anatomic site. The clinical course for MGH086 is summarized in Figure 3A. Following identification of an ALK rearrangement, MGH086 was sequentially treated with crizotinib and brigatinib. Despite prolonged clinical benefit on each agent, he ultimately developed recurrent left axillary adenopathy requiring local excision/biopsy on three separate occasions (post-crizotinib N=1, post-brigatinib N=2; Figure 3A–B). WES of the post-crizotinib specimen (MGH086-00) demonstrated an acquired ALK E1210K mutation. E1210 maps to the ribose-binding pocket of the ALK kinase (26). ALK E1210K confers resistance to crizotinib based upon in vitro mutagenesis screens, but this alteration has not been described in clinical NSCLC samples previously (26). We next performed WES on the first brigatinib-resistant excision specimen (MGH086-0), which revealed an ALK E1210K mutation and a new ALK S1206C mutation (Figure 3B). Notably, WES of the second brigatinib-resistant excision specimen (MGH086-1) also demonstrated the ALK E1210K mutation, but ALK S1206C was no longer observed. Instead, a new ALK D1203N mutation was found. Clonal analysis suggested that the E1210K mutation emerged as an early resistant clone after treatment with crizotinib. Subsequent treatment with brigatinib enriched for this clone, which eventually acquired a compound ALK S1206C mutation. It is possible that surgical excision of the patient’s left axillary node may have physically removed the E1210K+S1206C clone, but parental E1210K clones may have still been present microscopically. Ultimately, these clones may have acquired an ALK D1203N mutation with continued brigatinib exposure (Figure 3C).
Figure 3.
Clonal evolution of resistance to sequential ALK inhibitor therapy. A) Panel A depicts the treatment course of patient MGH086. Of note, the patient received several lines of therapy, including the EGFR inhibitor erlotinib, before identification of an ALK rearrangement. The points at which the patient underwent biopsies are indicated in red. B) Fused positron emission tomography (PET)-computed tomography (CT) images demonstrate a hypermetabolic, left axillary lymph node (white arrow) that developed at the time of disease progression on crizotinib. Whole-exome sequencing (WES) revealed an ALK E1210K mutation (cancer cell fraction [CCF] 0.82). This axillary lymph node initially responded to brigatinib but recurred after 12 months. WES of this brigatinib-resistant lesion (MGH086-0) demonstrated continued presence of the ALK E1210K mutation and a new ALK S1206C mutation. The patient remained on brigatinib. After an additional 9 months on brigatinib, he developed another recurrence in the left axilla (slightly more inferior than the prior lesion). Repeat WES (MGH086-1) revealed a new compound ALK E1210K+D1203N mutation. C) Panel C shows a model of clonal evolution of resistance to sequential ALK inhibitor therapy in patient MGH086. Using whole-exome sequencing, we determined that a founder ALK E1210K subclone was not present in a pre-crizotinib biopsy but later developed on crizotinib. When the patient was switched to brigatinib, the ALK E1210K subclone expanded and ultimately acquired a new ALK mutation, S1206C. Surgical excision of this site of progression may have depleted the compound mutant (ALK E1210K+S1206C), but microscopic parental E1210K clones may have persisted, ultimately acquiring ALK D1203N in combination with ALK E1210K.
These data demonstrate that compound ALK resistance mutations can arise in ALK-positive patients treated with sequential ALK inhibitors. Moreover, as will be discussed in greater detail below, such compound mutations can confer high levels of resistance to ALK inhibitors.
Broad Assessment of Genetic Mutations in Resistant Cancers
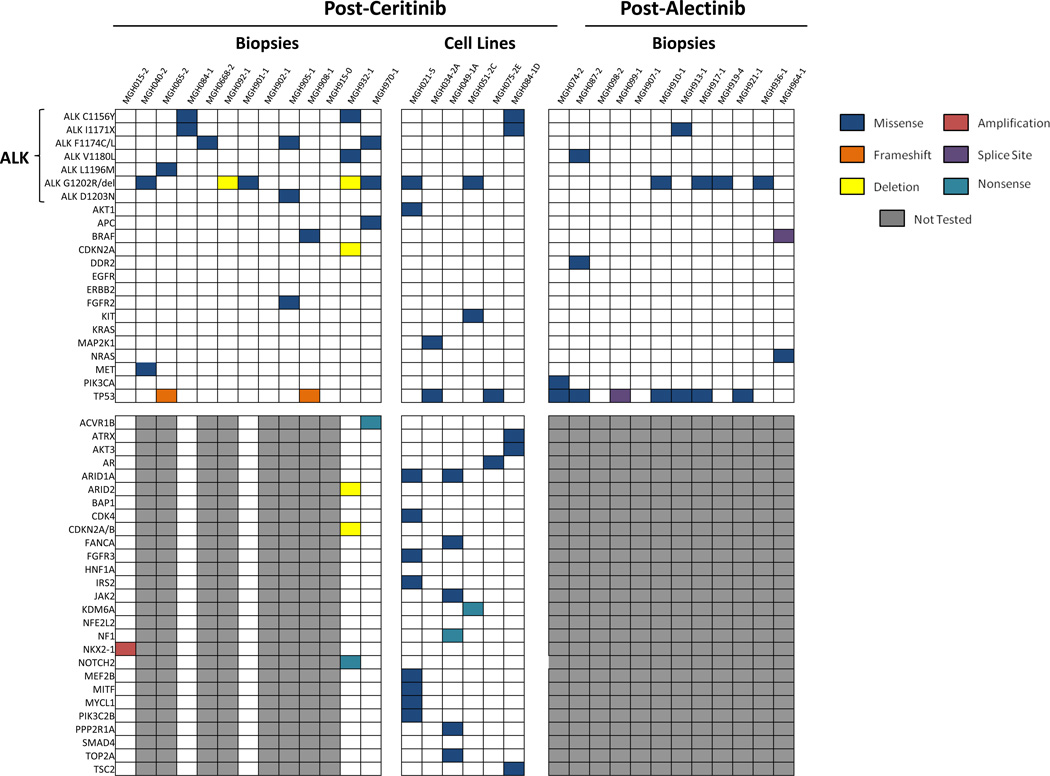
ALK resistance mutations appear to be the predominant mechanism of resistance to second-generation ALK inhibitors. Nonetheless, we observed that 44% of post-second-generation ALK TKI biopsies were negative for ALK mutations. To investigate the potential role of alternative mechanisms of resistance, such as up-regulation of bypass signaling pathways, we first performed targeted NGS on post-ceritinib, post-alectinib and post-brigatinib biopsy specimens using the MGH Snapshot NGS platform or FoundationOne™ NGS (Tables S8–10; Fig 4).
Figure 4.
Summary of genetic alterations in resistant biopsies among patients progressing on ceritinib or alectinib. Specimens underwent targeted, next generation sequencing (NGS) using the Massachusetts General Hospital (MGH) NGS assay or the FoundationOne platform. Only genes with at least one genetic alteration detected in resistant specimens are depicted. If a particular gene was not evaluated in a given specimen due to the type of sequencing platform used, it is represented in dark gray. Within this cohort, the most commonly mutated genes were ALK and TP53. Recurrent alterations in other genes were uncommon. In addition, six ceritinib-resistant, patient-derived cell lines underwent NGS using a 1000 gene panel (See Genotype Assessments in Methods). Genes from this panel are depicted in Figure 4 if the following criteria are met: (1) a genetic alteration was present in at least one of the patient-derived cell lines and (2) the gene was also included in either the MGH NGS or FoundationOne panels. Please see Table S11 for a comprehensive assessment of all genetic alterations identified within these cell lines.
Twenty-seven specimens had sufficient tissue for analysis. Beyond ALK resistance mutations, 15 (56%) specimens showed genetic alterations in at least one other gene. TP53 mutations were the most common, present in 9 (33%) specimens. Of note, we were unable to determine whether these mutations were present prior to treatment with second-generation ALK inhibitors due to a lack of baseline tissue for analysis. No KRAS or EGFR mutations were identified. Missense mutations in DDR2 (L610F), BRAF (G15V), FGFR2 (F645L), MET (T992I), NRAS (A155T) and PIK3CA (G106V) were each identified in one (3.7%) specimen, none of which were overlapping. Mutations in DDR2, BRAF, NRAS and FGFR2 did not occur in known hotspot residues, nor have they been observed in NSCLCs in the COSMIC database (COSMICv76); thus, the impact of these alterations on ALK inhibitor resistance is uncertain. MET T992I was observed in one specimen in this series (MGH040-2). MET T992I has been reported at low frequencies in various malignancies, but functional studies have demonstrated that this variant lacks transformative capacity and does not impact MET phosphorylation status (34). Thus, MET T992I was unlikely a major driver of resistance in this patient—particularly since this specimen harbored a concomitant ALK G1202R mutation. By contrast, the alectinib-resistant specimen MGH074-2 showed no ALK resistance mutations but harbored a PIK3CA G106V mutation. Previous studies have shown that PIK3CA G106V is a gain-of-function mutation that localizes to the p85/adaptor-binding domain of p110α and results in increased AKT phosphorylation (35). In addition, PIK3CA mutations have been associated with acquired resistance to EGFR inhibitors in EGFR-mutant NSCLC (36). Due to tissue availability, we were unable to assess this patient’s pre-alectinib specimen for the presence of PI3KCA G106V; thus, it remains unclear when this alteration arose. Among the remaining 26 cases that underwent NGS, no additional PIK3CA mutations were identified; however, we previously reported a PIK3CA H1047R mutation in a ceritinib-resistant specimen (MGH034-2) that was identified by the Snapshot allele-specific assay (10, 37).
To identify other potential off-target mechanisms of resistance to second-generation ALK inhibitors, we established six ceritinib-resistant, patient-derived cell lines. All samples underwent targeted NGS of 1000 known cancer genes (Table S11, Figure 4; ref 10). Samples without ALK resistance mutations (MGH034-2A, MGH049-1A, and MGH075-2E) were also evaluated using a combination drug screen to identify potential novel mechanisms of resistance (10). As we previously reported (10), MGH034-2A harbored an acquired MAP2K1 K57N mutation. Furthermore, treatment with a MEK inhibitor (AZD6244) re-sensitized these cells to ceritinib, suggesting that reactivation of the MAPK pathway promoted resistance in this specimen. In addition, for MGH049-1A, we previously observed that agents targeting SRC, EGFR and PI3K re-sensitized cells to ALK inhibition, implicating these signaling pathways as mediators of resistance in this model (10). Notably, no genetic alterations in these pathways were identified in our 1000-gene NGS analysis. Finally, we performed the same pharmacological screen on MGH075-2E, identifying dasatinib as the only hit (Figure S4A–B). In addition to ABL and Eph receptors, dasatinib potently inhibits SRC family kinases, which have been associated with resistance to ALK inhibitors previously (10). Of note, we could not identify a genetic basis for SRC activation using a 1000-gene NGS panel. Prior biochemical experiments using cell line models sensitive to combined ALK and SRC inhibition have suggested potential cross talk between ALK and SRC, with ALK inhibition leading to up-regulation of SRC signaling (10).
Collectively, these observations reinforce our finding that ALK resistance mutations are the predominant mechanisms of resistance to second-generation ALK inhibitors. While several potential bypass signaling tracts were identified in individual patients, no high frequency, recurring genetic alterations beyond TP53 mutations were observed.
Epithelial-Mesenchymal Transition Is Associated with ALK Inhibitor Resistance
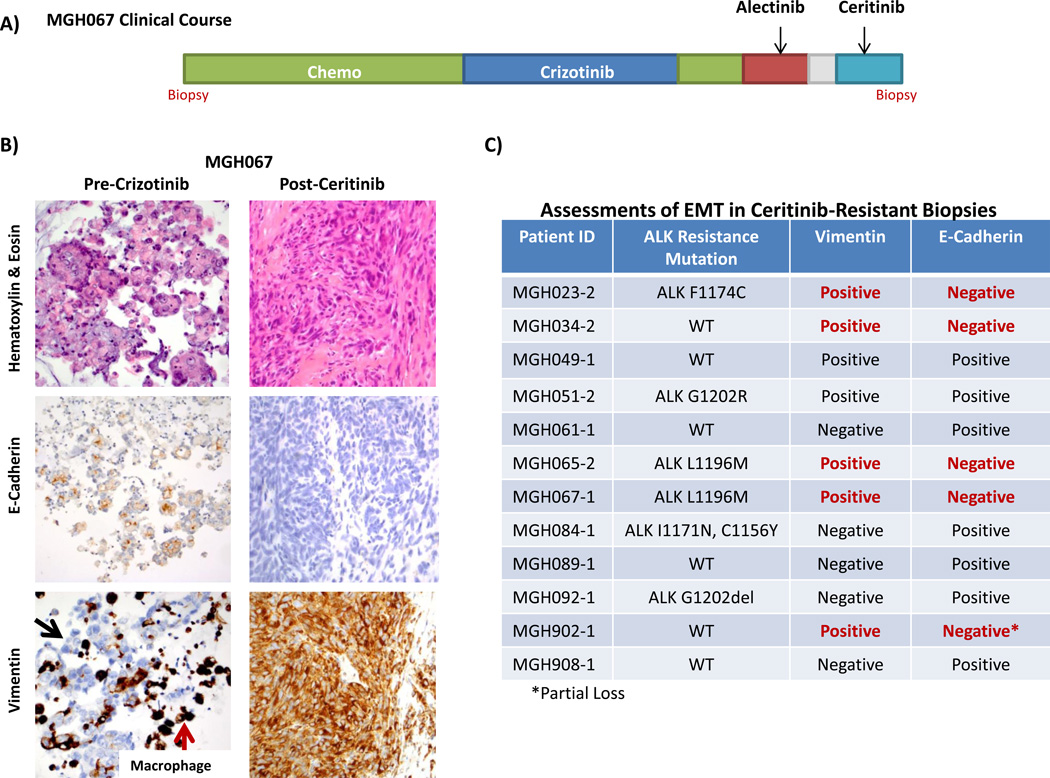
In preclinical models, ceritinib, alectinib and brigatinib have demonstrated activity against the ALK L1196M gatekeeper mutation (17, 18, 38). However, we identified three ALK-positive patients with ALK L1196M following treatment with second-generation ALK inhibitors (ceritinib N=2, alectinib N=1), suggesting that additional resistance mechanisms may be responsible. In one of these specimens (MGH067-1), obtained from an ALK-positive patient treated with crizotinib, alectinib, and ceritinib (Figure 5A), the post-ceritinib biopsy revealed a malignant spindle cell neoplasm with no morphological or immunohistochemical markers of epithelial differentiation (Figure 5B). In contrast to the patient’s initial diagnostic specimen, the post-ceritinib biopsy showed diffuse vimentin expression and loss of E-cadherin staining, consistent with epithelial-to-mesenchymal transition (EMT). Of note, ALK FISH was positive for a rearrangement (Figure S5A–C), but immunohistochemical staining for ALK showed negative to weak staining (Figure S5D–F). Post-crizotinib and/or post-alectinib biopsies were not performed in this patient; thus, we could not determine when the ALK L1196M mutation was acquired. However, we suspect that it was acquired before ceritinib. Indeed, it is possible that EMT, rather than the ALK L1196M mutation, may have contributed more significantly to ceritinib resistance in this patient.
Figure 5.
Epithelial-mesenchymal transition (EMT) is associated with ceritinib resistance. A) Panel A depicts the clinical course of patient MGH067. B) Pre-crizotinib and post-ceritinib biopsies (lung and subcutaneous lesions, respectively) from MGH067 underwent hematoxylin and eosin staining, and immunostaining for E-cadherin, and vimentin. The post-ceritinib biopsy shows a loss of E-cadherin staining and gain of vimentin expression, consistent with EMT. Black arrows indicate a lack of vimentin staining of tumor cells in the pre-crizotinib biopsy. Red arrows depict vimentin staining of alveolar macrophages in the same specimen. C) Twelve ceritinib-resistant biopsy specimens underwent E-cadherin and vimentin staining to assess for EMT. In total, five specimens demonstrated immunohistochemical features consistent with EMT.
Based upon the above observation, we performed immunohistochemical staining on 11 other ceritinib-resistant biopsy specimens with sufficient tissue for analysis to investigate the frequency of EMT in ALK-positive patients progressing on second-generation ALK inhibitors. EMT is generally associated with loss of E-cadherin staining and gain of vimentin expression (36). In total, we observed transition to a more mesenchymal phenotype in 5/12 (42%) specimens (Figures 5C). Interestingly, among these five specimens, three harbored ALK resistance mutations, including the two cases with ALK L1196M. Notably, post-crizotinib/pre-ceritinib biopsies were not available in these cases but baseline diagnostic specimens were available for comparison in all five cases (Table S12). Four of these were negative for EMT. In one patient (MGH065), the baseline biopsy showed strong vimentin expression, but preserved E-cadherin staining, which was lost in the ceritinib-resistant cancer. Of note, histological transformation to small cell lung cancer (SCLC) was not observed in these cases. Together, these findings suggest that EMT was acquired during ALK inhibitor therapy and may therefore play a role in resistance to second-generation ALK inhibitors. However, EMT may not be the sole driver of resistance in these patients, since several specimens with EMT had concomitant alterations that may also contribute to resistance.
Lorlatinib is Active Against ALK Resistance Mutations that Develop on Second-Generation ALK Inhibitors
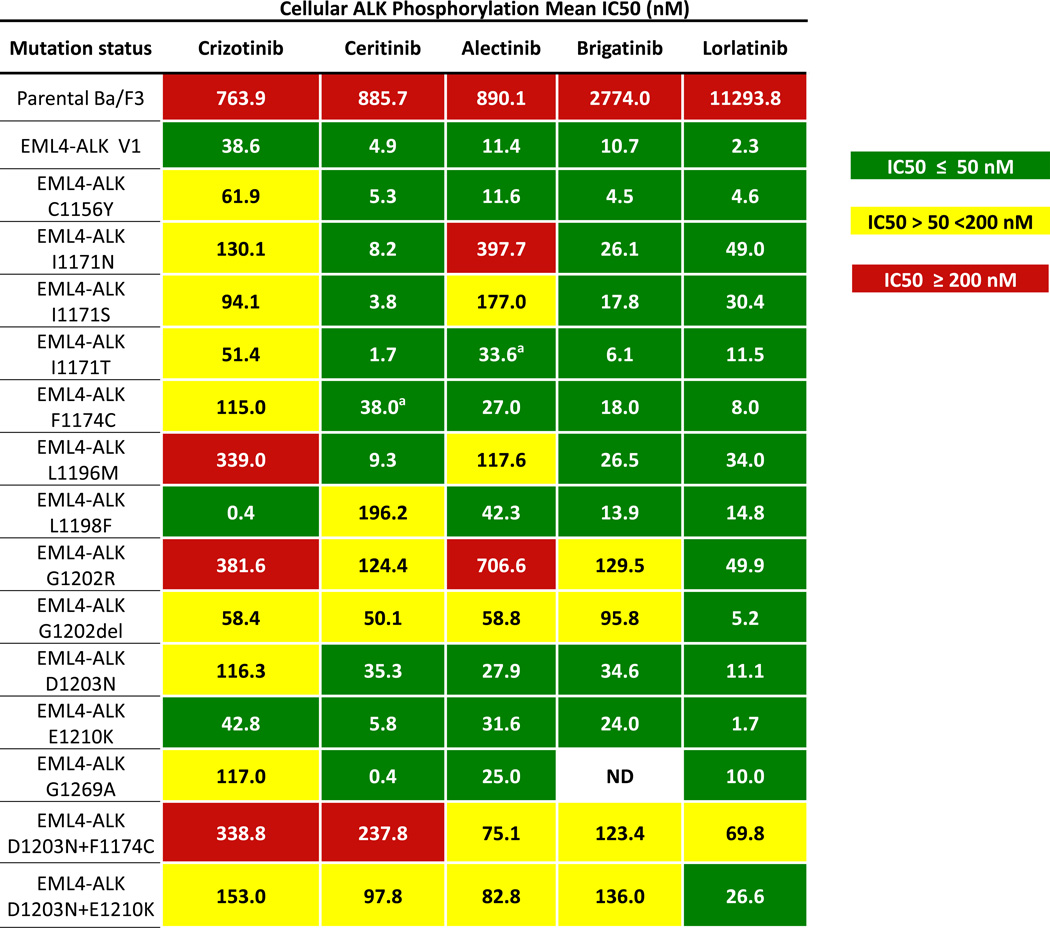
To investigate whether cancers that develop resistance to second-generation ALK inhibitors remain susceptible to continued ALK inhibition, we examined the activity of the third-generation ALK inhibitor, lorlatinib (PF-06463922). Lorlatinib is a potent, highly selective ALK/ROS1 inhibitor that is currently being evaluated in an ongoing phase 2 clinical trial (NCT01970865). We first engineered Ba/F3 cells to express wild-type EML4-ALK (E13;A20) or EML4-ALK harboring various ALK mutations (Figure 6). Ba/F3 cells were treated with crizotinib, ceritinib, alectinib, brigatinib and lorlatinib. Notably, lorlatinib was the only ALK inhibitor to potently inhibit ALK phosphorylation across all single ALK secondary mutations, including ALK G1202R (IC50 49.9 nM). Next, based upon our observations of compound ALK resistance mutations in a subset of patients, we also evaluated the activity of various ALK inhibitors against compound ALK resistance mutations. We found that crizotinib, ceritinib, alectinib and brigatinib were all inactive against D1203N+E1210K and F1174C+D1203N. Conversely, lorlatinib retained significant potency against the ALK double-mutant D1203N+E1210K and intermediate potency against D1203N+ F1174C.
Figure 6.
Lorlatinib potently inhibits ALK resistance mutations, including ALK G1202R. Absolute IC50 values of crizotinib, ceritinib, alectinib, brigatinib, and lorlatinib on cellular ALK phosphorylation in Ba/F3 cells harboring wild-type EML4-ALK variant 1 or various EML4-ALK resistance mutants are depicted. aIn Ba/F3 cells, ALK F1174C and ALK I1171T appear sensitive to ceritinib and alectinib, respectively; however, these mutations may not be susceptible to these agents in vivo based upon prior clinical reports.
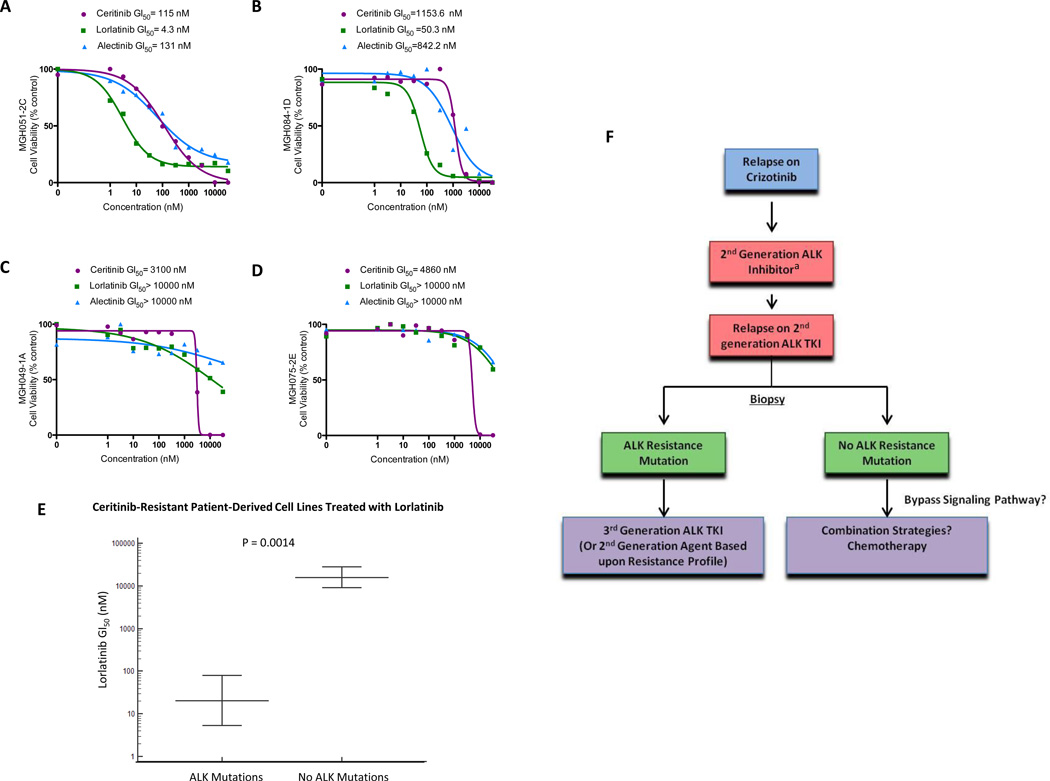
To further examine the activity of lorlatinib in ALK-resistant models, we next evaluated a series of ceritinib-resistant, patient-derived cell lines (N=6). Three cell lines harbored ALK resistance mutations (MGH021-5A, MGH051-2C, and MGH084-1D), while three cell lines (MGH034-2A, MGH049-1A, and MGH075-2E) were wild-type for ALK secondary mutations. Notably, alectinib and ceritinib had minimal effects on cell growth across all patient-derived cell lines (GI50 values 131 nM to >10,000 nM and 115 nM to >10,000 nM, respectively; Figures 7, S6A–B). By contrast, lorlatinib markedly inhibited cell growth in 3/6 patient-derived cell lines (Figure 7A–B, S6A). Interestingly, this activity was restricted to cell lines with ALK resistance mutations (MGH021-5A, MGH051-2C, MGH084-1D; Figure S7A–D). The remaining three cell lines that lacked ALK resistance mutations (MGH034-2A, MGH049-1A, and MGH075-2E) were insensitive to lorlatinib (GI50 values >10,000 nM; Figures 7C–D, S8A–D, S9A–C). These data suggest that in the setting of acquired resistance to second-generation ALK inhibitors, the presence of ALK resistance mutations indicates continued dependency on ALK signaling and susceptibility to the pan-inhibitory ALK inhibitor lorlatinib (Figure 7E–F). In contrast, the absence of ALK resistance mutations after failure of second-generation ALK inhibitors may indicate loss of ALK dependency and resistance to lorlatinib.
Figure 7.
ALK resistance mutations predict for sensitivity to lorlatinib in patient-derived cell line models of acquired resistance to ceritinib. A–B) Cell viability assays of two representative ceritinib-resistant, patient-derived cell lines harboring ALK resistance mutations (MGH051-2C [EML4-ALKG1202R] and MGH084-1D [EML4-ALKI1171N,C1156Y]) treated with ceritinib, alectinib and lorlatinib. The number of cells seeded and the duration of treatment were adjusted for each cell line in order to have a consistent proliferation index (3.5 to 5) at the end of treatment. Values are presented as means (N=3). C–D) Cell viability assays of two representative, ceritinib-resistant, patient-derived cell lines without ALK resistance mutations (MGH049-1A [EML4-ALKWT] and MGH075-2E [EML4-ALKWT]) treated with ceritinib, alectinib and lorlatinib. The number of cells seeded and the duration of treatment were adjusted for each cell line in order to have a consistent proliferation index (3.5 to 5) at the end of treatment. Values are presented as means (N=3). E) Comparison of cell viabilities of ceritinib-resistant, patient-derived cell lines treated with lorlatinib based upon ALK resistance mutation status. F) Proposed schema for the clinical approach to ALK-positive patients with acquired resistance. This paradigm incorporates repeat biopsies and decision-making based upon ALK resistance mutation status following disease progression on second-generation ALK inhibitors. aChoice of second-generation ALK inhibitors may be impacted by identification of specific ALK resistance mutations, such as G1202R and I1171N/S/T, which can be rarely seen after crizotinib.
DISCUSSION
In this study, we present the largest systematic analysis of ALK inhibitor resistance to date and the first study to evaluate mechanisms of resistance across a spectrum of first- and second-generation ALK inhibitors. Consistent with earlier reports (6, 7), we found that only a minority of ALK-positive patients (~20%) developed ALK resistance mutations on crizotinib. By contrast, ALK resistance mutations were present in over one-half of patients progressing on second-generation ALK inhibitors, likely reflecting the greater potency and selectivity of these agents compared to crizotinib. In parallel, we observed that the spectrum of ALK resistance mutations was different following progression on second-generation ALK inhibitors compared to crizotinib. Most notably, ALK G1202R, which was present in only 2% of crizotinib-resistant biopsies, emerged as the most common ALK resistance mutation after treatment with second-generation ALK inhibitors. These findings are consistent with data from Ba/F3 models, which demonstrate that ALK G1202R confers high levels of resistance to all currently available second-generation ALK inhibitors.
While ALK G1202R was a common shared resistance mutation in each second-generation ALK inhibitor cohort, it is noteworthy that the spectrum of other ALK resistance mutations appeared to differ across agents. For example, ALK F1174 mutations were observed in several post-ceritinib biopsy specimens (4/24; 16.7%), but were otherwise absent from post-alectinib and post-brigatinib biopsies. While this comparison is limited due to the relatively small sample size, such observations may reflect the structural differences between ALK inhibitors. Indeed, prior reports suggest that ALK F1174 mutations confer resistance to ceritinib but remain sensitive to alectinib (17, 23). Conversely, several investigators have described ALK I1171 mutations that mediate resistance to alectinib, while conferring sensitivity to ceritinib (23, 29, 39). Consistent with these reports, we observed ALK I1171 mutations to be the second-most common ALK resistance mutations in post-alectinib specimens (N=2; 12%). Of note, one (4.1%) post-ceritinib sample also contained an ALK I1171N mutation (in combination with ALK C1156Y), but this patient had previously received crizotinib, alectinib and ceritinib. Additional biopsies between therapies were unavailable; thus, it is unclear whether the ALK I1171N mutation was acquired as a result of ceritinib or alectinib. Nonetheless, our findings that different ALK resistance mutations impart differential sensitivities to second-generation ALK inhibitors may have important clinical implications. Specifically, this provides support for a new paradigm in which particular ALK resistance mutations inform the choice of subsequent ALK targeted therapies, especially after failure of two ALK inhibitors. Tailoring of ALK therapy after failure of crizotinib may also be important in the small proportion of cases with uncommon and refractory mutations like ALK G1202R.
From a therapeutic standpoint, overcoming resistance to ALK inhibitors may be further complicated by the emergence of compound ALK resistance mutations. In this study, we identified six specimens with ≥2 ALK resistance mutations. In all cases, patients had received multiple ALK inhibitors, suggesting that sequential use of ALK inhibitors may facilitate the development of compound ALK mutations. Furthermore, in Ba/F3 models, we demonstrated that compound ALK mutations conferred increased drug resistance. However, one important exception to this observation has emerged. We recently reported the case of an ALK-positive patient treated with crizotinib, ceritinib and lorlatinib, who ultimately acquired a dual ALK L1198F+C1156Y mutation at the time of disease progression (33). Interestingly, ALK L1198F paradoxically re-sensitized cells to crizotinib, again underscoring the importance of serial biopsies in ALK-positive NSCLC. Of note, the emergence of compound resistance mutations in ALK-positive NSCLC is analogous to the experience with other targeted therapies. For example, in CML, sequential use of different ABL inhibitors has been shown to select for more drug-resistant, compound BCR-ABL mutations (30, 40). Similarly, in EGFR-mutant NSCLC, compound drug-resistant T790M/C797S mutations have been described following sequential treatment with first- and third-generation EGFR inhibitors (31, 33, 41). Ultimately, new therapeutic strategies, such as up-front TKI combinations, may be needed to suppress the emergence of on-target resistance mechanisms, particularly compound resistance mutations.
We also investigated potential off-target resistance mechanisms using a combination of NGS, histological analyses, and functional drug screens. We previously identified up-regulation of bypass signaling tracts involving SRC and the MAPK pathway (10). We also observed PIK3CA mutations in two patients. PIK3CA mutations have been implicated in resistance to other targeted therapies, such as EGFR inhibitors (36). Nonetheless, outside of TP53 mutations, recurring genetic alterations were uncommon in this series. We did however observe evidence of EMT in some patients. EMT is believed to enhance cell motility and invasiveness (42). Preclinical studies have also suggested that EMT is associated with resistance to crizotinib and ceritinib (43–45), but clinical specimens were not examined in these reports. Of note, other phenotypic changes, such as transformation to SCLC, have been shown to mediate resistance in EGFR-mutant lung cancer and rarely in ALK-positive NSCLC (36, 46–48). However, we did not observe transformation to SCLC in any specimens in this series.
Beyond characterizing mechanisms of resistance to second-generation ALK inhibitors, this study also aimed to investigate the therapeutic impact of these findings. In particular, we evaluated the preclinical activity of the third-generation ALK inhibitor lorlatinib. Using a combination of Ba/F3 models and patient-derived cell lines, we demonstrated that lorlatinib was active against all single ALK resistance mutations. Moreover, lorlatinib was the only ALK inhibitor that retained significant activity against ALK G1202R. Such findings are consistent with preliminary phase I data in which lorlatinib has demonstrated an ORR of 44% among ALK-positive patients treated with two or more ALK inhibitors, with responses noted in ALK-positive patients harboring ALK G1202R mutations (49).
More broadly, our data suggest that therapeutic approaches to crizotinib resistance and resistance to second-generation ALK inhibitors will differ. In particular, we anticipate that repeat biopsies to identify ALK resistance mutations will play a larger role in guiding therapy decisions after progression on second-generation ALK TKIs compared to crizotinib. With respect to the latter, the absence of ALK resistance mutations has not been shown to impact ORRs or PFS to a second-generation ALK inhibitor among crizotinib-resistant patients (12, 50). Thus, such patients generally remain ALK-dependent, even in the absence of ALK resistance mutations—likely reflecting the relatively low potency of crizotinib against ALK. By contrast, data from our patient-derived cell line models suggest that ALK resistance mutation status after disease progression on second-generation ALK inhibitors is likely to be critically important in predicting sensitivity to the third-generation ALK inhibitor lorlatinib. Indeed, lorlatinib was only active in ceritinib-resistant, patient-derived cell lines harboring ALK resistance mutations in this study. Altogether, our findings support a new therapeutic paradigm in which clinicians tailor ALK inhibitor therapy based upon resistance mechanisms following disease progression on second-generation ALK inhibitors (Figure 7F).
This study has several important limitations. First, resistance to targeted therapies may be heterogeneous (10, 51), and a single biopsy may not adequately capture the full scope of resistance in a given patient. However, in general, it is not feasible to obtain biopsies of multiple sites in patients with NSCLC. In the future, circulating tumor DNA (ctDNA) platforms may enable us to evaluate tumor heterogeneity, while also allowing clinicians to readily monitor the evolution of resistance over time. A second limitation is that the sample sizes of several resistant-biopsy cohorts were relatively small, and patients did not always have adequate tissue for comprehensive molecular analysis or generation of patient-derived cell lines. As a result, we prioritized tissue for assessments of on-target mechanisms of resistance. Nonetheless, it is possible that our targeted ALK sequencing panels did not adequately capture low frequency variants or previously un-described ALK resistance mutations. Another limitation of this analysis is that pre-crizotinib and/or pre-second generation ALK inhibitor biopsies were generally not available; thus, we were unable to determine whether certain genetic alterations existed prior to the development of resistance to second-generation ALK inhibitors.
In summary, we demonstrate that the frequency and spectrum of ALK resistance mutations differs depending on the ALK inhibitor. Moreover, resistance profiles may evolve over time and in response to sequential ALK inhibitors. Moving forward, it will be important to incorporate repeat biopsies into clinical trials of next generation ALK inhibitors both before treatment and at progression. As biopsies are not always feasible and resistance may be heterogeneous, non-invasive techniques, such as ctDNA, will be crucial to develop and validate in parallel. Together, such efforts may facilitate discovery of novel mechanisms of resistance and new insights into the impact of heterogeneity on treatment response. On a practical level, this work will also allow clinicians to personalize ALK-targeted strategies based upon the presence or absence of specific ALK resistance mutations, which may ultimately translate into improved patient outcomes.
METHODS
Patients and Treatment
ALK-positive NSCLC patients underwent repeat biopsies of resistant tumors between January 2009 and June 2016. Tumor histology was classified according to World Health Organization criteria. Electronic medical records were retrospectively reviewed to obtain clinical data and treatment histories. All patients provided signed informed consent under an Institutional Review Board (IRB)-approved protocol. The study was conducted in accordance with the principles of the Declaration of Helsinki.
Genotype Assessments
All post-progression biopsies were analyzed for ALK resistance mutations. Testing methodologies included the MGH NGS platform, the FoundationOne NGS platform, and Sanger dideoxynucleotide sequencing of complementary DNA (cDNA) and genomic DNA (gDNA; Table S2). The MGH NGS platform (v1.1.4) uses anchored multiplex polymerase chain reaction (PCR) to detect single-nucleotide variants (SNVs) and insertions/deletions within 39 cancer-related genes, including ALK (exons 22, 23, and 25; ref 52). The FoundationOne platform (Foundation Medicine, Cambridge, MA) uses NGS to evaluate the entire coding sequence of 315 cancer-related genes as well select introns from 28 genes commonly altered in solid tumors (53). A subset of specimens underwent Sanger sequencing of the entire ALK kinase domain as previously described (6).
For whole exome sequencing, genomic DNA was extracted from formalin-fixed paraffin embedded (FFPE) samples. Whole-exome capture libraries were constructed from 100ng of extracted tumor and normal DNA. Ligated DNA was size-selected for lengths between 200–350bp and subjected to exonic hybrid capture using SureSelect v2 Exome bait (Agilent). Samples were multiplexed and sequenced on Illumina HiSeq flowcells (paired end 76bp reads) to an average on-target coverage depth ranging from 134 to 210× for all tumors and normal, respectively. Massively parallel sequencing data were processed using two consecutive pipelines as previously described (33).
Patient-derived cell lines (MGH021-5A, MGH034-2A, MGH049-1A, MGH051-2, MGH075-2E) were analyzed by NGS. RNA bait-based hybridization capture was performed to capture over 1000 known cancer genes (RightOn Cancer Sequencing Kit, developed in collaboration with Elim BioPharma), as previously described (10).
Fluorescence in situ Hybridization
ALK FISH was performed on FFPE tissue using dual color, break-apart rearrangement probes (Abbott-Vysis; ref 6). Multicolor FISH to assess for gene amplification was performed using a mix of custom FISH probes: Kreatech MET (7q31) blue, EGFR (7p11) green, and HER2/ERBB2 (17q12) red (Leica Biosystems).
Immunohistochemistry
ALK immunohistochemistry was performed using an anti-ALK monoclonal antibody (clone 5A4, Novocastra, Newcastle Upon Tyne, UK) at 1:50 dilution with Leica automation (Leica BOND-III, Leica Microsystems, Buffalo Groove, IL, USA). Immunohistochemical staining for vimentin and E-cadherin were performed as previously described (36).
Cell Lines and Reagents
Patient-derived cell lines were established as previously described (10). MGH075-2E and MGH049-1A were developed from malignant, ceritinib-resistant pleural effusions (March 2014 and July 2012, respectively). MGH084-1D, MGH034-2A and MGH051-2C were established from ceritinib-resistant, liver biopsies (April 2014, September 2012 and February 2013, respectively). MGH021-5A was derived from a malignant pleural effusion (June 2011). Cell lines were sequenced to confirm the presence of ALK rearrangements identified by clinical testing of biopsy specimens from the same patients. Additional authentication was not performed. Cells were grown either in RPMI-1640 or DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 1× Antibiotic-Antimycotic.
Ba/F3 immortalized murine bone marrow–derived pro-B cells were obtained from the RIKEN BRC Cell Bank (RIKEN BioResource Center) and cultured in DMEM supplemented with 10% FBS and IL3 (0.5 ng/mL). Cells were infected with lentiviral vectors (pLenti) expressing either wild type EML4–ALK variant 1 (E13;A20), or EML4-ALK harboring different ALK resistance mutations. Infected cells were selected in puromycin (0.8 µg/mL) for 2 weeks. After selection IL3 was withdrawn from the culture medium for at least 2 weeks before experiments.
Ceritinib, brigatinib and lorlatinib were purchased from Selleckchem. Alectinib was purchased from MedChem Express. Each compound was dissolved in DMSO for cell culture experiments.
Drug Screens
Patient-derived cell lines lacking known ALK resistance mutations underwent a combination drug screen consisting of 77 agents, as previously described (10). Cells were treated with vehicle or varying concentrations of drugs to be screened in the absence or presence of 0.3 µM ceritinib for 72 hours. Cell viability was determined as detailed below.
Antibodies and Immunobloting
5 × 105 cells were seeded in 6-well plates and treated with indicated agents for 6 hours. Lysates were prepared as previously described and equal volumes of total cell lysate were processed for immunoblotting (6). Antibodies against phospho-ALK (Y1282/1283), ALK, phospho-AKT (S473), AKT, Phospho-ERK (T202/Y204), ERK, phospho-S6, and S6 were obtained from Cell Signaling Technology. GAPDH was purchased from Millipore.
Survival Assays
Since the patient-derived cell lines used in this study have different growth kinetics, the number of cells seeded and the duration of treatment were adjusted for each cell line in order to have a consistent proliferation index (3.5 to 5) at the end of treatment (Table S13). Cells were seeded into 96-well plates and treated with serial dilutions of different ALK inhibitors over time. Each condition was performed in triplicate. For Ba/F3 cells, 2000 cells were plated into 96-well plates and treated for 48 hours.
At the end of treatment, cells were incubated with a CellTiter-Glo assay reagent (Promega) for 20 minutes, and luminescence was measured with a Centro LB 960 Microplate Luminometer (Berthold Technologies). GraphPad Prism version 5.0 were used to graphically display data. IC50 values were determined by nonlinear regression model utilizing a four-parameter analytical method.
Growth Assays
Indicated number of cells per cell line (Table S14) were seeded in six wells per condition in 96-well plates and treated with vehicle or indicated drugs at a concentration of 300 nM. Cell growth kinetics was measured over time using Real-time Glo reagent (Promega) according to the manufacture protocol.
Supplementary Material
Significance.
Secondary ALK mutations are a common resistance mechanism to second-generation ALK inhibitors and predict for sensitivity to the third-generation ALK inhibitor lorlatinib. These findings highlight the importance of repeat biopsies and genotyping following disease progression on targeted therapies, particularly second-generation ALK inhibitors.
Acknowledgments
Funding Sources: This work was supported by grants from the the National Cancer Institute (5R01CA164273, to ATS and JAE, C06CA059267 to JFG), by a National Foundation for Cancer Research (to ATS), by Be a Piece of the Solution, by the Evan Spirito Foundation, and by LungStrong.
JFG has served as a compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad, Loxo, Novartis, Merck, Clovis, Boehringer Ingelheim, Jounce Therapeutics, and Kyowa Hakko Kirin. SG has served as a compensated consultant for Pfizer, Bristol-Myers Squibb, Ariad, Genentech/Roche, AstraZeneca, and Boehringer Ingelheim. LVS has served as an uncompensated consultant for Boehringer Ingelheim, Clovis, Novartis, Merrimack, and Taiho Pharmaceuticals, and served as a compensated consultant or received honoraria from AstraZeneca, Genentech and Ariad. CHB has received research funding from Novartis and Amgen. MMK has served as a compensated consultant for Merrimack Pharmaceuticals. JAE is an employee of Novartis. JAE has served as a paid consultant for Novartis, Astra Zeneca, Chugai, and Genentech. JAE has received research funding from Novartis. ATS has served as a compensated consultant or received honoraria from Pfizer, Novartis, Genentech, Roche, Ignyta, Blueprint Medicine, Daiichi-Sankyo, Ariad, Chugai, Taiho Pharmaceuticals, and EMD Serono.
Footnotes
Disclosures:
The remaining authors have no financial interests to declare.
References
- 1.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 2.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 6.Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4:120ra117. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gainor JF, Shaw AT. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J Clin Oncol. 2013;31:3987–3996. doi: 10.1200/JCO.2012.45.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovly CM, McDonald NT, Chen H, Ortiz-Cuaran S, Heukamp LC, Yan Y, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med. 2014;20:1027–1034. doi: 10.1038/nm.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21:1038–1047. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, Camidge DR, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol. 2016;17:452–463. doi: 10.1016/S1470-2045(15)00614-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou SH, Ahn JS, De Petris L, Govindan R, Yang JC, Hughes B, et al. Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J Clin Oncol. 2016;34:661–668. doi: 10.1200/jco.2015.63.9443. [DOI] [PubMed] [Google Scholar]
- 15.Shaw AT, Gandhi L, Gadgeel S, Riely GJ, Cetnar J, West H, et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol. 2016;17:234–242. doi: 10.1016/S1470-2045(15)00488-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camidge D, Bazhenova L, Salgia R, Langer CJ, Gold KA, Rosell R, et al. Safety and efficacy of brigatinib (AP26113) in advanced malignancies, including ALK+ non-small cell lung cancer (NSCLC) J Clin Oncol. 2015;33 (suppl; abstr 8062) [Google Scholar]
- 17.Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–673. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami TA, et al. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19:679–690. doi: 10.1016/j.ccr.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Katayama R, Sakashita T, Yanagitani N, Ninomiya H, Horiike A, Friboulet L, et al. P-glycoprotein Mediates Ceritinib Resistance in Anaplastic Lymphoma Kinase-rearranged Non-small Cell Lung Cancer. EBioMedicine. 2016;3:54–66. doi: 10.1016/j.ebiom.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isozaki H, Ichihara E, Takigawa N, Ohashi K, Ochi N, Yasugi M, et al. Non-Small Cell Lung Cancer Cells Acquire Resistance to the ALK Inhibitor Alectinib by Activating Alternative Receptor Tyrosine Kinases. Cancer Res. 2016;76:1506–1516. doi: 10.1158/0008-5472.CAN-15-1010. [DOI] [PubMed] [Google Scholar]
- 21.Toyokawa G, Inamasu E, Shimamatsu S, Yoshida T, Nosaki K, Hirai F, et al. Identification of a Novel ALK G1123S Mutation in a Patient with ALK-rearranged Non-small-cell Lung Cancer Exhibiting Resistance to Ceritinib. J Thorac Oncol. 2015;10:e55–e57. doi: 10.1097/JTO.0000000000000509. [DOI] [PubMed] [Google Scholar]
- 22.Ignatius Ou SH, Azada M, Hsiang DJ, Herman JM, Kain TS, Siwak-Tapp C, et al. Next-generation sequencing reveals a Novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on crizotinib. J Thorac Oncol. 2014;9:549–553. doi: 10.1097/JTO.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 23.Ou SH, Milliken JC, Azada MC, Miller VA, Ali SM, Klempner SJ. ALK F1174V mutation confers sensitivity while ALK I1171 mutation confers resistance to alectinib. The importance of serial biopsy post progression. Lung Cancer. 2016;91:70–72. doi: 10.1016/j.lungcan.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canda. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 25.Zou HY, Friboulet L, Kodack DP, Engstrom LD, Li Q, West M, et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer Cell. 2015;28:70–81. doi: 10.1016/j.ccell.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Wang F, Keats J, Zhu X, Ning Y, Wardwell SD, et al. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chem Biol Drug Des. 2011;78:999–1005. doi: 10.1111/j.1747-0285.2011.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford D, et al. New response evaluation criteria in solid tumors: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Ou SH, Greenbowe J, Khan ZU, Azada MC, Ross JS, Stevens PJ, et al. I1171 missense mutation (particularly I1171N) is a common resistance mutation in ALK-positive NSCLC patients who have progressive disease while on alectinib and is sensitive to ceritinib. Lung Cancer. 2015;88:231–234. doi: 10.1016/j.lungcan.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Katayama R, Friboulet L, Koike S, Lockerman EL, Khan TM, Gainor JF, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res. 2014;20:5686–5696. doi: 10.1158/1078-0432.CCR-14-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah NP, Skaggs BJ, Branford S, Hughes TP, Nicoll JM, Paquette RL, et al. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J Clin Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niederst MJ, Hu H, Mulvey HE, Lockerman EL, Garcia AR, Piotrowska Z, et al. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin Cancer Res. 2015;21:3924–3933. doi: 10.1158/1078-0432.CCR-15-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Berggvist S, Brooun A, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med. 2016;374:54–61. doi: 10.1056/NEJMoa1508887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyner JW, Fletcher LB, Wang EQ, Yang WF, Rutenberg-Schoenberg ML, Beadling C, et al. MET receptor sequence variants R970C and T992I lack transforming capacity. Cancer Res. 2010;70:6233–6237. doi: 10.1158/0008-5472.CAN-10-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudd ML, Price JC, Fogoros S, Godwin AK, Sgroi DC, Merino MJ, et al. A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin Cancer Res. 2011;17:1331–1340. doi: 10.1158/1078-0432.CCR-10-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Wang F, Keats J, Ning Y, Wardwell SD, Moran L, et al. Abstract LB-298: AP26113, a potent ALK inhibitor, overcomes mutations in EML4-ALK that confer resistance to PF-02341066 (PF1066) Cancer Research. 2010;70:LB-298. [Google Scholar]
- 39.Toyokawa G, Hirai F, Inamasu E, Yoshida T, Nosaki K, Takenaka T, et al. Secondary mutations at I1171 in the ALK gene confer resistance to both Crizotinib and Alectinib. J Thorac Oncol. 2014;9:e86–e87. doi: 10.1097/JTO.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 40.Bauer RC, Sänger J, Peschel C, Duyster J, von Bubnoff N. Sequential inhibitor therapy in CML: in vitro simulation elucidates the pattern of resistance mutations after second- and third-line treatment. Clin Cancer Res. 2013;19:2962–2972. doi: 10.1158/1078-0432.CCR-13-0052. [DOI] [PubMed] [Google Scholar]
- 41.Yu HA, Tian SK, Drilon AE, Borsu L, Riely GJ, Arcila ME, et al. Acquired Resistance of EGFR-Mutant Lung Cancer to a T790M-Specific EGFR Inhibitor: Emergence of a Third Mutation (C797S) in the EGFR Tyrosine Kinase Domain. JAMA Oncol. 2015;1:982–984. doi: 10.1001/jamaoncol.2015.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gower A, Hsu WH, Hsu ST, Wang Y, Giaccone G. EMT is associated with, but does not drive resistance to ALK inhibitors among EML4-ALK non-small cell lung cancer. Mol Oncol. 2016;10:601–609. doi: 10.1016/j.molonc.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo F, Liu X, Qing Q, Sang Y, Feng C, Li X, et al. EML4-ALK induces epithelial-mesenchymal transition consistent with cancer stem cell properties in H1299 non-small cell lung cancer cells. Biochem Biophys Res Commun. 2015;459:398–404. doi: 10.1016/j.bbrc.2015.02.114. [DOI] [PubMed] [Google Scholar]
- 45.Kim HR, Kim WS, Choi YJ, Choi CM, Rho JK, Lee JC. Epithelial-mesenchymal transition leads to crizotinib resistance in H2228 lung cancer cells with EML4-ALK translocation. Mol Oncol. 2013;7:1093–1102. doi: 10.1016/j.molonc.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujita S, Masago K, Katakami N, Yatabe Y. Transformation to SCLC after Treatment with the ALK Inhibitor Alectinib. J Thorac Oncol. 2016;11:e67–e72. doi: 10.1016/j.jtho.2015.12.105. [DOI] [PubMed] [Google Scholar]
- 47.Cha YJ, Cho BC, Kim HR, Lee HJ, Shim HS. A Case of ALK-Rearranged Adenocarcinoma with Small Cell Carcinoma-Like Transformation and Resistance to Crizotinib. J Thorac Oncol. 2016;11:e55–e58. doi: 10.1016/j.jtho.2015.12.097. [DOI] [PubMed] [Google Scholar]
- 48.Miyamoto S, Ikushima S, Ono R, Awano N, Kondo K, Furuhata Y, et al. Transformation to small-cell lung cancer as a mechanism of acquired resistance to crizotinib and alectinib. Jpn J Clin Oncol. 2016;46:170–173. doi: 10.1093/jjco/hyv173. [DOI] [PubMed] [Google Scholar]
- 49.Solomon B, Bauer T, Felip E, Besse B, James LP, Clancy JS, et al. Safety and efficacy of lorlatinib (PF-06463922) from the dose-escalation component of a study in patients with advanced ALK+ or ROS1+ non-small cell lung cancer (NSCLC) J Clin Oncol. 2016;34 (suppl; abstr 9009) [Google Scholar]
- 50.Gainor JF, Tan DS, De Pas T, Solomon BJ, Ahmad A, Lazzari C, et al. Progression-Free and Overall Survival in ALK-Positive NSCLC Patients Treated with Sequential Crizotinib and Ceritinib. Clin Cancer Res. 2015;21:2745–2752. doi: 10.1158/1078-0432.CCR-14-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piotrowska Z, Niederst MJ, Karlovich CA, Wakelee HA, Neal JW, Mino-Kenudson M, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov. 2015;5:713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 53.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.