Abstract
CNDAC (2’-C-cyano-2’-deoxy-1-β-D-arabino-pentofuranosyl-cytosine, DFP10917) and its orally bioavailable prodrug, sapacitabine, are undergoing clinical trials for hematological malignancies and solid tumors. The unique action mechanism of inducing DNA strand breaks distinguishes CNDAC from other deoxycytidine analogs. To optimize the clinical potentials of CNDAC, we explored multiple strategies combining CNDAC with chemotherapeutic agents targeting distinct DNA damage repair pathways that are currently in clinical use. The ability of each agent to decrease proliferative potential, determined by clonogenic assays, was determined in paired cell lines proficient and deficient in certain DNA repair proteins. Subsequently each agent was used in combination with CNDAC at fixed concentration ratios. The clonogenicity was quantitated by median effect analysis, and a combination index was calculated. The c-Abl kinase inhibitor, imatinib, had synergy with CNDAC in HCT116 cells, regardless of p53 status. Inhibitors of PARP1 that interfere with homologous recombination (HR) repair or base excision repair (BER) and agents such as temozolomide that cause DNA damage repaired by the BER pathway were also synergistic with CNDAC. The toxicity of the nitrogen mustards, bendamustine and cytoxan, or of platinum compounds, which generate DNA adducts repaired by nucleotide excision repair and HR, was additive with CNDAC. An additive cell killing was also achieved by the combination of CNDAC with taxane mitotic inhibitors (paclitaxel and docetaxel). At concentrations which allow survival of the majority of wild type cells, the synergistic or additive combination effects were selective in HR-deficient cells. This study provides mechanistic rationales for combining CNDAC with other active drugs.
Keywords: sapacitabine, homologous recombination, synthetic lethality, clonogenicity
Introduction
Sapacitabine is an orally bioavailable prodrug of the deoxycytidine analog, CNDAC (2’-C-cyano-2’-deoxy-1-β-D-arabino-pentofuranosyl-cytosine). Sapacitabine has shown activity in AML and MDS (1, 2) and is currently in Phase III trial for older AML patients (www.clinicaltrials.gov identifier NCT01303796) and a Phase II trial for relapsed CLL/SLL with 11q22-23 deletion (NCT01253460). The parent nucleoside, CNDAC, formulated for parenteral infusion as DFP-10917, is in a Phase I/II trial for AML and ALL (NCT01702155) (3).
After being phosphorylated in vivo, CNDAC induces DNA damage by incorporation into replicating DNA with the subsequent formation of nicks through a β-elimination process that generates a 2’, 3’-dideoxy analog at the 3’-terminus which is not a substrate for ligation (4). These CNDAC-induced single-strand breaks (SSBs) may be repaired by a transcription-coupled nucleotide excision repair mechanism (5). Unrepaired SSBs can be converted into double-strand breaks (DSBs) when cells go through a second S-phase. The potentially lethal DSBs, resulting from unresolved SSBs, are repaired mainly by the homologous recombination (HR) pathway (6). We have demonstrated that deficiency in HR components, including ATM, RAD51, XRCC3, BRCA2, confer sensitivity to CNDAC. Preliminary studies reporting hypersensitivity of colon cancer cells lacking BRCA1 or BRCA2 to CNDAC (7) are in agreement with our findings.
CNDAC is distinguished from other structurally related nucleoside analogs (cytarabine, decitabine and gemcitabine) in its unique mechanism of action. To better understand and prepare for the next-step clinical applications, we exploited combination strategies of CNDAC with chemotherapeutic agents targeting different DNA repair pathways. Most of these agents are already in clinical use as first-line therapies. Imatinib, the first tyrosine-kinase inhibitor for the treatment of Ph+ CML as well as a number of other malignancies, inhibits the activity of c-Abl kinase in addition to the CML pathogenic Bcr-Abl kinase resulting from the t (9;22) translocation. c-Abl, activated by ATM kinase (8, 9), amplifies the DNA damage response in HR pathway. Inhibition of poly-(ADP-ribose) polymerase (PARP1), which facilitates gap filling in the BER pathway as well as increased activity of HR (10, 11), has shown promising therapeutic advantage in tumors deficient in HR function. Temozolomide, an oral alkylating agent used for brain tumors and melanoma, induces DNA lesions that are repaired in part by the base excision repair (BER) pathway (12, 13). Bendamustine and cytoxan, nitrogen mustards with wide-spread usage in solid tumors and hematologic malignancies, form bulky adducts repaired by the NER pathway (14). Adducts that escape this level of repair are capable of generating interstrand DNA cross links, which require HR repair. Cisplatin and oxaliplatin initially cause DNA mono-adducts and intra-strand crosslinks that are repaired by NER (15, 16), but the most toxic lesions are inter-strand crosslinks that are repaired by the Fanconi anemia and HR pathways (17–19). The last class of chemotherapeutic drug investigated in this study is the taxanes, which include paclitaxel and docetaxel. These “mitotic inhibitors” act by stabilizing tubulin and disrupting microtubule function, thereby inhibiting cell division (20).
Our investigations demonstrate that drugs that directly affect DSB repair (imatinib and inhibitors of PARP1) or which rely upon aspects of DSB repair (temozolomide), are synergistic with CNDAC. Combinations of CNDAC with agents that cause bulky adducts and crosslink DNA (platinum compounds or nitrogen mustards) or that affect the mitotic spindle (taxanes) produced loss of clonogenicity that were additive with that of CNDAC. In all cases, cells that were deficient in HR were selectively sensitized relative to those with normal repair capabilities. Considerations of the mechanisms that enable these positive interactions identify future paths of research and clinical opportunities.
Materials and Methods
Materials
Imatinib, temozolomide, bendamustine, cisplatin, oxaliplatin, paclitaxel, docetaxel, veliparib (ABT-888) and talazoparib (BMN 673) were purchased from Selleck Chemicals. Olaparib (AZD2281, KU-0059436) and CO-338 (PF-01367338-BW, rucaparib camsylate salt) were obtained from AstraZeneca and Clovis Oncology through MTAs, respectively. 4-HC was kindly provided by Drs. Michael Colvin and Susan Ludeman (Duke University, NC). Structures of PARP1 inhibitors used are shown in Suppl. Fig. 1.
Cell lines
The Chinese hamster ovary cell line AA8 (designated as WT1) and Chinese hamster lung cell line V79-4 (designated as WT2) were from ATCC (Manassas, VA). The AA8-derived mutant of XRCC3 (irs1SF) and the V79-derived mutant of BRCA2 (V-C8) were provided by Dr. Randy Legerski (M.D. Anderson Cancer Center, Houston, TX). 1SFwt8 (irs1SF complemented with human XRCC3 cDNA), 51D1 (RAD51D knockout line) and 51D1.3 (RAD51D complemented line) were gifts from Dr. Larry Thompson (Lawrence Livermore National Laboratory, Livermore, CA). All the above hamster cell lines, obtained in 2008 – 2010, were maintained in α-MEM supplemented with 10% fetal bovine serum (FBS) and Glutamax. In 2013, Dr. Wouter Wiegant (The Leiden University Medical Center, Leiden, Netherlands) also kindly provided hamster lung cell lines, V79 (designated as WT3) and paired V-C8 (BRCA2), which were maintained in Ham’s F-10 medium containing FBS (10%) and Glutamax. Colon carcinoma cell lines with wild type or knock-out p53 (HCT116 p53+/+ and p53−/−) were provided by Dr. Bert Vogelstein (Johns Hopkins Medical School, MD) in 2007, and were cultured in McCoy’s 5A with 10% FBS. HeLa RAD51C knockout line created by CRISPR technology and the RAD51C wild-type control line were obtained from Dr. Junjie Chen (M.D. Anderson Cancer Center, Houston, TX) in 2015, and were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum (FBS) and Glutamax. All cells were free of mycoplasma, as certified by Characterized Cell Line Core Facility at MDACC using the MycoAlert kit from Lonza (Switzerland). All hamster cell lines were functionally validated by clonogenic sensitivity to mitomycin C (21). STR DNA fingerprinting was used to authenticate human cell lines by the Characterized Cell Line Core at MDACC. Frozen aliquots of cells were used within 6 months of thawing.
Clonogenic assay
Attached cells were seeded in 6-well plates one day before cells were exposed to a range of concentrations of drugs for 24 hr. After washing into drug-free medium, cells were incubated for 3 to 8 days depending on growth rate of the particular cell line, and colonies (containing 50 or more cells) in triplicate wells were counted electronically (GelCount, Oxford Optronix, Oxford, UK).
Statistical analysis
Clonogenicity curves were plotted and IC50 values of single agents were calculated using the GraphPad Prism6 software (GraphPad Software, Inc. San Diego, CA) based on clonogenicity rates in triplicate.
Median-effect analysis of drug combinations
The sensitivity of individual cell lines to each single agent was determined as a basis for choosing fixed molar ratios of two agents to be combined (Suppl. Fig. 1–6). Clonogenicity of cells following treatment with two agents combined in fixed molar ratios over a range of concentrations for each agent that achieve 10% to 90% inhibition alone was analyzed by the median-effect method (22, 23) using the CalcuSyn software (Biosoft, Ferguson, MO). The calculated combination index (CI) <1, =1 and >1 indicate being synergistic, additive and antagonist, respectively. The dose-reduction index (DRI), calculated for a 90% decrease in clonogenicity, signifies when used in combination the fold dose-reduction for each drug to achieve a given lethal effect.
Results
Imatinib and CNDAC are synergistic regardless of p53 status
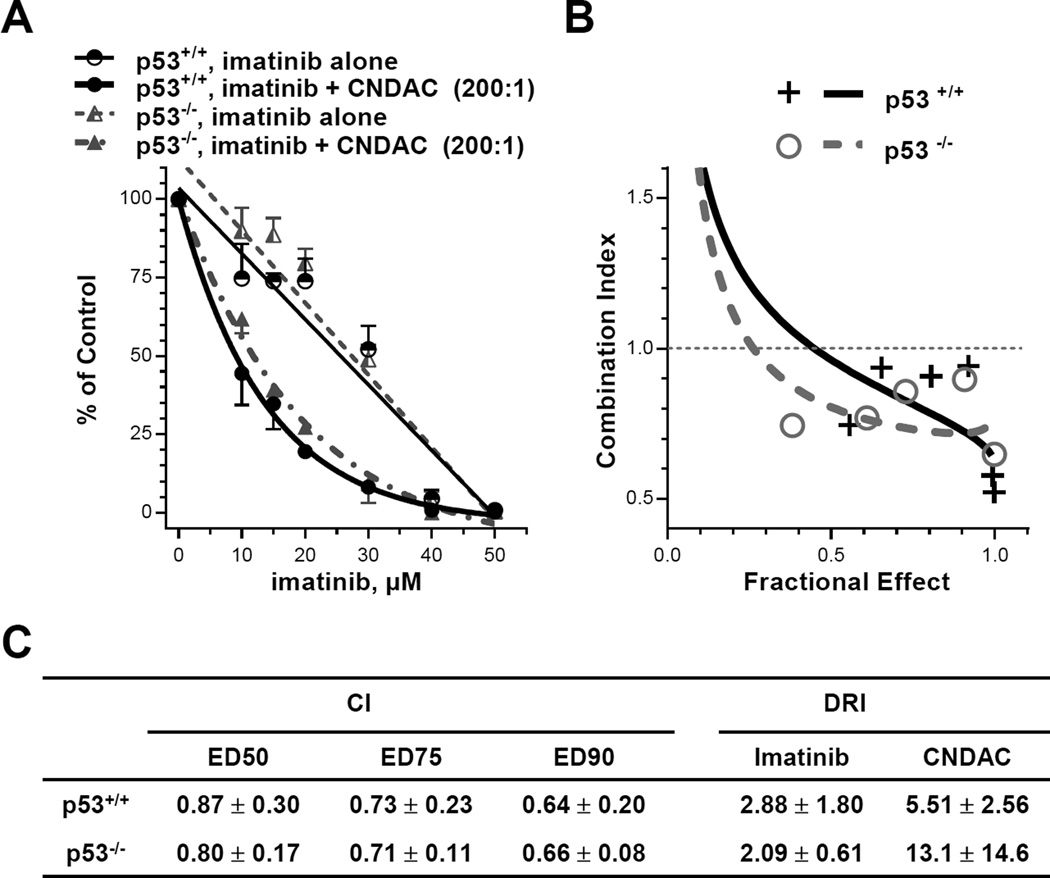
ATM-dependent homologous recombination is the major pathway to repair CNDAC-induced DNA double strand breaks (6), and cells lacking HR components are significantly sensitized to CNDAC. RAD51 recombinase, a key component of HR, forms filaments along 3’-overhanging single-stranded DNA and catalyzes annealing of resected DNA ends to homologous DNA sequences (24, 25). Because RAD51 is phosphorylated and activated by the c-Abl kinase in vitro (26), we hypothesized that inhibition of c-Abl by imatinib might down-regulate RAD51 activity and thereby suppress HR function. If this is the case, then imatinib may potentiate CNDAC toxicity. We studied the interaction between imatinib and CNDAC in human colon cancer cells and extended this to isogenic lines with wild-type p53 and knocked-out p53. p53 knockout cells were no more sensitive to imatinib or CNDAC alone compared to the wild type (Fig. 1A, Suppl. Fig. 2A and 2B). The IC50 values of imatinib and CNDAC in HCT116 cells are greater than 14 µM and 0.2–0.3 µM, respectively (Suppl. Fig. 1C). When these isogenic cells were exposed to the imatinib-CNDAC combination at fixed ratios (200:1 for Exp. 1, 50:1 for Exp. 2 and Exp. 3), loss of clonogenicity was greater than additive (Fig. 1B). Median effect analysis of the combination demonstrated that CI values of both lines were less than 1 in three independent experiments (Fig. 1C), indicating a synergistic interaction of these two agents. There was no significant difference between p53+/+ and p53−/− cells, indicating that the synergy between imatinib and CNDAC is independent of p53 status. The DRI values indicate that the same effect of either drug alone may be achieved at lesser concentrations in the presence of the second drug. In addition, c-Abl activity was equally inhibited by imatinib (50% inhibition at 7 – 8 µM, 70% - 75% inhibition at 40 µM) in both p53+/+ and p53−/− cells (Suppl. Fig. 2D).
Figure 1.
Imatinib and CNDAC are synergistic in HCT116 cells, regardless of p53 status. HCT116 cells with wild type p53 and p53 KO were pretreated with imatinib (5 – 50 µM) for 2 hours before co-incubation with CNDAC (0.05 – 0.4 µM). Cells were washed after 24 hours and incubated in medium with imatinib added at the same concentrations until colonies were fixed. A, comparison of clonogenicity of p53 wild type and p53 KO after treatment with imatinib alone vs imatinib-CNDAC combination at a fixed ratio (200:1), representative of 3 independent experiments (ratios 200:1 or 50:1). B, median effect analysis algebraic estimate of imatinib and CNDAC combination (ratio 200:1), representative of 3 independent experiments. C, values of combination index (CI) at 50%, 75% and 90% clonogenic inhibition and dose reduction index (DRI) at 90% clonogenic inhibition (mean ± S.D.).
CNDAC interacts synergistically with PARP1 inhibitors
PARP1 inhibitors target poly(ADP-ribose) polymerase 1, which is crucial for SSB repair by the base excision repair pathway and for repair of DSBs by either non-homologous end-joining or homologous recombination pathways, respectively. The actions of CNDAC have been shown to be independent of the BER proteins DNA polymerase β and Xrcc1, and cells lacking the NHEJ proteins DNA-PKcs and Ku-80 are not sensitized to CNDAC (6). Thus, interactions between CNDAC and PARP1 inhibitors are likely due to the actions of PARP1 inhibitors on HR (10, 11). Therefore we hypothesized that combination of CNDAC with a PARP1 inhibitor will create a synthetic lethal condition in HR-defective cells. To test this postulate we compared the sensitivity of paired BRCA2 lines to three PARP1 inhibitors, veliparib, olaparib and rucaparib. Rucaparib was shown to be the most potent agent among the three in sensitizing BRCA2 deficient cells (>2400-fold more sensitive than wild type), olaparib was less so (>400-fold) and veliparib was the least potent (>300-fold) (Suppl. Fig. 3A and 3D), consistent with earlier reports (27). Moreover, cells lacking XRCC3 were significantly more sensitive to olaparib compared to parental cells (>500-fold) but clonogenicity of cells lacking the NER component, XPF was minimally decreased (∼4-fold) (Suppl. Fig. 3C). Comparison of the lethality of PARP1 inhibition for 24 hr relative to prolonged incubation demonstrated that the potency of inhibition was increased 50-fold by continuous exposure (IC50 = 4 nM vs 200 nM) (Suppl. Fig. 3B).
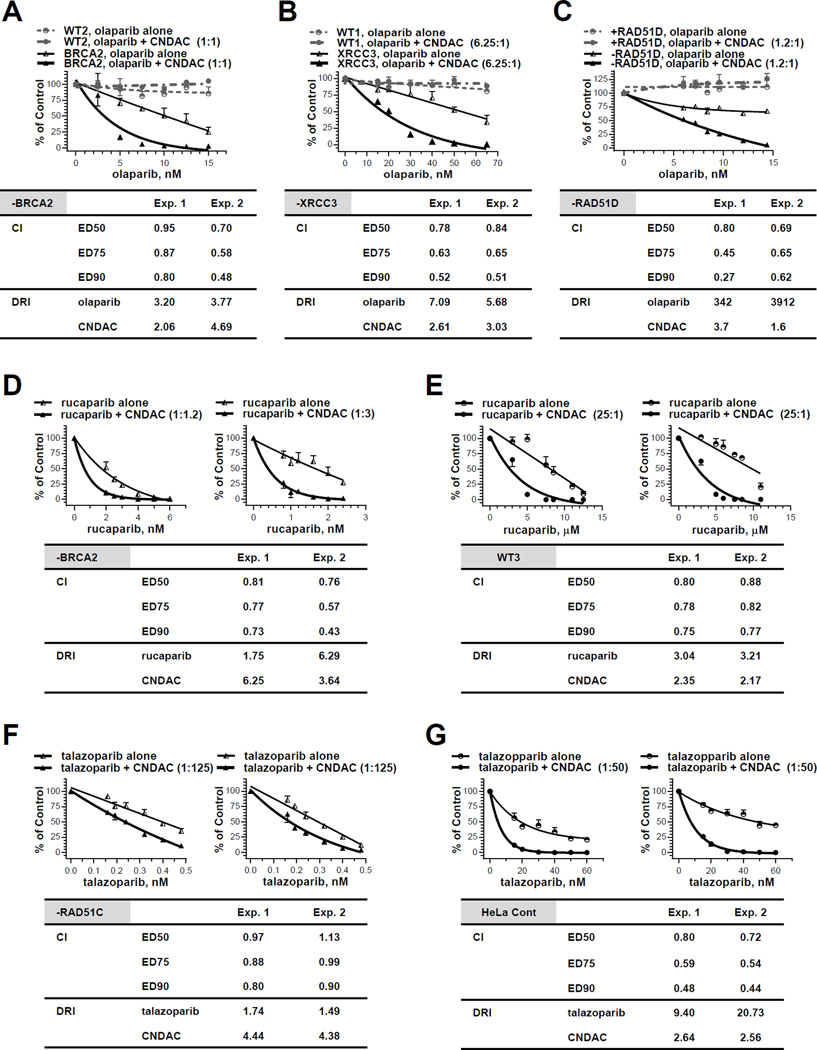
Combination of olaparib and CNDAC at a ratio of 1:1 (Exp. 1) or 1.3:1 (Exp. 2) achieved CI<1, indicating synergy in BRCA2 deficient cells (Fig. 2A). The olaparib-CNDAC combination in XRCC3 deficient cells (ratio 6.25:1 for both Exp. 1 and 2, Fig. 2B) and RAD51D deficient cells (ratio 1.2:1 for both Exp. 1 and 2, Fig. 2C) also achieved CI<1, suggesting this synergistic effect is common in HR defective cells. The concentrations of both agents tested were too low to kill the wild type cells, demonstrating aselectivity for the mutants. Similar to the olaparib-CNDAC combination, rucaparib and CNDAC (at nM concentration range) combined at a ratio of 1:1.2 (Exp. 1) or 1:3 (Exp. 2) was also synergistic with CI<1 in BRCA2 defective cells (Fig. 2D). However, this combination was also effective in the wild type cells (Fig. 2E), although at a higher concentration (µM) range of both rucaparib and CNDAC (25:1 for both Exp. 1 and 2, CI<1), suggesting selectivity is lost at greater concentrations.
Figure 2.
The PARP1 inhibitors olaparib, rucaparib and talazoparib have synergistic effect with CNDAC. A, Chinese hamster wild-type (WT2) and BRCA2 deficient (−BRCA2) cells were exposed to olaparib (2.5 – 19.5 nM) 1 hour before addition of CNDAC (2.5 – 15 nM). After 24 hours, cells were washed out of drugs and incubated in medium with olaparib added at the same concentrations until colonies were fixed. Upper panel, representative of two independent experiments, comparison of clonogenicity of wild type and BRCA2 deficient cells after treatment with olaparib alone vs olaparib-CNDAC combination at a fixed ratio (1:1 for Exp. 1 and 1.3:1 for Exp. 2). Lower panel, CIs and DRI at 90% clonogenic inhibition from median effect analysis of the two experiments in the BRCA2 deficient line. B, CHO wild-type (WT1) and XRCC3 deficient (−XRCC3) cells were exposed to olaparib (15 – 65 nM) 1 hour before addition of CNDAC (2.4 – 10.4 nM). After 24 hours, drugs were washed out and cells were incubated in medium with olaparib at the same concentrations until colonies were fixed. Upper panel, representative of two independent experiments, comparison of clonogenicity of wild type and XRCC3 deficient cells after treatment with olaparib alone vs olaparib-CNDAC combination at a fixed ratio (6.25:1). Lower panel, CIs and DRI at 90% clonogenic inhibition from median effect analysis of the two experiments in the XRCC3 deficient line. C, RAD51D deficient (−RAD51D) and complemented (+RAD51D) CHO cells were exposed to olaparib (6 – 14.4 nM) 1 hour before addition of CNDAC (5 – 12 nM). After 24 hours, drugs were washed out and cells were incubated in medium with olaparib at the same concentrations until colonies were fixed. Upper panel, representative of two independent experiments, comparison of clonogenicity of RAD51D deficient and repleted cells after treatment with olaparib alone vs olaparib-CNDAC combination at a fixed ratio (1.2:1). Lower panel, CIs and DRI at 90% clonogenic inhibition from median effect analysis of the two experiments in the RAD51D deficient line. D - E, BRCA2 deficient (−BRCA2, D) and wild-type (WT3, E) lines were exposed to rucaparib 1 hour before addition of CNDAC. Concentrations of rucaparib tested were 0.8 – 6 nM (−BRCA2) and 3 – 12.5 µM (WT3), while concentrations of CNDAC were 2.4 – 7.2 nM (−BRCA2) and 0.12 – 0.5 µM (WT3), respectively. After 24-hour incubation, drugs were washed out and medium with rucaparib at the same concentrations were added during colony formation. Upper panels, comparison of cell clonogenicity after treatment with rucaparib alone vs rucaparib-CNDAC combination at fixed ratios (1:1.2 and 1:3 for Exp. 1 and 2, respectively, in –BRCA2 line; 25:1 for both experiments in WT3). Lower panel, CIs and DRI at 90% clonogenic inhibition from median effect analysis of the two experiments in each line. F - G, HeLa RAD51C deficient (−RAD51C, F) and control line (G) and line were exposed to talazoparib 0.5 hour before addition of CNDAC. Concentrations of talazoparib tested were 0.16 – 0.48 nM (−RAD51C) and 15 – 60 nM (HeLa Cont), while concentrations of CNDAC were 20 – 60 nM (−RAD51C) and 0.75 – 3 µM (HeLa Cont), respectively. After 24-hour incubation, drugs were washed out and medium with talazoparib at the same concentrations were added during colony formation. Upper panels, comparison of cell clonogenicity after treatment with talazoparib alone vs talazoparib-CNDAC combination at fixed ratios (1:125 for Exp. 1 and 2 in –BRCA2 line; 1:50 for both experiments in WT3). Lower panel, CIs and DRI at 90% clonogenic inhibition from median effect analysis of the two experiments in each line.
In addition, HeLa RAD51C knockout cells have been shown to be sensitive to a newer and more potent PARP1 inhibitor, talazoparib (Suppl. Fig. 3E). Combination of talazoparib and CNDAC at fixed ratios presented synergy (CI<1) in both RAD51C deficient (ratio 1:125, at lower nM range, Fig. 2F) and proficient HeLa cells (ratio 1:50, at higher nM range, Fig. 2G). Thus, combinations of olaparib, rucaparib or talazoparib with CNDAC clearly demonstrated a synergistic interaction between PARP1 inhibitors and CNDAC in HR deficient cells. Although synergy with CNDAC is consistent or comparable to HR deficient cells, selectivity is lost at the higher rucaparib or talazoparib concentrations.
Temozolomide and CNDAC have synergy in HR defective cells
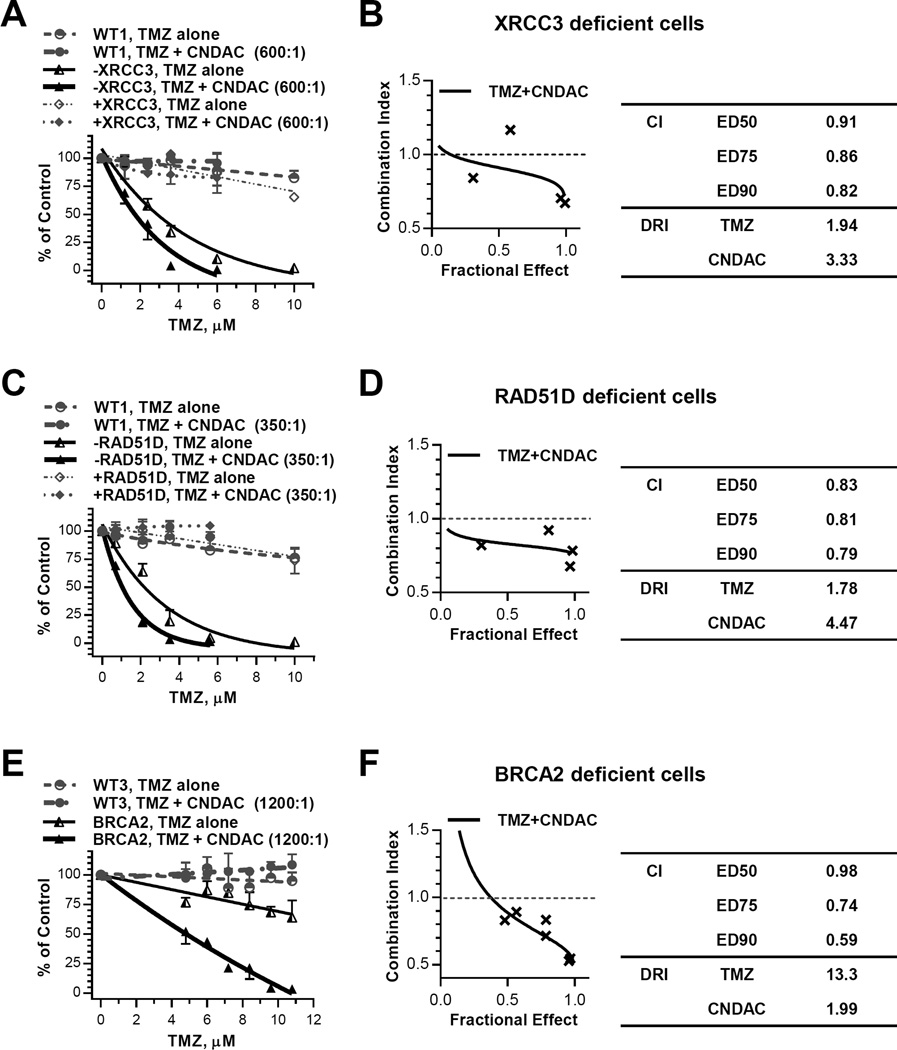
Temozolomide (TMZ) methylates DNA most often at the O6 or N7 positions of the guanine residues which cause mutations associated with tumorigenesis. The O6-MeG lesion can be recognized and directly repaired by O6-methylguanine-DNA-methyltranferase (28, 29), whereas N7-MeG, the major form of lesions, is recognized by methylpurine-DNA glycosylase and repaired via the BER pathway (12). We found that cells deficient in the HR proteins XRCC3 or RAD51D were more sensitive to TMZ (IC50 values 2.2 – 2.9 µM) relative to XRCC3 or RAD51D complemented cells (IC50 >>10 µM, Suppl. Fig. 3). This suggests that HR may be responsible, in part, for repair of DNA damage caused by TMZ, presumably due to DSBs arising from SSBs unrepaired by BER. The combination of TMZ and CNDAC at a fixed ratio of 600:1 indicated a dose response that was highly selective in XRCC3 deficient cells with respect to the parental and complemented lines (Fig. 3A). The results of median effect analysis (CI= 0.8–0.9) suggested a synergy (Fig. 3B). Similarly, the TMZ-CNDAC combination (ratio 350:1) in RAD51D defective cells was also selective (Fig. 3C) and indicative of synergy (CI ∼0.8) (Fig. 3D). In addition, selectivity and synergy of TMZ-CNDAC combination (ratio 1200:1) was also observed in BRCA2 deficient cells (Fig. 3E and 3F). Thus, TMZ and CNDAC have synergy in HR defective cells at concentrations insufficient to kill the wild type, XRCC3 or RAD51D complemented lines (Fig. 3A, 3C and 3E; Suppl. Fig. 4).
Figure 3.
Temozolomide and CNDAC are synergistic in HR deficient CHO lines. A and B, CHO wild-type (WT1), XRCC3 deficient (−XRCC3) and XRCC3 complemented (+XRCC3) cells were exposed to temozolomide (1.2 – 10 µM) and CNDAC (0.002 – 0.02 µM) concomitantly for 24 hours before washout and colony formation. A, the colonies were quantitated and clonogenicity of all three lines were compared for treatment with temozolomide alone vs temozolomide-CNDAC combination at a fixed ratio (600:1). B, median effect analysis algebraic estimate of temozolomide-CNDAC combination, CIs and DRI at 90% clonogenic inhibition of the XRCC3 deficient line. C and D, CHO wild-type (WT1), RAD51D deficient line (−RAD51D) and RAD51D complemented lines (+RAD51D) were treated as in A and B. Concentrations of temozolomide tested in all lines were 0.7 – 10 µM, while concentrations of CNDAC were 0.002 – 0.03 µM. C, clonogenicity of all three lines were compared for treatment with temozolomide alone vs temozolomide-CNDAC combination at a fixed ratio (350:1). D, median effect analysis algebraic estimate of temozolomide-CNDAC combination, CIs and DRI at 90% clonogenic inhibition of the RAD51D deficient line. E and F, Chinese hamster wild-type (WT3) and BRCA2 deficient (−BRCA2) cells were exposed to temozolomide (4.8 – 10.8 µM) and CNDAC (4 – 9 nM) concomitantly for 24 hours before washout and colony formation. E, clonogenicity of both lines were compared for treatment with temozolomide alone vs temozolomide-CNDAC combination at a fixed ratio (1200:1). F, median effect analysis algebraic estimate of temozolomide-CNDAC combination, CIs and DRI at 90% clonogenic inhibition of the BRCA2 deficient line (mean of two experiments).
Both bendamustine and 4-hydroperoxycyclophosphamide have an additive effect with CNDAC in HR-deficient cells
Bendamustine and 4-HC are nitrogen mustards, which form mono-adducts on DNA, activating the NER pathway. Notably, our preliminary studies demonstrated that cells defective in XPF, a key component of both NER and repair of interstrand crosslinks, were 88-fold more sensitive to 4-HC than the wild type (Suppl. Fig. 5B). Those adducts that are not repaired have the potential to form interstrand crosslinks. Although NER is the primary pathway for repair of damage due to bendamustine and 4-HC, HR has an important role in repairing secondary damage caused by these agents. For example, RAD51D deficiency rendered 12- and 13-fold sensitivity to bendamustine relative to RAD51-complemented cells and wild type cells. Deficiency in XRCC3 conferred 19-fold and 45-fold sensitivity to bendamustine relative to XRCC3 complemented and parental cells, respectively (Suppl. Fig. 5A). Similar to bendamustine, 4-HC sensitized RAD51D and XRCC3 deficient cells by approximately 50-fold compared to parental cells, whereas BRCA2 deficient cells were 20-fold more sensitive to 4-HC than the wild type (Suppl. Fig. 5B).
Bendamustine-CNDAC combination at molar ratio of 1000:1 resulted in CI ∼1 in both XRCC3 deficient (Fig. 4A) and RAD51D deficient cells (Fig. 4B). The combination of 4-HC and CNDAC led to CI ∼1 in both BRCA2 (ratio 4:1, Fig. 4C) and XRCC3 deficient cells (ratio 16:1, Fig. 4D). These results show that agents that cause lesions that are initially repaired by NER are additive to the action of CNDAC in HR-defective cells. All the concentrations tested in the above experiments were non-toxic to the wild type or complemented cell lines.
Figure 4.
Both bendamustine (BEN) and 4-hydroperoxycyclophophamide (4-HC) have additive effect with CNDAC in HR deficient cells. A, CHO cell lines wild-type (WT1), deficient in XRCC3 (−XRCC3) and complemented with XRCC3 (+XRCC3) were treated concomitantly with BEN (2 – 20 µM) and CNDAC (0.002 – 0.02 µM) for 24 hours before washout. Colonies formed were quantitated and clonogenicity of all three lines were compared for treatment with bendamustine alone vs bendamustine-CNDAC combination at a fixed ratio (1000:1). B, CHO cell lines wild-type (WT1), deficient in RAD51D (−RAD51D) and complemented with RAD51D (+RAD51D) were treated concomitantly with BEN (2 – 30 µM) and CNDAC (0.002 – 0.03 µM) as in A. Clonogenicity of all three lines were compared for treatment with bendamustine alone vs bendamustine-CNDAC combination at a fixed ratio (1000:1). C, Chinese hamster wild-type line WT2 and BRCA2 deficient line (−BRCA2) were exposed to CNDAC (0.002 – 0.02 µM) subsequent to addition of 4-HC (0.008 – 0.08 µM) and incubated for 24 hours before washout. Clonogenicity of all three lines were compared for treatment with bendamustine alone vs bendamustine-CNDAC combination at a fixed ratio (4:1). D, CHO cells deficient in XRCC3 (−XRCC3) and complemented with XRCC3 (+XRCC3) were treated concomitantly with CNDAC (0.002 – 0.012 µM) and 4-HC (0.032 – 0.192 µM) for 24 hours. Clonogenicity of all three lines were compared for treatment with bendamustine alone vs bendamustine-CNDAC combination at a fixed ratio (16:1). CIs of respective deficient cell lines in A - D are listed below the plots.
Combination of CNDAC with platinum compounds presents additive actions
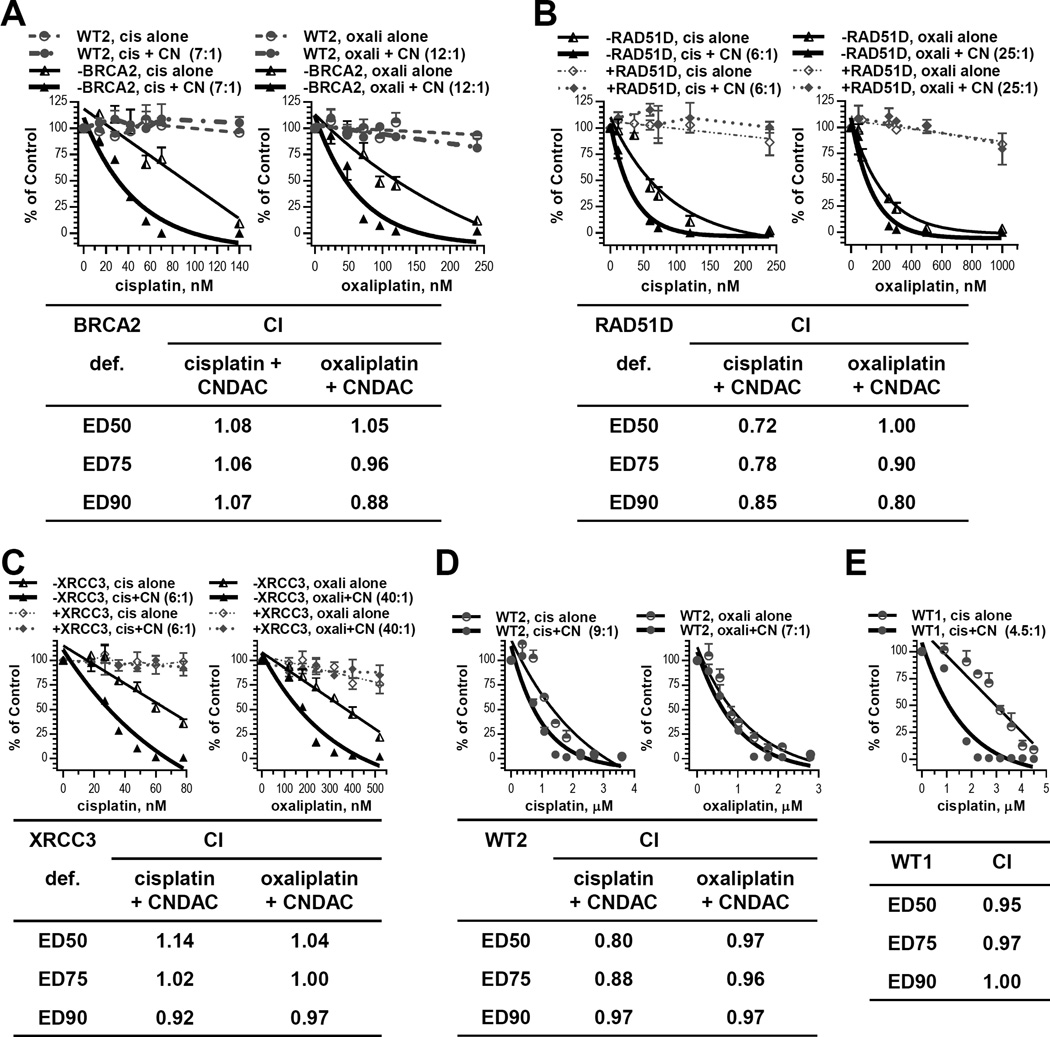
Cisplatin is a front-line therapy for several solid tumors. Oxaliplatin is used in combination strategies for both solid tumors and hematological malignancies. Both drugs act by intra- and inter-strand crosslinks in DNA. Preliminary investigations with single agents determined that BRCA2 deficient cells were 24- and 11-fold more sensitive to cisplatin and oxaliplatin, respectively, relative to wild type cells (Suppl. Fig. 6A). Furthermore, wild type cells were 78-, 39- and 55-fold more resistant to cisplatin than cells lacking XPF, RAD51D and XRCC3, respectively. Similarly, cells defective in XPF, RAD51D and XRCC3 were 29-, 13- and 8-fold more sensitive to oxaliplatin compared to parental cells (Suppl. Fig. 6B). Therefore, while NER is the initial and primary DNA repair pathway that acts on platinum adducts, HR also participates in the repair processes, presumably in resolving inter-strand crosslinks. The combination of cisplatin and CNDAC (ratio: 7:1, 6:1 and 6:1, respectively) resulted in CI ∼1, <1 and ∼1 in BRCA2-, RAD51D- and XRCC3-deficient cells, respectively (Fig. 5A, 5B and 5C, upper left panels). Replacement of cisplatin with oxaliplatin in the combination also achieved CI values ∼1 in the above deficient cell lines at oxaliplatin-CNDAC (ratio: 12:1, 25:1 and 4:1, respectively; Fig. 5A, 5B and 5C, upper right panels). These results demonstrate additive interaction between cisplatin or oxaliplatin and CNDAC at concentrations that were not toxic to HR proficient cells. Therefore, we combined cisplatin or oxaliplatin and CNDAC in greater concentration ranges to study if the additive effect applies to wild type cells as well. The cisplatin-CNDAC combination at ratios of 9:1 and 9:2 led to CI ∼ 1 in V79-4 and AA8 lines, respectively, while the oxaliplatin-CNDAC combination at 7:1 ratio (Fig. 5D and 5E) also confirmed that the combination effect of platinum compounds and CNDAC is additive in both HR-proficient and deficient cells.
Figure 5.
Platinum compounds have additive effect with CNDAC. A, Chinese hamster wild-type line V79-4 (WT2) and BRCA2 deficient line V-C8 (−BRCA2) were exposed to CNDAC (CN, 2 – 20 nM) subsequent to addition of cisplatin (cis, 14 – 140 nM) or oxaliplatin (oxali, 24 –240 nM) and incubated for 24 hours before washout. The colonies formed were quantitated and clonogenicity of both lines were compared for treatment with cisplatin alone vs cisplatin-CNDAC combination at a fixed ratio (7:1, left) or oxaliplatin vs oxaliplatin-CNDAC combination at a fixed ratio (12:1, right). B, CHO cell lines deficient in RAD51D (−RAD51D) and complemented with RAD51D (+RAD51D) were treated concomitantly with cisplatin (12 – 240 nM) or oxaliplatin (50 – 1000 nM) and CNDAC (2 – 40 nM) in clonogenic assays as in A. Clonogenicity of both lines were compared for treatment with cisplatin alone vs cisplatin-CNDAC combination at a fixed ratio (6:1, left) or oxaliplatin vs oxaliplatin-CNDAC combination at a fixed ratio (25:1, right). C, CHO cell lines deficient in XRCC3 (−XRCC3) and complemented with XRCC3 (+XRCC3) were treated concomitantly with cisplatin (18 – 78 nM) or oxaliplatin (120 – 520 nM) and CNDAC (3 – 13 nM) for 24 hours before washout in clonogenic assays. Clonogenicity of both lines were compared for treatment with cisplatin alone vs cisplatin-CNDAC combination at a fixed ratio (6:1, left) or oxaliplatin vs oxaliplatin-CNDAC combination at a fixed ratio (40:1, right). In A - C, CIs of the respective deficient cell lines are presented in the accompanying inserts. D, wild type hamster lung cell line (WT2) was exposed to cisplatin (0.36 – 3.6 µM) or oxaliplatin (0.28 – 2.8 µM) followed by CNDAC (0.04 – 0.4 µM) as in A-C. Clonogenicity was compared for treatment with cisplatin alone vs cisplatin-CNDAC combination at a fixed ratio (9:1, left) or oxaliplatin vs oxaliplatin-CNDAC combination at a fixed ratio (7:1, right). E, wild type CHO cells (WT1) were exposed to cisplatin (0.9 – 4.5 µM) followed by CNDAC (0.2 – 1 µM) as in D. Clonogenicity was compared for treatment with cisplatin alone vs cisplatin-CNDAC combination at a fixed ratio (4.5:1). CIs of the wild-type lines in D - E are presented in the accompanying inserts.
Taxane agents and CNDAC have additive effect in cells lacking HR function
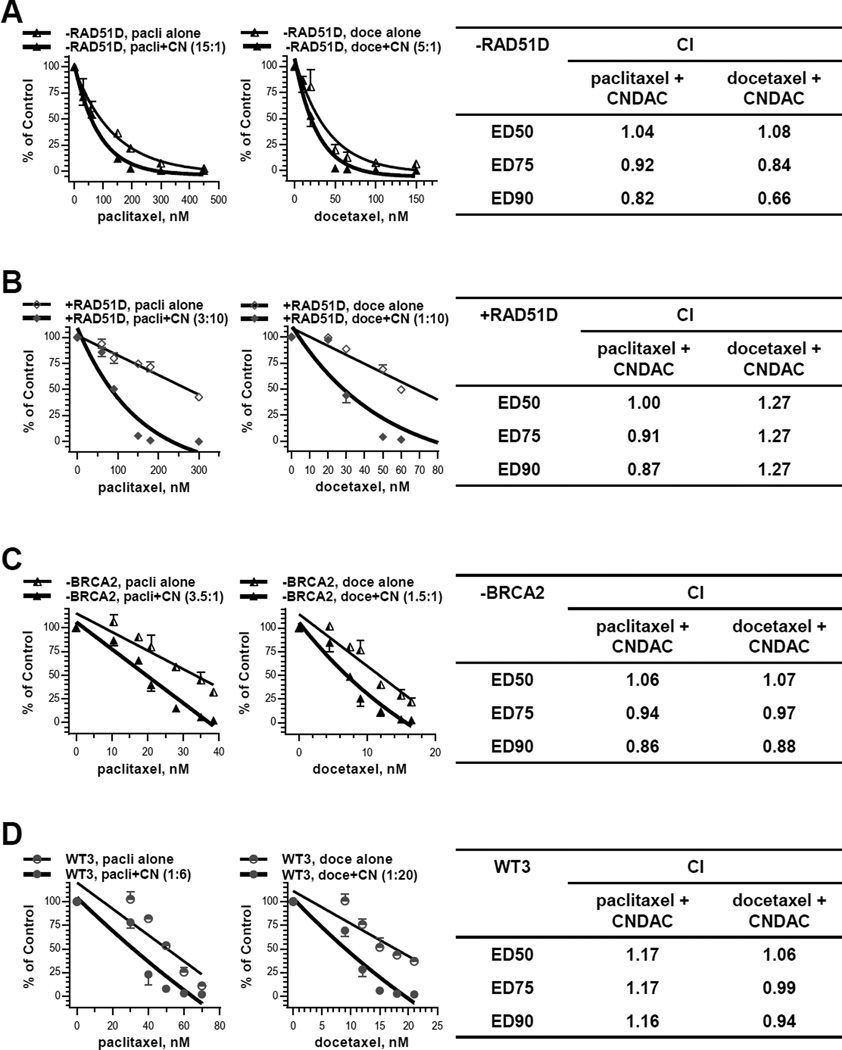
Paclitaxel and docetaxel are well-established tubulin-directed agents used against multiple cancer types. Because they impact specifically on mitosis, it is not a surprise that neither RAD51D nor BRCA2 deficient cells were significantly sensitive to these two agents relative to proficient cells (Suppl. Tab. 1). However, the paclitaxel-CNDAC (15:1) and docetaxel-CNDAC (5:1) combinations both achieved CI <= 1 in cells lacking RAD51D (Fig. 6A). Paclitaxel-CNDAC (3:10) and docetaxel-CNDAC (1:10) combinations achieved CI <= 1 and CI >1, respectively, in cells complemented with RAD51D (Fig. 6B). The concentrations of paclitaxel and docetaxel were in the same range for both cell lines. In contrast, the concentrations of CNDAC required to achieve a similar cell killing effect were 50-fold greater in RAD51D repleted than deficient cells (6). The interaction between taxane agent and CNDAC was also investigated in cells lacking BRCA2 and the wild type. Paclitaxel-CNDAC (ratios 10:1 for Exp. 1 and 7:2 for Exp. 2) and docetaxel-CNDAC (ratios 20:3 for Exp. 1 and 3:2 for Exp. 2) combinations both resulted in CI ∼1 in BRCA2 deficient cells (Fig. 6C). Paclitaxel-CNDAC (ratios 1:5 for Exp. 1 and 1:6 for Exp. 2) and docetaxel-CNDAC (ratios 1:6 for Exp. 1 and 1:20 for Exp. 2) combinations also led to CI >=1 in wild type cells (Fig. 6D). Again the concentrations of paclitaxel and docetaxel were similar for both cell lines. By contrast, the concentrations of CNDAC required to achieve similar cell killing effect were 20-fold greater in the wild type than BRCA2 deficient cells (6). Together, these results demonstrated an additive effect of taxanes and CNDAC in HR defective cells and proficient cells as well. This effect is selective in HR deficient cells at lower concentrations of CNDAC that allow survival of wild type cells.
Figure 6.
Taxane agents have additive effect with CNDAC in HR defective cells. A, CHO RAD51D deficient cells (−RAD51D) were treated concomitantly with paclitaxel (30 – 450 nM) or docetaxel (10 – 150 nM) and CNDAC (2 – 30 nM) for 24 hours in clonogenic assays. The colonies formed were quantitated and clonogenicity of both lines were compared for treatment with paclitaxel alone vs paclitaxel-CNDAC combination at a fixed ratio (representative 15:1, left) or docetaxel vs docetaxel-CNDAC combination at a fixed ratio (representative 5:1, middle). The mean CIs of paclitaxel-CNDAC combination (3 independent experiments) and docetaxel-CNDAC combination (2 independent experiments) are presented in the right panel. B, RAD51D complemented cells (+RAD51D) were treated concomitantly with paclitaxel (60 – 300 nM) or docetaxel (20 – 70 nM) and CNDAC (200 – 1000 nM, high concentration range) for 24 hours in clonogenic assays. Clonogenicity of both lines were compared for treatment with paclitaxel alone vs paclitaxel-CNDAC combination at a fixed ratio (3:10, left) or docetaxel vs docetaxel-CNDAC combination at a fixed ratio (1:10, middle). CIs of paclitaxel-CNDAC combination and docetaxel-CNDAC combination are presented in the right panel. C, BRCA2-deficient Chinese hamster lung cells (−BRCA2) were treated concomitantly with paclitaxel (10.5 – 75 nM) or docetaxel (4.5 – 50 nM) and CNDAC (2.4 – 11 nM, low concentration range) for 24 hours in clonogenic assays. Clonogenicity of both lines were compared for treatment with paclitaxel alone vs paclitaxel-CNDAC combination at a fixed ratio (representative 3.5:1, left) or docetaxel vs docetaxel-CNDAC combination at a fixed ratio (representative 1.5:1, middle). CIs (mean of 2 independent experiments) of paclitaxel-CNDAC combination and docetaxel-CNDAC combination are presented in the right panel. D, the BRCA2 wild-type hamster cells (WT3) were treated concomitantly with paclitaxel (24 – 80 nM) or docetaxel (9 – 40 nM) and CNDAC (120 – 420 nM, high concentration range) for 24 hours in clonogenic assays. Clonogenicity of both lines were compared for treatment with paclitaxel alone vs paclitaxel-CNDAC combination at a fixed ratio (representative 1:6, left) or docetaxel vs docetaxel-CNDAC combination at a fixed ratio (representative 1:20, middle). CIs of paclitaxel-CNDAC combination (mean of 2 independent experiments) and docetaxel-CNDAC combination are presented in the right panel.
Discussion
The increased DNA mutation rates that cause genome instability are a hallmark of cancer cells that allow them to generate resistance to single therapeutic agents. Given this limitation, combination chemotherapy has been the most effective approach to circumventing the emergence of resistance as well as affecting deeper responses. CNDAC has a novel mechanism of action that results in chemically defined, repair resistant SSBs, which are metabolized to DSBs upon DNA replication. Earlier work demonstrated that HR is principally responsible for the repair of this damage (6) with transcription-coupled nucleotide excision repair having a lesser role (5). In contrast, lack of either non-homologous end-joining, base excision repair or nucleotide mismatch repair had negligible effects on the sensitivity of cells to CNDAC (5, 30). Looking forward to the further development of CNDAC, this investigation sought to identify drugs that have the most advantageous level of cell killing activity when applied in combinations with CNDAC.
Taking advantage of colony formation assays and median-effect analyses, we evaluated the combinational effects of CNDAC with a wide spectrum of chemotherapeutic agents affecting different DNA repair pathways and targets. Drugs that affect DSB repair (imatinib and inhibitors of PARP1) or which may rely upon DSB repair (temozolomide), were synergistic with CNDAC. Combinations of CNDAC with agents that cause bulky adducts to DNA (platinum compounds or nitrogen mustards) or that affect the mitotic spindle (taxanes) produced loss of clonogenicity that was additive with that of CNDAC. In all cases, cells that were deficient in HR were selectively sensitized relative to those with normal repair capabilities. Considerations of the mechanisms that enable these positive interactions identify future paths of research and clinical opportunities.
Imatinib is one of several potent inhibitors of c-Abl that are currently used against a variety of cancers. c-Abl is activated by DNA damage, and subsequently amplifies the response by phosphorylating down-stream DNA damage proteins, including RAD51 the key enzyme for homology search in the HR process (31–33). In the context of synergy with CNDAC, inhibition of c-Abl by imatinib likely disables its contribution to this aspect of HR, thereby decreasing the possibility that CNDAC generated DSBs will be repaired. Imatinib has been shown to inhibit homologous recombination in cancer cell lines by downregulating Rad51 expression at both mRNA and protein levels (34, 35). This provides an alternative mechanism by which imatinib sensitizes HCT116 cells to CNDAC. However, this synergy between imatinib and CNDAC is likely specific to certain cell types, because the same synergy was not observed in HeLa Rad51C knockout line and its control line (data not shown).
Recent investigations have demonstrated mechanisms by which inhibition of PARP1 may block DNA repair, several of which may contribute to synergy with CNDAC. Upon DNA damage, PARP1 quickly adds poly (ADP-ribose) polymers (PARylation) to proteins, including itself, that are adjacent to DNA breaks. Such modifications are recognized by BARD1, which is responsible for bringing its binding partner BRCA1 to the vicinity of the damage (36). BRCA2 has its own role in HR by enabling RAD51 to relocate to form the single strand filament that is required for homology search (37, 38). In addition, the attraction of BRCA2 to DSB has been shown to be dependent upon PARylation, a process that is blocked by PARP1 inhibitors (39). Thus, inhibiting PARylation may diminish HR repair of CNDAC-induced DSB. Also, self-PARylation by PARP1 is thought to assist its exit from DNA damage to permit repair proteins to act (40). Inhibition of such self-PARylation is thought to be the cause of PARP1 trapping onto DNA, thereby inhibiting the HR repair processes (41). In addition, a study from Kaufmann’s group has highlighted the contribution of NHEJ to PARP1 inhibitor lethality in HR-deficient contexts (42), supporting the “NHEJ activation” model (43). Although NHEJ does not appear to participate in the repair of CNDAC (30), microhomology-mediated end joining, an alternative to NHEJ, has a PARP1-dependent step that may be inhibitory to HR (44). Finally, PARP1 inhibitors are thought to interfere with BER at steps following removal of a damaged nucleotide, thereby permitting endogenous gap to be converted to a DSB upon subsequent DNA replication (45, 46). These additional breaks may serve to compete for HR proteins that otherwise would be directed to repair of breaks generated by CNDAC. Each of these mechanisms of PARP1 inhibitors may serve to enable synergy in combination with CNDAC.
The interactions of temozolomide with CNDAC produced a lesser synergistic loss of clonogenicity. The single strand gap generated by CNDAC is terminated on the 3’-end by a 2’, 3’-dideoxy nucleotide β-elimination product that is not a substrate for ligation by BER (5). It is possible that this structure attracts the BER proteins (XRCC1, DNA polβ, DNA ligase 4) that normally insert and ligate a cognate nucleotide, thus diminishing the capacity for BER. Under such conditions, repair of temozolomide may be diminished and not completed effectively. Thus, DNA with gaps may be replicated generating DSBs that compete for repair with those caused by CNDAC.
The taxanes are commonly used to treat both breast cancers and ovarian cancer, each of which include significant cohorts of patients that are deficient in HR repair due to loss or mutation of BRCA1 or BRCA2 (47). Several cells lines lacking different elements of HR repair were selectively sensitized to combinations of CNDAC, but no evidence of synergy between CNDAC and paclitaxel or docetaxel was observed. The repair of interstrand DNA crosslinks associated with the mechanisms of action of platinum compounds and nitrogen mustards requires components of the HR process. Although cells lacking HR repair were clearly sensitized to these agents alone and in combination, it was surprising that synergy with CNDAC was not observed under the conditions tested with the combinations. Nevertheless, the consistent additive actions of CNDAC and these active agents provide a rationale for their use in combinations, particularly against tumors that lack HR capacity.
Supplementary Material
Acknowledgments
Grant Support
This work was supported in part by grants CA28596 (W. Plunkett), CA100632 (W. Plunkett), and Cancer Center Support grant P30 CA16672 from the National Cancer Institute, Department of Health and Human Services.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflict of Interest
The authors have no potential conflict of interest to disclose.
References
- 1.Kantarjian H, Garcia-Manero G, O’Brien S, Faderl S, Ravandi F, Westwood R, et al. Phase i clinical and pharmacokinetic study of oral sapacitabine in patients with acute leukemia and myelodysplastic syndrome. J Clin Oncol. 2010;28:285–291. doi: 10.1200/JCO.2009.25.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kantarjian H, Faderl S, Garcia-Manero G, Luger S, Venugopal P, Maness L, et al. Oral sapacitabine for the treatment of acute myeloid leukaemia in elderly patients: A randomised phase 2 study. Lancet Oncol. 2012;13:1096–1104. doi: 10.1016/S1470-2045(12)70436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kantarjian HM, Jabbour E, Garcia-Manero G, Kadia TM, Dinardo CD, Daver NG, et al. First report of a phase i/ii study of dfp-10917, a nucleoside analog, given by continuous infusion (ci) in patients (pts) with relapsed or refractory acute leukemia. J Clin Oncol. 2015;33(suppl) abstr 7077. [Google Scholar]
- 4.Azuma A, Huang P, Matsuda A, Plunkett W. 2’-C-cyano-2’-deoxy-1-beta-D-arabino-pentofuranosylcytosine: A novel anticancer nucleoside analog that causes both DNA strand breaks and G(2) arrest. Mol Pharmacol. 2001;59:725–731. doi: 10.1124/mol.59.4.725. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Liu X, Matsuda A, Plunkett W. Repair of 2’-C-cyano-2’-deoxy-1-beta-D-arabino-pentofuranosylcytosine-induced DNA single-strand breaks by transcription-coupled nucleotide excision repair. Cancer Res. 2008;68:3881–3889. doi: 10.1158/0008-5472.CAN-07-6885. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Wang Y, Benaissa S, Matsuda A, Kantarjian H, Estrov Z, et al. Homologous recombination as a resistance mechanism to replication-induced double-strand breaks caused by the antileukemia agent cndac. Blood. 2010;116:1737–1746. doi: 10.1182/blood-2009-05-220376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapiro GI, Hilton J, Cleary JM, Tolaney SM, Ghandi L, Kwak EL, et al. Responses to sequential sapacitabine and seliciclib in patients with brca-deficient solid tumors. Proceedings AACR 104th annual meeting. 2013;73(8 suppl) abstr# LB-202. [Google Scholar]
- 8.Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, et al. Ataxia telangiectasia mutant protein activates c-abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 9.Shafman T, Khanna KK, Kedar P, Spring K, Kozlov S, Yen T, et al. Interaction between atm protein and c-abl in response to DNA damage. Nature. 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 10.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor ag14361. J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 11.Guillot C, Favaudon V, Herceg Z, Sagne C, Sauvaigo S, Merle P, et al. PARP inhibition and the radiosensitizing effects of the parp inhibitor ABT-888 in in vitro hepatocellular carcinoma models. BMC Cancer. 2014;14:603. doi: 10.1186/1471-2407-14-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res. 2005;65:6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 13.Goellner EM, Grimme B, Brown AR, Lin YC, Wang XH, Sugrue KF, et al. Overcoming temozolomide resistance in glioblastoma via dual inhibition of NAD+ biosynthesis and base excision repair. Cancer Res. 2011;71:2308–2317. doi: 10.1158/0008-5472.CAN-10-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheson BD, Leoni L. Bendamustine: Mechanism of action and clinical data. Clin Adv Hematol Oncol. 2011;9:1–11. [PubMed] [Google Scholar]
- 15.Reardon JT, Vaisman A, Chaney SG, Sancar A. Efficient nucleotide excision repair of cisplatin, oxaliplatin, and bis-aceto-ammine-dichloro-cyclohexylamine-platinum(iv) (jm216) platinum intrastrand DNA diadducts. Cancer Res. 1999;59:3968–3971. [PubMed] [Google Scholar]
- 16.Basu A, Krishnamurthy S. Cellular responses to cisplatin-induced DNA damage. J Nucleic Acids. 2010;2010:1–16. doi: 10.4061/2010/201367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Andrea AD. The fanconi anemia/brca signaling pathway: Disruption in cisplatin-sensitive ovarian cancers. Cell Cycle. 2003;2:290–292. [PubMed] [Google Scholar]
- 18.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the fanconi anemia-brca pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 19.Chirnomas D, Taniguchi T, de la Vega M, Vaidya AP, Vasserman M, Hartman AR, et al. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol Cancer Ther. 2006;5:952–961. doi: 10.1158/1535-7163.MCT-05-0493. [DOI] [PubMed] [Google Scholar]
- 20.Abal M, Andreu JM, Barasoain I. Taxanes: Microtubule and centrosome targets, and cell cycle dependent mechanisms of action. Curr Cancer Drug Targets. 2003;3:193–203. doi: 10.2174/1568009033481967. [DOI] [PubMed] [Google Scholar]
- 21.Liu XJ, Nowak B, Wang YQ, Plunkett W. Sapacitabine, the prodrug of cndac, is a nucleoside analog with a unique action mechanism of inducing DNA strand breaks. Chin J Cancer. 2012;31:373–380. doi: 10.5732/cjc.012.10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 23.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 24.Aylon Y, Kupiec M. New insights into the mechanism of homologous recombination in yeast. Mutat Res. 2004;566:231–248. doi: 10.1016/j.mrrev.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 26.Chen G, Yuan SS, Liu W, Xu Y, Trujillo K, Song B, et al. Radiation-induced assembly of RAD51 and RAD52 recombination complex requires atm and c-ABL. J Biol Chem. 1999;274:12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 27.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of brca2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 28.Jacinto FV, Esteller M. Mgmt hypermethylation: A prognostic foe, a predictive friend. DNA Repair (Amst) 2007;6:1155–1160. doi: 10.1016/j.dnarep.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Kanzawa T, Bedwell J, Kondo Y, Kondo S, Germano IM. Inhibition of DNA repair for sensitizing resistant glioma cells to temozolomide. J Neurosurg. 2003;99:1047–1052. doi: 10.3171/jns.2003.99.6.1047. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Matsuda A, Plunkett W. Ataxia-telangiectasia and rad3-related and DNA-dependent protein kinase cooperate in G2 checkpoint activation by the DNA strand-breaking nucleoside analogue 2’-C-cyano-2’-deoxy-1-beta-D-arabino-pentofuranosylcytosine. Mol Cancer Ther. 2008;7:133–142. doi: 10.1158/1535-7163.MCT-07-0416. [DOI] [PubMed] [Google Scholar]
- 31.Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, et al. Regulation of rad51 function by c-ABL in response to DNA damage. J Biol Chem. 1998;273:3799–3802. doi: 10.1074/jbc.273.7.3799. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu H, Popova M, Fleury F, Kobayashi M, Hayashi N, Sakane I, et al. c-ABL tyrosine kinase stabilizes RAD51 chromatin association. Biochem Biophys Res Commun. 2009;382:286–291. doi: 10.1016/j.bbrc.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 33.Meltser V, Ben-Yehoyada M, Shaul Y. C-abl tyrosine kinase in the DNA damage response: Cell death and more. Cell Death Differ. 2011;18:2–4. doi: 10.1038/cdd.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell JS, Brady K, Burgan WE, Cerra MA, Oswald KA, Camphausen K, et al. Gleevec-mediated inhibition of RAD51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed] [Google Scholar]
- 35.Choudhury A, Zhao H, Jalali F, Al Rashid S, Ran J, Supiot S, et al. Targeting homologous recombination using imatinib results in enhanced tumor cell chemosensitivity and radiosensitivity. Mol Cancer Ther. 2009;8:203–213. doi: 10.1158/1535-7163.MCT-08-0959. [DOI] [PubMed] [Google Scholar]
- 36.Li M, Yu X. Function of brca1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The brc repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U S A. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of RAD51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 39.Zhang F, Shi J, Bian C, Yu X. Poly(adp-ribose) mediates the BRCA2-dependent early DNA damage response. Cell Rep. 2015;13:678–689. doi: 10.1016/j.celrep.2015.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muthurajan UM, Hepler MR, Hieb AR, Clark NJ, Kramer M, Yao T, et al. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc Natl Acad Sci U S A. 2014;111:12752–12757. doi: 10.1073/pnas.1405005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical parp inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-ribose) polymerase inhibitors: Recent advances and future development. J Clin Oncol. 2015;33:1397–1406. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, et al. Homologous-recombination-deficient tumours are dependent on poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iglehart JD, Silver DP. Synthetic lethality--a new direction in cancer-drug development. N Engl J Med. 2009;361:189–191. doi: 10.1056/NEJMe0903044. [DOI] [PubMed] [Google Scholar]
- 46.Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA Cancer J Clin. 2011;61:31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- 47.Petrucelli N, Daly MB, Feldman GL. BRCA1 and BRCA2 hereditary breast and ovarian cancer. GeneReviews® [Internet] Initial Posting. 1998 Sep 4; Last Update: September 26, 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.